Abstract
The cerebral cortex is a cellularly complex structure comprising a rich diversity of neuronal and glial cell types. Cortical neurons can be broadly categorized into two classes—excitatory neurons that use the neurotransmitter glutamate, and inhibitory interneurons that use γ-aminobutyric acid (GABA). Previous developmental studies in rodents have led to a prevailing model in which excitatory neurons are born from progenitors located in the cortex, whereas cortical interneurons are born from a separate population of progenitors located outside the developing cortex in the ganglionic eminences1,2,3,4,5. However, the developmental potential of human cortical progenitors has not been thoroughly explored. Here we show that, in addition to excitatory neurons and glia, human cortical progenitors are also capable of producing GABAergic neurons with the transcriptional characteristics and morphologies of cortical interneurons. By developing a cellular barcoding tool called ‘single-cell-RNA-sequencing-compatible tracer for identifying clonal relationships’ (STICR), we were able to carry out clonal lineage tracing of 1,912 primary human cortical progenitors from six specimens, and to capture both the transcriptional identities and the clonal relationships of their progeny. A subpopulation of cortically born GABAergic neurons was transcriptionally similar to cortical interneurons born from the caudal ganglionic eminence, and these cells were frequently related to excitatory neurons and glia. Our results show that individual human cortical progenitors can generate both excitatory neurons and cortical interneurons, providing a new framework for understanding the origins of neuronal diversity in the human cortex.
Similar content being viewed by others
Main
The neocortex is responsible for carrying out many higher-order cognitive functions, such as decision making, language comprehension and sensory perception. During neural development, progenitors in the cortex and ganglionic eminences produce the diverse array of neurons and glia that comprise the neocortex. Extensive developmental studies in rodents have demonstrated that the two principal types of cortical neuron—glutamatergic excitatory neurons and GABAergic inhibitory interneurons—are produced by two distinct groups of progenitors1,2,3,4,5. Excitatory neurons are generated by progenitors located dorsally in the developing cortex and migrate radially to occupy their terminal positions in the cortical plate. By contrast, cortical interneurons are generated by progenitors located in the ganglionic eminences of the ventral forebrain and then migrate dorsally into the developing neocortex.
A few limited studies have suggested the possibility that human cortical progenitors might also give rise to cortical interneurons, but the findings have been conflicting6,7,8,9,10. Short-term clonal labelling of progenitors in the human cortex revealed the local generation of newborn GABAergic neurons7, but did not determine whether they were cortical interneurons or another type of interneuron such as those of the olfactory bulb, which have previously been shown to derive from cortical progenitors in mice11,12,13. Similarly, in vitro cultures derived from human cortical progenitors have been shown to generate GABAergic inhibitory neurons10. Conversely, a later study that labelled dividing cells in short-term cultures of human organotypic tissue slices did not find a substantial fraction of newborn inhibitory neurons in the cortex8. The overwhelming consensus remains that human cortical progenitors give rise to excitatory neurons but not cortical inhibitory neurons; however, this has not been thoroughly examined and the developmental potential of individual human cortical progenitors remains largely unknown.
Design and validation of lineage tracer
In order to carry out high-throughput clonal lineage tracing of primary human neural progenitors, we developed STICR—an ultra-high-complexity barcoding strategy that allows us to permanently label cells and their progeny. This approach uses a lentivirus encoding a heritable, transcribed molecular barcode within the 3′-untranslated region of the enhanced green fluorescent protein (EGFP) reporter gene (Fig. 1a). The combinatorial, single-molecule barcode design of STICR allows for a predefined, error-correctable barcode library with a maximum diversity of 125 million sequences (Extended Data Fig. 1a; see Methods). Deep sequencing of STICR plasmid and lentiviral libraries confirmed their ultra-high barcode diversity (approximately 50 million to 65 million unique barcodes per library) without any overrepresented barcodes (Extended Data Fig. 1b). Using the observed barcode diversity and frequency from each STICR library, we modelled the rate of barcode ‘collision’—the event in which two different cells are independently labelled with the same barcode—and found that STICR could be used to label more than 250,000 cells before reaching an estimated barcode collision rate of around 0.5% (Extended Data Fig. 1c). To confirm the accuracy of STICR barcode recovery from single cells, we carried out a cell-mixing, or ‘barnyard’, experiment in which we labelled mouse and human cells with different STICR libraries that were readily distinguishable by a constant ‘viral index’ sequence unique to each library (Extended Data Fig. 1d). Following single-cell RNA sequencing (scRNA-seq), recovered STICR barcodes were 100% concordant with the STICR library used to infect each population (Extended Data Fig. 1e, f), indicating that our method is accurate and can be applied robustly to carry out high-throughput clonal lineage tracing.
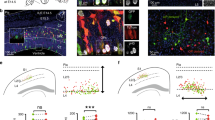
a, Design of the STICR lentiviral vector. CAG, cytomegalovirus/chicken β-actin promoter; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element; pA, polyadenylation signal. b, The experimental design used to label and capture samples. GW, gestational week; Rep, replicate. c, Uniform manifold approximation and projection (UMAP) embedding and Leiden clustering of STICR-labelled cells following scRNA-seq. IPC, intermediate progenitor cell. d, Feature plots showing the principal-cell-trajectory marker genes DLX2 (GABAergic inhibitory neurons and DLX2+ IPCs), NEUROD2 (excitatory neurons), GFAP (glia), and EOMES (EOMES+ IPCs). e, Histogram showing clone sizes within each sample. Left, clone sizes of 1–75 cells. Right, clone sizes of more than 75 cells, in 25-cell bins.
Lineage tracing of human neural progenitors
To determine the developmental potential of individual human cortical progenitors, we derived primary human cell cultures from the cortical germinal zone of three different specimens at stages of peak neurogenesis (gestational weeks 15 and 18, GW15 and GW18) (Fig. 1b). Before clonal labelling, one of the specimens (GW18) was further dissected on the basis of known anatomical landmarks, allowing us to generate region-specific cultures from the germinal zones of the prefrontal cortex (PFC), primary visual cortex (V1) and medial ganglionic eminence (MGE). We then labelled cells with STICR and cultured them for six weeks in vitro before carrying out scRNA-seq. Transcriptome analysis of 121,290 cells identified three principal cortical cell type trajectories— excitatory neurons, GABAergic inhibitory neurons and glia—on the basis of differential gene expression, including that of marker genes NEUROD2, DLX2 and GFAP, respectively (Fig. 1c, d and Extended Data Figs. 2, 3). We identified intermediate progenitor cells (IPCs) within both the inhibitory and the excitatory neuron trajectories, which we refer to as DLX2+ IPCs (inhibitory trajectory) and EOMES+ IPCs (excitatory trajectory) (Extended Data Fig. 3). Cluster correlation analysis of STICR datasets with an scRNA-seq atlas of the developing primary human cortex at comparable developmental time points and regions14 further supported these cell-type designations (Extended Data Fig. 2d).
We recovered STICR barcodes in 63 ± 23% (mean ± s.d., n = 5 libraries) of cells per culture, including cells of each principal cell type (Supplementary Table 1 and Extended Data Fig. 3g). In total, we identified 1,461 unique clonal barcodes, 1,324 of which belonged to multicellular clones with a median size of 23 cells per clone (Fig. 1e). Although there is very little known about the output of human cortical progenitors over this time scale, we observe a maximum clone size of 1,209 cells, which is congruent with a prior study that measured the output from three individual human outer radial glia15. Notably, no STICR barcodes were shared between experimental groups (Extended Data Fig. 3h), further indicating that cells identified as members of multicellular clones here are not the result of barcode collision. Although all three principal cortical cell types (excitatory glutamatergic neurons, GABAergic interneurons and glia) were found in multicellular clones in both GW15 samples and the GW18 V1 sample, GW18 PFC clones contained relatively more GABAergic inhibitory neurons than excitatory neurons or glia (Extended Data Fig. 3i). Consistent with previous studies, the GW18 MGE culture completely lacked excitatory neurons and instead comprised almost entirely interneurons (Extended Data Fig. 3i). Thus, STICR reveals clonal lineage relationships of all principal cell types derived from human forebrain progenitors.
Clonal relationship of human cortical neurons
In order to determine the lineage relationships of cells born from cortical progenitors, we first analysed the relative proportions of principal cell types in multicellular clones. The majority (66%; 829 of 1,252) of multicellular clones from cortical cultures contained at least 1 excitatory neuron, with these clones containing a median of 3 excitatory neurons per clone (Extended Data Fig. 4a, b and Supplementary Table 1). Although our study of regional cortical progenitors is limited to a single GW18 sample, we found that PFC clones contained proportionally fewer excitatory neurons than clones in the V1 sample (Extended Data Fig. 4b, c). This is consistent with the neurodevelopmental gradient in the cortex, where developmental milestones are reached in the PFC several weeks before the V1 region16. Across all samples, only 1.5% (19 of 1,252) of cortical clones were composed entirely of excitatory neurons, and 51% (635 of 1,252) contained a combination of excitatory neurons, inhibitory neurons and glia (Fig. 2a–c, Extended Data Fig. 4c and Supplementary Table 1). Owing to extensive aggregation of excitatory neurons within in vitro cultures and the gentle dissociation used (Extended Data Fig. 4d, e), production of excitatory neurons may be underestimated in the dataset derived from cell culture experiments. Nevertheless, across GW15 samples, the coincidence of excitatory and inhibitory neurons within the same clone occurred across the range of clone sizes (Fig. 2b). Together, these results suggest that individual human cortical progenitors reproducibly generate both excitatory and inhibitory neurons.
a, Stacked boxplot depicting the average (mean ± s.d.) proportion of GABAergic inhibitory neurons, DLX2+ IPCs, EOMES+ IPCs, excitatory neurons and glia in different sized clones. b, Bar graph depicting the proportions of clones of different sizes that contain both excitatory and inhibitory neurons. c, Representative 6-cell (GW15, replicate 1, clone 113) and 61-cell (GW15, replicate 1, clone 732) clones depicted in UMAP space. Cells within each clone are coloured in red. Dashed lines depict borders of principal cell types from Fig. 1c. d, Pseudotime transcriptional trajectories of subclustered inhibitory neurons. Three different interneuron transcriptional trajectories (IN.1, IN.2 and IN.3) are indicated with arrows. e, Violin plots depicting expression of IN.1 marker genes (SST, NPY, TAC3 and NXPH1) and general CGE marker genes (SCGN, SP8, PCDH9 and BTG1) in IN.1, IN.2 and IN.3 trajectories. f, Volcano plot comparing differences in gene expression between IN.2 and IN.3 trajectory cells. Not sig, genes with log2 fold-change less than 0.125 and/or adjusted P value greater than 0.1. g, Venn diagram depicting the number of multicellular cortical clones that contain IN.3 cells or excitatory neurons.
