Abstract
Protein ubiquitination at sites of DNA double-strand breaks (DSBs) by RNF168 recruits BRCA1 and 53BP11,2, which are mediators of the homologous recombination and non-homologous end joining DSB repair pathways, respectively3. Non-homologous end joining relies on 53BP1 binding directly to ubiquitinated lysine 15 on H2A-type histones (H2AK15ub)4,5 (which is an RNF168-dependent modification6), but how RNF168 promotes BRCA1 recruitment and function remains unclear. Here we identify a tandem BRCT-domain-associated ubiquitin-dependent recruitment motif (BUDR) in BRCA1-associated RING domain protein 1 (BARD1) (the obligate partner protein of BRCA1) that, by engaging H2AK15ub, recruits BRCA1 to DSBs. Disruption of the BUDR of BARD1 compromises homologous recombination and renders cells hypersensitive to PARP inhibition and cisplatin. We further show that BARD1 binds nucleosomes through multivalent interactions: coordinated binding of H2AK15ub and unmethylated H4 lysine 20 by its adjacent BUDR and ankyrin repeat domains, respectively, provides high-affinity recognition of DNA lesions in replicated chromatin and promotes the homologous recombination activities of the BRCA1–BARD1 complex. Finally, our genetic epistasis experiments confirm that the need for BARD1 chromatin-binding activities can be entirely relieved upon deletion of RNF168 or 53BP1. Thus, our results demonstrate that by sensing DNA-damage-dependent and post-replication histone post-translation modification states, BRCA1–BARD1 complexes coordinate the antagonization of the 53BP1 pathway with promotion of homologous recombination, establishing a simple paradigm for the governance of the choice of DSB repair pathway.
Similar content being viewed by others
Main
The equilibrium between accurate DNA DSB repair by homologous recombination and error-prone DSB repair by non-homologous end joining is controlled by the BRCA1 and 53BP1 proteins and their interplay with two states of histone post-translational modification (PTM). DNA damage recognition by both proteins involves ubiquitination of H2A-type histones at DSB sites by the DNA-damage-responsive E3 ubiquitin ligase RNF1681,2,3,6. Chromatin engagement of 53BP1 and BRCA1 complexes also requires binding to histone H4 tails, through recognition of distinct methylation states of lysine 20 that undergo DNA replication-dependent oscillations. Essential to its promotion of non-homologous end joining, 53BP1 binds nucleosomes that carry mono- and dimethylated lysine 20 of histone H4 (H4K20me1 and H4K20me2, respectively); these two histone PTMs are highly abundant on old histones in pre- and post-replicative chromatin7,8,9. Conversely, BRCA1 complexes recognize H4 histones specifically when they are unmethylated at lysine 20 (H4K20me0), a state which is restricted to newly synthesized histones incorporated into chromatin during DNA replication10. H4K20me0 thereby recruits BRCA1 to post-replicative chromatin, where its promotion of homologous recombination is essential for genome stability and tumour suppression3,10.
Specialized histone-binding domains in BARD1 (the obligate interaction partner of BRCA1) and 53BP1 mediate histone H4 interactions. It has previously been shown that the ankyrin repeat domain (ARD) in BARD1 binds several residues in the H4 tail, and specifically lysine 20 in its unmethylated state10; the tandem-Tudor domain (TTD) of 53BP1 mediates the converse methylation-dependent interaction with H4K20me1 and H4K20me2 (hereafter, H4K20me1/2)7. To achieve specificity for chromatin that is proximal to DSBs, 53BP1 couples the binding of widespread H4K20me1/2 with recognition of the RNF168-dependent H2AK15ub PTM in DSB-proximal chromatin using its ubiquitin-dependent recruitment motif (UDR), which is a TTD-proximal sequence that binds H2AK15ub and features of the nucleosome surface4,5. The equivalent dependence of BRCA1 recruitment on RNF168 activity1,2 similarly implicates H2AK15ub recognition; however, the mechanism that links BRCA1 complexes to this modification during homologous recombination has remained unknown.
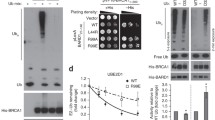
Given that 53BP1–nucleosome interactions involve simultaneous binding to H4K20me1/2 and H2AK15ub, we considered whether the BRCA1–BARD1 complex might also possess sequences that bind H2AK15ub and couple this to H4K20me0 recognition by the ARD of BARD1. BARD1 comprises an N-terminal RING domain, a central ARD and two tandem BRCA1 C-terminal domain (BRCT) repeats at its C terminus (Fig. 1a). Tandem BRCTs are frequently present in DNA-damage-responsive proteins and typically bind phosphoserine-containing peptide ligands in partner proteins11,12,13,14. Putative phosphopeptide-binding residues are conserved in the BRCTs of BARD1, but reportedly bind to poly-ADP-ribose (PAR) chains induced at sites of DNA damage15. Despite this, it has been shown that a PAR-binding-defective point mutant of BARD1 (BARD1(K619A)) is fully proficient in repairing olaparib-induced DNA lesions10. In agreement, mice that are homozygous for equivalent mutations in the BRCTs of BARD1 were not tumour-prone and displayed a cellular proficiency for homologous recombination, which dismisses a role for BRCT-dependent interactions with PAR or phosphoproteins in tumour suppression16. Nevertheless, when BARD1AID/AID HCT-116 cells—which are engineered to encode biallelic auxin-dependent degron tags in the BARD1 C terminus10,17—that had been treated with auxin (hereafter, BARD1∆/∆ cells) were reconstituted with a BARD1 transgene in which the BRCTs are deleted (BARD1∆BRCT), their hypersensitivity to olaparib (Fig. 1b) was consistent with their previously identified importance for homologous recombination18. Notably, BARD1(∆BRCT) protein was expressed at endogenous levels and stabilized BRCA1 (Extended Data Fig. 1a), which prompted us to consider a specific and undescribed function for the BRCTs of BARD1 in homologous recombination.
a, BARD1 domain map, with ARD 3A and K619A mutations indicated. b, Survival of the indicated BARD1∆/∆ cells grown in the presence of olaparib. Cultures were supplemented with doxycycline (2 μg ml−1) for 24 h before addition of 1 mM auxin (indole-3-acetic acid (IAA)). Olaparib (500 nM) was added 1 h after IAA. Cells were stained following a 10-day treatment with olaparib. Representative data, n = 3 biological experiments. WT, wild type. c, Space-filling (top) and ribbon (bottom) models of the BARD1 tandem BRCT crystal structure (Protein Data Bank code 2NTE), pseudocoloured to indicate amino acid conservation. Red dashed ovals indicate the inter-β2′–β3′ loop. d, As in b, for additional BARD1 mutants with and without IAA and/or olaparib treatment. Representative data, n = 3 biological experiments. e, The inter-β2′–β3′ loop, with amino acid side chains represented.
To identify putative functional surfaces in the BRCTs of BARD1, we mapped sequence conservation onto a crystal structure of this domain19, and used this to prioritize highly conserved solvent-exposed residues for mutagenesis (Fig. 1c). We then stably integrated BARD1 transgenes bearing neutral or disruptive amino acid substitutions at nine positions into BARD1AID/AID cells, and assayed for olaparib sensitivity following auxin-induced depletion of endogenous BARD1 (Fig. 1d, Extended Data Fig. 1b). Only mutations within a focused cluster of three conserved residues (Arg705, Asp712 and Gln715) conferred olaparib sensitivity (Fig. 1d). These all mapped to the loop formed between β-sheets 2 and 3 of the second BRCT of BARD1 (inter-β2′–β3′ loop of BRCT2) (Fig. 1e), a protruding feature that comprises three 310 helices previously noted to be unique among BRCTs19. The observation that all three mutant BARD1 proteins were stable (Extended Data Fig. 1b, c) but potentiated olaparib hypersensitivity indicated a direct role for the inter-β2′–β3′ loop in homologous recombination.
