Abstract
Systemic insulin sensitivity shows a diurnal rhythm with a peak upon waking1,2. The molecular mechanism that underlies this temporal pattern is unclear. Here we show that the nuclear receptors REV-ERB-α and REV-ERB-β (referred to here as ‘REV-ERB’) in the GABAergic (γ-aminobutyric acid-producing) neurons in the suprachiasmatic nucleus (SCN) (SCNGABA neurons) control the diurnal rhythm of insulin-mediated suppression of hepatic glucose production in mice, without affecting diurnal eating or locomotor behaviours during regular light–dark cycles. REV-ERB regulates the rhythmic expression of genes that are involved in neurotransmission in the SCN, and modulates the oscillatory firing activity of SCNGABA neurons. Chemogenetic stimulation of SCNGABA neurons at waking leads to glucose intolerance, whereas restoration of the temporal pattern of either SCNGABA neuron firing or REV-ERB expression rescues the time-dependent glucose metabolic phenotype caused by REV-ERB depletion. In individuals with diabetes, an increased level of blood glucose after waking is a defining feature of the ‘extended dawn phenomenon’3,4. Patients with type 2 diabetes with the extended dawn phenomenon exhibit a differential temporal pattern of expression of REV-ERB genes compared to patients with type 2 diabetes who do not have the extended dawn phenomenon. These findings provide mechanistic insights into how the central circadian clock regulates the diurnal rhythm of hepatic insulin sensitivity, with implications for our understanding of the extended dawn phenomenon in type 2 diabetes.
Similar content being viewed by others
Main
REV-ERB-α and REV-ERB-β (also known as NR1D1 and NR1D2, respectively) constitute a key druggable component of the circadian clock machinery. REV-ERB-α is the primary form and is highly enriched in the SCN, which is mainly composed of GABAergic neurons5 (Fig. 1a). REV-ERB-α and REV-ERB-β display a robust diurnal rhythm in the SCN, with peak expression at Zeitgeber time (ZT)6–96 (Extended Data Fig. 1a). To assess the function of REV-ERB-α and REV-ERB-β in GABAergic neurons, we generated mice with a GABAergic-neuron-specific knockout of REV-ERB-α and REV-ERB-β (hereafter, knockout mice) by crossbreeding Nr1d1loxPNr1d2loxP double-floxed mice7,8 with Vgat-ires-cre (vgat-cre) mice. The REV-ERB-α allele generates a truncated protein without a DNA-binding domain8 upon Cre recombination and, therefore, could lead to a functionally hypomorphic knockout and/or have effects on transcription that are independent of DNA binding. vgat-cre mice alone, or floxed mice that were not crossbred with vgat-cre mice, served as wild-type controls. Knockout mice were born at a Mendelian ratio and did not show any developmental defects. They displayed a normal diurnal rhythm of wheel-running activities under regular 12-h light/12-h dark (light–dark) conditions (Fig. 1b, Extended Data Fig. 1b–f). Under constant darkness, knockout mice showed a shorter period but maintained overall rhythmicity (Extended Data Fig. 1g–j). Knockout mice also exhibited normal diurnal patterns of food intake (Fig. 1c, d, Supplementary Table 1), total daily food intake (Extended Data Fig. 2a) and body weight (Fig. 1e) on a chow diet in regular light–dark conditions.
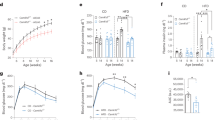
a, RNAscope analysis of REV-ERB-α gene expression in 3-month-old mice at ZT6–9. Scale bars, 500 μm. b, Wheel-running activity in light–dark conditions in 5-month-old wild-type (WT) and knockout (KO) mice (n = 7 mice). c, d, Food intake in light–dark conditions in 4-month-old mice under continuous monitoring (c) or recorded every 12 h (d) (n = 5 mice). e, Body weight (n = 10 mice). f–h, GTTs in 4-month-old mice at ZT0–2 (f), ZT6–8 (g) and ZT12–14 (h) (n = 8 mice). i, Serum insulin levels in 5-month-old mice at ZT12–14 (n = 8 mice). j–m, Insulin clamp analyses in 5-month-old mice (n = 4 mice). j, Glucose infusion rate (GIR). k, Hepatic glucose production rate (HGP). l, Glucose disposal rate (GDR). m, Basal hepatic glucose production rate. Data are mean ± s.e.m. Box plot centre lines, box limits and whiskers represent median, quartiles and minimum and maximum values, respectively. *P < 0.05 by two-way ANOVA or two-sided t-test; NS, not significant. Statistical details are in Supplementary Table 1.
