Abstract
The voltage-dependent motor protein prestin (also known as SLC26A5) is responsible for the electromotive behaviour of outer-hair cells and underlies the cochlear amplifier1. Knockout or impairment of prestin causes severe hearing loss2,3,4,5. Despite the key role of prestin in hearing, the mechanism by which mammalian prestin senses voltage and transduces it into cellular-scale movements (electromotility) is poorly understood. Here we determined the structure of dolphin prestin in six distinct states using single-particle cryo-electron microscopy. Our structural and functional data suggest that prestin adopts a unique and complex set of states, tunable by the identity of bound anions (Cl− or SO42−). Salicylate, a drug that can cause reversible hearing loss, competes for the anion-binding site of prestin, and inhibits its function by immobilizing prestin in a new conformation. Our data suggest that the bound anion together with its coordinating charged residues and helical dipole act as a dynamic voltage sensor. An analysis of all of the anion-dependent conformations reveals how structural rearrangements in the voltage sensor are coupled to conformational transitions at the protein–membrane interface, suggesting a previously undescribed mechanism of area expansion. Visualization of the electromotility cycle of prestin distinguishes the protein from the closely related SLC26 anion transporters, highlighting the basis for evolutionary specialization of the mammalian cochlear amplifier at a high resolution.
Similar content being viewed by others
Main
Mammals have evolved a highly sophisticated sense of hearing6 that is characterized by extraordinary sensitivity and the ability to process high-frequency sounds7,8. This evolutionary outcome is the result of a mechanism of amplification that relies on the specialized electromotility of the mammalian outer-hair cells (OHCs)9,10,11. The cochlear amplifier depends on the voltage-dependent longitudinal contractions or elongations of OHCs triggered by the concerted action of millions of fast ‘motor’ proteins in the basolateral membrane12. This molecular motor was identified as prestin (SLC26A5), a piezoelectric member of the SLC26 family of anion transporters1. Importantly, the absence or dysfunction of prestin is associated with non-syndromic hearing loss in mammals2,3,4. Despite the intense attention to prestin since its identification as the OHC motor1, fundamental questions remain unanswered, including its oligomeric state13,14,15,16, its underlying voltage-sensing mechanisms, the molecular basis of electromotility, and the evolutionary relationship between prestin and other non-electromotive albeit closely-related SLC26 family members17.
We used single-particle cryo-electron microscopy (cryo-EM) to determine the structure of prestin under various ionic conditions and in complex with the reversible inhibitor salicylate. These structures, together with site-directed mutagenesis, functional data and electrostatic calculations show that prestin adopts a defined set of states as part of its electromotility cycle. These states point to mechanisms of voltage-dependent changes in area, highlight the evolutionary differences with SLC26 transporters and constrain explicit models that help to explain prestin’s behaviour as a piezoelectric motor.
The structure of the prestin homodimer in Cl−
An initial screening using fluorescence-detected size-exclusion chromatography (FSEC) led to the identification of a prestin candidate from the bottlenose dolphin (Tursiops truncatus). Dolphin prestin has around 94% identity (around 97% similarity) to human prestin. HEK293 cells expressing dolphin prestin display the characteristic bell-shaped nonlinear capacitance (NLC) with a V1/2 (the voltage at maximum NLC) of −61 ± 0.5 mV (mean ± s.e.m., n = 20) (Fig. 1a) and similar voltage sensitivity values (1/α = 37±3 mV, mean ± s.e.m., n = 20) to those reported for other mammalian systems13,16,18. Importantly, HEK293 cells overexpressing prestin at high levels undergo voltage-driven cell movements under whole-cell patch-clamp configuration, consistent with robust levels of electromotility (Fig. 1b, Extended Data Fig. 1a and Supplementary Video 1). Expressed in baculovirus-infected mammalian cells, dolphin prestin is characterized by a main gel-filtration peak of around 160 kDa (predicted molecular mass of around 80 kDa), suggesting a stable dimer in solution (Extended Data Fig. 1b, c). We report no evidence of larger oligomeric assemblies.
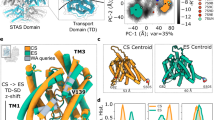
a, Normalized change in NLC (NLC = C − Clin) (top) and charge movement (bottom) of dolphin prestin expressed in HEK293 cells and using patch-clamp electrophysiology. Compared with mock-transfected (GFP only) cells, dolphin prestin has a bell-shaped change in capacitance with a V1/2 of −61 ± 0.5 mV. n = 20. Data are mean ± s.e.m. The NLC curve of prestin (grey) versus transfected cells with GFP alone (blue, n = 8) in whole-cell patches is shown. Prestin’s NLC follows a Boltzmann distribution (fit with solid lines). b, The electromotility of HEK293 cells transfected with dolphin prestin in the whole-cell configuration. n = 7. Cells were lifted off the surface to avoid any possible effect of cell-surface adhesion forces on the cellular movement. After changing the membrane potential (the amplitude was increased in each step), the overexpression of dolphin prestin in HEK293 cells can visibly deform the cells. By tracking the change in the two-dimensional projected area of the cell (red and yellow traces), the cell expands after hyperpolarization. Cells transfected with GFP only do not show such a response (blue solid line). n = 5. Cell movement has been normalized to the largest diameter of the cell, d0. Scale bars, 5 µm (left), 1 s (right, horizontal) and 0.05 (right, vertical). c, Cryo-EM density map and the overall structure of the dolphin prestin homodimer at 3.3 Å nominal resolution when Cl− is the main anion (Methods); the subunits are coloured violet and green. Scale bar, 25 Å. d, Side and top views of the prestin structure coloured according to the different domains, with the core domain in red, the gate domain in blue, and the N and C termini and STAS domain in grey. UCSF ChimeraX was used for illustration.
We determined the structure of full-length dolphin prestin at a resolution of 3.3 Å using single-particle cryo-EM, with Cl− as the main anion (Fig. 1c, d and Extended Data Fig. 1d). It shows that prestin is a symmetric homodimer (C2) with 14 α-helical transmembrane (TM) segments (Extended Data Fig. 2), divided into two overlapping domains: a gate domain (TM segments 5–7 and 12–14) and a core domain (TM segments 1–4 and 8–11) as part of the individual subunit. The TM regions are domain-swapped with the STAS domain containing N- and C-terminal cytoplasmic domains (Fig. 1d). This putative Cl−-bound conformation is structurally closest to that of the intermediate state in SLC26A9 (Protein Data Bank (PDB) accession 6RTF)19 (Extended Data Fig. 3), in which a large cavity is observed that is accessible to the intracellular face of the molecule between the gate and core modules. We observed no explicit density associated with a bound Cl−, as is the case with other SCL26 structures19,20.
SO4 2− binding reveals new prestin states
Replacing Cl− with SO42− as the main anion reduces dolphin prestin’s NLC by around 75% and shifts it approximately 80 mV towards positive potentials (Fig. 2a). This is in agreement with previous studies of prestin in other species18,21,22,23,24,25. Thus, at 0 mV, almost half of its total charge has moved across the electric field and SO42−-bound prestin would display maximal conformational heterogeneity. Indeed, the cryo-EM structural determination of SO42−-bound prestin revealed three distinct new states: two conformations with solvent access to the anion-binding pocket (down I and II) and an intermediate conformation preceding the ‘up’ state (Fig. 2b–d and Extended Data Fig. 4). When compared with Cl−-bound prestin (up), the major differences among these states were quantitatively described on the basis of two measurements between the core and gate domains: z, the position of residue Arg399 (TM10, core domain) relative to Val499 in TM14 in the gate domain25; and d, the gap that separates the TM3–TM10 helical dipole (Fig. 2b).
a, Normalized NLC and charge transfer measurement of HEK293 cells that were transiently transfected with dolphin prestin in Cl− and SO42−. In SO42− as the main internal and external anion, the NLC amplitude is attenuated and V1/2 is shifted to around +20 ± 3 mV compared with in Cl−. Data are mean ± s.e.m. n = 10. Statistical analysis was performed using one-sided Student’s t-tests; *P = 0.002. b, The wild-type prestin structure in a SO42−-based medium corresponds to three distinct configurations. Two reaction coordinates (z and d) determine the structural landscape of prestin: z is the distance between the Arg399 backbone carbon and Val499 backbone carbon; d is the distance between Arg399 Cα and Phe137 Cα, equivalent to the diameter of the binding pocket. All three residues are shown as sticks. On the basis of the z and d values, we defined the following prestin states: down I, down II, intermediate (intermed.) (all SO42−) and up (Cl−), shown in red, yellow, blue and violet, respectively. Right, cross-sectional area expansion in the down (expanded) versus up (compact) state. c, Movement of the anion-binding site from the down state to the up state: as TM3 moves towards TM10, the electrical field becomes more focused (red arrow); this is followed by an elevator-like movement of the binding site from down to up, shown by the cartoons and the yellow arrow. The structures were aligned on the basis of residues 460–550 (TM13–TM14, the least mobile part of prestin). For clarity, only TM14 is shown. d, All-atom molecular dynamics simulation of these states in a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipid bilayer reveals the dynamics of the anion-binding pocket and the degree of water hydration (cyan). The number of water molecules in the intracellular cavity of prestin reduces from the down state to the up state (left to right). Furthermore, the pocket is the widest in the down I state (weak anion binding) and most confined in the up (Cl−) state. For clarity, only water molecules within eight of the residues, Gln97, Phe101, Phe137 Val397, Ser398, Arg399, Glu280 and Glu404, were screened and illustrated.