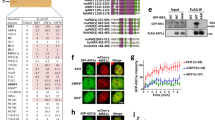
We noted that cells with mutations that affect the inter-β2′–β3′ loop of BRCT2 exhibited olaparib-sensitivity profiles equivalent to those of cell lines in which the ARD of BARD1 is mutated (N470A/E467A/D500A; hereafter, ARD 3A) (Fig. 2a, Extended Data Fig. 2a), and considered that the ARD and BRCTs of BARD1 might be functionally interconnected. Consistent with this notion, ARD 3A mutations did not synergize with the D712A mutation (which is in the inter-β2′–β3′ loop) in increasing cellular hypersensitivity to olaparib (Fig. 2b, Extended Data Fig. 2b) or cisplatin (Fig. 2c, Extended Data Fig. 2c). We therefore used BARD1∆/∆ cells complemented with wild-type, BARD1ARD 3A, BARD1D712A or BARD1ARD 3A/D712A double-mutant BARD1 transgenes to assess whether cooperation between the ARD and tandem BRCTs in BARD1 was necessary for BRCA1–BARD1 recruitment to DSB sites. We combined high-content imaging of BRCA1 ionizing-radiation (IR)-induced foci (IRIF) with immunofluorescence intensity-labelling of H4K20me0 to quantify BRCA1 recruitment in H4K20me0high cell populations in which IR-induced BRCA1 recruitment is highest10. BARD1-deficient cells complemented with a glutathione S-transferase (GST) control exhibited marked BRCA1 recruitment defects, which were suppressed upon complementation with wild-type BARD1 (Fig. 2d, e, Extended Data Fig. 2d, e). However, only very low frequencies of BRCA1 IRIF were observed in cells complemented with the BARD1ARD 3A, BARD1D712A or BARD1ARD 3A/D712A transgenes (Fig. 2d, e, Extended Data Fig. 2d, e). This confirmed a requirement for ARD–BRCT cooperation in the recruitment of BRCA1. We suspected that residual BRCA1 IRIF detected in ARD- and BRCT-mutant-complemented cells were dependent on the BRCA1-A complex, a protein complex composed of BRCC36, ABRAXAS, BRE, MERIT40 and RAP80 that recruits BRCA1–BARD1 to DNA damage sites via RAP80-mediated interactions with lysine-63-linked polyubiquitin chains20,21,22,23,24 but is dispensable for BRCA1-dependent homologous recombination25. Consistent with previous reports25,26, BRCA1 IRIF frequencies in BARD1∆/∆ cells complemented with wild-type BARD1 were only modestly reduced by deletion of RAP80 (Fig. 2f, g). By contrast, BRCA1 IRIF were ablated in RAP80−/−BARD1∆/∆ cells complemented with the BARD1(D712A), BARD1(ARD 3A) and BARD1(ARD 3A/D712A) double mutants (Fig. 2f, g, Extended Data Fig. 2f, g). Thus, the ARD and BRCTs of BARD1 recruit BRCA1 to DSBs independently of RAP80 and BRCA1-A. Notably, however, BARD1(D712A) expression substantially improved the survival of olaparib-treated RAP80+/+BARD1∆/∆ cells but offered no survival benefit when expressed in RAP80−/−BARD1∆/∆ cells (Fig. 2h). Thus, our results suggest that in the absence of BARD1-dependent recruitment of BRCA1–BARD1, residual RAP80-dependent recruitment of BRCA1 makes a significant—albeit partial—positive contribution to DNA repair.
a–c, Survival of the indicated BARD1AID/AID cell lines grown for 7 days in the presence of IAA (1 mM), doxycycline (2 μg ml−1) and the indicated doses of olaparib or cisplatin. Resazurin cell viability assay, n = 3 biological experiments, mean ± s.d. d, Immunofluorescent microscopy of BRCA1 IRIF in H4K20me0+ BARD1AID/AID cell lines. Cultures that were supplemented with doxycycline (2 μg ml−1 for 24 h) before addition of IAA (1 mM) were irradiated (5 Gy) after 2 h, and fixed 2 h later. Scale bar, 5 μm. Representative of n = 2 biological experiments. e, Top, quantification of BRCA1 IRIF from d. Boxes, median ± 25th–75th percentiles; whiskers, 10th–90th percentiles. BRCA1 foci measurements are made for nuclei in the top quartile of H4K20me0 integrated staining intensity (≥171 nuclei per condition). Integrated intensity and foci quantifications were made using CellProfiler. Significance was determined by two-sided Kruskal–Wallis H test with Dunn’s correction for multiple comparisons. ****P ≤ 0.0001. Bottom, mean number of BRCA1 foci per cell from two independent experiments. f, g, Same as in d, e, respectively, in BARD1∆/∆RAP80−/− cells (≥ 178 nuclei per condition). h, Survival of RAP80+/+ (left) and RAP80−/− (right) BARD1∆/∆ cell lines grown for 7 days in the presence of olaparib. Cultures were supplemented with doxycycline (2 μg ml−1) for 24 h before addition of IAA (1 mM) and olaparib. Resazurin cell viability assay, n = 3 biological experiments, mean ± s.d. i, Immunofluorescent microscopy of RAD51 IRIF in BARD1∆/∆ cells expressing BARD1D712A, BARD1ARD 3A or BARD1ARD 3A/D712A transgenes. Cultures that were supplemented with doxycycline (2 μg ml−1, 24 h) before addition of IAA (1 mM, 2 h) were irradiated (5 Gy) and fixed 2 h later. Scale bar, 5 μm. Representative of n = 3 biological experiments. j, Top, quantification of RAD51 foci per cell from i. Nuclei per condition, ≥255. Boxes, median ± 25th–75th percentiles; whiskers, 10th–90th percentiles. Significance was determined by two-sided Kruskal–Wallis H test with Dunn’s correction for multiple comparisons. ****P ≤ 0.0001. Representative of n = 3 biological experiments. Bottom, mean number of RAD51 IRIF from three independent biological experiments ± s.d.
Next, to examine the contribution of the ARD and BRCTs of BARD1 to homologous recombination, we quantified the effect of the BARD1D712A and BARD1ARD 3A mutations on RAD51 recruitment into IRIF. In contrast to BARD1∆/∆ cells complemented with wild-type BARD1 (in which RAD51 frequencies were fully restored), complementation with BARD1(D712A), BARD1(ARD 3A) or BARD1(ARD 3A/D712A) did not improve RAD51 recruitment into IRIF when compared to GST-complemented control cells (Fig. 2i, j). These results collectively confirm that the ARD–tandem BRCT architecture in BARD1 functions as the primary recruitment module of BRCA1 during homologous recombination.
Interdependence between the ARD and BRCTs of BARD1 suggested their cooperation in chromatin binding at DSB sites. We therefore speculated that the BARD1 C-terminal domain architecture might couple H4K20me0 and H2AK15ub binding in a manner analogous to the TTD–UDR domains of 53BP1. If the BRCTs of BARD1 interacted with H2AK15ub, we reasoned that they might rescue the recruitment of a UDR-mutated IRIF-forming fragment of 53BP127 (amino acids 1220–1711) that cannot bind H2AK15ub4,5. We tested this hypothesis by expressing chimeric proteins in which wild-type or D712A-mutant versions of the BRCTs of BARD1 were fused C-terminal to wild-type or UDR-mutated fragments of 53BP1(1220–1711), and examined their ability to form IRIF in 53BP1−/−BARD1∆/∆ cells. The 53BP1(1220–1711)–BARD(BRCT1–BRCT2) fusion proteins readily formed IRIF that were completely ablated when recruitment-neutralizing UDR mutations in 53BP1 (L1619A)4,27 and the BRCTs of BARD1 (D712A) were both present (Extended Data Fig. 3a, b). However, the ability of equivalent proteins bearing either the wild-type UDR or the wild-type BRCTs of BARD1 to form IRIF (+D712A and +L1619A panels, respectively, in Extended Data Fig. 3a) strongly suggested that the BRCTs of BARD1—akin to the 53BP1 UDR4—might recognize H2AK15ub at DSB sites.