Knockout mice showed ZT-dependent abnormalities in glucose metabolism, with a slight impairment of glucose tolerance at ZT0–2 (Fig. 1f), normal glucose tolerance at ZT6–8 (Fig. 1g) and robust impairment at ZT12–14 (Fig. 1h). The differential glucose intolerance phenotype of knockout mice between ZT6–8 and ZT12–14 was not affected by differential fasting severities (Extended Data Fig. 2b, c) and was not associated with different basal glucose levels (Extended Data Fig. 2d). Knockout mice did not show an altered rhythmicity in their levels of blood insulin or glucagon (Extended Data Fig. 2e, f), but had different levels of corticosterone, glucagon-like peptide 1 and growth hormone at certain ZTs, compared to wild-type mice (Extended Data Fig. 2g–i). The ZT-dependent glucose-tolerance phenotype in knockout mice was not caused by the vgat-cre mouse line (Extended Data Fig. 2j, k). Knockout mice showed a normal increase of blood insulin levels after a glucose injection at ZT12–14 (Fig. 1i), suggesting impaired insulin sensitivity.
Insulin clamp revealed a notable reduction in the glucose infusion rate in knockout mice compared to wild-type mice at ZT12–14 and to a lesser degree at ZT0–2, but not at ZT6–8 (Fig. 1j, Extended Data Fig. 2l–o), providing evidence of ZT-dependent insulin resistance in knockout mice. The low glucose infusion rate in knockout mice at ZT12–14 was mainly driven by a high rate of hepatic glucose production (Fig. 1k), and not by a low glucose disposal rate (Fig. 1l). By contrast, the low glucose infusion rate in knockout mice at ZT0–2 was caused by both a high hepatic glucose production rate and a low glucose disposal rate, which probably accounts for the different shapes of blood glucose curves during glucose-tolerance tests (GTTs) at ZT0–2 versus ZT12–14 in knockout mice. Notably, wild-type mice showed diurnal rhythms in systemic insulin sensitivity (Fig. 1j), hepatic sensitivity to insulin-mediated suppression of glucose production (Fig. 1k) and basal hepatic glucose production (Fig. 1m). All three rhythms peaked at waking (ZT12–14). The insulin sensitivity rhythm—but not the basal glucose production rhythm—was disrupted in knockout mice. These data show that neural REV-ERB has an essential role in regulating the hepatic insulin sensitivity rhythm, independent of eating behaviours or basal glucose production.
In line with the glucose phenotype that was observed on a normal chow diet, knockout mice on a high-fat diet (HFD) displayed increased glucose intolerance and dampened responses to insulin at ZT12–14 (Extended Data Fig. 3a–e). Knockout mice also showed a more marked increase in blood glucose levels than wild-type mice in a mouse model of diabetes induced by the combination of HFD and streptozotocin (Extended Data Fig. 3f–i).
REV-ERB regulates the firing of SCNGABA neurons
Quantitative PCR with reverse transcription (RT–qPCR) analyses at ZT6—a time point corresponding to the peak level of REV-ERB expression—revealed that the most prominent differences in the expression of clock genes Bmal1 (also known as Arntl) and Npas2 between knockout and wild-type mice occurred in the SCN, as compared to the liver and other brain regions (Extended Data Fig. 4 a–i). Whole-cell patch-clamp recordings showed that the firing activity of SCNGABA neurons in knockout mice was higher than that in wild-type mice at ZT12–14 and was also slightly increased at ZT0–2, but there was no difference between knockout and wild-type mice at ZT6–8 (Fig. 2a, b, Extended Data Fig. 5a). The temporal pattern of differences in the firing activity of SCNGABA neurons between knockout and wild-type mice aligns well with the glucose phenotype (Fig. 1). The high firing activity at ZT12–14 in knockout versus wild-type mice was mainly associated with a higher amplitude of miniature excitatory postsynaptic currents (mEPSCs), with less change in the frequency of mEPSCs (Fig. 2c, d, Extended Data Fig. 5b) or in the frequency or amplitude of miniature inhibitory postsynaptic currents (mIPSCs) between knockout and wild-type mice (Fig. 2e, f, Extended Data Fig. 5c). These data suggest that a change in postsynaptic excitatory neurotransmission is a potential mechanism for the difference in SCNGABA firing activity between knockout and wild-type mice, although other mechanisms cannot be excluded.