When linked as part of a possible trajectory of conformational rearrangements, these states comprise the overall working cycle of prestin, from the down to up conformations. In the down states, the core domain is positioned towards the intracellular face of the membrane (z of 8–10 Å), whereas, in the up state, the core domain moves around 6 Å towards the extracellular face of the membrane (Fig. 2b, c). The movement (Fig. 2c and Supplementary Video 2) is reminiscent of a partial ‘elevator’ rearrangement that is seen in other SLC transporters whereby the uppermost conformation inprestin goes beyond SLC26 transporters in an occluded (intermediate) state19. Moreover, the core domain movements associated with the substitution of Cl− by SO42− also trigger a unique widening of the anion-binding pocket (parameter d). Molecular dynamics simulations confirm that these structural changes lead to substantial changes in water penetration, whereby the anion-binding pocket is fully water accessible in states down I and II, but water accessibility is substantially reduced in the intermediate and up states (Fig. 2d). Even when the prestin voltage sensor is in a maximally up conformation, the anion-binding site is still not exposed to the extracellular side, preventing the release of anions to the extracellular space. Even though further up states are yet to be discovered, we speculate that prestin is unable to reach a completely upward-facing state (in a canonical elevator cycle), which rationalizes why mammalian prestin does not efficiently transport anions, in contrast to other members of the SLC26 family.
As proposed, force transmission through an area motor model26,27,28 requires prestin to populate at least two distinct conformations characterized by different intramembrane cross-sectional areas. Indeed, the movement of the binding pocket from down to up is accompanied by an eccentric contraction of the intermembrane cross-sectional area of prestin, whereby the Cl−-bound up conformation corresponds to a compact state and the SO42−-bound down conformations represent expanded states. These two sets of conformations differ in cross-sectional area by around 10% (Fig. 2b and Extended Data Fig. 5), indicating a mechanism whereby prestin might exert lateral forces that directly influence the physical state of the bilayer. Moreover, this feature seems to be unique to the motor function when compared with the Cl− transporter SLC26A9 (ref. 19; Extended Data Fig. 5).
Salicylate locks prestin in an intermediate state
The amphiphilic drug salicylate is known to cause tinnitus29,30, abolishing the NLC and electromotility in mammalian OHCs1,31,32, while also inhibiting Cl− transport in its non-mammalian homologues33. Whole-cell patch-clamp recordings show that 10 mM Na-salicylate flattens the NLC curve of dolphin prestin across the physiological voltage range1,31,32 (Fig. 3a and Extended Data Fig. 6). However, the molecular basis of the inhibitory effects of salicylate remains unclear. We determined the structures of the prestin–salicylate complex in the presence of either Cl− (360 mM) or SO42− (120 mM) at a resolution of 3.8 Å and 3.7 Å, respectively (Fig. 3b and Extended Data Fig. 7). In each case, 50 mM Na-salicylate was present.
a, Salicylate (NaSal; 10 mM) inhibits prestin function, flattening NLC and charge transfer when expressed in HEK293 cells. n = 7. Statistical analysis was performed using one-sided Student’s t-tests; P = 0.005. b, The resolved structural densities point to a salicylate moiety (shown in red) bound to the prestin anion pocket (at the core–gate interface). c, A magnification of the space between the TM1 helix and the micro-dipole of TM3 and TM10 reveals the anion-binding site formed by residues Gln97, Phe101, Phe137, Leu397, Ser398 and Arg399 (yellow) (left). The salicylate molecule and density are shown in salmon. Middle left, the relative positions of the TM3 and TM10 densities in Cl− (up, green) and in Cl− + salicylate (inhibited I, blue). Middle right, an overlay of the EM densities for sensor up (Cl−, green) versus inhibited I (Cl− + salicylate, blue). Right, cartoon illustrating the relative positions of TM3 and TM10 between the up (green) and inhibited I (blue) conformations. d, Overlap of the inhibited I (Cl− + salicylate) and inhibited II (Cl− + salicylate) structures at the anion-binding site (TM13–TM14 helices were aligned). Notably, the TM6 helix is kinked in the inhibited II state (yellow) causing this conformation to be further expanded. e, Sequence alignment of prestin TM6 across different species. We focused on the mammalian-only glycine residues that probably underlie helix flexibility during its motor function. f, Mutating the mammalian-conserved glycine residues (Gly274 and Gly275; GG) has major consequences for NLC and charge transfer. Double mutation diminishes the NLC, the GG to GI mutation largely removed the NLC and the GG to IG mutation left-shifted the NLC to −114 ± 3. n = 16. Statistical analysis was performed using one-way analysis of variance; P = 0.005. Mutation of the other conserved glycine residues along the TM6 helix shows their effects on the prestin NLC and V1/2. Statistical analysis was performed using one-sided Student’s t-tests; *P = 0.01. Mutation of the residue that is conserved only in mammals (G270V) left shifts the NLC to −75 ± 0.6 (n = 12), whereas G263V and G265C greatly diminishes the NLC (n = 11 and n = 19, respectively).
Salicylate probably outcompetes any other bound anion at the binding pocket and, given that no anion can be resolved even under saturating Cl− or SO42− concentrations (Fig. 3b), the salicylate density helps to confirm the overall nature of ion coordination at the anion-binding pocket. In both structures, the salicylate density fits snuggly into the pocket formed by the residues Gln97, Phe101, Phe137, Leu397, Ser398 and Arg399 (Fig. 3c). The majority of these residues are conserved across SLC transporters and other prestin homologues (Extended Data Fig. 6c). Comparing the anion-binding site in the up conformation with that of the Cl− + salicylate structure (inhibited I) shows how the core domain TM10 moves approximately 3 Å downwards, creating enough space to accommodate the larger salicylate (Fig. 3c). The salicylate-inhibited structures (Cl− and SO42−) appear to converge into an intermediate state that displays common conformations at the anion-binding site (Fig. 3d (left)). The major differences are observed at the TM6–TM7 helices, in the periphery of the gate domain, facing the lipid bilayer (Fig. 3d (right)). On the basis of these results, we suggest that salicylate inhibits the motor function of prestin by immobilizing the relative displacement of the core domain around the anion-binding site.
The hybrid nature of prestin’s voltage dependence
A large body of experimental evidence points to the complex nature of the voltage sensor in prestin21,23,31,34, whereby intrinsic charges or dipoles interact with bound anions to generate the NLC, ultimately driving electromotility. Indeed, neutralizing the residue Arg399 completely eliminates the NLC (Extended Data Fig. 8a, e) in mammalian prestin13, whereas neutralization of a series of positively charged residues (Lys276, Lys359, Lys56 and Lys449) have a partial effect on unitary charge transfer35,36. Yet, the voltage dependence of prestin shows an acute dependence on the binding and occupancy of anions (Fig. 2a and Extended Data Fig. 8b). The major effect of anions on prestin’s function is to shift prestin’s V1/2 along the voltage axis (Fig. 2a). This simple experimental manipulation enables a structure-based estimate of prestin’s electrostatic properties (Extended Data Fig. 8b–d) and, by extension, offers a glimpse of the mechanism of voltage dependence. Owing to the presence of a relatively large intracellular water-filled cavity, the electric field displays fairly limited focussing (in contrast to voltage sensing domains in ion channels and enzymes37,38). Within this profile, it is clear that the anion-binding pocket is sensing only around 30–35% of the field. As a result, positive fields at the anion-binding pocket are only partially neutralized in the presence of a bound anion, regardless of their valence. In principle, this would explain the apparent paradox posed by the non-correlated effects of anion valence with the elementary charges estimated from NLC values21,23. Voltage dependence can be estimated from the movement of the charged residues within a defined electric field39. Gating charge calculations carried out on the basis of the two extreme conformations (down I to the up state) estimate that approximately 0.4 unitary charges move across the electric field (Extended Data Fig. 8d). These estimates are approximately consistent with earlier evaluations of charge movement in prestin40,41. Thus, in addition to the TM3–TM10 helical dipole, the positive charges located at the bilayer mid-plane (particularly Arg399) create an attracting field for anion binding and together probably define prestin’s voltage sensor.
TM6 flexibility defines an electromotility elbow
In all of our conformations, TM6–TM7 at the interface of prestin with the lipid bilayer has been the most dynamic entity, as evidenced by the diffused cryo-EM density in this region. Compared with a relatively straight helical configuration in the up state, TM6 helix bends around 65° outwardly, most noticeably in the inhibited II state (Fig. 3d). Around this region, prestin appears to have a strong influence in the shape of its surrounding micelle (Extended Data Fig. 5d). In the intermediate and up conformations, the micelle surrounding prestin is shaped as an elongated oblate that thickens towards the longitudinal ends of the dimer (viewed extracellularly). By contrast, the micelle in the inhibited II state is not only wider and thinner along the sides, but it also displays a ‘notch’ that induces a considerable thinning of the micelle. Molecular dynamics simulations show that TM6 bending substantially thinned the lipid bilayer by up to around 25% at the lipid–prestin interface (Extended Data Fig. 5b, c).