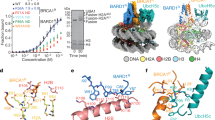
To directly test whether the BRCTs of BARD1 bind H2AK15ub-labelled nucleosomes, we recombinantly expressed and purified a GST-fusion protein fragment encoding the ARD and BRCTs of BARD1 (GST–BARD1(ARD–BRCT)) (Extended Data Fig. 3c). We then performed GST pull-down assays after incubation with recombinant nucleosomes that were unmodified, chemically methylated at H4K20 (H4Kc20me2), chemically ubiquitinated at H2AK15 (H2AKc15ub) or modified at both positions (Extended Data Fig. 3d, e). In these experiments, interactions between the BARD1(ARD–BRCT) fragment and nucleosomes were strongly stimulated by the presence of H2AKc15ub, but inhibited when H4Kc20me2 was also present (Fig. 3a). By contrast, the presence of both histone PTMs was required when we performed nucleosome binding experiments using a control recombinant GST-fusion polypeptide that encodes the TTD–UDR of 53BP1 (GST–53BP1(TTD–UDR)) (Fig. 3a), consistent with previous observations4,5,28. We confirmed the inhibitory effect of H4K20 methylation on the binding of BARD1 to H2AK15ub-labelled nucleosomes using an electrophoretic mobility shift assay (EMSA), in which a recombinant monomeric 6×His–maltose-binding protein (MBP)–BARD1(ARD–BRCT) fragment exhibited over fourfold-higher affinity for H2AK15ub-labelled nucleosomes when they were not methylated on H4K20 (Fig. 3b, Extended Data Fig. 3f). The inverse was seen for a GST–53BP1(TTD–UDR) fragment: H4K20 dimethylation stimulated binding to H2AKc15ub-labelled nucleosomes by EMSA (Extended Data Fig. 3f), as expected4,5,7. Lastly, interactions between H2AKc15ub-modified nucleosomes and GST–BARD1(ARD–BRCT) fragments were sensitive to the ARD 3A and D712A mutations alone and in combination (Fig. 3c, Extended Data Fig. 3g). These results collectively confirm that the ARD and BRCT domains cooperate in nucleosome binding, and imply a specificity for RNF168-dependent H2AK15ub. H2AK15ub-directed specificity was furthermore confirmed in nucleosome pull-down experiments in which ubiquitin was instead conjugated to H2A lysine 119 (H2AKc119ub), a Polycomb repressive complex 1 (PRC1)-dependent modification that is present on 5–15% of all H2A in vertebrate cells29. In contrast to its high affinity for H2AKc15ub, we did not detect an interaction between recombinant GST–BARD1(ARD–BRCT) polypeptides and H2AKc119ub-containing nucleosomes, indicating a selectivity for H2AK15ub (Fig. 3d). Thus, the BRCTs of BARD1 are a reader of DNA-damage-dependent H2AK15ub. It was because of the analogous functioning UDR in 53BP1 (which also binds H2AK15ub4,5,28) that we gave the inter-β2′–β3′ loop of BARD1 the name BUDR (for BRCT-associated ubiquitin-dependent recruitment motif).
a, Immunoblots from pull-down assay using GST–BARD1(ARD–BRCT) and GST–53BP1(TTD–UDR) fragments immobilized on glutathione affinity beads and incubated with recombinant nucleosome variants. Nucleosomes were either modified with dimethyl-lysine analogues at H4 position 20 and/or chemically ubiquitinated at H2A position 15. Representative of n = 3 independent experiments. b, Quantification of EMSA experiments in which H2AKc15ub-modified or H2AKc15ub- and H4Kc20me2-modified nucleosomes (or control DNA) were incubated with increasing concentrations of 6×His–MBP–BARD1(ARD–BRCT). Complexes were resolved by native PAGE and visualized using Diamond DNA stain. Data from n = 3 independent experiments ± s.e.m. c, Immunoblots from pull-down assay using tandem GST (GST2), GST–BARD1(ARD–BRCT) wild type, and the indicated point mutants. Pull-down was performed on immobilized GST-tagged proteins incubated with recombinant nucleosomes chemically ubiquitinated at lysine 15 on H2A. Representative of n = 3 independent experiments. d, Immunoblots from pull-down assay using GST–BARD1(ARD–BRCT) fragments immobilized on glutathione affinity beads, and incubated with recombinant nucleosome variants. Nucleosomes were chemically ubiquitinated at H2A position 15 or 119. Representative of n = 3 independent experiments.
The RNF168 E3 ubiquitin ligase catalyses ubiquitination of H2AK15 at sites of DSBs6, promoting BRCA1 and 53BP1 recruitment into IRIF1,2,6. Consistently, RNF168-deletion in BARD1AID/AID cells blocked BRCA1 and 53BP1 recruitment near completely (Extended Data Fig. 4a–d). This effect was specific, as cells deleted of the PRC1 E3 ubiquitin ligases (RING1A and RING1B) were fully proficient in supporting 53BP1 and BRCA1 IRIF, despite ablating H2AK119ub (Extended Data Fig. 4e, f). Likewise, BARD1∆/∆ cells complemented with the BARD1(R99E) mutant defective for H2AK127 ubiquitination30,31 supported normal frequencies of BRCA1 and 53BP1 IRIF (Extended Data Fig. 4g). Thus, BRCA1–BARD1 recruitment to DNA damage sites does not involve Polycomb- and BRCA1–BARD1-directed H2A ubiquitination events.
In cell viability assays performed in the presence of increasing doses of olaparib, RNF168 deletion markedly increased the survival of BARD1∆/∆ cells (Fig. 4a). However, olaparib resistance in RNF168−/−BARD1∆/∆ cells was fully rescued to levels comparable to those of non-auxin-treated RNF168−/−BARD1AID/AID control cells only when the former were additionally complemented with wild-type BARD1 (Fig. 4a, b). An equivalent rescue also occurred when RNF168−/−BARD1∆/∆ cells were complemented with BARD1(ARD 3A) or BARD1(D712A) expression (Fig. 4a, b). This indicates a genetic epistasis between loss of RNF168-dependent H2AK15ub and mutations that affect the ability of BARD1 to recognize this mark. In complete agreement, IRIF formed by a fragment of BARD1 encompassing its minimal chromatin-binding domains fused to the monomeric GFP variant mClover2 (mClover2–BARD1(ARD–BRCT)) were equally disrupted by deletion of RNF168, or ARD 3A or BUDR mutations in live-cell imaging experiments (Fig. 4c). However, it is noteworthy that mClover2–BARD1(ARD–BRCT) IRIF did not withstand fixation procedures, suggesting other nucleosome-binding features of the BRCA1–BARD1 complex32 may stabilize ARD–BRCT-dependent chromatin interactions.
a, b, Survival of the indicated BARD1AID/AID cell lines grown for 7 days in the presence of the indicated doses of olaparib. Cultures were grown in doxycycline (2 μg ml−1) for 24 h before addition of IAA (1 mM) (a) or dimethyl sulfoxide (DMSO) (b) (carrier control), and subsequent addition of olaparib (at 25 h). Seven days later, cell viability was measured by resazurin assay. n = 3 biological experiments, mean ± s.d. c, BARD1AID/AID cells expressing an mClover2-BARD1ARD-BRCT transgene or derivatives with ARD 3A or D712A mutations were grown in doxycycline (2 μg ml−1, 24 h) before addition of IAA (1 mM). Two h later, cultures were irradiated (10 Gy); live imaging began 60 min after irradiation. Scale bar, 10 μm. Representative of n = 2 independent experiments. BARD1(ARD–BRCT) corresponds to amino acids 425–777. NLS, nuclear localization signal. d–g, Survival of the indicated 53BP1+/+BARD1∆/∆ (left panels) or 53BP1−/−BARD1∆/∆ (right panels) cell lines grown for 7 days in the presence of the indicated doses of olaparib or cisplatin. Cultures were grown in doxycycline (2 μg ml−1) for 24 h before addition of IAA (1 mM), and subsequent addition of olaparib or cisplatin (at 25 h). Seven days later, cell viability was measured by resazurin assay. n = 3 biological experiments, mean ± s.d. Data in d and Fig. 2a were collected from common experiments, with the indicated cell lines displayed separately for clarity. h, Proposed model for bivalent nucleosome recognition by BARD1. i, Logic gate depicting how combinatorial H2AK15 and H4K20 PTM states govern the choice of DSB repair pathway. HR, homologous recombination; NHEJ, non-homologous end joining.