Analysis of cortically born inhibitory neurons
We next sought to determine the transcriptional identities of cortically born GABAergic neurons. In addition to DLX2 (Fig. 1d), inhibitory-trajectory cells were broadly enriched for general markers of interneurons, including GAD1, ARX, SLC32A1 and DLX6AS-1 (Extended Data Fig. 3e). Iterative subclustering and transcriptional trajectory analysis of the inhibitory cells along with the DLX2+ IPCs revealed three distinct subgroups of GABAergic inhibitory neurons that we termed IN.1, IN.2 and IN.3 (Fig. 2d and Extended Data Fig. 5a–c). IN.1 cells were enriched for markers of SST+ cortical interneurons, including SST, NPY, TAC3 and NXPH1 (Fig. 2e). Consistent with developmental studies in mice which showed that SST+ cortical interneurons derive primarily from the MGE17, 73% (492 of 671 cells) of IN.1 cells are produced by MGE progenitors (Extended Data Fig. 5d). By contrast, IN.1 cells comprised only 0.3% (179 of 56,244 cells) of progeny born from cortical progenitors (Extended Data Fig. 5e). Furthermore, MGE-derived IN.1 cells expressed canonical MGE-born interneuron genes, including LHX6, NKX2-1, ACKR3 (CXCR7), PDE1A and MAF, while cortically born IN.1 cells did not (Extended Data Fig. 5f). Together, these data suggest that IN.1 cells are transcriptionally similar to SST+ cortical interneurons and that the majority of IN.1 cells derive from the MGE. In contrast to IN.1 trajectory cells, IN.2 and IN.3 trajectory cells were transcriptionally similar to cells born from the caudal ganglionic eminence (CGE) based on their expression of marker genes such as SCGN, SP8, PCDH9 and BTG1 (Fig. 2e)18. Furthermore, IN.2 and IN.3 cells differed from IN.1 cells in that they were derived entirely from cortical progenitors, with no contribution from MGE progenitors (Extended Data Fig. 5d). Top IN.2 markers included TSHZ1, PBX3, MEIS2, CALB2, CDCA7L, SYNPR and ETV1, which are enriched in mouse olfactory bulb interneurons (Fig. 2f)19,20. By contrast, top IN.3 marker genes included NR2F1, NFIX, PROX1 and NR2F2,which are enriched within the CGE, as well as SOX6 and CXCR4, which are enriched in cortical interneurons (Fig. 2i), suggesting that these cells are transcriptionally similar to CGE-derived cortical interneurons14,21,22,23,24,25. Comparison of IN.2 and IN.3 marker genes with orthogonal datasets, including those from the Allen Brain Atlas26, similarly support this distinction (Extended Data Fig. 5g, h). Thus, although there are currently no marker genes that can unequivocally distinguish cortical interneurons from olfactory bulb interneurons, our transcriptome-wide data suggest that IN.2 cells resemble olfactory bulb interneurons while IN.3 cells are similar to CGE-born cortical interneurons.
Previous fate-tracing studies in mice have shown that cortical progenitors can produce a subset of embryonically born olfactory bulb interneurons11. In agreement with these findings, we found that multicellular cortical clones frequently contain olfactory-bulb-like (IN.2 branch) GABAergic inhibitory neurons (Extended Data Fig. 5d, e). Notably, many clones containing olfactory-bulb-like GABAergic inhibitory neurons also contained excitatory neurons (61%; 321 of 530 clones) (Extended Data Fig. 5j). Such clones were found in all cortical samples but were especially common in both GW15 samples (Supplementary Table 1). GW18 PFC clones contained the highest proportion of olfactory-bulb-like GABAergic neurons (41%; 1,637 of 3,949 cells) out of any sample analysed, and although relatively few of the GW18 PFC clones contained excitatory neurons (9 of 141 clones), 8 of these also contained at least one olfactory-bulb-like interneuron (Extended Data Fig. 5d and Supplementary Table 1). These results suggest that, similar to mice13, human cortical progenitors can generate olfactory bulb interneurons during embryogenesis, and that some excitatory neurons and embryonically born olfactory bulb interneurons are clonally related.
Surprisingly, we found that most (79%; 655 of 829) multicellular clones that contained excitatory neurons also included putative cortical interneurons (IN.3 cells) (Fig. 2g), indicating that some human cortical progenitors can generate both excitatory neurons and cortical interneurons. Many of these clones contained multiple cells of both types, and mixed clones were especially abundant in GW15 and GW18 V1 samples. Further subclustering analysis of excitatory neurons revealed both deep-like and upper-like excitatory neuron subgroups, both of which were frequently clonally related to IN.3 cells (Extended Data Fig. 6). Notably, every cortical sample we analysed contained clones with both excitatory and inhibitory cortical neurons (Fig. 2g). These mixed clones also frequently contained olfactory bulb interneurons as well as glial-trajectory cells (Extended Data Fig. 5i, j). Taken together, our results suggest that human cortical progenitors cultured in vitro are unexpectedly multipotent in their ability to generate a wide variety of principal neural cell types, including both excitatory neurons and putative cortical interneurons—two cell types previously thought to be produced by different pools of spatially restricted progenitors in the developing forebrain.
Mixed excitatory–inhibitory clones in xenografts
To confirm that the observed lineage relationship between excitatory neurons and cortical interneurons was not an artefact of in vitro culture, we STICR-labelled cortical germinal zone cells from three additional GW15 specimens as above, and transplanted them into the cortex of early postnatal immunodeficient mice, where we allowed them to develop for six weeks before analysis (Fig. 3a). Immunohistochemistry revealed xenografted human cells at the injection site itself as well as distributed throughout the adjacent tissue (Figs. 3b and Extended Data Fig. 7a–c). To quantify the proportion of the principal cell types in the xenografts, we carried out immunohistochemistry for the excitatory-neuron marker NEUROD2, the interneuron marker GABA, and the glia markers SOX9 and OLIG2. Excitatory neurons were the most common cell type, accounting for 75.7 ± 8.4% of xenograft-derived cells, followed by glia at 16.8 ± 6.3%, and interneurons at 7.9 ± 3.2% (mean ± s.d.; n = 7 recipient mice; Extended Data Figs. 7a–c). In parallel, we isolated EGFP+ xenograft cells by fluorescence-activated cell sorting (FACS), and carried out scRNA-seq to determine their transcriptional identities and clonal relationships. Similar to our in vitro cultures, xenograft cells formed distinct transcriptional clusters of GABAergic neurons, excitatory neurons and glia (Fig. 3c and Extended Data Fig. 7d–f). The proportions of excitatory and inhibitory neurons within multicellular clones from xenograft experiments differed from those observed in our in vitro cultures (Fig. 2g). This was likely to be due to the specific loss of excitatory neurons from in vitro culture during dissociation, as these cells frequently associated in tightly formed masses that were difficult to dissociate (Extended Data Figs. 4d, e). Quantification of principal cell types by scRNA-seq closely matched the cell proportions observed by immunohistochemistry, and was reproducible across biological replicates (Extended Data Fig. 7g). These data suggest that xenograft-derived STICR-labelled cells analysed by scRNA-seq accurately reflected the cell proportions produced within the xenograft.
a, Experimental design and analysis of STICR-labelled progenitors by immunohistochemistry (IHC) and scRNA-seq following transplantation into the postnatal murine cortex. b, Representative image of transplanted human cortical cells. EGFP expression from STICR is in green. Scale bar, 500 μm. c, UMAP embedding and Leiden clustering of xenografted cells following scRNA-seq. d, Histogram of clone sizes within each xenograft sample. Left, clone sizes from 1–25 cells. Right, clone sizes of greater than 25 cells, in 25-cell bins. e, Stacked barplot depicting the relative proportions of principal cell types within multicellular clones of each sample. f, UMAP embedding of both cultured and xenograft-derived inhibitory neurons. Xenograft-derived cells that are members of multicellular clones are highlighted in red. IN.2 and IN.3 trajectories are depicted with arrows. g, Venn diagrams showing the number of multicellular clones containing excitatory neurons and/or IN.3 neurons.