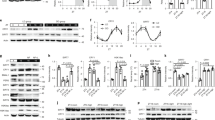
a, b, Spontaneous firing frequency (a) and resting membrane (RM) potential (b) of SCNGABA neurons in 4-month-old mice (n = 12–17 neurons). c, d, Amplitude (c) and frequency (d) of mEPSCs in SCNGABA neurons in 4-month-old mice (n = 10–15 neurons). e, f, Amplitude (e) and frequency (f) of mIPSCs in SCNGABA neurons in 4-month-old mice (n = 11 or 12 neurons). g, Differentially expressed genes (DEGs) in knockout versus wild-type mice identified by RNA-seq of the SCN of 4-month-old mice at ZT12–14. LOC100861615 is also known as Gm3411. Fold change refers to log2-transformed fold change in gene expression in knockout versus wild-type mice. h–k, RT–qPCR analysis of the mRNA levels of Nr1d1 (REV-ERB-α) (h), Nr1d2 (REV-ERB-β) (i), Rgs16 (j) and Gm10406 (α7-Takusan) (k) in the SCN of 4-month-old mice (n = 6 mice). Primers for Nr1d1 and Nr1d2 span the floxed exons. Data are mean ± s.e.m. *P < 0.05 by two-way ANOVA or two-sided t-test. Statistical details are in Supplementary Table 1.
RNA sequencing (RNA-seq) identified differentially expressed genes (knockout versus wild type) in the SCN at ZT12–14 (Fig. 2g). These differentially expressed genes were enriched for genes that encode cell-surface proteins related to neurotransmission (Fig. 2g). For example, Rgs16 (regulator of G-protein signalling 16) is required for circadian production of cyclic AMP in the SCN9. Notably, 13 out of the top 30 upregulated genes belong to the Takusan gene family. Members of this poorly annotated gene family encode proteins that are known to increase the amplitude of mEPSCs10. Rgs16 and several other genes of the Takusan family are known to have oscillatory patterns of expression6,11 with a peak at ZT0–6 (Extended Data Fig. 5d, e), immediately preceding the peak expression of REV-ERB at ZT6–9 (Extended Data Fig. 5f, g). RT–qPCR confirmed the disrupted rhythmicity of expression of Rgs16 and the Takusan gene α7-Takusan (Gm10406) in knockout versus wild-type mice (Fig. 2h–k, Extended Data Fig. 5h–k, Supplementary Table 2). REV-ERB can suppress its own transcription12, leading to artificially upregulated REV-ERB gene expression in the knockout mice when the primers do not span the floxed exons (Extended Data Fig. 5h, i). In situ hybridization confirmed that the rhythmicity of Rgs16 and the Takusan gene Gm3500 in the SCN was disrupted in knockout mice (Extended Data Fig. 5l–o). Chromatin immunoprecipitation revealed that REV-ERB-α binds near to these genes in a temporal pattern that is consistent with the levels of REV-ERB-α in the hypothalamus (Extended Data Fig. 5p, q). These data suggest that the rise in REV-ERB levels late in the day accounts for the repression of Rgs16 and Takusan gene expression in the early stages of the night, although indirect regulatory mechanisms cannot be excluded. Overexpression of Rgs16 or α7-Takusan (Gm10406) in SCNGABA neurons reduced glucose tolerance and impaired glucose responses to insulin at ZT12–14, but to a lesser degree at ZT6–8 (Extended Data Fig. 6a–f). The expression level and the temporal pattern of the exogenous Rgs16 or Takusan genes need to be further fine-tuned to fully address whether they are the de facto cause of the glucose phenotype. These results nonetheless indicate that Rgs16 and/or members of the Takusan family have a role in glucose metabolism.
SCNGABA firing regulates glucose metabolism
We next investigated how the firing activity of SCNGABA neurons per se regulates glucose metabolism. The engineered G-protein-coupled receptor hM3Dq can activate neurons in response to clozapine-N-oxide (CNO). CNO injection in mice expressing hM3Dq at the SCNGABA neurons caused glucose intolerance at ZT12–14 (Fig. 3a, b, Extended Data Fig. 7a, b). The effect was not observed at ZT12–14 in the absence of CNO (Fig. 3c), or at ZT6–8 in the presence of CNO when the baseline firing activity was already high (Fig. 3d). These results suggest that hyperactivity of SCNGABA neurons is sufficient to cause glucose intolerance at waking.