Given the vital function H2AK15ub also has in recruiting 53BP1 to chromatin at DSB sites and the importance of BRCA1 for antagonizing 53BP1 interactions with post-replicative chromatin10,33,34, we hypothesized that the chromatin-reader activities of BARD1 may have evolved to counteract 53BP1 pathway activity. In agreement with this notion, RAD51 IRIF were diminished in BARD1∆/∆ cells and restored in 53BP1−/−BARD1∆/∆ cells, and no further increases accompanied reconstitution of wild-type or mutant BARD1 protein expression (Extended Data Fig. 5d). However, despite this rescue of RAD51 recruitment, 53BP1−/−BARD1∆/∆ cells retained substantial sensitivity to both olaparib and cisplatin treatment (half-maximal inhibitory concentration (IC50) of about 63 nM for olaparib) (Fig. 4d–g), a result that suggested incomplete restoration of homologous recombination. By contrast, resistance to both cisplatin and olaparib treatment was fully restored in 53BP1−/−BARD1∆/∆ cells upon reconstitution with BARD1(D712A) and BARD1(ARD 3A) mutant proteins (IC50 for olaparib of >1 μM), which was comparable to cells reconstituted with wild-type BARD1 (Fig. 4d–g, Extended Data Fig. 5e–h). In summary, the equal effect of 53BP1 and RNF168 loss in restoring homologous recombination functionality to cells proficient or deficient in BARD1–chromatin interactions is consistent with 53BP1 pathway inhibition representing the primary role for the interactions of BARD1 with H4K20me0- and H2AK15ub-marked nucleosomes. Our results also suggest that BRCA1–BARD1 complexes exert important functions that do not require BARD1-mediated chromatin binding, which perhaps partly explains the incomplete phenotypic suppression by 53BP1 deletion in mice that bear severe Brca1 loss-of-function alleles35,36.
Altogether, our results answer the long-standing question of how the DNA-damage-associated H2AK15ub histone modification promotes recruitment of BRCA1–BARD1 complexes, and how these complexes coordinate the promotion of homologous recombination with the inhibition of 53BP1-dependent non-homologous end joining. In identifying the BRCTs of BARD1 as a specific receptor for H2AK15ub, we also reveal a conserved and simple principle that governs the equilibrium between competing DSB repair pathways, in which two histone PTM states—one of which is cell-cycle-regulated (H4 with or without mono- or dimethylation at K20) and one of which is DNA-damage-dependent (H2A with or without ubiquitination at K15)—specify bivalent interactions with the reader domains of the mediator proteins of distinct repair pathways (Fig. 4h–i). The shared affinity of the ARD–BUDR architecture of BARD1 and the TTD–UDR architecture of 53BP1 for H2AK15ub-modified nucleosomes, but their inverse affinities for H4K20 methylation, explains the respective preferences of these proteins for DSB-associated chromatin in post- and pre-replicated regions of the genome, and the establishment of DSB repair pathway choice.
Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized, and investigators were not blinded to allocation during experiments and outcome assessment.
Cell lines and culture conditions
BARD1AID/AID cells lines were generated by biallelic knock-in of auxin-inducible degron tags at the C terminus of the endogenous BARD1 loci in the adult male HCT116 colorectal carcinoma cells (parental cell line was a gift from I. Tomlinson; RRID: CVCL_0291) carrying doxycycline-inducible copies of Oryza sativa TIR1 integrated at the AAVS1 loci, as previously described8. All BARD1AID/AID and derivative cell lines were maintained in Dulbecco’s modified Eagle medium (DMEM)–high glucose (Sigma-Aldrich, D6546) supplemented with 10% FBS, penicillin–streptomycin and 2 mM l-glutamine. Cultures were maintained at 37 °C with 5% CO2.
To generate lentivirus for stable transgene complementation, HEK293T female embryonic kidney cells (obtained from Francis Crick Institute Cell Services; RRID: CVCL_0063) were co-transfected with a lentiviral vector encoding the transgene of interest, pHDM-tat1b, pHDM-G, pRC/CMV-rev1b and pHDM-Hgpm2 using 1.29 μg polyethylenimine per μg of DNA in Opti-MEM (Thermo Fisher, 31985062). Viral supernatants were collected at 48 h and 72 h after transfection, syringe-filtered (0.45 μm), and immediately used to transduce target cells populations in the presence of 4 μg ml−1 polybrene. Transduced populations were selected with antibiotic beginning 24 h after the last round of transduction until a non-transduced control population was completely dead. Stably transduced cell lines were maintained in the presence of selective antibiotic.
All knockout cell lines were generated by CRISPR–Cas9. Gene-specific gRNAs were integrated into pSpCas9(BB)-2A-GFP (PX458) (Addgene no. 48138) and 2 μg of plasmid was electroporated into 106 cells using a Lonza 4D-Nucleofector according to the manufacturer’s protocol for HCT116 cells. GFP-positive cells were sorted 24 h after electroporation using a Sony SH800 cell sorter with the brightest 5% being pooled for recovery in medium containing 50% FBS for 4 days. Sorted populations were then seeded at low density and individual clones were isolated after 10 days of outgrowth. Individual clones were validated by western blot and sequencing. All cell lines were validated as described in the reporting checklist, and tested for mycoplasma contamination upon entering the laboratory.
Survival experiments
To generate survival curves for BARD1AID/AID and derivative cell lines, 300 cells per well were seeded in the presence of doxycycline (2 μg ml−1) in triplicate for each drug concentration in a 96-well plate. Each cell line was plated in duplicate for plus and minus IAA conditions. After 24 h, IAA (1 mM) or carrier (DMSO) was added. One hour after IAA addition, olaparib or cisplatin was added to the indicated final concentrations. Seven days after drug addition, the medium was replaced with phenol red-free DMEM (Thermo Fisher, 21063-029) supplemented with 10% FBS, penicillin–streptomycin, 2 mM l-glutamine and 10 μg ml−1 resazurin (Sigma-Aldrich, R7017). Plates were then returned to the incubator for 2–4 h or until the growth medium in untreated control wells began to develop a pink colour. Relative fluorescence was measured with a BMG LABTECH CLARIOstar plate reader. The mean of three technical repeats after background subtraction was taken as the value for a biological repeat and three biological repeats were performed for each experiment. All survival curves presented in this Article represent the mean of three biological repeats ± s.d.
For survival experiments analysed by crystal violet staining, 104 cells were seeded per well of a 6-well plate in triplicate for each cell line in the presence of doxycycline (2 μg ml−1). After 24 h, IAA (1 mM) or carrier (DMSO) was added. One hour after IAA addition, olaparib was added to the indicated final concentrations. Ten days after plating, the growth medium was removed and the cells were washed briefly with PBS before the addition of crystal violet stain (0.5% crystal violet in 25% methanol). Cells were stained for 5 min, washed with ddH2O and dried before scanning. Representative wells were selected for display.
Immunofluorescence
For experiments analysing BRCA1 foci, 106 cells were passed through a 70-μm mesh cell strainer (Thermo Fisher, 22363548) and seeded in a single well of a 6-well plate in the presence of doxycycline (2 μg ml−1). After 24 h, IAA was added to a final concentration of 1 mM. Cells were irradiated (5 Gy) 2 h after IAA addition, trypsinized 2 h after irradiation, and 105 cells were plated on fibronectin-coated glass coverslips (13 mm) using a cytospin. Coverslips were immediately moved to ice-cold cytoskeletal buffer (10 mM PIPES pH 6.8, 300 mM sucrose, 50 mM NaCl, 3 mM EDTA, 0.5% Triton X-100, protease inhibitor cocktail (cOmplete EDTA-free; Roche, 27368400)) for 5 min before fixation in 2% PFA. BARD1AID/AID 53BP1−/− cells stably transduced with 53BP1–BARD1 fusion protein were prepared identically as described for BRCA1 foci, but were immediately fixed in 2% PFA after cytospin. After fixation, these cells were permeabilized in PBS containing 0.2% Triton X-100. For analysis of BRCA1 foci in RNF168−/−, PRC1−/− and BARD1R99E cells (Extended Data Fig. 4c-g), 2 × 105 cells were passed through a 70-μm mesh cell strainer and seeded on 3 fibronectin-coated glass coverslips (13 mm) in a single well of a 6-well plate in the presence of doxycycline (2 μg ml−1). After 24 h, IAA was added to a final concentration of 1 mM. Two h after IAA, cultures were treated with 40 μM EdU for 10 min. EdU was washed out with fresh medium and the cells were immediately irradiated (5 Gy), then fixed in 2% PFA 2 h after irradiation.