We recovered STICR barcodes from 76 ± 10% (n = 3 libraries) of xenografted cells and identified 660 multicellular clones that ranged in size from 2 to 101 cells, with a median clone size of 5 cells (Fig. 3d). Within multicellular clones, the proportions of principal cortical cell types were highly similar across biological samples (Fig. 3e). We then further analysed cells categorized as either inhibitory neurons or DLX2+ IPCs in order to determine their transcriptional identities and clonal relationships. Subclustering analysis revealed several distinct groups of GABAergic cells (Extended Data Fig. 8a), including one that was enriched for genes found in cortical interneuron-like IN.3 cells—such as NR2F1, KLHL35, NFIX and SCGN—and was similar to reference GABAergic cortical inhibitory neurons (Extended Data Fig. 8b, c). In order to directly compare the transcriptomic identities of GABAergic neurons from xenografts to those observed following in vitro culture, we integrated scRNA-seq data of GABAergic neurons from both sets of experiments, using the in vitro cultured cells as a reference. GABAergic neurons from the xenografts clustered well with their in vitro counterparts (Fig. 3f, Extended Data Fig. 8d) and integrated entirely within the previously defined transcriptional trajectories (Fig. 2f). Consistent with their marker-gene expression, 85% (211 of 249) of GABAergic xenograft cluster-1 cells clustered within the IN.3 trajectory (Extended Data Fig. 8d), suggesting that they had the transcriptional identity of cortical interneurons. Few (4%; 9 of 249 cells) GABAergic xenograft cluster-1 cells clustered within the IN.2 trajectory (Extended Data Fig. 8d), consistent with previous transplantation studies which found that the cortical environment is not conducive to the generation of olfactory bulb interneurons27. Xenograft-derived GABAergic IN.3 neurons were found in 56 multicellular clones, 43 (77%) of which also contained excitatory neurons (Fig. 3g, Extended Data Fig. 8e–g and Supplementary Table 2). Mixed IN.3/excitatory-neuron clones were found in all three xenografted samples, including GW15 replicate 5, which was transplanted immediately after transduction with STICR and was never cultured (see Methods). Thus, as we had previously observed in vitro, cortical progenitors can generate mixed excitatory/inhibitory-neuron clones when xenografted into the perinatal mouse brain, and these inhibitory neurons are transcriptionally similar to cortical interneurons.
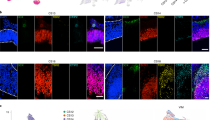
Finally, to determine whether human cortical-progenitor-derived GABAergic inhibitory neurons had additional features consistent with cortical interneurons, we characterized their morphology and distribution in two additional xenograft experiments from GW17 samples. For these experiments, we used a FACS-based isolation strategy to enrich for human progenitor cells before transducing them with STICR (Fig. 4a, Extended Data Fig. 9). Given the protracted time scale of human brain development, we waited for 12 weeks after transplantation before analysing the xenografts by immunohistochemistry. We observed STICR-labelled cells with well-elaborated processes distributed throughout the cortical layers and also within the olfactory bulbs (Fig. 4b and Extended Data Fig. 9). Of STICR-labelled cells located in the cortex, 8.3% (89 of 1,071) expressed GABA, and these were also broadly dispersed across the cortical layers (Fig. 4c, d and Extended Data Fig. 10). Although most STICR-labelled GABA+ cells were located at a similar rostral–caudal level as the transplantation site, we did observe cells more distant from the transplantation site in both directions. Together, these morphological characteristics of cortically derived GABAergic inhibitory neurons are consistent with their classification as cortical interneurons. Thus, individual human cortical progenitors can give rise to both excitatory neurons and cortical GABAergic inhibitory neurons (Fig. 4e).
a, Experimental design and analysis of PTPRZ1-enriched, STICR-labelled progenitors by IHC following transplantation into the postnatal murine cortex. b, Representative images of transplanted human cortical cells analysed by IHC. EGFP expression from STICR is depicted in green, NEUROD2 in red, and DAPI counterstain in blue. Arrows indicate NEUROD2+/STICR+ double-positive cells. Scale bar, 50 μm. c, Representative images of STICR-labelled GABA+ cells throughout the cortical plate. Red arrows depict soma of two cells in the same image. Scale bars, 10 μm. d, Relative laminar positions of GABA+/STICR+ cells in the cortical plate from the corpus callosum (value of 0.0) to the pial surface (value of 1.0) in each host mouse brain. Host brains 1 and 2 were transplanted with GW17 (replicate 1); host brains 3 and 4 were transplanted with GW17 (replicate 2). e, Schematic showing the differences in the developmental potential of human and mouse cortical progenitors. OB, olfactory bulb.
Discussion
By carrying out high-throughput lineage tracing of approximately 1,900 human cortical progenitors, we have been able to demonstrate a lineage relationship between human excitatory neurons and putative cortical interneurons—two populations that had been widely believed to arise from distinct progenitor populations. Although existing scRNA-seq informatic tools have been used to describe the transcriptional trajectories between progenitors and mature cell types (that is, pseudotime analysis28), extensive analysis of the developing human brain at this time has not uncovered this lineage relationship29, nor would further analysis be likely do so given the degree to which the transcriptional trajectories of these cell types differ. Furthermore, pseudotime analysis itself does not provide direct evidence of clonal relationships, and the lineages of transcriptionally similar progenitors can differ. At present, approaches that retrospectively infer developmental lineage relationships from somatic mutations predicted to have arisen during development30,31,32 lack the resolution to confidently assign daughter cells to individual progenitors, and thus would not have revealed this relationship.
Previous studies suggested that cortical progenitors generate cortical interneurons in primates, but were limited by their use of a small set of putative ‘marker’ genes that do not directly link progenitors to their progeny6,7. In these studies, differences in the expression of GABAergic marker genes between different germinal zones were presumed to persist across differentiation from progenitors to inhibitory neurons. These markers were thus used to retrospectively infer the origin of cortical interneurons. However, this type of approach cannot account for potential changes in gene expression that can occur over development, or for potentially unknown sources of cells expressing the same putative ‘marker’ genes. Additionally, although one study identified cortically born interneurons using a gammaretorviral labelling approach7, their approach did not determine whether these cells were cortical interneurons or olfactory bulb interneurons, which our work and others have found to be produced from progenitors in the cortex11,12,13. Our prospective approach of clonal labelling by STICR combined with scRNA-seq-based transcriptome-wide analysis has allowed us to distinguish between these two types of interneurons, and we have found, surprisingly, that both types can be clonally related to excitatory neurons (Figs. 2–4). As with any prospective labelling study carried out in humans, we cannot definitively rule out the possibility that our methodologies influenced progenitor behaviour. However, the observation of clonally related excitatory and inhibitory neurons in all of our experimental contexts provides strong evidence that human cortical progenitors have the capacity to generate both types of neuron.
In this study, we investigated the clonal relationships of cells born from GW15–GW18 human cortical progenitors over a six-week developmental window. A previous study in which newborn neurons were labelled with 5-bromo-2′-deoxyuridine (BrdU)8 quantified BrdU+;DLX2+ cells in organotypic slice culture derived from GW17.5–GW20.5 human cortex. After roughly eight to ten days of BrdU labelling, that work8 did not find a substantial amount of BrdU+;DLX2+ cells from cortical progenitors. Given the longer window of our experiments, these data suggest that the production of interneurons by cortical progenitors does not occur throughout the entire duration of corticogenesis, but instead begins at some point after midgestation. Consistent with this interpretation, six weeks after labelling we observed a higher proportion of inhibitory neurons within clones derived from GW18 progenitors than within those from GW15 progenitors (Supplementary Table 1). Furthermore, extensive chains of migrating interneurons have been observed in the perinatal human cortex33. Thus, production of cortical interneurons from cortical progenitors may extend beyond the period in which excitatory neurons are born. It is also possible that the different experimental systems used in these studies (that is, in vitro cell culture and xenograft here, and ex vivo organotypic slice culture in ref. 8) might contribute to some of the observed incongruencies between these three systems. Future studies aimed at detailing the output of cortical progenitors over a broader developmental period and throughout different cortical regions will help to further elucidate the contribution of this phenomenon to human brain development.
In a companion study34, STICR was used to perform in vivo clonal labelling of embryonic mouse forebrain progenitors, and STICR-labelled cells were analysed postnatally by scRNA-seq. Although both glutamatergic excitatory neurons and cortical GABAergic inhibitory neurons were recovered, they did not occur within the same clone. The lineage relationship that we observe between cortical excitatory and inhibitory neurons herein thus raises new questions regarding the development of the human cerebral cortex. First, what are the implications of a single progenitor producing both excitatory neurons and cortical interneurons? Evolutionary expansion of the primate neocortex has been attributed to the increased proliferative capacity of cortical neural progenitors. Adaptations in cortical progenitor competence to produce both principal types of cortical neuron could help to ensure the appropriate inhibitory/excitatory balance, despite the dramatic increase in the pool of cortical excitatory neurons35. Recent studies have revealed that although the inhibitory/excitatory balance increases from mice to humans, the relative composition of cortical interneuron types remains relatively constant across evolution. Although existing technical limitations prevent us from confidently estimating the precise cellular contributions of cortical progenitors to the mature human brain, future studies that quantify the relative contributions of progenitors from the cortex and ganglionic eminences will be helpful in understanding the cellular basis of how normal human cortical function is achieved.
Our study opens many avenues for future investigation. The molecular mechanisms that regulate the production of locally born cortical inhibitory neurons are unknown at present. In mice, signalling through the Sonic hedgehog pathway36 is required for individual cortical progenitors to undergo a GABAergic ‘switch’ and to generate inhibitory neurons37 that migrate to the olfactory bulb13. Does a similar molecular mechanism govern the production of cortically-derived cortical interneurons in humans? Furthermore, what molecular markers, if any, can distinguish them from cortical interneurons born in the CGE or MGE? Previous studies have found that NR2F1 and NR2F2 are expressed not only in the CGE but also in cortical progenitors9,10. Given the transcriptional similarity of cortically-derived IN.3 cells to CGE-derived interneurons observed here, it is possible that a similar developmental program is used. These are just some of the questions raised by our new understanding of the human cortical lineage, and addressing these will help to further decipher the origins and mechanisms underlying human brain development.
Methods
STICR barcode design
STICR barcode fragment sequences were generated using the Barcode Generator script written by L. Comai and T. Howell (http://comailab.genomecenter.ucdavis.edu/index.php/Barcode_generator), with a sequence length of 15 base pairs (bp) and a minimum hamming distance of 5. Sequences containing the restriction-enzyme sites matching STICR’s multicloning site (MCS) or homopolymer repeats longer than 4 bp were excluded. In total, 3 non-overlapping sets of 500 sequences meeting these design criteria were selected (Supplementary Table 3).