We identified a series of glycine residues that seem to endow TM6 with its unusual flexibility. Although Gly265 appears to be highly conserved across all vertebrates42, a series of three additional Glycine residues appears only among mammals (Gly263, Gly274 and Gly275; Fig. 3e) and helps to define the phylogenetic relationships between bona fide mammalian electromotive OHCs with those of other vertebrates. Two of these residues, Gly274 and Gly275, are located precisely at the hinge that allows TM6 bending in the inhibited II state. Notably, the C-terminal end of the TM6 is also capped by a cluster of charged residues (Fig. 3f) a fact that probably contributes to the membrane thinning. To assess the importance of the TM6 glycine residues in prestin function, we systematically mutated these residues, and the individual mutants were subsequently evaluated using patch–clamp electrophysiology. Mutating Gly274 and Gly275 to leucine and valine (GG to LV) abolishes prestin NLC (Fig. 3f and Extended Data Fig. 9) without changing the levels of expression. The potential role of helical flexibility has been suggested previously43. Aligned with these observations, electromotility and NLC are strongly influenced by the thickness of the bilayer44,45 as well as by membrane tension generated from changes in surface area41,46,47.
The structural rearrangements that cause changes in the cross-sectional area between these states are not limited to TM6–TM7, as there are substantial conformational changes along other peripheral helices: TM5b (gate) and TM8 (core) (Extended Data Fig. 6a). Thus, the present data clearly indicate a model in which TM6 helical hinge-bending, the ‘electromotility elbow’, together with localized rearrangements on TM5, TM7 and TM8, underlie the putative transition between the compact and expanded prestin states (Fig. 4). This is fully consistent with the process of OHC electromotility, whereby cell elongation coincides with an expanded prestin state (hyperpolarization), while depolarization-driven contractions are correlated with a compact prestin state27,41,48,49,50,51,52.
a, A complete set of prestin structures either stabilized by using anion binding as a proxy for shifts in the midpoint of the NLC (V1/2) or stabilizing the overall conformational dynamics by binding salicylate, namely: up (in high Cl−; PDB: 7S8X), inhibited I (Cl− + salicylate; PDB: 7S9A), down I (SO42−; PDB: 7S9B), down II (SO42−; PDB: 7S9C), intermediate (SO42−; PDB: 7S9D) and inhibited II (SO42− + salicylate; PDB: 7S9E). Each experimental condition is represented by a cartoon, with its core domain (blue) adjacent to the gate and intracellular domains. Transitions between states depend on the binding/displacement of anions or salicylate. The TM3–TM10 macrodipole gap is represented by two yellow cylinders encompassing the anion-binding pocket. b, The structural transition of prestin monomer from the down to up conformation after membrane depolarization (top, from left to right). Anions are attracted to the focused positive potential field of the anion-binding pocket, which acts as voltage sensor. After depolarization, the voltage sensor moves from down to up, which is accompanied by an eccentric contraction of the intermembrane cross-sectional area of prestin as well as a major increase in the hydrophobic thickness of the protein (exaggerated cartoon). If the plasma membrane follows the footprint of prestin during the process of prestin charge transfer, that means that the plasma membrane would thicken and the cell would contract after membrane depolarization. This is consistent with the OHC movement (contraction) after applying depolarizing potentials56. Moreover, salicylate physically occludes the pocket and inhibits the voltage-sensor movement, and therefore inhibits the charge transfer and electromotility of prestin.
A molecular model for electromotility
Here we have determined a complex set of interrelated conformations that define the prestin electromotility cycle at the molecular level (Fig. 4a). Similar prestin structures have recently been reported in Cl− (refs. 53,54) as well as SO42− and salicylate53. Although these structures were derived from different species, they are in overall agreement with some of the conformational highlights described here and point to a conserved structural basis of prestin function. Under physiological conditions, in which Cl− is the major intracellular anion, prestin populates its most compact state with its voltage sensor in the up conformation at 0 mV. Displacement of Cl− by SO42− shifts the V1/2 by around 100 mV in the depolarizing direction, enabling us to sample both the expanded and intermediate conformations. Additional inhibited conformations (stabilized with bound salicylate) show expansion at TM6 in the membrane–protein interface. On the basis of these structures, we have been able to delineate the intricate sequence of conformational changes that take place in each prestin monomer (Fig 4b) and, by doing so, we have been able propose an explicit molecular mechanism of OHC electromotility. Transitions between the compact (up) and expanded (down) conformations are accompanied by changes in the TM3–TM10 helical macrodipole (putatively affecting anion binding) and a displacement of the core relative to the gate domain. These movements appear to be transmitted allosterically to the helices in the gate domain (TM6–TM7) such that, during the transitions from the down to up conformation, the intrahelical spaces of the core domain contract, leading to the reorientation of the TM6 electromotility elbow and TM1 helix.
Prestin is present in the lateral wall of OHCs at very high densities (around 5,600–7,000 per µm2)12 in which they may interact cooperatively, directly or through adaptor proteins that are yet to be identified. The efficiency of force transfer to the OHC lateral membrane depends on how individual motors are arranged in the plasma membrane, whether prestin is vectorially or randomly oriented. On the basis of a model of the OHC lateral membrane with fully polarized prestins along the longitudinal direction of the area change, the maximum expected change in cell length would be around 2.6 µm, or 5% elongation relative to the initial length (Methods and equation (3)). This estimate is fully consistent with experimental values at which the OHC elongates or contracts within a length of approximately 4% relative to the initial length48,52,55. This estimate suggests that electromotility in OHCs is probably intrinsic to the changes in the physical state of the plasma membrane. However, a definitive understanding of the molecular and cellular basis of OHC electromotility will necessitate consideration of additional structural rearrangements and potentially other molecular actors, such as cytoplasmic elements.
Methods
Cell lines
The HEK293S GnTI- cells in suspension that were used for protein expression and purification were obtained from ATCC (CRL-3022). GnTI- cells expressing dolphin prestin were grown at 37 °C and 7.8% CO2 in FreeStyle 293 expression medium (Gibco, Thermo Fisher Scientific) supplemented with 2% heat-inactivated fetal bovine serum (FBS) and 10 µg ml−1 penicillin–streptomycin. Adherent HEK293T cells were obtained from ATCC (CRL-1573) and were grown in Dulbecco’s modified Eagle’s medium (Gibco, Thermo Fisher Scientific) supplemented with 10% FBS at 37 °C and 5% CO2. Sf9 cells (Thermo Fisher Scientific, 12659017) were cultured in SF-900 II SFM medium (Gibco, Thermo Fisher Scientific) supplemented with 10% FBS and 10 µg ml−1 gentamicin at 28 °C.
Generation of the prestin construct
The primary dolphin (T. truncatus) prestin construct was a gift from the Shi laboratory18. Dolphin prestin was subcloned into a modified pEG BacMam vector containing a C-terminal HRV 3C protease site, an eGFP tag and an 8×-His tag with 5′ NotI and 3′ XbaI restriction sites. Furthermore, the full-length coding sequence of dolphin prestin was subcloned into the same vector without the protease site, the eGFP tag and the 8×-His tag for electrophysiology recordings. Our eGFP tag alone coding sequence was subcloned into a pcDNA3 vector. The P0 baculovirus was generated using the Bac-to-Bac method (Invitrogen) using Cellfectin II as the transfection reagent (Thermo Fisher Scientific, 10362100). The QuikChange site-directed mutagenesis method (Agilent) was used to introduce the mutations of Gly274 and Gly275—GG to GI, GG to IG, GG to LV, respectively—and G265C, G263V, G270V, R399Q and F101Y into dolphin prestin using KOD DNA polymerase (71085, EMD Millipore). All of the mutant and wild-type constructs were confirmed by DNA sequencing before structural and electrophysiological experiments.