We found RAD51 foci staining to be disrupted by cytospin plating. For RAD51 foci quantification, 2 × 105 cells were passed through a 70-μm mesh cell strainer and seeded on 3 fibronectin-coated glass coverslips (13 mm) in a single well of a 6-well plate in the presence of doxycycline (2 μg ml−1). After 24 h, IAA was added to a final concentration of 1 mM. Cells were irradiated (5 Gy) 2 h after IAA addition and fixed in 2% PFA 2 h after irradiation.
Staining of all fixed cells began with 15 min blocking (3% BSA, 0.1% Triton X-100 in PBS), followed by 1 h incubation with primary antibody in a humidity chamber. For experiments in which cells were treated with EdU, the Click-iT EdU Cell Proliferation Kit, Alexa Fluor 647 (Thermo Fisher, C10340) was used to label EdU-positive cells according to the manufacturer’s protocol between blocking and primary antibody incubation. The following primary antibodies were used at the indicated concentrations: mouse anti-HA (1:200, HA.11 901501 Biolegend), mouse anti-BRCA1 D-9 (1:40, sc-6954 Santa Cruz), rabbit anti-H4K20me0 (1:250, ab227804 Abcam), rabbit anti-RAD51 (1:1,000, 70-001 BioAcademia), mouse anti-γH2AX (1:500, 05-636 Millipore), rabbit anti-53BP1 (1:250, NB100-304) and rabbit anti-γH2AX (1:500, 2212-1 Epitomics). Following primary, coverslips were washed 3 times with PBS containing 0.1% Triton X-100 before incubation with secondary antibody for 1 h in a humidity chamber. Secondary antibodies used in this study were: goat anti-mouse Alexa Fluor 488 (1:500, A-11001 Invitrogen) and goat anti-rabbit Alexa Fluor 568 (1:500, A-11011 Invitrogen). Coverslips were then washed 3 more times with PBS containing 0.1% Triton X-100, once with PBS, and mounted on glass microscope slides using a drop of ProLong Gold antifade reagent with DAPI (Life Technologies, P36935).
Live imaging of BARD1AID/AID cells expressing mClover2-BARD1ARD-BRCT fusion transgenes was performed using Leica SP8-X SMD confocal microscope. For each cell line, 105 cells were seeded in duplicate on 35-mm glass-bottomed dishes in the presence of doxycycline (2 μg ml−1). IAA (1 mM) was added after 24 h and cells were irradiated (10 Gy) 2 h later. Imaging began 1 h after irradiation and continued for 30 min.
Immunofluorescence images for BRCA1 quantification were acquired on a Leica DMi8 widefield microscope (Fig. 2d–g, Extended Data Fig. 2d–g) or Leica SP8-X SMD confocal microscope (Extended Data Fig. 4c–g). 53BP1–BARD1 fusion protein experiments were visualized on a Leica SP8-X SMD confocal microscope. CellProfiler (Broad Institute) was used for foci quantification. Images were visualized and saved in Fiji and assembled into figures in Adobe Illustrator.
Protein extraction and western blotting
Cells were washed once with PBS and lysed by resuspension in ice-cold benzonase cell lysis buffer (40 mM NaCl, 25 mM Tris pH 8.0, 0.05% SDS, 2 mM MgCl2, 10 U ml−1 benzonase, and cOmplete EDTA-free protease inhibitor cocktail (Roche, 27368400)). Extracts were then incubated on ice for 10 min before protein concentration was calculated by Bradford assay (Bio-Rad, 500-0006). Extracts were then mixed with Laemmli buffer and boiled at 95 °C for 5 min before loading on SDS–PAGE gels.
Protein samples were fractionated on NuPAGE 4–12% 1.0 mm Bis-Tris polyacrylamide gels (Life Technologies, NP0322) before transferring to 0.45-μm nitrocellulose membranes (GE Healthcare, 10600003). After transfer, membranes with blocked with 5% milk in PBST for at least 30 min and then incubated overnight with primary antibody in PBST supplemented with 0.03% NaN3 and 3% BSA. Primary antibodies used for western blot in this study include: rabbit anti-53BP1 (1:2,500, Novus Biological, NB100-304), mouse anti-BRCA1 D-9 (1:400, sc-6954 Santa Cruz), rabbit anti-BARD1 (1:500, ab64164 Abcam), mouse anti-β-actin (1:2,000, A1978 Sigma-Aldrich), rabbit anti-RING1B D22F2 (1:1,000, 5694 Cell Signaling), rabbit anti-H2A-K119-Ub D27C4 (1:2,000, 8240S Cell Signaling), rabbit anti-CHK2-Phospho-Thr-68 C13C1 (1:1,000, 2197 Cell Signaling), rabbit anti-H2AX (1:1,000, ab124781 Abcam; also recognizes H2A), and mouse anti-HA (1:2,000, HA.11 901501 Biolegend). Following primary, membranes were incubated with either HRP-conjugated goat anti-mouse (1:20,000, Thermo Fisher, 62-6520) or HRP-conjugated goat anti-rabbit (1:20,000, Thermo Fisher, 65-6120) secondary antibodies. Membranes were developed with Clarity Western ECL Substrate (Bio-Rad, 170-5061) and imaged using a Gel Doc XR System (Bio-Rad).
For nucleosome pull-down assays, proteins were separated using 4–20% Tris glycine gradient gels (BioRad) before transfer onto PVDF membranes. All blocking and antibody incubations were performed in Tris-buffered saline containing either 5% (w/v) BSA or 5% (w/v) skimmed milk powder. For western blotting the following commercial primary antibodies were used: rabbit anti-H2A (Abcam, ab18255), rabbit anti-H3 (Abcam, ab1791), mouse anti-GST (Santa Cruz, sc-138). HRP-conjugated goat anti-rabbit IgG (Vector Laboratories, PI-1000) and HRP-conjugated horse anti-mouse IgG (Vector Laboratories, PI-2000) secondary antibodies were used with enhanced chemiluminescence solution (ECL supersignal, Thermo Scientific) was used for protein detection.
Protein purification
GST2, GST–BARD1(ARD–BRCT) and 6×His–MBP–BARD1(ARD–BRCT) (corresponding to residues 425–777) variants were expressed using 200 μM IPTG in BL-21 DE3 RIL Escherichia coli overnight cultures grown at 16 °C in 2YT broth. Cell pellets were resuspended in lysis buffer (25 mM Tris pH 8, 300 mM NaCl, 0.1% Triton (v/v), 10% glycerol (v/v), 5 mM β-mercaptoethanol, 1× protease inhibitor mix (284 ng ml−1 leupeptin, 1.37 μg ml−1 pepstatin A, 170 μg ml−1 phenylmethylsulfonyl fluoride and 330 μg ml−1 benzamindine), 1 mM AEBSF and 5 μg ml−1 DNaseI). Cells were lysed by sonication and lysozyme treatment and spun at 39,000g for 30 min. For GST-tagged proteins, clarified lysate was applied to a glutathione sepharose 4B column (GE Healthcare). After extensive washing, bound protein was eluted using 30 mM reduced glutathione and concentrated using a 30K MWCO centrifugation device (Amicon). The 6×His–MBP–BARD1(ARD–BRCT) lysates were treated the same, with the addition of 15 mM imidazole to the buffer after lysis. The 6×His–MBP–BARD1(ARD–BRCT) was applied to a chelating HP column (GE Healthcare) preloaded with nickel ions. After extensive washing, protein was eluted using a gradient of imidazole (15 column volumes, final imidazole concentration 300 mM). Proteins were further purified by size-exclusion chromatography using a Superdex 200 Increase 10/300 in SEC buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1 mM DTT, 5% glycerol) and the main mono-disperse protein-containing peak was collected, concentrated, flash frozen in liquid nitrogen and stored at −80 °C. GST–53BP1(TTD–UDR) (residues 1484–1631) was expressed and purified as previously described5.