STICR library creation
STICR barcode libraries were created using a modified pSico lentiviral plasmid (Addgene, catalogue number 11578) in which the sequence between the central polypurine tract (cPPT)/central termination sequence (CTS) and 3′ long terminal repeat (LTR) was replaced with a DNA fragment containing the hybrid cytomegalovirus/chicken β-actin promoter (CAG) promoter, EGFP transgene, WPRE, multiple cloning site (MCS), and bGH polyadenylation signal. The MCS consisted of three adjacent pairs of restriction-enzyme sites (EcoRI–BamHI–NheI–XhoI) between which the STICR barcode fragments would be added. Each double-stranded DNA (dsDNA) STICR barcode fragment was synthesized as a pair of single-stranded DNA (ssDNA) oligonucleotides (Genewiz), annealed, and then pooled together with all the other barcode fragments belonging to its set. Each set of barcode fragments were added into the STICR MCS individually over three rounds of restriction-enzyme cloning. After each round of barcode fragment addition, a ‘negative selection’ digestion was performed using a restriction enzyme that targeted the stuffer sequence in the MCS that should have been replaced with a barcode fragment, so that undigested/barcoded molecules were removed from the library. Following each ‘negative selection’, the resulting STICR barcode plasmid library was amplified by transformation into MegaX DH10B electrocompetent Escherichia coli (Thermo, C640003) and grown overnight on LB agar plates at 37 °C. In order to maintain even sequence distribution, transformed bacteria were plated at high density (roughly 75 million colonies per large-format plate), which helped to restrict colony size and make them grow more uniformly. In order to maximize the diversity of barcode sequences, we transformed enough barcoded plasmid to get at least ten times as many colonies as there were potential barcode sequences. Additionally, the STICR plasmid contained a sequencing primer site upstream of the STICR barcode. Each STICR library contained a 3-bp viral ‘index’ immediately downstream of the sequencing primer binding site. The viral index is unique to each library and allowed us to differentiate STICR barcodes from different libraries.
Generation of STICR NGS libraries for modelling
To generate a STICR plasmid library for next-generation sequencing (NGS), we first digested 1 μg of STICR plasmid library with XhoI and then ligated a polymerase chain reaction (PCR) adaptor containing a unique molecular identifier (UMI) to this site (Supplementary Table 4). Ligation products were amplified by PCR using a Q5 Hot Start high fidelity 2× master mix (NEB, catalogue number M0494) using primers targeted to the STICR sequencing primer site and the adaptor sequence (Supplementary Table 4), using the following program: 1, 98 °C, 30 s; 2, 98 °C, 10 s; 3, 62 °C, 10 s; 4, 72 °C, 10 s; 5, repeat steps 2–4 15 times; 6, 72 °C, 2 min; 7, 4 °C, hold. Following PCR amplification, a 0.8–0.6× dual-sided size selection was performed using Ampure XP beads (Beckman Coulter, catalogue number A63881). The resulting libraries were sequenced to the depth of roughly 30 million reads.
To generate a STICR lentiviral library for NGS, we carried out an RNA extraction from 1/20 of the total lentiviral prep using 300 μl Trizol (Thermo Fisher, catalogue number 15596026). After incubating for 5 min, 60 μl chloroform was added, incubated for 3 min, then centrifuged at 12,000g at 4 °C for 15 min. The aqueous phase was extracted and mixed with an equal volume of 100% ethanol, then loaded onto a Zymo Direct-zol RNA Microprep (Zymo, R2061) column. The Zymo protocol was followed from there to bind and rinse the RNA, which was eluted in a final volume of 7 μl. Complementary DNA (cDNA) was generated from 5 μg of template RNA with the SuperScript IV kit (Thermo Fisher, catalogue number 18090010) using 1 μl of 2 μM STICR viral library reverse transcription (RT) primer (Supplementary Table 4). To add a UMI and a primer-binding handle to the individual cDNA molecules, one-cycle PCR was performed with cDNA and STICR viral library cDNA UMI primer (Supplementary Table 4) with 25 μl Q5 high fidelity 2× MasterMix (NEB, catalogue number M0492S), 2.5 μl primer (10 μM), 2.5 μL H2O, and 20 μl cDNA with the following program: 1, 98 °C, 40 s; 2, 62 °C, 20 s; 3, 72 °C, 2 min; 4, 4 °C, hold. Primers were removed with a left-sided 0.8× SPRISelect cleanup (Beckman Coulter, catalogue number B23318). Finally, cDNA was amplified using the same methods as for the lentiviral library above, and the library was sequenced to a depth of roughly 100 million reads.
Diversity and collision modelling of STICR libraries
STICR barcode sequences were extracted from fastq files with custom scripts that removed PCR duplicate reads using the UMI (for a general description, see the section ‘scRNA-seq analysis and STICR barcode analysis’ below). As it is prohibitively expensive to sequence high-diversity libraries to saturation, we extrapolated the total number of unique STICR barcodes using the Preseq38 command lc_extrap using default settings. Together with the measured relative barcode abundances, we used the extrapolated STICR barcode library size to model barcode collisions with the R (v4.0.1) programming language. Using base R functions, we simulated the labelling of a starting population of cells with a range of sizes from 101 to 106 and repeated each simulation 20,000 times. We then quantified the mean number of unique barcodes chosen for each starting cell population size. The difference between the starting cell population size and the number of unique barcodes present represented the number of collisions that happened at that population size.
Cell lines
We used NIH/3T3 cells (ATCC) and Lenti-X HEK293T cells (Takara Bio) here; we did not test them for mycoplasma or authenticate them.
Lentivirus production
STICR lentivirus was produced using a third-generation lentivirus packaging system: pMDLg/pRRE (Addgene, catalogue number 12251), pRSV-Rev (Addgene, 12253) and VSVG envelope (Addgene, 12259). Plasmids were transfected into Lenti-X HEK293T cells (Takara Bio, 632180) using jetPRIME (Polyplus, 114-15). In order to improve the viral titre, which is reduced in part because of the reverse orientation of the STICR EGFP-barcode transcript relative to the external lentiviral promoter, we also co-transfected pcDNA3.1 puro Nodamura B2 plasmid (Addgene, 17228) along with the other plasmids. Lenti-X 293T cells were grown and transfected in Dulbecco’s modified Eagle’s medium (DMEM) (Fisher, MT10017CV) supplemented with 10% fetal bovine serum (FBS) (Hyclone, SH30071.03) and 1% penicillin streptomycin (Fisher, 15070063). Twenty-four hours after transfection, media was replaced with Ultraculture media (Lonza, BE12-725F) supplemented with sodium pyruvate (final concentration 0.11 mg ml−1, Sigma P2256-25G) and sodium butyrate (0.005 M final molarity, Sigma B5887-1G) and penicillin streptomycin. Seventy-two hours after transfection, media were collected, passed through a 0.45 μM filter (Corning, 431220), and then ultracentrifuged at 22,000g for 2 h. Pellets were resuspended in 100 μl of sterile phosphate-buffered saline (PBS) (Thermo, 14190250) overnight at 4 °C and then aliquoted and stored at −80 °C.
Tissue procurement and STICR transduction
Deidentified tissue samples were collected with previous patient consent in strict observance of the legal and institutional ethical regulations. Protocols were approved by the Human Gamete, Embryo, and Stem Cell Research Committee (Institutional Review Board) at the University of California, San Francisco. In order to visualize tissue for microdissection, samples were embedded in 3% low-melting-point agarose (Fisher, catalogue number BP165-25) and then cut into 300-μm sections perpendicular to the ventricle on a Leica VT1200S vibrating blade microtome in oxygenated artificial cerebrospinal fluid containing 125 mM NaCl, 2.5 mM KCl, 1 mM MgCl2, 1 mM CaCl2 and 1.25 mM NaH2PO4. The germinal zone was then isolated by microdissection using a scalpel and fine forceps under a Leica MZ10F dissecting microscope. In order to dissociate cells into a single-cell suspension, microdissected tissue was incubated in 200 μl of 0.25% trypsin (Fisher, catalogue number 25200056) and 2,000 units ml−1 of DNase I (NEB, M0303) for 20 min at 37 °C and then gently mechanically triturated with a 1,000 μl pipetteman 10 times. We then added 800 μl of DMEM supplemented with 10% FBS to the sample to neutralize the papain, and removed the trypsin/DNase/FBS solution by centrifuging the sample for 5 min at 300g. The sample was rinsed in Dulbecco’s PBS (Thermo, 14190250) and then centrifuged again for 5 min at 300g to remove the rinse.
Cells were then immediately resuspended in NES media (defined in ref. 39), supplemented with ROCK inhibitor (Tocris, 1254) at a final concentration of 10 μM to reduce cell death and 1% penicillin streptomycin (Fisher, 15070063), and then plated on a 24-well tissue culture dish coated with 0.01% poly-l-ornithine (Sigma, P4957), 5 μg ml−1 laminin (Invitrogen, 23017-015) and μg ml−1 fibronectin (Corning, 354008) at a density of 500,000 cells per well. STICR lenitivirus was added to culture media at a roughly 1:250 to 1:500 dilution so that approximately 30% of cells were infected. After 24 h, virus-containing medium was removed and replaced with fresh medium. Seventy-two hours after infection, cultures were dissociated using papain (Worthington, LK003163), and EGFP+ cells were isolated by FACS and then used in a barnyard assay or in vitro culture assay. In order to maximize cell viability in the in vivo mouse transplantation assay, EGFP+ cells were not FACS-isolated before transplantation, but instead the entire culture (containing both EGFP− and EGFP+ cells) was used. See below for descriptions of each assay. In an effort to label subclones, the GW15 replicate-2 in vitro sample was initially infected with a STICR viral library derived from a fully barcoded STICR plasmid encoding a GFP-T2a-TVA transgene and a viral index ‘E’ and then plated on mouse astrocytes as above. This culture was infected two and four weeks later with EnvA-pseudotyped (Addgene, 74420) STICR libraries with viral index ‘1’ (two weeks) and ‘3’ (four weeks) with a 1:100 dilution of virus:medium. During analysis, we found that labelling with EnvA-pseudotyped libraries was poor, so STICR barcodes with a ‘1’ or ‘3’ index were not considered during analysis. Therefore, clonal analysis of GW15 replicate 2 was carried out using only the initial ‘E’ index STICR barcodes transduced six weeks before analysis, as for all the other in vitro libraries.