Expression and purification of dolphin prestin
To access a biochemically stable prestin preparation suitable for single-particle cryo-EM, a variety of orthologues were examined. We hypothesized that those adapted to detect high-frequency signals (that is, bats and whales) might exemplify particularly robust systems. An initial screening by FSEC led to the identification of a prestin candidate from the bottlenose dolphin (T. truncatus). P0 virus was amplified once to yield P1 baculovirus, which was used to infect HEK293S GnTI- cells at a ratio of 1:10 (v/v). After 20–24 h of incubation at 37 °C, 10 mM sodium butyrate was added to the cells and the culture was transferred to 30 °C. Cells were collected 50–56 h after infection by low-speed centrifugation. The cell pellets were washed with phosphate-buffered saline pH 7.4, collected by low-speed centrifugation, flash-frozen and stored at −80 °C for later purification. For purification, all of the steps were performed at 4 °C. The flash-frozen cell pellets (1 l) were thawed in a water bath at room temperature, and were resuspended and dounce homogenized in Cl−- or SO42−-based buffer (buffer A and buffer B, respectively). Buffer A contained 360 mM NaCl, 20 mM Tris-HCl, 3 mM dithiothreitol and 1 mM EDTA at pH 7.5, buffer B contained 125 mM Na2SO4, 5 mM MgSO4, 20 mM Tris-OH and 10–15 mM methanesulfonic acid to adjust the pH to 7.5. A protease inhibitor cocktail of 1 µg ml−1 leupeptin, 1 µg ml−1 aprotinin, 1 µg ml−1 pepstatin, 100 µg ml−1 soy inhibitor, 1 mM benzamidine, 0.2 mM phenylmethylsulfonyl fluoride, 0.1 mg ml−1 AEBSF and 10 µg ml−1 DNase as well as a cOmplete Protease Inhibitor Cocktail tablet (Roche) was added to the solutions. Protein was extracted with a final concentration of 1% n-dodecyl-β-d-maltopyranoside (DDM; Anatrace) and 0.2% cholesteryl hemisuccinate (Anatrace) for 90 min. Solubilized supernatant was isolated by ultracentrifugation and the supernatant was incubated for 2 h with 2 ml CNBR-activated Sepharose beads (GE Healthcare) coupled with 4 mg high-affinity GFP nanobodies57. Beads were collected by low-speed centrifugation and washed in batch with main buffer containing 0.05% DDM (Anatrace) and 0.01% cholesteryl hemisuccinate. The beads were transferred to a plastic column and further washed (each wash step was four column volumes), exchanging step-wise to buffer containing buffer A (or buffer B) with 0.02% GDN. Additional protease inhibitors were added in each wash step (1 µg ml−1 leupeptin, 1 µg ml−1 aprotinin, 1 µg ml−1 pepstatin, 100 µg ml−1 soy inhibitor). Protein was cleaved by HRV 3C protease58 for 2–4 h, concentrated and analysed using SEC on a Superose 6, 10/300 GE column (GE Healthcare), with the running buffer including buffer A (or buffer B), 0.02% GDN, 1 µg ml−1 aprotinin and 1 µg ml−1 pepstatin. Peak fractions were collected and concentrated using a 100 kDa molecular mass cut-off centrifugal filter (Millipore concentrator unit) to 2–3 mg ml−1. The concentrated protein was used immediately for the cryo-EM grid-freezing step. For samples with salicylate, 50 mM Na-salicylate was added to the purified protein before freezing the cryo-EM grids.
Cryo-EM sample preparation and imaging
Quantifoil 200-mesh 1.2/1.3 grids (Quantifoil) were plasma-cleaned for 30 s in an air mixture in a Solarus Plasma Cleaner (Gatan). Purified prestin samples were applied onto the grids and frozen in liquid-nitrogen-cooled liquid ethane using a Vitrobot Mark IV (FEI) and the following parameters: 3.5 µl sample volume; 2.5 s, 3.5 s, 5 s blot times (blot time varied from sample to sample); blot force 3; 100% humidity; temperature of 22 °C, and double filter papers on each side of the vitrobot. Grids were screened on a 200 kV Talos side-entry microscope (FEI) equipped with a K2 summit direct detector (Gatan) using a Gatan 626 single-tilt holder. Replicate grids from the same preparation were either imaged at our own facility (University of Chicago) or shipped to the National Cryo-Electron Microscopy Facility at the National Cancer Institute (NCI) and Case Western Reserve University (CWRU). Grids were imaged on a Titan Krios with a K3 detector (in super-resolution mode) and GIF energy filter (set to 20 eV) at a nominal magnification of ×130,000, corresponding to a super-resolution pixel size of 0.5315 Å, 0.55 Å or 0.56 Å per pixel depending on the default set-up at the above-mentioned EM facilities, respectively. Videos were acquired at 1 e− A−2 per frame for 50 frames.
Single-particle cryo-EM analysis
All of the structure determination steps were performed using Relion59. All the movies were binned by 2 and motion-corrected using Motioncor2 (ref. 60). Contrast transfer function (CTF) estimation was performed using CTFFIND4.1 (ref. 61). A total of 2,000 particles was manually picked and classified in 2D to generate templates for autopicking. We used either the SPHIRE-crYOLO package62 or Relion’s built-in reference-based auto picker for particle picking and the coordinates were fed into Relion for particle extraction. For each dataset, we picked between 1,000,000-5,000,000 initial particles, which were processed for 2D classification. Around 150,000–400,000 particles were selected from good classes depending on the dataset. Between 130,000 to 250,000 of these particles were used to generate an initial model with C1 symmetry (for only the very first obtained density for prestin) and, in other cases, C2 symmetry was imposed. All of the particles were then processed for 3D refinement with C2 symmetry, yielding a 4.8 Å nominal resolution map for the up (Cl−) state. Classification of the particles with C1 symmetry resulted in a map that closely resembled the overall architecture of the C2-symmetry-imposed map, albeit with a lower resolution. Postprocessing of the focused TM map was performed using the star file of the K3 detector at 300 kV and a masked nominal resolution of 3.3 Å was calculated for up (Cl−) state according to the gold-standard 0.143 Fourier shell correlation criterion63,64. The nominal resolution for other states was 3.8, 3.7, 4.2, 6.7 and 4.6 Å for the inhibited I, inhibited II, down I, down II and intermediate states, respectively. After a subset of particles (between 110,000 to 180,000 depending on the state) was identified for the final refinement, the particles underwent per-particle CTF refinement followed by Bayesian polishing. A final 3D refinement step was performed, followed by a postprocessing step using a tighter mask and by imposing C2 symmetry. Local resolution was calculated using ResMap65.
Model building and molecular visualization
For the very first dolphin prestin model, Swiss-Model66 was used to generate a homology model based on murine and human SLC26A9 template structures19,20. The homology model was then mutated to poly(alanine) using Chainsaw67, and all of the loops were subsequently deleted. The secondary structural elements were then rigid-body fit to the density. The 3.3 Å density map (up) was used for the initial model building. The rest of model building was pursued manually and in Coot68,69,70 by registering secondary structural elements using bulky residues (such as Phe, Arg) and building loops where appropriate. The density was of sufficient quality to assign rotamers for key residues. Side chains of residues that could not be assigned even tentative rotamers were stubbed at the Cβ. Models were refined in real space without secondary structure restraints using phenix.real_space_refine71,72. Strong non-crystallographic symmetry constraints in phenix.real_space_refine were used to immobilize the domain that was not currently being refined (that is, the cytosolic domain during the TM-domain-focused map refinement). Several iterations of manual refinement and global refinement using Phenix and Coot were performed after visual screening. The dimer model was generated by applying C2 symmetry operations to the monomer in UCSF Chimera and ChimeraX73,74,75. The initially built model was used as template for building the models for our other five EM densities. A primary fit was made using Cryofit2 tool in Phenix. The resulting structures were next processed for several rounds of refinement in Phenix and Coot. The EM density map used in modelling the TM6–TM7 kink in the inhibited II state was not postprocessed or sharpened, as the kink is more defined before any postprocessing step. The sidechains and other parts of this model were fit to the postprocessed map. For models containing salicylate, a PDB file of salicylate was imported into Coot and fit to the density as a ligand. Molecular visualization and analyses were performed using UCSF Chimera, ChimeraX and VMD76.
Patch-clamp electrophysiology
Adherent HEK293 cells were used for all heterologous expression experiments. HEK293T cells were plated for 24 h before transient transfection. Then, 3–3.5 µg prestin plasmids (without eGFP) and 0.4 µg eGFP plasmid were transfected into HEK293T cells using 10 µl of Lipofectamine 3000 (reagent 2:1 ratio; Thermo Fisher Scientific) in 500 ml Opti-MEM (Life Technologies). Cells were transferred from 37 °C to 30 °C after 20–24 h incubation to boost the expression. After 24 h to 48 h of incubation, successfully transfected cells were used for NLC measurements.
The membrane capacitance was measured in whole-cell configuration, using a sine wave stimulus with a frequency of 1 kHz and an amplitude of 10 mV, applied during voltage steps 10 ms after the transient response. Voltage steps varied from −140 mV to 140 mV with a holding potential of −70 mV. The admittance (Y(\(\omega \))) of the system was calculated by spectral analysis and the DC conductance (b) was obtained from the steady state current before the sine wave stimulus. The circuit components—the capacitance (C), membrane resistance (Rm) and series resistance (Rs)—were calculated as follows77:
where FFT is fast Fourier transform; ℜ and ℑ are the real and imaginary components of the admittance, respectively. The membrane capacitance was fitted to the derivative of a Boltzmann function plus a lineal component:
where Qmax is the maximum charge transfer in response to voltage stimulation; V1/2 is the voltage at which the maximum charge is equally distributed across the membrane or, in other words, the peak of the voltage-dependent capacitance; Clin is the linear capacitance, which is proportional to the surface area of the cell; and α is the slope factor of the voltage dependence of the charge transfer ,
The unit is mV−1. k is Boltzmann’s constant, T is the absolute temperature, z is the valence of charge movement, and e is the electron charge. The NLC was obtained by subtracting the linear component (NLC = Cm − Clin) divided by Clin (NLC/Clin) to compare the magnitude of NLC obtained from different cells with different levels of prestin expression. The charge movement was also normalized to Clin and therefore has the units of femtocoulomb per picofarad (Qmax/Clin, fC pF−1). We used our custom written MATLAB codes for data processing and fitting. For salicylate inhibition recordings, first, the NLC was measured and 10 mM Na-salicylate was then added to the bath; after 3 min, the NLC measurement was repeated.