Protein concentrations were determined via absorbance at 280 nm using a Nanodrop 8000 (Thermo Scientific), followed by SDS–PAGE and InstantBlue (Expedeon) staining with comparison to known amounts of control proteins (Extended Data Fig. 3c, f).
Human histone proteins including site-specific cysteine mutations were expressed in BL-21 DE3 RIL cells and purified from inclusion bodies, essentially as previously described5,37. The 6×His-TEV-ub G76C was expressed in E. coli BL-21 DE3 CodonPlus cells, lysed in 1× Recom-500 buffer (25 mM Na-phosphate buffer pH 7.4, 300 mM NaCl, 0.1% (v/v) Triton, 10% (v/v) glycerol, 4 mM β-mercapthoethanol, 1× protease inhibitor mix, 5 μg ml−1 DNaseI) and treated with lysozyme and sonication. Clarified cell lysate was loaded onto a HiTrap chelating column (GE Healthcare) pre-loaded with Ni2+ ions. After extensive washing, 6×His-TEV-ub was eluted using a gradient of imidazole and peak-protein-containing fractions were concentrated using a 3K MWCO centrifugation device (Amicon). The 6×His-TEV-ub was further purified on a S75 10/300 column in SEC buffer. Protein-containing fractions were dialysed into water supplemented with 1 mM acetic acid before lyophilization.
H4 methyl lysine analogue preparation
H4K20C was expressed and purified as described for other histones. Cysteine-engineered histone H4K20C protein was alkylated essentially as previously described38. In brief, pure histone H4 was reduced with DTT before addition of a 50-fold molar excess of (2-chloroethyl) dimethylammonium chloride (Sigma-Aldrich). The reaction was allowed to proceed for 4 h at room temperature before quenching with 5 mM β-mercaptoethanol. The H4 protein was separated and desalted using a PD-10 desalting column (GE Healthcare), pre-equilibrated in water supplemented with 1 mM acetic acid and lyophilized. After incorporation of alkylation agents was assessed by 1D intact weight ESI mass spectrometry, roughly 85% was found to be modified. Lyophilized H4 was subject to a second round of alkylation as described above with the final reaction proceeding to near completion (about 95%).
H2A chemical ubiquitylation
Mutant human histone H2A engineered with a single cross-linkable cysteine (H2A K15C or K119C) was chemically ubiquitylated essentially as previously described5,39. In brief, an alkylation reaction was assembled with an H2A cysteine mutant (700 μM), 6×His-TEV-ubiquitin G76C (700 μM) and 1,3-dibromoacetone (4.2 mM, Santa Cruz) in 250 mM Tris-Cl pH 8.6, 8 M urea and 5 mM TCEP and allowed to react for 16 h on ice. The reaction was quenched by the addition of 10 mM β-mercaptoethanol and pH adjusted to 7.5. Chemically ubiquitylated H2A (H2AKc15ub or H2AKc119ub) was purified using a HiTrap SP HP column (GE Healthcare) and 6×His-TEV-H2AKc15ub-containing fractions were pooled and enriched over a HiTrap chelating column (GE Healthcare) pre-loaded with Ni2+ ions. The 6×His tag was removed by TEV cleavage and subsequent Ni2+ column subtraction. The resulting flow-through was dialysed against a 2 mM β-mercaptoethanol and dH20 solution and lyophilized. H2AKc15ub was refolded and wrapped into nucleosomes as described in ‘Nucleosome reconstitution’.
Nucleosome reconstitution
Nucleosomes were reconstituted essentially as previously described5,37. Biotinylated 175-bp Widom-601 DNA fragments for wrapping nucleosomes were generated by PCR-based amplification, essentially as previously described40. For PCR amplification, 384 100-μl reactions PCR reactions using Pfu polymerase and HPLC pure oligonucleotides (IDT) were pooled, filtered and purified using a ResourceQ column and salt gradient.
For octamer formation, 4 core histones were mixed at equimolar ratios in unfolding buffer (7 M guanidine HCl, 20 mM Tris pH 7.5, 5 mM DTT) before dialysis to promote refolding into 2 M NaCl, 15 mM Tris pH 7.5, 1 mM EDTA, 5 mM β-mercaptoethanol. Octamers were selected by gel filtration chromatography and assembled into nucleosomes via salt gradient dialysis. Soluble nucleosomes were partially precipitated with 9% polyethylene glycol (PEG) 6000 and resuspended in 10 mM HEPEs pH 7.5, 100 mM NaCl, 1 mM EDTA, 1 mM DTT. Nucleosome formation and quality was checked by native gel electrophoresis and used within one month of wrapping (Extended Data Fig. 3e).
Nucleosome pull-down assays
Pull-down assays were performed essentially as previously described5. In brief, 2.5 μg of GST-tagged 53BP1(TTD–UDR) or 8.5 μg of GST–BARD1(ARD–BRCT) or GST2 was immobilized on BSA-blocked glutathione sepharose beads. Beads were separated and incubated with 2.2 μg of nucleosome variant in pull-down buffer (50 mM Tris-HCL pH 7.5, 150 mM NaCl, 0.02% NP40, 0.1 mg ml−1 BSA, 10% glycerol, 1 mM EDTA, 2 mM β-mercaptoethanol) for 2 h with rotation at 4 °C. Pull-downs were washed three times in pull-down buffer and resuspended directly in 2× SDS loading buffer. All pull-down assays were repeated at least two times, with a single representative immunoblot displayed.
Modified nucleosomes binding assays (EMSA)
Twenty nM of either H2AKc15ub-modified nucleosomes, H4Kc20me2- and H2AKc15ub-modified nucleosomes, or 175-bp Widom-601 DNA (the same DNA used to wrap nucleosomes) was incubated with serial dilutions of recombinant 6×His–MBP–BARD1(ARD–BRCT) to a final volume of 20 μl in EMSA buffer (15 mM Tris pH 7.5, 75 mM NaCl, 0.05 mg ml−1 BSA, 0.5 mM EDTA, 10% glycerol, 0.05% triton X-100, 1 mM DTT, 8% sucrose, 0.01% bromophenol blue). Samples were incubated at 4 °C for 45 min to ensure end point of binding was reached. Products were separated on a native 5% polyacrylamide gel with 0.5× TBE as the running buffer for 2 h at 4 °C. Gels were stained using Diamond DNA stain (Promega). Binding was estimated on the basis of the disappearance of the band corresponding to nucleosome or free DNA and quantified in Image Lab (BioRAD, version 6.1). Binding curves and apparent binding affinity were determined in GraphPad Prism (version 9), using nonlinear regression analysis. EMSA assays were repeated at least in triplicate.
Statistics
Prism 9 (Graphpad Software) was used for graphing and statistical analysis. Relevant statistical methods for individual experiments are detailed within figure legends.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
All data are available in the Article and its Supplementary Information. Source data are provided with this paper.
References
Doil, C. et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136, 435–446 (2009).
Stewart, G. S. et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136, 420–434 (2009).
Hustedt, N. & Durocher, D. The control of DNA repair by the cell cycle. Nat. Cell Biol. 19, 1–9 (2017).
Fradet-Turcotte, A. et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499, 50–54 (2013).
Wilson, M. D. et al. The structural basis of modified nucleosome recognition by 53BP1. Nature 536, 100–103 (2016).
Mattiroli, F. et al. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 150, 1182–1195 (2012).
Botuyan, M. V. et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373 (2006).
Bothmer, A. et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol. Cell 42, 319–329 (2011).