Barnyard experiment
To confirm that transcribed STICR barcodes can be accurately recovered using scRNA-seq, we carried out a ‘barnyard experiment’ in which we infected separate cultures of human cortical cells (GW18 sample) and mouse 3T3 cells (ATCC) with different STICR libraries. These libraries could be distinguished from each other by a constant sequence unique to each library (‘viral index’) (Fig. 1a). After three days, we dissociated cultures with papain and FACS-isolated EGFP+ cells. EGFP+ cells from both species were then mixed together and processed using a Chromium Single Cell 3ʹ GEM, Library & Gel Bead Kit v3 (10X Genomics, PN-1000075). Following sequencing (see below), transcript libraries were aligned with Cell Ranger (version 3.0.2) to a hybrid mouse/human genome and droplets were determined to be either a mouse cell, a human cell, or a multiplet. Quantification of recovered STICR viral index (see below) for mouse, human and multiplate droplets is included in Extended Data Fig. 1f.
In vitro culture assay
Long-term in vitro experiments were performed using an astrocyte co-culture system. Primary mouse cortical astrocytes were isolated from CD-1 mice at postnatal day 1 and cultured as described40, but with additional subdissection to remove the subventricular zone surrounding the lateral ventricles. Astrocytes were plated at a density of 400,000 cells per 3.5 cm2 in 12-well cell-culture plates containing DMEM (Fisher, MT10017CV) supplemented with 10% FBS (Hyclone, SH30071.03) and 1% penicillin streptomycin (Fisher, 15070063) 3–5 days before the addition of human cells.
For each experiment, roughly 1,000–2,000 STICR-labelled (EGFP+) cells were added to a 12-well plate already containing mouse cortical astrocytes and cultured in DMEM, 1% B-27 supplement (Invitrogen, 12587-010), 1% N-2 supplement (Invitrogen, 17501-048) and 1% penicillin streptomycin. Cell culture medium was half-changed every three to four days for six weeks. Cultures were then dissociated into single-cell solution using papain, and EGFP+ cells were isolated by FACS. Following FACS isolation, EGFP+ cells were concentrated by centrifugation (300g for 10 min), and prepared for scRNA-seq.
PTPRZ1-based FACS
For enrichment of progenitors before xenograft transplantation, we used an adaptation of the protocol in ref. 41 in order to isolate cells that express the cell-surface protein PTPRZ1, which is enriched on the surface of cortical progenitors15,42. Cortical tissue was dissociated to a single-cell suspension as above, then resuspended in a solution of 1% bovine serum albumin (BSA) (Sigma Aldrich, A7979-50ML) and 0.1% glucose in Hanks’ balanced salt solution (HBSS) (Life Technologies, 14175-095) for staining. Cells were incubated with mouse anti-PTPRZ1 primary antibody (Santa Cruz Biotechnology, sc-33664) at 1:50 dilution for 20 min on ice, washed with HBSS/BSA/glucose, incubated in goat anti-mouse IgM 488 secondary antibody (Thermo Fisher Scientific, A-21042) at 1:500 dilution for 20 min on ice, washed, and resuspended in HBSS/BSA/glucose. PTPRZ1+ cells were then isolated via FACS, plated, labelled with STICR, and cultured as above. Representative FACS plots are shown in Extended Data Fig. 9a.
Xenograft transplantation assay
Mouse transplantation assays were performed in CB17.Cg-PrkdcscidLystbg-J/Crl mice (Envigo) at postnatal day 3–5. STICR-labelled cultures were dissociated with papain, centrifuged at 300g for 5 min, rinsed once with DPBS, and then resuspended in ice-cold L15 media (Fisher, 11-415-064) with 180 Kunitz per millilitre of DNase (Fisher, 50-100-3290). Following anaesthetization, 100 nl of cell mixture (roughly 40,000 to 80,000 cells) was injected through a bevelled glass needle using a stereotactic rig into L −1, A 2.5, D −0.8 mm from lamda. At least five mice were injected with cells from each human specimen. Both male and female mice were used. In order to minimize clumping of xenograft cells, 20 mM EGTA (Sigma, E4378) was added to the cell mixture for GW17 replicate-1 and replicate-2 samples as well as GW15 replicate 3.
After 6 weeks, mice were euthanized and one brain from each set of transplantations was cut into 1-mm coronal sections using a brain mould (Stoelting, 51386). EGFP+ regions of the cortex were dissected from slices using a fluorescent dissecting scope and then dissociated into a single-cell solution using papain. EGFP+ cells were then isolated by FACS, concentrated by centrifugation (300g for 10 min), and prepared for scRNA-seq. Mice were housed in a barrier facility with a 12-h light/12-h dark cycle and temperature and humidity control (70 °F, 50% rack humidity). All protocols and procedures followed the guidelines of the Laboratory Animal Resource Center at the University of California, San Francisco, and were conducted with approval of the Institutional Animal Care and Use Committee (IACUC).
IHC of in vitro cultures and xenografts
At the experimental endpoint, transcardiac perfusion of sterile PBS followed by 4% PFA (Fisher, 50-980-487) was used to rinse and then fix the specimens. Brains were dissected out and drop-fixed overnight in 4% PFA at 4 °C. Brains used for cryosections (Fig. 3 and Extended Data Fig. 7) were then cryopreserved in a 1:1 solution of OCT (VWR, 25608-930) and 30% sucrose, embedded in cryomolds containing the same solution, frozen on dry ice, and stored at −80 °C. Brains were then cryosectioned at 12 μm onto glass slides and stored at −80 °C. Blocking and permeabilization were performed using a blocking solution consisting of 10% normal donkey serum, 1% Triton X-100 and 0.2% gelatin in PBS for 1 h. Primary and secondary antibodies were diluted and incubated in this same blocking solution. Cryosections were incubated with primary antibodies at 4 °C overnight, washed 3 × 10 min with washing buffer (0.1% Triton X-100 in PBS), incubated with secondary antibodies for 2 h at room temperature, and washed 3 × 10 min with washing buffer; coverslips (Azer Scientific, 1152460) were then mounted using Prolong Gold Antifade reagent (Invitrogen, P36930).
Brains used for morphological analysis of GABAergic cells (Fig. 4 and Extended Data Figs. 9, 10) were fixed as above but stored in PBS at 4 °C. Brains were then sectioned on a Leica VT1000 S vibrating blade microtome to 40 μm and slices were stored in PBS. Slices were incubated in blocking solution composed of 10% normal donkey serum and 0.1% Triton-X in PBS at room temperature for 2 h and then incubated in primary antibodies diluted in blocking solution overnight at 4 °C, washed 5 × 30 min in 0.1% PBST, incubated in secondary antibodies in blocking solution overnight at 4 °C, washed 5 × 30 min in 0.1% PBST, mounted on glass slides, and coverslipped as above using ProLong Gold Antifade reagent.
In vitro cultures comprising human cortical cells co-cultured with mouse astrocytes were prepared as described above, but without FACS isolation of EGFP+ STICR-labelled cells, and plated on eight-well chamber slides (Thermo Scientific, 154534PK). After 6 weeks, cultures were fixed with 4% PFA for 1 h at 4 °C. Cultures were then washed three times and stored in PBS. Immunohistochemistry was performed as described for the 40-μm mouse brain sections but with 10-min washes. Slides were coverslipped with ProLong Gold Antifade reagent.
The antibodies used include: chicken anti-GFP (Aves, GFP-1020; 1:1,000), mouse anti-human nuclear antigen (Novus, NBP2-34342; 1:100), rabbit anti-GABA (Millipore Sigma, A2052-100ul; 1:250), rabbit anti-NEUROD2 (Abcam, ab104430; 1:500), guinea pig anti-DCS (Millipore Sigma, AB2253; 1:200), rabbit anti-GFAP (abcam, ab7260; 1:1,500), rabbit anti-SOX9 (Abcam, ab104430; 1:250) and mouse anti-OLIG2 (Millipore Sigma, MABN50; 1:200). Secondary antibodies used include AlexaFluor anti-chicken 488 (Jackson Immunoresearch 703-545-155; 1:500), anti-mouse 488 (ThermoFisher A-21042; 1:500), anti-rabbit 594 (ThermoFisher A-21207; 1:500), anti-guinea pig 647 (Jackson Immunoresearch 706-605-148; 1:500), anti-mouse IgG1 488 (ThermoFisher A-21121; 1:500), and anti-mouse IgG2a 647 (ThermoFisher A-21241; 1:500).
Confocal imaging was performed using a Leica SP8 confocal microscope with either a 10× or a 20× air objective; a 2-μm optical z-step was used for all images. Images were processed using ImageJ/Fiji. For quantification of the major cell types in xenografted mice (Fig. 3 and Extended Data Fig. 7), tilescans of the transplanted region were z-projected with average intensity; channel intensity was normalized across images; and cells expressing EGFP and/or human nuclear antigen in addition to cell-type markers were counted manually using the CellCounter plugin for ImageJ/Fiji.
GABA+ STICR-labelled cells in the cortex of host mice (Fig. 4) were quantified by imaging four 40-μm-thick brain slices derived from within 400 μm of the transplantation site of each animal. Two mice for each of the two GW17 specimens were analysed in total. GFP+ cells in the cortex were counted using the CellCounter plugin for ImageJ/Fiji. The relative laminar positions of GABA+/GFP+ double-positive cells were measured using the Measure tool from ImageJ/Fiji to draw a line from the top of the corpus callosum straight up to the pial surface through the soma of each cell. The relative location of the cell’s soma to the top of the corpus callosum was then divided by the total length of the line drawn from the corpus callosum to pial surface.
scRNA-seq library preparation
Preparation of scRNA-seq libraries was carried out using a Chromium Single Cell 3ʹ GEM, Library & Gel Bead Kit v3 (10x Genomics, #PN-1000075). Libraries were prepared according to the manufacturer’s protocol.