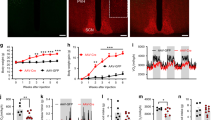
a, Expression of hM3Dq in SCNGABA neurons after bilateral stereotaxic injection of adeno-associated virus (AAV) expressing hM3Dq (Methods). Blue, DAPI; red, mCherry. Scale bar, 100 μm. b–d, GTTs in control (mCherry)- and hM3Dq-injected 3–4-month-old wild-type mice at ZT12–14 treated with CNO (b), ZT12–14 treated with saline (c) or ZT6–8 treated with CNO (d) (n = 11 mice). e, f, GTTs in 3–4-month-old wild-type (n = 12) and knockout (n = 14) mice that were bilaterally injected with AAV expressing hM4Di at the SCN and treated with saline (e) or CNO (f). g, h, Pyruvate-tolerance tests (PTTs) in hM4Di-injected 4-month-old wild-type (n = 6) and knockout (n = 7) mice treated with saline (g) or CNO (h). i, AAV vectors for inducible re-expression of REV-ERB-α. j, Immunostaining for Flag-tagged REV-ERB-α 9 h and 21 h after injection of doxycycline (Dox). Blue, DAPI; red, Flag. Scale bars, 200 μm. k, GTT in 3-month-old mice at ZT12–14, 12 h after injection of doxycycline at ZT0 (n = 9 mice). l, GTT in 3.5-month-old mice at ZT12–14, 24 h after injection of doxycycline at ZT12 (n = 9 mice). Data are mean ± s.e.m. *P < 0.05 by two-way ANOVA or two-sided t-test. Statistical details are in Supplementary Table 1.
Conversely, chemogenetic suppression of SCNGABA neurons with hM4Di blunted the difference between knockout and wild-type mice at ZT12–14 in glucose tolerance and pyruvate tolerance (Fig. 3e–h, Extended Data Fig. 7c, d). Knockout mice did not differ from wild-type mice in GTTs at ZT6–8, regardless of CNO (Extended Data Fig. 7e, f). These results suggest that hyperactivity of SCNGABA neurons at ZT12–14 is required for glucose intolerance and for increased hepatic glucose production in knockout mice. Therefore, the decrease in SCNGABA neuron firing activity in wild-type mice at waking promotes glucose tolerance, probably in anticipation of upcoming feeding behaviours.
REV-ERB rhythmicity regulates metabolism
To address whether the glucose phenotype of knockout mice is due to developmental disruption or clock-independent functions of REV-ERB, we performed inducible re-expression of REV-ERB-α (iREV-ERB) in adult mice specifically in SCNGABA neurons (Fig. 3i, Extended Data Fig. 8a, b). Injection of doxycycline led to robust re-expression of REV-ERB-α after 9 h, followed by a substantial decline after 21 h (Fig. 3j). By timing the injection of doxycycline, we were able to control the oscillatory pattern of exogenous REV-ERB to be either in-phase or anti-phase with the endogenous REV-ERB. The in-phase re-expression improved glucose tolerance in knockout mice at ZT12–14 (Fig. 3k), but had no significant effect at ZT6–8 (Extended Data Fig. 8c). Injection of doxycycline 24 h before GTTs at ZT12–14 (Fig. 3l) or 12 h before GTTs at ZT6–8 (Extended Data Fig. 8d) had no effect on glucose tolerance. Re-expression of REV-ERB-α also suppressed the expression of Rgs16 and Gm10406 (α7-Takusan) in the SCN at ZT12–14 (Extended Data Fig. 8e). Collectively, these results demonstrate that the oscillatory temporal pattern of REV-ERB expression in SCNGABA neurons is critical for systemic glucose metabolism.