Borosilicate glass pipettes were pulled using the Sutter micropipette puller (P-1000, Flaming/Brown). The resistance of the capillary pipettes ranged from 1.6 MΩ to 2.4 MΩ. The current was amplified using an Axopatch 200B amplifier (Molecular Devices) and the data were acquired at 5 kHz with a Digidata 1440A (Axon instruments) interface using the pCLAMP 10 acquisition software (Molecular Devices). The internal and external (bath) solutions were chosen such that the endogenous ionic current from HEK293 cells was minimal. Here we used almost identical solutions to those that were used in other prestin studies9,18.
For the Cl−-based experiments, the internal solution contained 140 mM CsCl, 2 mM MgCl2, 10 mM EGTA and 10 mM HEPES. The external solution contained 115 mM NaCl, 20 mM TEA-Cl, 5 mM CsCl, 2 mM CoCl, 2 mM MgCl2, 10 mM HEPES and 5 mM glucose. The osmolarities of the internal and external solutions were adjusted with glucose to 310 mOsm l−1 and 320 mOsm l−1, respectively, and were both adjusted to pH 7.4. For sulfate-based measurements, the internal solution contained 10 mM CsCl, 115 mM Cs2SO4, 2 mM MgCl2, 10 mM EGTA and 10 mM HEPES. The external solution contained 10 mM CsCl, 115 mM Na2SO4, 5 mM MgSO4 and 20 Tris-OH. The pH was adjusted to 7.4 using 10–15 mM methanesulfonic acid. All data were acquired from at least four independent cell transfections.
Electromotility measurements
For testing dolphin prestin electromotility, HEK293T cells transiently expressing dolphin Prestin were first tested for their level of NLC while bathed in the Cl−-based extracellular solution. The electromotility was followed only when the NLC was more than 0.8 pF (indicative of high prestin expression). To track the cell movement better, cells were first lifted off the substrate using a uMp piezoelectric manipulator (Sensapex). To evoke prestin-mediated electromotility, either voltage steps of 0.4 s, going from −140 to +150 mV in 10 mV increasing steps (Fig. 1b) or, in some cases, constant voltage steps of −120 to +120 mV were applied. Simultaneously, the video of the cell movement was recorded using a CS2100M-USB Quantalux sCMOS camera (Thorlabs) and using ×60 magnification. The videos were analysed using a custom Python code to track the cell movement. The maximum cellular displacement normalized to the largest diameter of the corresponding cell, d0, as shown in Fig. 1b. The normalized values in Extended Data Fig. 1a are the corresponding values at a change of membrane potential from +120 mV to −120 mV. The holding potential was set at −70 mV, and all the recordings were performed in the whole-cell configuration at room temperature. All data were acquired from at least three independent cell transfections.
Phylogeny analyses and sequence alignments
Selected metazoan prestin protein sequences were extracted from the complete proteomes in the NCBI Assembly database. From each proteome, one protein showing the highest BLAST78 bit score to the human prestin protein query was extracted. For generation of the phylogenetic tree, sequences were aligned using MUSCLE (v.3.5)79, and the ML phylogeny was inferred using RAxML (v.8.2.11)80 (best-fit model of evolution: LG + G + X). The schematic representation of the phylogeny was generated using iTOL81. Clustal Omega was used for all other sequence alignments82.
Molecular dynamics calculations
All-atom systems were constructed using the six atomic cryo-EM structures of prestin determined in different conditions. The orientation and position of the prestin structures in membranes were calculated online using the web server of the Orientations of Proteins in Membranes (OPM) database83. The protein was inserted into a POPC lipid bilayer and solvated in 100 mM KCl using the VMD program76. Titratable residues were assigned their default protonation state at pH 7. One additional chloride ion, sulfate ion or salicylate molecule was added into one of the two ion-binding sites of prestin. The two ion-binding sites were further hydrated by water molecules according to prediction using the Dowser++ program84. The resulting systems contain ~300,000 atoms with orthorhombic periodic box dimensions of ~180 × 120 × 150 Å3 and were electronically neutral.
All of the simulation systems were initially energy-minimized for 5,000 steps, and then equilibrated for 20 ns with gradually decreasing positional restraints being applied to the protein heavy atoms. Each system was further simulated for 150 ns under the NPT ensemble to investigate the morphology of lipid molecules surrounding the protein, the backbone atoms of which were harmonically restrained with a force constant of 1 kcal mol−1 Å−2. Two representative systems (occluded (Cl−) and inhibited II (SO42− and salicylate)) were simulated for another 1 µs to increase the statistical significance using the special-purpose supercomputer ANTON2 (ref. 85). To calculate the electrostatic potential of the central ion-binding sites, we performed an extra 50 ns simulation under the NVT ensemble for each system after the 150 ns NPT run, with the same positional restraint applied to the backbone atoms of the protein. To calculate the fraction of membrane potential, the same simulations were performed under an additional TM voltage of 500 mV or −500 mV. To estimate the gating charge corresponding to the conformational change of apo-prestin, we constructed five new systems based on the occluded (Cl−) system by changing the protein from up (Cl−) to the other five conformations using 20 ns targeted MD simulations. Thus, the final systems have exactly the same compositions, and without any ions in the central binding sites. Each system was then simulated for 50 ns under the NVT ensemble at −500 mV, −100 mV, 100 mV and 500 mV, with the backbone atoms of the protein being harmonically restrained with a force constant of 1 kcal mol−1 Å−2 and the ions in the solution were prevented from entering the cavity of the protein using the tcl force plugin.
The MD simulations other than the ANTON2 simulation were performed using the NAMD program86 with a time step of 2 fs. The CHARMM36 force field with torsional backbone corrections87,88,89 was used for protein, lipids and ions and the TIP3P model90 for water in all of the simulations. The GAMMP program was used to parameterize the all-atom force field for salicylate and sulfate anions91. In NAMD, the temperature and pressure were controlled at 300 K and 1 atm, respectively, using the Langevin dynamics and the Nose−Hoover Langevin piston method92,93. The van der Waals interactions were smoothly switched off at 10−12 Å. The long-range electrostatic interactions were calculated using the particle mesh Ewald method94. In ANTON2, the Nose–Hoover thermostat and the semi-isotropic MTK barostat95 were used to control the temperature and pressure, respectively. The k-space Gaussian split Ewald method96,97 was used to calculate the long-range electrostatic interactions.
Electrostatic potential and fraction of membrane potential calculations
The electrostatic potential maps were calculated using the PMEPOT plugin of VMD98. Snapshots (n = 4,000) from the last 40 ns trajectory of each system run under the NVT ensemble were used for the calculation. The time-averaged three-dimensional electrostatic potential maps were then used to compute the two-dimensional potential in the x–z plane87,99,100 crossing the two central ion-binding sites. Snapshots (n = 4,000) from the last 40 ns trajectories of the systems run under a TM voltage of +500 mV and −500 mV were used for the calculation of the fraction of membrane potential drop. The time-averaged three-dimensional electrostatic potential maps were then used to compute the fraction of membrane potential in the x–z plane crossing the two central binding sites.
Lipid bilayer morphology and cross-section areas of the protein
For area calculations, PDB models were aligned such that their z axis aligns with the symmetry axis using the PPM server. On the basis of local resolution maps, residues 460–505 always had a sub-3 Å resolution and were the least mobile. We therefore aligned all of the structures based on this region, unless otherwise specified. We kept the same criteria for SLC26A9 for comparison. The CHARMM-GUI87 membrane builder tool was used for prestin and SLC26A9 area calculation across the membrane thickness.
Charge displacement calculations
The gating charges of protein conformational change were estimated by calculating the average displacement charge using \({Q}_{d}=\sum _{i}{q}_{i}\frac{{z}_{i}}{{L}_{z}}\), where qi and zi are the partial charge and the unwrapped z coordinate of atom i, respectively, and Lz is the length of the simulation box along the z direction98,100. Snapshots (n = 4,000) from the last 40 ns trajectories at different TM voltages of each system were used to calculate the average displacement charges. The calculated average displacement charges of each system were then linearly fitted together, and the offset constants correspond to the gating charge changes between different conformations.