STICR barcode library recovery
STICR barcodes were subamplified from each 10X cDNA library using the Q5 Hot Start High Fidelity 2× master mix (NEB, M0494). In brief, 10 μl of cDNA was used as template in a 50 μl PCR reaction containing STICR barcode read 1 and 2 primers (0.5 μM, each) targeting the region immediately upstream of the STICR viral index/barcode as well as the partial Illumina Read1 sequence added during cDNA library preparation (Supplementary Table 4), using the following program: 1, 98 °C, 30 s; 2, 98 °C, 10 s; 3, 62 °C, 20 s; 4, 72 °C, 10 s; 5, repeat steps 2–4 11 times; 6, 72 °C, 2 min; 7, 4 °C, hold. Following PCR amplification, a 0.8–0.6 dual-sided size selection was performed using Ampure XP beads (Beckman Coulter, #A63881).
Sequencing
10X transcriptomic libraries and STICR barcode libraries were sequenced using Illumina Novaseq 6000 or Illumina HIseq 4000 machines. 10x transcriptomic libraries were sequenced to the average depth of approximately 50,000 reads per cell (2,200 genes per cell). STICR barcode libraries from lineage-tracing experiments were sequenced to a depth of roughly 30 million reads per library. STICR plasmid libraries for diversity and collision modelling were sequenced to a depth of approximately 30 million reads per library, while a single STICR lentiviral library was sequenced to a depth of approximately 100 million reads.
scRNA-seq analysis and STICR barcode analysis
10X transcriptomic libraries were aligned to the hg38 genome using CellRanger (version 3.0.2). Aligned cell/transcript counts were processed by Seurat43 (version 3.2.0.9014 for initial in vitro cultures and version 4.0 for subsequent xenografted cultures and integration of xenograft data into in vitro data) to remove cells containing fewer than 1,000 genes, fewer than 1,250 transcripts, or a high abundance of mitochondrial reads (greater than 7% of total transcripts). Cells passing these thresholds were then processed with Cellbender44(version 0.1) in order to identify and remove background reads and instances of barcode swapping. We then identified and remove multiplets that arose during cell capture using Solo45(version 0.1). Additionally, xenograft libraries were aligned to a chimaeric hg38/mm10 genome using CellRanger (version 3.0.2) in order to identify potential cross-species multiplets; cells identified as such were removed from analysis. Libraries were integrated using Seurat’s SCTransform and FindIntegrationAnchors functions, to identify integration features. First, in vitro cultured STICR experiments were integrated together. Subsequently, transcriptomic libraries form xenograft experiments were integrated with in vitro cells using integration anchors identified from in vitro cultures. Leiden cell clustering, pseudotime analysis and data visualizations (that is, creation of UMAP images) were performed using Monocle346 (version 0.2.1.9). Pseudotime analysis of cells of the inhibitory-neuron trajectory was performed by setting the root node within mitotic inhibitory neuron clusters. IN.1, IN.2 and IN.3 trajectories were defined as major branches of the principal graph that led to distinct sets of clusters. Within subclustered inhibitory neurons, cluster 17 appeared to be at the beginning of both IN.2 and IN.3 trajectory cells in pseudotime analysis and was thus termed ‘IN.early’.
Differential gene expression analysis was conducted using Seurat FindMarkers/FindAllMarkers functions. With the exception of cluster 34 from the in vitro culture data (Fig. 1c), we identified marker-gene expression consistent with previously described cell types. Cluster 34 expressed markers of multiple cell types and did not show strong cluster correlation with reference data (Extended Data Fig. 2d). Thus, we refer to cells in this cluster as ‘Unknown’ in Supplementary Table 1.
Iterative subclustering of excitatory neuron and EOMES+ IPC trajectory cells from in vitro STICR cultures revealed ‘upper-like’ (subclusters 3, 4, 7, 9), ‘deep-like’ (subclusters 1, 5), ‘newborn excitatory neurons’ (subcluster 6), ‘EOMES+ IPCs’ (subcluster 2), and ‘EN.other’ (subcluster 8) subgroups based on gene expression. The EN.other group did not show a strong correlation with excitatory neurons, and thus cells in this category were removed from further clonal analysis. Within xenografted excitatory-neuron and EOMES+ IPC trajectory cells, iterative subclustering revealed ‘excitatory neuron’ and ‘EOMES.IPC’ subclusters.
STICR barcode analysis was performed using custom scripts. First, BBMap (BBMap, Bushnell B; sourceforge.net/projects/bbmap/) was used to remove low-quality reads and then extract reads containing STICR barcode sequences. Then, BBMap was used to extract individual STICR barcode fragments that were then aligned to our predefined fragment reference sets using Bowtie47 (version 5.2.1), allowing for up to two mismatches per fragment. Aligned STICR barcodes were compiled into a file containing their corresponding 10x cell barcode and 10x UMI sequences using Awk. Finally, UMI-tools48 (version 0.5.1) was used to remove duplicate STICR barcode/cell barcode (CBC) reads by UMI, allowing for 1-bp mismatches in the UMI. STICR barcodes/CBC) pairings with at least five distinct UMIs were retained. Cells with a single STICR barcode meeting this criterion were retained for clonal analysis. Possible instances of STICR barcode superinfection (multiple STICR barcodes per starting progenitor) were identified by calculating jaccard similarity indexes of all STICR barcodes pairings found to co-occur within a single cell. Those pairings with a jaccard similarity index of 0.55 or greater that occurred in 10 cells or more were considered to be a valid superinfection clone and retained for clonal analysis. Cells that contained multiple STICR barcodes that were not determined to be valid superinfections were further analysed for the relative abundance of individual STICR barcodes. Cells that contained a ‘dominant’ STICR barcode with five or more times the number of barcode counts compared with the next most abundant STICR barcode, as determined by UMI, were retained and assigned that dominant barcode. Those cells that did not contain barcodes meeting these criteria were not considered for clonal analysis.
Validation of marker genes with Allen Brainspan data
Expression data (Z-score) for IN.2 and IN.3 marker genes were downloaded from the Allen Brainspan database (https://www.brainspan.org). Biological samples from the cortex, basal ganglia and rostral migratory stream/olfactory bulb were retained for further analysis. Hierarchical clustering was performed using the R package pheatmap(v.1.0.12), with the ward.D clustering method. For visualization purposes, the data ranges were divided into 20 quantiles.
Transcriptional cluster correlation
Marker genes from each cluster in the Nowakowski primary human brain reference atlas14 were calculated using Seurat FindAllMarkers, restricting genes to those present in at least 25% of cells of that cluster. The top 100 marker genes by fold-expression for each cluster were then retained for further analysis. The average expression for each cluster’s top marker genes was calculated for each cluster in the Nowakowski reference atlas as well as in each cluster within the STICR transcriptional datasets. We then calculated pairwise Pearson correlations between every reference and STICR cluster, and depicted the result using a heatmap.
Statistics and reproducibility
Images shown in figures are representative of results found in multiple replicates: Fig. 3b (two specimens each xenografted into three mice), Fig. 4b, c (two specimens each xenografted into two mice), Extended Data Fig. 4d, e (four specimens), and Extended Data Figs. 9, 10 (two specimens each xenografted into two mice).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
scRNA-seq transcriptomic data and STICR barcode data are available at the database of Genotypes and Phenotypes (dbGAP; https://www.ncbi.nlm.nih.gov/gap/) under accession number phs002624.v1.p1; and at the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE187875. An interactive browser of single-cell data can be found at the University of California, Santa Cruz (UCSC) cell browser49( https://human-cortical-lineage.cells.ucsc.edu). Publicly available reference genomes hg38 and mm10 were used for analysis. Source data are provided with this paper.
Code availability
Custom codes used in this study are available at the following GitHub repository: https://github.com/NOW-Lab/STICR.
References
Anderson, S. A., Eisenstat, D. D., Shi, L. & Rubenstein, J. L. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science 278, 474–476 (1997).
Sussel, L., Marin, O., Kimura, S. & Rubenstein, J. L. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development 126, 3359–3370 (1999).
Gorski, J. A. et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 22, 6309–6314 (2002).
Xu, Q., Tam, M. & Anderson, S. A. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J. Comp. Neurol. 506, 16–29 (2008).
Anderson, S. A., Marín, O., Horn, C., Jennings, K. & Rubenstein, J. L. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development 128, 353–363 (2001).
Petanjek, Z., Berger, B. & Esclapez, M. Origins of cortical GABAergic neurons in the cynomolgus monkey. Cereb. Cortex 19, 249–262 (2009).
Letinic, K., Zoncu, R. & Rakic, P. Origin of GABAergic neurons in the human neocortex. Nature 417, 645–649 (2002).
Hansen, D. V. et al. Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat. Neurosci. 16, 1576–1587 (2013).
Alzu’bi, A. et al. Distinct cortical and sub-cortical neurogenic domains for GABAergic interneuron precursor transcription factors NKX2.1, OLIG2 and COUP-TFII in early fetal human telencephalon. Brain Struct. Funct. 222, 2309–2328 (2017).
Alzu’bi, A. et al. The transcription factors COUP-TFI and COUP-TFII have distinct roles in arealisation and GABAergic interneuron specification in the early human fetal telencephalon. Cereb. Cortex 27, 4971–4987 (2017).
Kohwi, M. et al. A subpopulation of olfactory bulb GABAergic interneurons is derived from Emx1- and Dlx5/6-expressing progenitors. J. Neurosci. 27, 6878–6891 (2007).
Young, K. M., Fogarty, M., Kessaris, N. & Richardson, W. D. Subventricular zone stem cells are heterogeneous with respect to their embryonic origins and neurogenic fates in the adult olfactory bulb. J. Neurosci. 27, 8286–8296 (2007).