Circadian clock and the extended dawn phenomenon
The glucose intolerance observed in knockout mice at waking resembles the ‘dawn phenomenon’—an increase in the levels of blood glucose in the early morning, which affects about 50% of patients with diabetes13. The dawn phenomenon has mostly been studied in type 1 diabetes, and was attributed to waning levels of exogenous insulin at night in patients with type 1 diabetes13,14. The altered sensitivity to insulin in our mouse model suggests that disruption of the circadian clock might be particularly related to the dawn phenomenon in type 2 diabetes. A prominent feature of the dawn phenomenon is persistent hyperglycaemia after breakfast, which is known as the ‘extended dawn phenomenon’3,4. Knockout mice exhibited glucose intolerance in the absence of fasting hyperglycaemia, which suggests that circadian disruption contributes to the extended dawn phenomenon. To address whether the extended dawn phenomenon in type 2 diabetes is associated with the disrupted circadian clock, we used continuous glucose monitoring (CGM) (Extended Data Fig. 9) to study hospitalized patients with type 2 diabetes whose blood glucose levels were already under control through a fixed medication regimen. Patients with the Somogyi phenomenon or who had irregular patterns of sleep or meals before hospitalization were excluded (Supplementary Table 3). The dawn phenomenon was defined by the absolute increment from the lowest level of glucose during the night to the values before or after breakfast (Fig. 4a, b, Methods). There were no differences in age, body mass index, medication use or times of meals or sleeping during hospitalization between patients with and without the dawn phenomenon (Supplementary Tables 4–6), and no differences in the circulating levels of cortisol, insulin and melatonin (Fig. 4c–e). Patients with the dawn phenomenon exhibited a significant increase in the levels of growth hormone in the early morning (Fig. 4f). There was no difference in sleep duration, breathing, blood oxygen or heart rate, except that patients with the dawn phenomenon woke up less during sleep and had less-severe central sleep apnoea (Supplementary Table 5). The temporal pattern of expression of circadian clock genes in the blood and in other peripheral tissues correlates well with biomarkers of the central clock and responds rapidly to entraining signals in humans15,16. Therefore, we inferred the molecular clock functionality through an RT–qPCR analysis of peripheral blood mononuclear cells (PBMCs). The mRNA levels of NR1D1 and NR1D2 (encoding REV-ERB-α and REV-ERB-β, respectively) were significantly different between patients with and without the dawn phenomenon (Fig. 4g, h). BMAL1 (also known as ARNTL) also showed a trend towards significance, whereas PER2 and PER3 showed no difference between patients with and without the dawn phenomenon (Fig. 4i–l). These data suggest that disruption of the molecular clock is associated with the extended dawn phenomenon in type 2 diabetes.
a, b, CGM data from patients with type 2 diabetes over a 24-h period (a) and at different times before and after breakfast (b) (n = 16 patients without the dawn phenomenon (DP−); n = 11 patients with the dawn phenomenon (DP+)). c–f, Oscillation of the levels of cortisol (c), insulin (d), melatonin (e) and growth hormone (f) in patients with type 2 diabetes (n = 16 DP−; n = 11 DP+). g–l, RT–qPCR analysis of the mRNA levels of NR1D1 (g), NR1D2 (h), PER2 (i), PER3 (j), CLOCK (k) and BMAL1 (l) in PBMCs collected at the indicated times. Data were normalized to the average value of all time points for each patient (n = 12 DP−; n = 10 DP+). Data are mean ± s.e.m. *P < 0.05 by two-way repeated-measures ANOVA with Holm-Sidak’s test. Statistical details are in Supplementary Table 1.
Discussion
Our work suggests that the increase of REV-ERB in SCNGABA neurons in the late sleep cycle contributes to a reduction in the firing rate of SCNGABA neurons upon waking, which increases insulin sensitivity by enhancing the insulin-mediated suppression of hepatic glucose production. This model is supported by several lines of evidence: (1) depletion of REV-ERB in GABAergic neurons causes glucose intolerance, particularly at waking; (2) depletion of REV-ERB disrupts the diurnal rhythm of SCNGABA neuron firing and insulin sensitivity, and affects hepatic glucose production but not blood glucose disposal; (3) chemogenetic manipulation of SCNGABA neuron firing can mimic or rescue the glucose intolerance that is caused by REV-ERB depletion; and (4) inducible re-expression of REV-ERB in knockout mice—only at the correct phase—rescues glucose intolerance at waking. This diurnal regulation of insulin sensitivity is independent of eating or locomotor behaviours. In line with our findings, systemic insulin sensitivity, particularly insulin-mediated suppression of hepatic glucose production, was shown to peak at waking1,2. Surgical lesion of the SCN or pharmacological silencing of SCN neural activity can either improve glucose tolerance17 or impair insulin-mediated suppression of hepatic glucose production18,19. The specificity and duration of these manipulations, as well as the timing of the assay, may contribute to the discrepancy. How sub-SCN populations of neurons contribute to the glucose phenotype warrants further investigation.