Dimitrova, N., Chen, Y.-C. M., Spector, D. L. & de Lange, T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456, 524–528 (2008).
Nakamura, K. et al. H4K20me0 recognition by BRCA1–BARD1 directs homologous recombination to sister chromatids. Nat. Cell Biol. 21, 311–318 (2019).
Manke, I. A., Lowery, D. M., Nguyen, A. & Yaffe, M. B. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302, 636–639 (2003).
Yu, X., Chini, C. C. S., He, M., Mer, G. & Chen, J. The BRCT domain is a phospho-protein binding domain. Science 302, 639–642 (2003).
Glover, J. N. M., Williams, R. S. & Lee, M. S. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem. Sci. 29, 579–585 (2004).
Wu, Q., Jubb, H. & Blundell, T. L. Phosphopeptide interactions with BRCA1 BRCT domains: more than just a motif. Prog. Biophys. Mol. Biol. 117, 143–148 (2015).
Li, M. & Yu, X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell 23, 693–704 (2013).
Billing, D. et al. The BRCT domains of the BRCA1 and BARD1 tumor suppressors differentially regulate homology-directed repair and stalled fork protection. Mol. Cell 72, 127–139 (2018).
Natsume, T., Kiyomitsu, T., Saga, Y. & Kanemaki, M. T. Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep. 15, 210–218 (2016).
Laufer, M. et al. Structural requirements for the BARD1 tumor suppressor in chromosomal stability and homology-directed DNA repair. J. Biol. Chem. 282, 34325–34333 (2007).
Birrane, G., Varma, A. K., Soni, A. & Ladias, J. A. A. Crystal structure of the BARD1 BRCT domains. Biochemistry 46, 7706–7712 (2007).
Sobhian, B. et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316, 1198–1202 (2007).
Wang, B. et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316, 1194–1198 (2007).
Kim, H., Chen, J. & Yu, X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316, 1202–1205 (2007).
Sims, J. J. & Cohen, R. E. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol. Cell 33, 775–783 (2009).
Sato, Y. et al. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 28, 2461–2468 (2009).
Hu, Y. et al. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 25, 685–700 (2011).
Shao, G. et al. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 23, 740–754 (2009).
Zgheib, O., Pataky, K., Brugger, J. & Halazonetis, T. D. An oligomerized 53BP1 Tudor domain suffices for recognition of DNA double-strand breaks. Mol. Cell. Biol. 29, 1050–1058 (2009).
Hu, Q., Botuyan, M. V., Cui, G., Zhao, D. & Mer, G. Mechanisms of ubiquitin-nucleosome recognition and regulation of 53BP1 chromatin recruitment by RNF168/169 and RAD18. Mol. Cell 66, 473–487 (2017).
Cao, J. & Yan, Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front. Oncol. 2, 26 (2012).
Densham, R. M. et al. Human BRCA1–BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat. Struct. Mol. Biol. 23, 647–655 (2016).
Kalb, R., Mallery, D. L., Larkin, C., Huang, J. T. & Hiom, K. BRCA1 is a histone-H2A-specific ubiquitin ligase. Cell Rep. 8, 999–1005 (2014).
Witus, S. R. et al. BRCA1/BARD1 site-specific ubiquitylation of nucleosomal H2A is directed by BARD1. Nat. Struct. Mol. Biol. 28, 268–277 (2021).
Chapman, J. R., Sossick, A. J., Boulton, S. J. & Jackson, S. P. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J. Cell Sci. 125, 3529–3534 (2012).
Pellegrino, S., Michelena, J., Teloni, F., Imhof, R. & Altmeyer, M. Replication-coupled dilution of H4K20me2 guides 53BP1 to pre-replicative chromatin. Cell Rep. 19, 1819–1831 (2017).
Nacson, J. et al. BRCA1 mutation-specific responses to 53BP1 loss-induced homologous recombination and PARP inhibitor resistance. Cell Rep. 24, 3513–3527 (2018).
Chen, J. et al. 53BP1 loss rescues embryonic lethality but not genomic instability of BRCA1 total knockout mice. Cell Death Differ. 27, 2552–2567 (2020).
Dyer, P. N. et al. in Methods in Enzymology Vol. 375 (eds Allis, C. D. & Wu, C.) 23–44 (Academic, 2003).
Simon, M. D. et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128, 1003–1012 (2007).
Long, L., Furgason, M. & Yao, T. Generation of nonhydrolyzable ubiquitin-histone mimics. Methods 70, 134–138 (2014).
Wilson, M. D. et al. Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer. Nat. Commun. 10, 4189 (2019).
Acknowledgements
We thank all members of the laboratory of J.R.C. for discussions; D. Durocher for plasmid reagents; N. Johnson for discussions regarding unpublished data; and T. Humphrey for comments on the manuscript. This work was funded by a Cancer Research UK (CRUK) Career Development Fellowship (C52690/A19270 to J.R.C.), which previously provided salary support to J.R.B., and CRUK Oxford Centre funding (C5255/A18085 to J.R.B. and J.R.C.). J.R.B.’s salary is provided by a Ruth L. Kirschstein NRSA Individual Postdoctoral Fellowship (F32) (NIH/NCI - F32CA239339). The laboratory of J.R.C. is also supported by the Medical Research Council (MRC - MR/R017549/1), the MRC Molecular Haematology Unit (MRC MHU, UK) and the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre (BRC). J.R.C. holds a Lister Institute Research Prize Fellowship. The Wellcome Centre for Human Genetics (WHG) is supported by Wellcome core award 090532/Z/09/Z. C.B. was sponsored by an ERASMUS+ internship. M.D.W.’s work is supported by the Wellcome Trust (210493), the MRC (T029471/1) and the University of Edinburgh. The Wellcome Centre for Cell Biology is supported by core funding from the Wellcome Trust (203149). A.G.’s research is supported by the Lundbeck Foundation (R198-2015-269 and R313-2019-448), the European Research Council (ERC CoG no. 724436) and Independent Research Fund Denmark (7016-00042B and 4092-00404B). Research at CPR is supported by the Novo Nordisk Foundation (NNF14CC0001). We also acknowledge the BHF Centre of Research Excellence, Oxford (RE/13/1/30181), and the WHG Cellular Imaging core for equipment access and support.
Author information
Authors and Affiliations
Contributions
J.R.B. designed and analysed the majority of experiments, and supervised experiments performed by C.B. M.D.W. and G.C. undertook and analysed all in vitro nucleosome binding experiments. The project was initiated in collaboration with A.G. J.R.C. conceived and supervised the project, and designed and analysed experiments. J.R.B. and J.R.C. co-wrote the manuscript, with author input.
Corresponding author
Ethics declarations
Competing interests
A.G. is co-founder and CSO in Ankrin Therapeutics. No other authors have competing interests.
Additional information
Peer review information Nature thanks Daniel Durocher, Joanna Morris and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 BARD1 BRCT-mutated transgenes are stably expressed in BARD1AID/AID HCT-116 cells.
Related to Fig. 1. a, Immunoblots of whole-cell lysates collected at the indicated time points after IAA addition. Expression of the auxin-degron-targeting SCF-complex E3 ligase Oryza sativa TIR1 was induced using doxycycline (2 μg ml−1), 24 h before the depletion of endogenous BARD1–AID protein with IAA (1 mM). Representative of two biological repeats. b, Immunoblot from whole-cell lystes of BARD1 BRCT mutants screened for olaparib sensitivity. c, Immunoblot of whole cell lysates from BARD1AID/AID cells expressing the indicated transgenes. Cells were seeded in the presence of doxycycline (2 μg ml−1) and IAA (1 mM) was added after 24 h. Lysates were collected at the indicated time points after IAA addition. Representative of two biological repeats.
Extended Data Fig. 2 ARD and BRCTs in BARD1 cooperate in recruiting BRCA1 to post-replicative chromatin during homologous recombination.