Calculation of cell motility based on prestin’s cross-sectional expansion
We assumed that all of the prestin dimers are aligned along the lateral line of the OHCs and that they are cooperative. It was further assumed that the OHC membrane fully follows the footprint of prestin and only the lateral wall of OHC extends due to cross-sectional expansion of prestin. For simplicity, it was also assumed that OHCs have a perfect cylindrical shape.
where ΔLOHC is the maximum possible somatic motility of the OHC based solely based on collective cross-sectional expansion of prestin molecules in the lateral wall; AOHC is the average lateral area of an OHC; ρ is the prestin density (prestin counts per µm2); ΔAprestin is the average cross-sectional area expansion of a prestin dimer from down to up (Cl−); ROHC is the average circumferential radius of OHC; and LOHC is the average length of OHC. Typical values used for the above variables were as follows: AOHC = average length of ρ = 7,000 prestin per µm2, ΔAprestin = 700 Å2, ROHC = 4 µm and LOHC = 50 µm (refs. 9,48,52,55).
Note that the OHC dimensions (length and diameter) vary along tonotopically defined frequency segments of the cochlea and are also species dependent. Thus, the calculations provided represent rough estimates of whether prestin in-plane area expansion at the molecular scale could underlie somatic motility in the OHCs.
Statistical analysis
Statistical significance (criteria, *P < 0.005) was determined using unpaired Student’s t-tests. In the electrophysiology data, this was determined by a comparison of data obtained from wild-type dolphin prestin in Cl− with wild-type prestin in SO42−. To compare electrophysiology data of the mutants with wild-type prestin, we used one-way analysis of variance with Tukey’s post hoc test. The software used in this project was curated by SBGrid101.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
The atomic structure coordinates have been deposited at the RCSB PDB under accession numbers 7S8X, 7S9A, 7S9B, 7S9C, 7S9D and 7S9E; and the EM maps have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-24928, EMD-24930, EMD-24931, EMD-24932, EMD-24933 and EMD-24934. All materials generated during the current study are available from the corresponding author under a materials transfer agreement with The University of Chicago.
References
Zheng, J. et al. Prestin is the motor protein of cochlear outer hair cells. Nature 405, 149–155 (2000).
Liu, X. Z. et al. Prestin, a cochlear motor protein, is defective in non-syndromic hearing loss. Hum. Mol. Genet. 12, 1155–1162 (2003).
Liberman, M. C. et al. Prestin is required for electromotility of the outer hair cell and for the cochlear amplifier. Nature 419, 300–304 (2002).
Dallos, P. et al. Prestin-based outer hair cell motility is necessary for mammalian cochlear amplification. Neuron 58, 333–339 (2008).
Cheatham, M., Huynh, K., Gao, J., Zuo, J. & Dallos, P. Cochlear function in Prestin knockout mice. J. Physiol. 560, 821–830 (2004).
Masterton, B., Heffner, H. & Ravizza, R. The evolution of human hearing. J. Acoust. Soc. Am. 45, 966–985 (1969).
Heffner, H. & Masterton, B. Hearing in glires: domestic rabbit, cotton rat, feral house mouse, and kangaroo rat. J. Acoust. Soc. Am. 68, 1584–1599 (1980).
Fettiplace, R. Diverse mechanisms of sound frequency discrimination in the vertebrate cochlea. Trends Neurosci. 43, 88–102 (2020).
Ashmore, J. A fast motile response in guinea‐pig outer hair cells: the cellular basis of the cochlear amplifier. J. Physiol. 388, 323–347 (1987).
Brownell, W. E., Bader, C. R., Bertrand, D. & De Ribaupierre, Y. Evoked mechanical responses of isolated cochlear outer hair cells. Science 227, 194–196 (1985).
Kachar, B., Brownell, W. E., Altschuler, R. & Fex, J. Electrokinetic shape changes of cochlear outer hair cells. Nature 322, 365–368 (1986).
He, D. Z. et al. Changes in plasma membrane structure and electromotile properties in prestin deficient outer hair cells. Cytoskeleton 67, 43–55 (2010).
Gorbunov, D. et al. Molecular architecture and the structural basis for anion interaction in prestin and SLC26 transporters. Nat. Commun. 5, 3622 (2014).
Hallworth, R. & Nichols, M. G. Prestin in HEK cells is an obligate tetramer. J. Neurophysiol. 107, 5–11 (2012).
Zheng, J. et al. Analysis of the oligomeric structure of the motor protein prestin. J. Biol. Chem. 281, 19916–19924 (2006).
Navaratnam, D., Bai, J.-P., Samaranayake, H. & Santos-Sacchi, J. N-terminal-mediated homomultimerization of prestin, the outer hair cell motor protein. Biophys. J. 89, 3345–3352 (2005).
He, D. Z., Lovas, S., Ai, Y., Li, Y. & Beisel, K. W. Prestin at year 14: progress and prospect. Hear. Res. 311, 25–35 (2014).
Liu, Z., Qi, F.-Y., Xu, D.-M., Zhou, X. & Shi, P. Genomic and functional evidence reveals molecular insights into the origin of echolocation in whales. Sci. Adv.4, eaat8821 (2018).
Walter, J. D., Sawicka, M. & Dutzler, R. Cryo-EM structures and functional characterization of murine Slc26a9 reveal mechanism of uncoupled chloride transport. eLife 8, e46986 (2019).
Chi, X. et al. Structural insights into the gating mechanism of human SLC26A9 mediated by its C-terminal sequence. Cell Discov. 6, 55 (2020).
Rybalchenko, V. & Santos-Sacchi, J. Anion control of voltage sensing by the motor protein prestin in outer hair cells. Biophys. J. 95, 4439–4447 (2008).
Tan, X. et al. From zebrafish to mammal: functional evolution of prestin, the motor protein of cochlear outer hair cells. J. Neurophysiol. 105, 36–44 (2011).
Santos-Sacchi, J., Song, L., Zheng, J. & Nuttall, A. L. Control of mammalian cochlear amplification by chloride anions. J. Neurosci. 26, 3992–3998 (2006).
Rybalchenko, V. & Santos‐Sacchi, J. Cl− flux through a non‐selective, stretch‐sensitive conductance influences the outer hair cell motor of the guinea‐pig. J. Physiol. 547, 873–891 (2003).
Homma, K., Duan, C., Zheng, J., Cheatham, M. A. & Dallos, P. The V499G/Y501H mutation impairs fast motor kinetics of prestin and has significance for defining functional independence of individual prestin subunits. J. Biol. Chem. 288, 2452–2463 (2013).
Homma, K. & Dallos, P. Evidence that prestin has at least two voltage-dependent steps. J. Biol. Chem. 286, 2297–2307 (2011).
Iwasa, K. A two-state piezoelectric model for outer hair cell motility. Biophys. J. 81, 2495–2506 (2001).
Ludwig, J. et al. Reciprocal electromechanical properties of rat prestin: the motor molecule from rat outer hair cells. Proc. Natl Acad. Sci. USA 98, 4178–4183 (2001).
Cazals, Y. Auditory sensori-neural alterations induced by salicylate. Prog. Neurobiol. 62, 583–631 (2000).
Chen, G.-D. et al. Salicylate-induced cochlear impairments, cortical hyperactivity and re-tuning, and tinnitus. Hear. Res. 295, 100–113 (2013).
Oliver, D. et al. Intracellular anions as the voltage sensor of prestin, the outer hair cell motor protein. Science 292, 2340–2343 (2001).
Kakehata, S. & Santos-Sacchi, J. Effects of salicylate and lanthanides on outer hair cell motility and associated gating charge. J. Neurosci. 16, 4881–4889 (1996).
Schaechinger, T. J. & Oliver, D. Nonmammalian orthologs of prestin (SLC26A5) are electrogenic divalent/chloride anion exchangers. Proc. Natl Acad. Sci. USA 104, 7693–7698 (2007).
Song, L. & Santos-Sacchi, J. Conformational state-dependent anion binding in prestin: evidence for allosteric modulation. Biophys. J. 98, 371–376 (2010).
Bai, J.-P. et al. Current carried by the Slc26 family member prestin does not flow through the transporter pathway. Sci. Rep. 7, 46619 (2017).
Bai, J.-P. et al. Prestin’s anion transport and voltage-sensing capabilities are independent. Biophys. J. 96, 3179–3186 (2009).
Jogini, V. & Roux, B. Dynamics of the Kv1.2 voltage-gated K+ channel in a membrane environment. Biophys. J. 93, 3070–3082 (2007).
Starace, D. M. & Bezanilla, F. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature 427, 548–553 (2004).
Bezanilla, F. How membrane proteins sense voltage. Nat. Rev. Mol. Cell Biol. 9, 323–332 (2008).
Dong, X.-X., Ehrenstein, D. & Iwasa, K. Fluctuation of motor charge in the lateral membrane of the cochlear outer hair cell. Biophys. J. 79, 1876–1882 (2000).
Dong, X.-X. & Iwasa, K. Tension sensitivity of prestin: comparison with the membrane motor in outer hair cells. Biophys. J. 86, 1201–1208 (2004).
Beurg, M., Tan, X. & Fettiplace, R. A prestin motor in chicken auditory hair cells: active force generation in a nonmammalian species. Neuron 79, 69–81 (2013).