There seem to be two opposing actions at play in normal chronophysiology. On the one hand, basal glucose production peaks around waking20,21,22, to prevent hypoglycaemia during sleep and to provide fuel for the neurocognitive and locomotor activities that ensue after waking. On the other hand, hepatic sensitivity to insulin-mediated suppression of glucose production also peaks at waking1,2, in anticipation of the upcoming intake of food, so as to efficiently assimilate dietary nutrients for replenishing the energy reservoir that dwindles during sleep. These two mechanisms, combined with oscillatory insulin levels, keep blood glucose levels stable throughout the day in healthy individuals. A prominent increase in hepatic glucose production in the morning is associated with type 2 diabetes23,24. Here we show that the SCN circadian clock regulates the rhythm of hepatic insulin sensitivity. The association of changes in REV-ERB expression with the extended dawn phenomenon suggests that the clock-mediated anticipatory regulation of hepatic insulin sensitivity may account for the uncontrolled rise of hepatic glucose production at waking and the pathogenesis of type 2 diabetes with the extended dawn phenomenon.
Our patch-clamp data suggest that the firing rhythmicity of SCNGABA neurons is dependent on REV-ERB. REV-ERB-regulated genes—such as Rgs16 and members of the Takusan family—have a diurnal expression rhythm and can modulate synaptic activity9,10, which might regulate the amplitude of mEPSCs. GABA can have excitatory roles in the SCN25,26,27 and might contribute to the higher spontaneous firing frequency in knockout mice at higher mEPSC amplitudes, given the intra-SCN connections between SCNGABA neurons.
The extended dawn phenomenon in the patients in our study is unlikely to be due to the waning effects of medications because (1) blood glucose levels were controlled with a fixed medication regimen throughout the study; (2) oral antidiabetic drugs do not affect the dawn phenomenon4; and (3) the extended dawn phenomenon was not associated with changes in insulin levels. It is intriguing that REV-ERB was upregulated in human PBMCs in the afternoon, rather than being downregulated in the morning. One potential explanation is that the disrupted function of REV-ERB could lead to the upregulation of its own gene transcription owing to negative feedback12. The opposite phases between nocturnal mice and diurnal humans in clock outputs could be another contributing factor. It is unclear what caused the disrupted REV-ERB rhythmicity in patients with the extended dawn phenomenon. In summary, our findings reveal a REV-ERB-mediated, SCNGABA-dependent pathway that underlies the diurnal rhythm of hepatic insulin sensitivity in mice, with potential applications for further studying the extended dawn phenomenon in type 2 diabetes in humans.
Methods
Mice
Mice with floxed alleles at the Nr1d1 and Nr1d2 (REV-ERB-α and REV-ERB-β) loci (Nr1d1loxP/loxP and Nr1d2loxP/loxP) were obtained from PHENOMIN - ICS (Institut Clinique de la Souris, Illkirch-Graffenstaden; http://www.phenomin.fr/), as previously reported7,8. vgat-cre mice28 and Rosa26-tdTomato29 mice were obtained from The Jackson Laboratory. All mice were of the C57BL/6 genetic background. Female mice showed a more robust and reproducible phenotype during the initial characterization of the metabolic phenotype. Therefore, we used female mice for all experiments. The age of the mice is shown in the figure legends. Littermate Nr1d1loxP/loxP and Nr1d2loxP/loxP mice were used as the wild-type control for most studies unless otherwise indicated. vgat-cre mice or Rosa-CAG-LSL-tdTomato-WPRE;vgat-cre (tdTomato/vgat-cre) mice were used as wild-type controls for electrophysiology, chemogenetics, REV-ERB re-expression rescue and RGS16 and Takusan overexpression studies, as indicated in the figure legends. All vgat-cre mice used are heterozygous for the cre allele. Normal chow diet is composed of 23% kcal from protein, 14.8% kcal from fat and 62.1% kcal from carbohydrates (3002906-203, PicoLab). HFD is composed of 20% from protein, 60% from fat and 20% kcal from carbohydrates, with a total of 5.21 kcal g−1 energy density (D12492, Research Diet). Four mice were housed in each cage at room temperature (21 °C) and 40% humidity in a 12 h-light/12-h dark (light–dark, 07:00 light on, 19:00 light off) condition with free access to water and food for all experiments except for the wheel-running experiment under constant darkness. Some mice were housed in a reverse light–dark schedule (07:00 light off, 19:00 light on) for more than two weeks for convenience for the experimenters to analyse the phenotype in the dark cycle. In this case, the experiments were performed under dim red light to minimize the effects of the light on the central circadian clock. Wheel running was conducted in circadian cabinets (Actimetrics) for 10 days in light–dark conditions followed by constant darkness for another 10 days. The actogram, chi-square periodogram, phase angle and period length were generated and analysed by ClockLab software (Actimetrics). The phase angle of entrainment reflects the difference between the onset of activity and the onset of the dark period. It was determined by performing a linear regression of activity onset for the first eight days in constant darkness and extrapolating this line to the last day of the light–dark cycle30. Mice were group-housed except temporary single housing during the wheel-running experiment or the post-surgery recovery period. Mice of the same age and genotypes were randomized into different surgery groups or treatment groups. Streptozotocin was solubilized in freshly made 0.1 mmol l−1 sodium citrate buffer (pH 4.5) at 4 mg ml−1 and intraperitoneally injected at 40 mg kg−1 for 5 consecutive days. All of the animal procedures were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of Baylor College of Medicine.