Fuentealba, L. C. et al. Embryonic origin of postnatal neural stem cells. Cell 161, 1644–1655 (2015).
Nowakowski, T. J. et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323 (2017).
Pollen, A. A. et al. Molecular identity of human outer radial glia during cortical development. Cell 163, 55–67 (2015).
Bystron, I., Blakemore, C. & Rakic, P. Development of the human cerebral cortex: Boulder Committee revisited. Nat. Rev. Neurosci. 9, 110–122 (2008).
Bandler, R. C., Mayer, C. & Fishell, G. Cortical interneuron specification: the juncture of genes, time and geometry. Curr. Opin. Neurobiol. 42, 17–24 (2017).
Mayer, C. et al. Developmental diversification of cortical inhibitory interneurons. Nature 555, 457–462 (2018).
Li, J. et al. Transcription factors Sp8 and Sp9 coordinately regulate olfactory bulb interneuron development. Cereb. Cortex 28, 3278–3294 (2018).
Guo, T. et al. Dlx1/2 are central and essential components in the transcriptional code for generating olfactory bulb interneurons. Cereb. Cortex 29, 4831–4849 (2019).
Batista-Brito, R. et al. The cell-intrinsic requirement of Sox6 for cortical interneuron development. Neuron 63, 466–481 (2009).
Stumm, R. K. et al. CXCR4 regulates interneuron migration in the developing neocortex. J. Neurosci. 23, 5123–5130 (2003).
López-Bendito, G. et al. Chemokine signaling controls intracortical migration and final distribution of GABAergic interneurons. J. Neurosci. 28, 1613–1624 (2008).
Tripodi, M., Filosa, A., Armentano, M. & Studer, M. The COUP-TF nuclear receptors regulate cell migration in the mammalian basal forebrain. Development 131, 6119–6129 (2004).
Azim, E., Jabaudon, D., Fame, R. M. & Macklis, J. D. SOX6 controls dorsal progenitor identity and interneuron diversity during neocortical development. Nat. Neurosci. 12, 1238–1247 (2009).
Lein, E. S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007).
Herrera, D. G., Garcia-Verdugo, J. M. & Alvarez-Buylla, A. Adult-derived neural precursors transplanted into multiple regions in the adult brain. Ann. Neurol. 46, 867–877 (1999).
Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017).
Bhaduri, A. et al. An atlas of cortical arealization identifies dynamic molecular signatures. Nature 598, 200–204 (2021).
Lodato, M. A. et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science 350, 94–98 (2015).
Ludwig, L. S. et al. Lineage tracing in humans enabled by mitochondrial mutations and single-cell genomics. Cell 176, 1325–1339.e22 (2019).
Lareau, C. A. et al. Massively parallel single-cell mitochondrial DNA genotyping and chromatin profiling. Nat. Biotechnol. 39, 451–461 (2021).
Paredes, M. F. et al. Extensive migration of young neurons into the infant human frontal lobe. Science 354, aaf7073 (2016).
Bandler, R. C. et al. Single-cell delineation of lineage and genetic identity in the mouse brain. Nature https://doi.org/10.1038/s41586-021-04237-0 (2021).
Rakic, P. Evolution of the neocortex: a perspective from developmental biology. Nat. Rev. Neurosci. 10, 724–735 (2009).
Zhang, Y. et al. Cortical neural stem cell lineage progression is regulated by extrinsic signaling molecule Sonic hedgehog. Cell Rep. 30, 4490–4504.e4 (2020).
Cai, Y., Zhang, Y., Shen, Q., Rubenstein, J. L. & Yang, Z. A subpopulation of individual neural progenitors in the mammalian dorsal pallium generates both projection neurons and interneurons in vitro. Stem Cells 31, 1193–1201 (2013).
Daley, T. & Smith, A. D. Modeling genome coverage in single-cell sequencing. Bioinformatics 30, 3159–3165 (2014).
Onorati, M. et al. Zika virus disrupts phospho-TBK1 localization and mitosis in human neuroepithelial stem cells and radial blia. Cell Rep. 16, 2576–2592 (2016).
Schildge, S., Bohrer, C., Beck, K. & Schachtrup, C. Isolation and culture of mouse cortical astrocytes. J. Vis. Exp. 71, 50079 (2013).
Crouch, E. E. & Doetsch, F. FACS isolation of endothelial cells and pericytes from mouse brain microregions. Nat. Protocols 13, 738–751 (2018).
Bhaduri, A. et al. Outer radial blia-like cancer stem cells contribute to heterogeneity of glioblastoma. Cell Stem Cell 26, 48–63.e6 (2020).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e1821 (2019).
Fleming, S. J., Marioni, J. C. & Babadi, M. CellBender remove-background: a deep generative model for unsupervised removal of background noise from scRNA-seq datasets. bioRxiv https://doi.org/10.1101/791699 (2019).
Bernstein, N. J. et al. Solo: doublet identification in single-cell RNA-seq via semi-supervised deep learning. Cell Syst. 11, 95–101.e5 (2020).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Smith, T., Heger, A. & Sudbery, I. UMI-tools: modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res. 27, 491–499 (2017).
Speir, M. L. et al. UCSC cell browser: visualize your single-cell data. Bioinformatics https://doi.org/10.1093/bioinformatics/btab503 (2021).
Acknowledgements
We thank A. Bhaduri for discussions regarding scRNA-seq analysis; B. Rabe and C. Cepko for discussions regarding viral vectors and sharing of reagents; C. Cadwell and M. Paredes for discussions regarding interneuron morphology; J. Rubenstein and R. Andersen for reading the manuscript; and M. Speir and B. Wick for data wrangling at the UCSC single-cell browser. This study was supported by the Psychiatric Cell Map Initiative Convergence Neuroscience award U01MH115747; an Innovation Award from the Broad Foundation (to T.J.N.); a New Frontiers Research Award from the Sandler Program for Breakthrough Biomedical Research (PBBR) (to T.J.N.); a National Science Foundation (NSF) Graduate Research Fellowship Program (GRFP) award (to D.E.A.); an Autism Speaks Predoctoral Fellowship (11874 to R.S.Z.); NIH K08 NS116161 and UCSF Physician Scientist Scholars Program to EEC; and gifts from Schmidt Futures and the William K. Bowes Jr Foundation (to T.J.N.). Work in the Alvarez-Buylla laboratory is supported by National Institutes of Health (NIH) grants R01NS028478 and R01EY025174, and a gift from the John G. Bowes Research Fund. A.A.-B. is the Heather and Melanie Muss Endowed Chair and Professor of Neurological Surgery at UCSF.
Author information
Authors and Affiliations
Contributions
R.N.D conceived of the project, designed and generated the STICR barcode library, designed and conducted experiments, analysed the data, and wrote the manuscript. D.E.A. helped to design experiments, conducted experiments, analysed data, and helped to write the manuscript. M.G.K helped to design experiments, conducted experiments and helped to write the manuscript. W.R.M.L carried out xenograft transplantations. R.S.Z. helped to construct the STICR library. E.E.C. carried out PTPRZ1 FACS. A.A.-B. helped to supervise the research. T.J.N conceived of the project, helped to design experiments, assisted in the interpretation of data, and helped to write the manuscript.
Corresponding authors
Ethics declarations
Competing interests
A.A.-B. is co-founder and on the Scientific Advisory Board of Neurona Therapeutics.
Additional information
Peer review information Nature thanks Zoltan Molnar and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Validation of the STICR barcode design.
a, Histogram showing pairwise hamming distances between every sequence in each STICR fragment pool. b, Barcode diversity extrapolations derived from sequencing a representative STICR plasmid or lentiviral library. Mean ± 95% confidence range for each library is shown. c, Simulated barcode collision frequencies (mean ± s.d.) for a range of starting cell numbers, based on the barcode diversity estimated in b. Barcode sampling was performed with replacement using measured proportions of barcodes within the representative plasmid and lentiviral libraries depicted in b. Each simulation was performed 20,000 times. Most error bars (depicting standard deviations) are not visible as they are smaller than the dots (depicting mean values). d, The ‘barnyard’ species-mixing experiment. e, Plot depicting species-specific transcript counts from barnyard experiment. Each dot depicts a single cell and the dot colour indicates whether the cell was determined to be a 3T3 cell (mouse), cortex cell (human), or mixed droplet (multiplet). f, Violin plots depicting the number of unique STICR barcode molecules recovered from droplets identified as either mouse, human, or multiplet. ND, not detected.
Extended Data Fig. 2 Cluster analysis of in vitro STICR data sets.
a, UMAP plots of each individual biological sample highlighted. b, Top marker-gene expression for each cluster. The size of each dot corresponds to the proportion of cells in the cluster that express the gene, while the colour of the dot corresponds to the average expression level per cluster. c, d, Heatmap depicting pairwise transcriptional cluster correlation of in vitro cultured cells with itself (c) and the 2017 Nowakowski scRNA-seq atlas (d)14. The principal cell-type designation is depicted next to each column and row. The dendrogram depicts hierarchical clustering distance.
Extended Data Fig. 3 Transcriptional analysis of in vitro STICR data sets.
a–f, Feature plots depicting expression of genes corresponding to cell cycle (a), glia (b), oligodendroglia (c), excitatory neurons (d), interneurons (e), and regional markers (f). g, Bar plot depicting the proportion of cells within each cluster with a recovered STICR barcode. h, Heatmap depicting the percentage of STICR barcodes shared between biological samples. GW15 (Rep1) and all GW18 samples were labelled with the same viral stock, while GW15 (Rep2) was labelled with a different stock (see Methods). i, Stacked barplot depicting relative proportions of principal cell types within each sample, restricted to cells that are members of multicellular clones.