Dallos, P. & He, D. Z. Two models of outer hair cell stiffness and motility. J. Assoc. Res. Otolaryngol. 1, 283–291 (2000).
Izumi, C., Bird, J. E. & Iwasa, K. H. Membrane thickness sensitivity of prestin orthologs: the evolution of a piezoelectric protein. Biophys. J. 100, 2614–2622 (2011).
Fang, J., Izumi, C. & Iwasa, K. H. Sensitivity of prestin-based membrane motor to membrane thickness. Biophys. J. 98, 2831–2838 (2010).
Santos‐Sacchi, J., Shen, W., Zheng, J. & Dallos, P. Effects of membrane potential and tension on prestin, the outer hair cell lateral membrane motor protein. J. Physiol. 531, 661–666 (2001).
Kakehata, S. & Santos-Sacchi, J. Membrane tension directly shifts voltage dependence of outer hair cell motility and associated gating charge. Biophys. J. 68, 2190–2197 (1995).
Ashmore, J. Cochlear outer hair cell motility. Physiol. Rev. 88, 173–210 (2008).
He, D. Z., Evans, B. N. & Dallos, P. First appearance and development of electromotility in neonatal gerbil outer hair cells. Hear. Res. 78, 77–90 (1994).
Dallos, P. The active cochlea. J. Neurosci. 12, 4575–4585 (1992).
Santos-Sacchi, J. Asymmetry in voltage-dependent movements of isolated outer hair cells from the organ of Corti. J. Neurosci. 9, 2954–2962 (1989).
Santos-Sacchi, J. & Dilger, J. Whole cell currents and mechanical responses of isolated outer hair cells. Hear. Res. 35, 143–150 (1988).
Ge, J. et al. Molecular mechanism of prestin electromotive signal amplification. Cell 184, 4669–4679 (2021).
Butan, C. et al. Single particle cryo-EM structure of the outer hair cell motor protein prestin. Preprint at bioRxiv https://doi.org/10.1101/2021.08.03.454998 (2021).
Santos-Sacchi, J. On the frequency limit and phase of outer hair cell motility: effects of the membrane filter. J. Neurosci. 12, 1906–1916 (1992).
Holley, M. & Ashmore, J. F. On the mechanism of a high-frequency force generator in outer hair cells isolated from the guinea pig cochlea. Proc. R. Soc. Lond. B Biol. Sci. 232, 413–429 (1988).
Kirchhofer, A. et al. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 17, 133–138 (2010).
Clark, M. D., Contreras, G. F., Shen, R. & Perozo, E. Electromechanical coupling in the hyperpolarization-activated K+ channel KAT1. Nature 583, 145–149 (2020).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Wagner, T. et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019).
Scheres, S. H. & Chen, S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Biasini, M. et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 (2014).
Stein, N. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J. Appl. Crystallogr. 41, 641–643 (2008).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D 71, 136–153 (2015).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D 74, 531–544 (2018).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Pettersen, E. F. et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Lindau, M. & Neher, E. Patch-clamp techniques for time-resolved capacitance measurements in single cells. Pflügers Arch. 411, 137–146 (1988).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinform. 10, 421 (2009).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019).
Sievers, F. et al. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I. & Lomize, A. L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012).
Morozenko, A. & Stuchebrukhov, A. Dowser++, a new method of hydrating protein structures. Proteins Struct. Funct. Bioinform. 84, 1347–1357 (2016).
Shaw, D. E. et al. Anton, a special-purpose machine for molecular dynamics simulation. Commun. ACM 51, 91–97 (2008).
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM‐GUI: a web‐based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
MacKerell, Jr A. D., Feig, M. & Brooks, C. L. Improved treatment of the protein backbone in empirical force fields. JACS 126, 698–699 (2004).
MacKerell, Jr A. D. et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Huang, L. & Roux, B. Automated force field parameterization for nonpolarizable and polarizable atomic models based on ab initio target data. J. Chem. Theory Comput. 9, 3543–3556 (2013).
Feller, S. E., Zhang, Y., Pastor, R. W. & Brooks, B. R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 103, 4613–4621 (1995).
Martyna, G. J., Tobias, D. J. & Klein, M. L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 101, 4177–4189 (1994).
Essmann, U. et al. A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 (1995).
Martyna, G. J., Klein, M. L. & Tuckerman, M. Nosé–Hoover chains: the canonical ensemble via continuous dynamics. J. Chem. Phys. 97, 2635–2643 (1992).
Shan, Y., Klepeis, J. L., Eastwood, M. P., Dror, R. O. & Shaw, D. E. Gaussian split Ewald: a fast Ewald mesh method for molecular simulation. J. Chem. Phys. 122, 054101 (2005).
Aksimentiev, A. & Schulten, K. Imaging α-hemolysin with molecular dynamics: ionic conductance, osmotic permeability, and the electrostatic potential map. Biophys. J. 88, 3745–3761 (2005).
Roux, B. The membrane potential and its representation by a constant electric field in computer simulations. Biophys. J. 95, 4205–4216 (2008).
Castillo, J. P. et al. Mechanism of potassium ion uptake by the Na+/K+-ATPase. Nat. Commun. 6, 7622 (2015).
Khalili-Araghi, F. et al. Calculation of the gating charge for the Kv1. 2 voltage-activated potassium channel. Biophys. J. 98, 2189–2198 (2010).
Morin, A. et al. Cutting edge: Collaboration gets the most out of software. eLife 2, e01456 (2013).
Acknowledgements
We thank M. Zhao for discussions and assistance with imaging; K. Poole, C. Cox, T. Ngo, O. Bavi, R. Hulse, C. Bassetto, Y. Nikolaev, Y. Krishnan, P. Bezanilla, H. Mchaourab, P. Gueorguieva, S. Zhong, M. Karami, P. Haller and F. Galan, and the members of the Perozo laboratory for exchanging ideas and comments on the manuscript; P. Shi for sharing the Tursiops prestin plasmid; J. Fuller, J. Austin II and T. Lavoie at the University of Chicago Advanced Electron Microscopy Facility for microscope maintenance and training; and U. Baxa and T. J. Edwards at NCEF for cryo-EM data collection. Anton 2 computer time was provided by the Pittsburgh Supercomputing Center (PSC) through grant R01GM116961 from the National Institutes of Health. The Anton 2 machine at PSC was made available by D.E. Shaw Research. N.B. acknowledges the Biology of Inner Ear course (BIE2019) and Gordon Conference (Auditory System Gordon Research Conference) for inspiring him to study hearing and prestin. This work was supported by NIDCD grant R01 DC019833 to E.P. N.B. was the recipient of a Chicago Fellowship. M.D.C. was supported by F30MH116647 and T32GM007281. This research was in part supported by the National Cancer Institute’s National Cryo-EM Facility at the Frederick National Laboratory for Cancer Research under contract HSSN261200800001E.
Author information
Authors and Affiliations
Contributions
N.B., M.D.C. and E.P. conceived the project. N.B. expressed and purified the protein. N.B. and M.D.C. prepared cryo-grids. N.B. and B.G.R. performed EM data collection. N.B., B.G.R. and M.D.C. processed the cryo-EM data and built and refined the atomic models. N.B. and G.F.C. performed and analysed the electrophysiological experiments. R.S. carried out MD simulations and electrostatic calculations. N.B. and W.M. carried out molecular cloning, mutagenesis and created all of the expression constructs. N.B. and W.M. managed cell culture. All of the authors analysed the data. N.B. and E.P. wrote the manuscript with input from all of the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Rachelle Gaudet and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Function, biochemistry and structural features of electromotile prestin.
a, Electromotility analysis of HEK293 cells transfected with wild-type dolphin prestin compared to GFP-only transfected cell (Mock-GFP). The cellular displacement was normalized based on the cell largest diameter, d0 (Fig. 1b). The normalized electromotility was 0.05±02 (n = 6) versus 0.008±0.002 (n = 5) for wild-type prestin and Mock-GFP, respectively. These values were measured at the depolarizing voltage step changing from +120 mV to −120 mV (mean ± SD, is the number of independent cells. One-sided Student t-test, unpaired, P=0.005). b, Size-exclusion chromatography (SEC) curves of the full-length dolphin prestin purified in GDN, run on a Superose 6 column, in high Cl− (red) and SO42− (blue) based solution. The fractions indicated by black dotted lines in both represent purified proteins that were used for cryo-EM imaging. c, Purified dolphin prestin cryo-EM samples, run on a Stain-free SDS-PAGE gel, indicating size of ~80 kDa for the full-length prestin monomer (representation of n = 3).d, Topology of dolphin prestin. Different domains are indicated by color; the gate domain is colored in blue, the core domain in red and the C- and N-termini as well as the STAS domain in grey. The transmembrane helices are numbered from 1 to 14. The N- and C-termini as well as the STAS domain are oriented towards the cytoplasm.