Metabolic testing and insulin clamp
For GTTs and PTTs, mice were handled daily for a week before the test to allow them to get used to the handling to minimize stress. On the day of the test, mice fasted for 6 h for GTT or 8 h for PTT. Baseline blood glucose levels were taken using a glucometer (OneTouch Ultra2) from tail bleeding, followed immediately by intraperitoneal injection of glucose (1.5 or 2.5 g per kg body weight) or pyruvate (2 g per kg body weight). Blood glucose levels at 15, 30, 60, 90 and 120 min after injection were monitored using the glucometer. Tests at ZT12–14 were performed under dim red light. Serum insulin, glucagon, corticosterone, glucagon-like peptide 1 (GLP-1) and growth hormone levels were measured using the ultra-sensitive mouse insulin ELISA kit (90080, Crystal Chem), glucagon ELISA kit (10-1281-01, Mercodia), corticosterone ELISA kit (ADI-900-097, Enzo Life Sciences), mouse GLP-1 ELISA kit (81508, Crystal Chem) and growth hormone ELISA kit (EZRMGH-45K, Sigma), respectively. For insulin clamp, mice were catheterized at a single jugular vein and were allowed to recover for a week. Human insulin was infused at 2.5 mU kg−1 min−1 into unrestrained mice, and blood glucose levels were maintained between 120–140 mg dl−1 by infusing 20% glucose at various rates. 3H-glucose was used to trace hepatic glucose production. The clamp assay lasts about two hours from the start of the insulin infusion to the end. Tests at ZT12–14 were performed under dim red light.
Virus, stereotaxic injection and chemogenetics
We integrated the flip-excision (FLEX) system31 with the Tet-ON system32 to achieve inducible re-expression of REV-ERB-α specifically in SCNGABA neurons. AAV-TRE-Flag-REV-ERB-α and AAV-FLEX-rtTAM2 plasmids were constructed based on the pAAV-FLEX-GFP plasmid (Addgene 28304, from E. Boyden), pMA3211 (Addgene 46879)33, and FUW-M2rtTA (Addgene 20342)34. Adult knockout mice were co-injected with AAV-FLEX-rtTAM2 and AAV-TRE-Flag-REV-ERB-α for inducible REV-ERB re-expression (iREV-ERB) at the SCN bilaterally. As the control, knockout mice were co-injected with the rtTAM2 vector and an empty vector. For overexpressing RGS16 and α7-Takusan, the ORF clones for Rgs16 (NM_002928.4) and Gm10406 were purchased from GenScript and cloned into the pAAV-FLEX plasmid to generate AAV-FLEX-RGS16-2A-GFP or AAV-FLEX-α7-Takusan-2A-GFP. The AAV vector expressing GFP alone served as the control vector. The recombinant AAV virus was pseudotyped with an AAV9 serotype and injected at the SCN bilaterally in adult mice. For chemogenetic studies, we used the designer receptors exclusively activated by designer drugs (DREADDs) tool with engineered G-protein-coupled receptors to activate or silence specific neurons in response to CNO35. For chemogenetic activation, we injected AAV-hSyn-FLEX-hM3Dq-mCherry36 into the SCN of adult vgat-cre (wild-type) mice with AAV-hSyn-FLEX-mCherry as the control. We performed GTTs at 120 min after intraperitoneal injection of CNO. For chemogenetic suppression, we injected AAV-hSyn-FLEX-hM4Di37 at the SCN of adult knockout mice. vgat-cre (wild-type) mice injected with the same hM4Di vector served as the control. AAV5-FLEX-hM4Di-mCherry and AAV5-FLEX-mCherry were obtained from B. Roth37 through the Vector Core at the University of Northern Carolina at Chapel Hill. AAV5-FLEX-hM3Dq-mCherry was obtained from Addgene (44361-AAV-5)36. All virus was titred at around 1013 GC ml−1. For stereotaxic injection, adult mice were anaesthetized with ketamine and xylazine (100 mg kg−1 and 10 mg kg−1), and