Extended Data Fig. 4 Clonal analysis of cortical clones containing excitatory neurons.
a, Histogram of excitatory neuron (EN) counts within each multicellular cortical clone. Left, clone sizes from 1–25 cells in single-cell bins. Right, clone sizes of more than 25 cells in the indicated bin sizes. b, Box-and-whisker plot depicting the proportion of EN cells within individual multicellular clones for each biological sample. Maxima and minima of boxes depict third and first quartiles, while box centres depict medians. Whiskers depict 1.5× the interquartile distance. Individual clone values are shown as dots. The number of clones is listed below each sample group. c, Ternary plots depicting the relative proportions of inhibitory neurons, excitatory neurons and all other cell types (‘Other’) within individual clones. d, e, Immunohistochemistry of in vitro cultures derived from GW15 germinal zone cells labelled with STICR. d, Low-magnification image to show distribution; scale bar, 25 μm. e, High-magnification image showing a cluster of ENs; scale bar, 250 μm.
Extended Data Fig. 5 Clonal and transcriptional analysis of inhibitory neurons and DLX2+ IPCs in vitro.
a, UMAP embedding and Leiden subclustering of GABAergic inhibitory neuron (IN) trajectory cells. b, Feature plots depicting expression of MKI67, STMN2, CENPF and ERBB4. c, Heatmap depicting pairwise transcriptional cluster correlation of this data set with itself. d, Stacked barplot depicting relative proportion of multicellular clones from each sample that comprise each IN trajectory. e, Stacked barplot depicting the relative proportions of different IN trajectory cells within multicellular clones of each sample. f, Feature plots depicting MGE-derived cells (red) and expression of NKX2-1, LHX6, ACKR3, MAF and PDE1A. The enlarged insets below show IN.1 trajectory cells. g, Heat plot depicting differential expression of IN.2 and IN.3 marker genes in the developing human cortex, olfactory bulb/rostral migratory stream and basal ganglia. Data are derived from the Allen BrainSpan Laser Capture Microdissection database. Dendrograms reflect hierarchical clustering of genes and samples while colours represent quantile-normalized z-scores. h, Paired violin plots and in situ hybridization (ISH) images of P60 mouse brains from the Allen Brain Atlas for select genes. The log2 fold difference between IN.2 (olfactory-bulb-like) and IN.3 (cortical-interneuron-like) cells is depicted above each violin plot. i, Stacked barplots depicting relative proportions of IN.1, IN.2, IN.3, EN and glia trajectory cells within multicellular clones. The number of clones is listed below each sample. j, Venn diagram depicting the number of EN-containing multicellular cortical clones that also contain IN.2 and/or IN.3 cells.
Extended Data Fig. 6 Clonal and transcriptional analysis of excitatory neurons and EOMES+ IPCs in vitro.
a, UMAP embedding and Leiden subclustering of excitatory neuron (EN) trajectory cells. b, c, Heatmap depicting pairwise transcriptional cluster correlation of subclustered EN trajectory cells with self (b) and with the 2017 Nowakowski developing human brain scRNA-seq atlas (c)14. d, Feature plots depicting the expression of genes corresponding to labelled subclustered EN trajectory subtypes. e, Stacked barplot depicting relative proportions of EN subtypes within EN trajectory cells of multicellular clones. f, Venn diagram depicting the number of multicellular cortical clones containing deep-like ENs, upper-like ENs, and IN.3 cells.
Extended Data Fig. 7 Characterization of human cortical progenitor xenografts at six weeks.
a, b, Representative images of transplanted human cortical cells analysed by IHC for principal cell-type markers six weeks after transplantation. EGFP expression from STICR is in green, with NEUROD2 or GABA expression in red. Scale bars: a, 50 μm; b,10 μm. c, Barplot depicting the proportion (mean ± s.d.) of transplanted cells expressing principal cell-type markers as assessed by IHC. n = 7 sections derived from 6 xenografted mice, 3 of which were transplanted with donor cells from GW15 Rep1 and 3 with cells from GW15 Rep2. d, Top marker-gene expression for each cluster from xenografted cells. Sizes of dots correspond to the proportion of cells in the cluster expressing the gene, while dot colours correspond to the average expression level per cluster. e, UMAP embedding of xenografted cells and feature plots depicting expression of NEUROD2, EOMES, DLX2, MKI67 and GFAP. f, Heatmap depicting pairwise transcriptional cluster correlation of subclustered excitatory neuron (EN) trajectory cells with the 2017 Nowakowski developing human cortex scRNA-seq atlas14. g, Comparison of principal cell-type quantification (mean) in transplanted cells by analysis method (IHC versus scRNA-seq) and biological replicate.
Extended Data Fig. 8 Transcriptional analysis of excitatory and inhibitory neurons from xenografts.
a, UMAP embedding and Leiden subclustering of inhibitory neuron (IN) trajectory cells from xenografts. b, Feature plots depicting expression of CENPF, MKI67, ERBB4, NR2F1, NFIX, SP8, SCGN and KLHL35. c, Heatmap depicting pairwise transcriptional cluster correlation of subclustered xenograft IN and DXL2+ IPC trajectory cells with the 2017 Nowakowski developing human cortex scRNA-seq atlas14. d, UMAP embedding depicting cells in multicellular clones from xenograft IN subclusters 1 (salmon) and 2 (lime), integrated with an in vitro cultured STICR IN subset. e, UMAP embedding depicting individual interneuron trajectory cells from multicellular clones from xenograft experiments integrated with interneuron trajectory cells from in vitro cultures, split by biological replicate. Members of such clones are highlighted in red. f, UMAP embedding and Leiden subclustering of excitatory neuron (EN) and EOMES+ IPC trajectory cells from xenografts. g, Heatmap depicting pairwise transcriptional cluster correlation of subclustered xenograft EN and EOMES+ IPC trajectory cells with the 2017 Nowakowski developing human cortex scRNA-seq atlas14.
Extended Data Fig. 9 Analysis of PTPRZ1-sorted STICR+ cells in the cortex, subventricular zone, rostral migratory stream and olfactory bulb at 12 weeks.
a, Representative FACS plots depicting isolation of PTPRZ1+ cells from the cortical germinal zone. b, Representative image of transplanted human cortical cells in the cortex of a 12-week-old host mouse. EGFP expression from STICR is in green, with DAPI in blue. Scale bar, 50 μm. CC, corpus callosum. c, Representative images of PTPRZ1-sorted, STICR-labelled cells in the dorsolateral corner of the lateral ventricle of a 12-week-old host mouse analysed by IHC. EGFP from STICR is in green, DCX in red, and DAPI in blue. Scale bar, 100 μm. d, High-magnification inset of the region boxed in c. Scale bar, 10 μm. SVZ, subventricular zone. e, Representative images of PTPRZ1-sorted, STICR-labelled cells in the rostral forebrain, analysed by IHC. The rostral migratory stream (RMS) is outlined by the white box labelled f. GFP expression is in green, DCX expression in red, and DAPI in blue. Scale bar, 100 μm. f, High-magnification insets of the RMS depicted in the white box in e. Scale bar, 10 μm. The cell outlined by the white box is magnified below. g, Representative images of PTPRZ1-sorted, STICR-labelled cells that migrated from the transplantation site in the cortex to the olfactory bulb, analysed by IHC. EGFP expression is in green and DAPI in magenta. GCL, granule cell layer; MCL, mitral cell layer. Scale bar, 10 μm.
Extended Data Fig. 10 Immunohistochemistry of STICR-labelled cortical INs from xenografts at 12 weeks.
Representative images of STICR-labelled GABA+ cells throughout the cortical plate, analysed by IHC. EGFP from STICR is in green, GABA in red, and DAPI in blue. Same cells from Fig. 4d. Arrows point to STICR-labelled GABA+ cells. Scale bar, 10 μm.
Supplementary information
Supplemental Table 1
Table describing clone composition from in vitro culture experiments. Columns: Clone Barcode (“Full_VBC”), clone size (“Total Cells”), sample of origin (“Library”), as well as the number of cells within that clone assigned to principal cell types (“EN”, “IN”, “glia”, “DLX.IPC”, “EOMES.IPC”), cluster 34 (“Unknown”), the excitatory neuron subclustering cell types (“Upper”, “Deep”, “EN.Newborn”, “EN.other”), and the inhibitory neuron/DLX.IPC subclustering cell types (“DLX2.IPC”, “IN.early”, “IN.1”,”IN.2”,”IN.3”). Principal cell type designations reflect subclustering results.
Supplemental Table 2
Table describing clone composition from xenograft experiments (GW15 Reps 3, 4, and 5). Columns: Clone Barcode (“Full_VBC”), clone size (“Total Cells”), sample of origin (“Library”), as well as the number of cells within that clone assigned to principal cell types (“EN”, “IN”, “glia”, “DLX.IPC”, “EOMES.IPC”), “microglia”, “Undetermined” and the inhibitory neuron/DLX.IPC subclustering cell types (“DLX2.IPC”, “IN.early”, “IN.2”,”IN.3”). Principal cell type designations reflect subclustering results.
Supplemental Table 3
Table of STICR ssDNA oligomers used to create barcode fragment sequences.
Supplemental Table 4
Table of primers used to amplify STICR barcodes from plasmid and lentiviral libraries as well as from 10X genomics cDNA libraries.
Rights and permissions
About this article
Cite this article
Delgado, R.N., Allen, D.E., Keefe, M.G. et al. Individual human cortical progenitors can produce excitatory and inhibitory neurons. Nature 601, 397–403 (2022). https://doi.org/10.1038/s41586-021-04230-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-04230-7
- Springer Nature Limited
This article is cited by
-
Genetics of human brain development
Nature Reviews Genetics (2024)
-
ATF3 coordinates the survival and proliferation of cardiac macrophages and protects against ischemia–reperfusion injury
Nature Cardiovascular Research (2024)
-
Cell-type-resolved mosaicism reveals clonal dynamics of the human forebrain
Nature (2024)
-
Generation of ‘semi-guided’ cortical organoids with complex neural oscillations
Nature Protocols (2024)
-
Protracted neuronal recruitment in the temporal lobes of young children
Nature (2024)