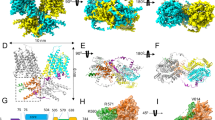
Extended Data Fig. 2 Flow chart for the cryo-EM data processing and structure determination of the dolphin prestin in high Cl− condition.
a, The final reconstruction has a nominal resolution of 3.3 Å (at FSC=0.143). The yellow scale bar on the micrograph represents 200 Å. All the images in this figure were created in UCSF ChimeraX.
Extended Data Fig. 3 Structure of prestin in high Cl− and comparison with the Intermediate.
a, Comparison between prestin (high Cl−) (blue and Red) and SLC26A9 Intermediate state (6RTF, grey). The structures are aligned based on residues 460 to 505 of one subunit (TM13-TM14, dotted box). ChimeraX was used for illustration. b, Electrostatic potential and surface charge distribution of SLC26A9 intermediate state19 compared with that of prestin in high Cl− panel c. The electrostatic charge distribution ranges from −5 to 5 kT from negative to positive charge. ChimeraX was used for illustration.
Extended Data Fig. 4 Flow chart for the cryo-EM data processing and structure determination of the dolphin prestin in SO42−.
a, b Cryo-EM data processing and structure determination of the dolphin prestin in Down I (SO42−) and Down II (SO42−) states. A was obtained from Dataset I, which was combined with Class B from Dataset II. The final reconstruction yielded two structures, Down I (SO42−) and Down II (SO42−), which have nominal resolutions of 4.2 and 6.7 Å, respectively (at FSC=0.143). See Supplementary Figure 5 for the steps on how Class A and B were further processed. Evidence of both states was found in dataset II, however merging of datasets was required to improve resolution of states. c, Flow chart for the cryo-EM data processing and structure determination of the dolphin prestin in the Intermediate state (SO42−) (See Methods for details). The final reconstruction has a nominal resolution of 4.6 Å (at FSC=0.143). UCSF ChimeraX was for illustration of all the structures. The yellow scale bar on all the micrographs represents 200 Å.
Extended Data Fig. 5 Prestin’s cross-sectional area changes upon transition from Down to Up states.
a, Upon the transition from Down to Up state and the movement of the anion-binding site, the most obvious changes are seen in the peripheral helices TM5b, TM6-TM7, and TM8. b, MD simulation of prestin in Up state is compared with the Inhibited II state (Cl− and Salicylate) equilibrated in POPC lipid bilayers. The cross-sectional area of outer and inner monolayers with mapped leaflet coordinate in the Z direction (across the membrane thickness) using all-atom molecular dynamics simulations (1µs). Δz shows movement of the phosphate group of the lipids in the Z (thickness) direction. The comparison was made between Up (Cl−) and Inhibited II (SO42−) states. The largest difference was observed at the location of the TM6 helix. c, Cross-sectional area calculations of the transmembrane domain of SLC26A9(12) along the hydrophobic thickness using CHARMM-membrane builder. Cross-sectional area change of SLC26A9 from Inward-facing to Intermediate states (6RTC and 6RTF) per monomer19. Note that prior to area calculation, the spatial arrangements of all the structures with respect to the hydrocarbon core of the lipid bilayer were first adjusted using the PPM server(30). The structures were aligned based on residues 460 to 505 (TM13-TM14). d, Comparison of the change in the micelle morphology between two salicylate-inhibited structures Inhibited I (Cl−) and Inhibited II (SO42− + Salicylate) states. The overlay of the two states shows drastic changes in the micelle thickness especially around TM6 region in addition to the overall changes in the micelle in-lane direction, both indicative of major structural rearrangements between the two states. ChimeraX was used for illustration.
Extended Data Fig. 6 Salicylate outcompetes SO42− in binding to anion-binding pocket.
a, The NLC measurements of HEK293T cells transfected with dolphin prestin in SO42− (0.15±0.06; n = 6). The NLC of these cells were completely abrogated (0.01±0.01) by 10 mM Na-Salicylate (mean ± s.e.m.; n, is the number of independent cells. One-sided student’s t-test, unpaired, P=0.01) b, Density of Salicylate (orange) in the anion-binding site (blue) was resolved in the Inhibited II (SO42−) state of dolphin prestin. c, Sequence alignment of prestin and close SLC transporters across different species. Residues forming the anion-binding site are largely conserved (e.g. Q97, F101, F137). Putative voltage-sensing residue R399 in dolphin prestin is replaced by a valine in murine SLC26A9. Clustal Omega was used for the sequence alignments. ChimeraX was used for illustration.
Extended Data Fig. 7 Flow chart for the cryo-EM data processing and structure determination of the dolphin prestin in the Salicylate-Inhibited states.
Flow chart of the dolphin prestin in the a, Inhibited I state (Cl− + Salicylate) and b, (SO42− + Salicylate) The final reconstructions have a nominal resolution of 3.8 Å and 3.7 Å, respectively (at FSC=0.143). All the images in this figure were created in UCSF ChimeraX. The yellow scale bar on all the micrographs represents 200 Å.
Extended Data Fig. 8 Electrostatic calculations and charge transfer of prestin across the membrane.
a, Mutation of the key residues in the anion binding pocket either completely abolishes the NLC (R399Q) or right shifts the V1/2 by more than 80 mV (F101Y) to around +25 ± 5 mV (mean ± s.e.m.; n, is the number of independent cells. One-sided Student t-test, P=0.001); a similar effect has been observed in other prestin homologues using patch-clamp electrophysiology (51). b, Snapshots from the MD trajectories of the systems, and calculation of the electrostatic potential across the membrane at two states, the Down I state (with SO42− in the left cavity, and without SO42− in the right cavity) versus Up (with Cl− in the left cavity and without any Cl− in the right cavity). The x-z plane is crossing the two central anion-binding sites. In both models, the positive field is mainly focused around the transmembrane mid-plane and around the anion-binding site, creating an attractive (blue) field for the binding of the anion. However, in the Up state the field is more positive around the mid-plane compared to the corresponding region in the Intermediate state. In both cases, the presence of the anion only partially neutralizes (~35%) the positive field around the bilayer mid-plane. Note that the actual size of the simulation box is larger than what is illustrated here (see Methods). c, Averaged 1-D fraction of membrane potential in the z direction along the two central binding sites (shown as dashed blue lines in panel A with the central binding sites highlighted using the red cross symbols). The 1-D and 2-D maps were directly extracted from the ensemble averaged 3-D fraction of membrane potential map. The location of the phosphate atoms of the outer and inner lipid leaflets along the z axis was highlighted with dashed gray lines). d, Displacement of charge for prestin in the Up and Down I conformations at different transmembrane potentials. The gating charge between the two states is 0.38 +0.25 e calculated as the offset constant between the linear fits. (n = 3; data are mean ± SD; One-sided Student’s t-test; P=0.05). e, R399 in both monomers have been mutated to Q, S and E in different systems to see the contribution of R399 residue to the positive charge at the bilayer mid-plane using electrostatic calculations. R399 mutation to polar residues shows that R399 has almost ~40% contribution the positive charge of the field at the bilayer mid-plane. The remainder likely comes from the TM3-TM10 helical dipole and other positive charges in this area.
Extended Data Fig. 9 Whole cell patch-clamp electrophysiology of the mutations of different glycine residues along the TM6 helix.
All the individual data points, that has been averaged in Fig. 3f, has been presented here. Compared to wild-type prestin, mutation of evolutionary conserved glycine residues, a, G274 and G275 and b, G263, G265 and G270 largely affects the NLC.
Supplementary information
Supplementary Information
This file contains further supportive results for the findings in this study, including Supplementary Figs. 1–10 (which include the uncropped gel source data).
Supplementary Video 1
Electromotility measurements of HEK293 cells transfected with dolphin prestin using whole-cell patch-clamp electrophysiology. To evoke prestin-mediated electromotility, the membrane potential was held at −70 mV; 10 mV increase-in-amplitude voltage steps were applied up to the final steps, which was from +150 mV to −140 mV (Fig. 1b). The magenta square indicates the area that was chosen in our custom-written code to track the cellular displacements.
Supplementary Video 2
Structural changes from the expanded (down I) to the compact (up) conformation as a linear interpolation. The side front and top views of the dimer have been shown in one single frame. The anion-binding site is highlighted in red and Arg399 is shown in stick representation and the backbone has been coloured yellow. The videos were made in UCSF ChimeraX.
Rights and permissions
About this article
Cite this article
Bavi, N., Clark, M.D., Contreras, G.F. et al. The conformational cycle of prestin underlies outer-hair cell electromotility. Nature 600, 553–558 (2021). https://doi.org/10.1038/s41586-021-04152-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-04152-4
- Springer Nature Limited
This article is cited by
-
Mechanism of anion exchange and small-molecule inhibition of pendrin
Nature Communications (2024)
-
Activation mechanisms of dimeric mechanosensitive OSCA/TMEM63 channels
Nature Communications (2024)
-
Interdomain-linkers control conformational transitions in the SLC23 elevator transporter UraA
Nature Communications (2024)
-
Substrate binding plasticity revealed by Cryo-EM structures of SLC26A2
Nature Communications (2024)
-
A mechanical-coupling mechanism in OSCA/TMEM63 channel mechanosensitivity
Nature Communications (2023)