Related to Fig. 2. a–c, Survival of the indicated BARD1AID/AID cell lines grown for 7 days without IAA in the presence of the indicated doses of olaparib or cisplatin. Cell lines were seeded in doxycycline (2 μg ml−1) for 24 h before olaparib or cisplatin addition. Resazurin cell viability assay, n = 3 biological experiments, mean ± s.d. d, High-content immunofluorescent microscopy of BRCA1 IRIF in H4K20me0+ BARD1AID/AID cells expressing the indicated transgenes. Cultures were grown in the presence of doxycycline (2 μg ml−1) for 24 h before IAA (1 mM) addition, irradiated 2 h later, and fixed following irradiation. Scale bar, 5 μm. Representative of n = 2 biological experiments. e, Top, quantification of BRCA1 IRIF from d. Boxes indicate the 25th–75th percentiles with the median denoted, and whiskers indicate the 10th–90th percentiles. BRCA1 foci measurements are made for nuclei in the bottom quartile of H4K20me0 integrated staining intensity (≥172 nuclei per condition). Foci quantification and H4K20me0 integrated intensity measurements were performed with CellProfiler. Bottom, mean number of BRCA1 foci per cell from two independent experiments ± s.d. f, g, Same as in d, e, respectively, in RAP80−/− cells. ≥ 179 nuclei per condition. h, i, Survival of the indicated BARD1AID/AID cell lines grown for 7 days without IAA in the presence of olaparib. Cell lines were seeded in doxycycline (2 μg ml−1) for 24 h before olaparib addition. Resazurin cell viability assay, n = 3 biological experiments, mean ± s.d
Extended Data Fig. 3 Purification of BARD1 and 53BP1 fragments and assembly of modified nucleosomes.
Related to Fig. 3. a, Western blot of HA-tagged 53BP1–BARD1 fusion proteins used in b stably expressed in BARD1AID/AID 53BP1−/− cells. b, Top, model depicting the 53BP1–BARD1 fusion protein. The fusion is a chimera composed of the 53BP1 minimal focus-forming region (amino acids 1220–1711) and BARD1 BRCTs (amino acids 555–777). Expressed form includes an N-terminal 2×HA–Flag epitope tag. Bottom, confocal immunofluorescent microscopy of 53BP1–BARD1 chimeric fusion proteins in irradiated BARD1AID/AID53BP1−/− cells. Cultures were grown in the presence of doxycycline (2 μg ml−1) for 24 h before IAA (1 mM) addition and irradiated (5 Gy) 2 h later. Cells were fixed 2 h following irradiation. Scale bar, 10 μm. Representative of n = 2 biological experiments. c, SDS–PAGE gel, stained with InstantBlue protein stain of proteins used in Fig. 3a. d, SDS–PAGE gel, stained with InstantBlue protein stain of nucleosomes used in this study. e, Native gel electrophoresis of Widom 601 DNA in isolation and wrapped with nucleosomes used in this study. f, Representative gel images from EMSA experiments quantified in Fig. 3b. H2AKc15ub-modified, or H2AKc15ub- and H4Kc20me2-modified, nucleosomes (or control DNA) were incubated with increasing concentrations of 6×His–MBP–BARD1(ARD–BRCT) or GST–53BP1(TTD–UDR). Complexes were resolved by native PAGE and visualized using Diamond DNA stain. g, SDS–PAGE gel, stained with InstantBlue protein stain of BARD1 variants used in Fig. 3c. Neighbouring lanes were loaded with two different concentrations.
Extended Data Fig. 4 BRCA1 recruitment to IRIF is RNF168-dependent, but independent of PRC1 and BRCA1–BARD1 ubiquitin ligase activity.
Related to Fig. 4. a, Model indicating the three major known sites of ubiquitin attachment on histone H2A and the genetic manipulations used to block each individually in our experiments. PRC1−/− indicates RING1A−/−RING1B−/− double knockout. b, Immunoblot of whole-cell lysates from BARD1AID/AID parental cells and RNF168−/− derivatives. Cells were seeded in the presence of doxycycline (2 μg ml−1) and IAA (1 mM) was added after 24 h. Lysates were collected 8 h after IAA addition. Representative of two biological repeats. c, Immunofluorescent microscopy of BARD1AID/AID parental cells and RNF168−/− derivatives. Cultures were seeded 24 h before irradiation (5 Gy), and fixed 2 h later. Representative of n = 3 biological experiments. Scale bar, 10 μm. d, Quantification of BRCA1 and 53BP1 IRIF from c. Boxes indicate the 25th–75th percentiles with the median denoted, and whiskers indicate the 10th–90th percentiles. BRCA1 foci measurements are made for EdU-positive nuclei. Foci quantification was performed using using CellProfiler. Significance was determined by two-sided Kruskal–Wallis H test with Dunn’s correction for multiple comparisons. ****P ≤ 0.0001. Representative of n = 3 biological replicates. e, Immunoblot of whole-cell lysates from BARD1AID/AID parental cells and RING1A−/−, RING1B−/− and RING1A−/−RING1B−/− (denoted as PRC1−/−) derivatives. Cultures were seeded 24 h before irradiation (10 Gy), and collected 2 h later. f, g, The indicated cell lines were treated as in d. Representative of n = 3 biological replicates
Extended Data Fig. 5 The β2′–β3′ loop and ARD counteract toxic 53BP1-dependent non-homologous end joining.
Related to Fig. 4. a, Immunofluorescent microscopy of RAD51 IRIF in BARD1AID/AID cells expressing the indicated transgenes. Cultures were grown in the presence of doxycycline (2 μg ml−1) for 24 h before IAA (1 mM) addition, irradiated 2 h later (5 Gy), and fixed with PFA 2 h following irradiation. Data were collected from the same experiment as Fig. 2i, j. Scale bar, 5 μm. Representative of n = 3 biological experiments. b, Top, quantification of RAD51 foci per cell from a. Per condition, ≥255 nuclei. Boxes indicate the 25th–75th percentiles with the median denoted, and whiskers indicate the 10th–90th percentiles. Significance was determined by two-sided Kruskal–Wallis H test with Dunn’s correction for multiple comparisons. ****P ≤ 0.0001, ***P = 0.0003. Data are from same experiment presented in Fig. 2i, j. BARD1AID/AID cells expressing GST and BARD1 are displayed in both for comparison. Representative of n = 3 biological experiments. Bottom, mean number of RAD51 IRIF from three independent biological experiments ± s.d. c, d, Immunoblot of whole-cell lysates from BARD1AID/AID (top) or BARD1AID/AID53BP1−/− (bottom) cells expressing the indicated transgenes. Cells were seeded in the presence of doxycycline (2 μg ml−1) and IAA (1 mM) was added after 24 h. Lysates were collected at the indicated time points after IAA addition. Representative of two biological repeats. e–h, Survival of the indicated BARD1AID/AID cell lines grown without IAA for 7 days in the presence of the indicated doses of olaparib or cisplatin. Cultures were seeded in doxycycline (2 μg ml−1) and olaparib or cisplatin was added 24 h later. Survival was measured after 7 days by resazurin cell viability assay (n = 3 biological experiments) mean ± s.d
Supplementary information
Supplementary Figure
This file contains the uncropped blots shown in Fig. 3 and Extended Data Figs 1, 3, 4 and 5.
Rights and permissions
About this article
Cite this article
Becker, J.R., Clifford, G., Bonnet, C. et al. BARD1 reads H2A lysine 15 ubiquitination to direct homologous recombination. Nature 596, 433–437 (2021). https://doi.org/10.1038/s41586-021-03776-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03776-w
- Springer Nature Limited
This article is cited by
-
Evolved histone tail regulates 53BP1 recruitment at damaged chromatin
Nature Communications (2024)
-
DNMT3B PWWP mutations cause hypermethylation of heterochromatin
EMBO Reports (2024)
-
Shieldin and CST co-orchestrate DNA polymerase-dependent tailed-end joining reactions independently of 53BP1-governed repair pathway choice
Nature Structural & Molecular Biology (2024)
-
Recent advances in chemical protein synthesis: method developments and biological applications
Science China Chemistry (2024)
-
Genetic separation of Brca1 functions reveal mutation-dependent Polθ vulnerabilities
Nature Communications (2023)