the head was fixed on the surgery platform by a stereotaxic system (Stoelting). A blur hole was drilled on each side of the skull (posterior 0.00 mm and lateral 0.14 mm to the bregma; depth 6.00 mm for the SCN), and 200 nl of each virus per side per mouse was injected. Mice were allowed to recover from surgery for at least two weeks before tests. The accurate injection was validated in each mouse by post-mortem histological examination. Mis-injected mice were excluded from the final data analysis. Mice that did not recover to their pre-surgery body weight were also excluded. For doxycycline-induced REV-ERB-α re-expression, Dox was intraperitoneally. injected at 30 mg kg−1. For chemogenetic manipulation, mice were intraperitoneally injected with 1 mg kg−1 CNO at 2 h before the injection of glucose or pyruvate during GTT or PTT analyses.
Histology
Mice were anaesthetized for transcardiac perfusion with cold PBS and 4% paraformaldehyde. Overnight post-fixed brains were immersed in 30% sucrose, embedded in optimal cutting temperature compound (OCT) and frozen in precooled isopentane. Coronal brain sections (around 12 μm) were prepared on the Leica CM1850 cryostat slicer. The coronal sections corresponding to bregma −0.34 mm to −0.70 mm were collected. REV-ERB-α (Nr1d1) and Rgs16 were assayed using RNAscope following the standard protocol from ACD with minor modifications. In brief, brain sections were rinsed with PBS to remove OCT. The brain sections were incubated at 60 °C for 30 min. Then the brain sections were post-fixed in 4% PFA at 4 °C for 15 min. After the post-fixation, the brain sections were dried in ethanol. The brain sections were then incubated with hydrogen peroxide at room temperature for 10 min. The sections were rinsed for 2 min three times in distilled water, and then the brain sections were retrieved in RNAscope 1× target retrieval reagent at 100 °C for 5 min. The slides were then rinsed in distilled water for 2 min three times and re-dried in 100% alcohol for storage. The pretreated brain sections were incubated with protease III for 30 min at 40 °C. The protease III was removed, and the brain sections were rinsed in distilled water for 2 min three times. The brain sections were hybridized with the mixed probes of Nr1d1 (ACD, 529151-C2), Rgs16 (ACD, 539201) and Vgat (also known as Slc32a1) (ACD, 319191-C3) for 2 h at 40 °C. After that, the brain sections were rinsed for 2 min three times in wash buffer to remove the excessive probes. The RNAscope Multiplex FL v2 Amp1 was added to the brain sections and incubated at 40 °C to amplify the signal for one probe. The brain sections were rinsed with wash buffer after 30 min. After the incubation of Amp1, the other two probes were amplified by Amp2 and Amp3 under the same condition. The probe signals were detected using the RNAscope Multiplex Fluorescent Detection Reagents V2 (ACD 323110). The in situ hybridization analysis of α-Takusan was performed with a probe for Takusan Gm3500 (Supplementary Table 2). For detecting the re-expression of Flag–REV-ERB-α in the SCN, the brain sections were incubated with rat anti-Flag antibodies (Novus, nbp1-06712ss, dilution 1:50) at 4 °C overnight. The sections were then washed three times in PBS at room temperature and further incubated with goat anti-rat antibodies tagged with a fluorescence dye (Abcam, A-21247, dilution 1:1,000) for 1 h at room temperature. These sections were then washed three times in PBS at room temperature, counterstained with Hoechst and sealed with the coverslip. The immunofluorescence of brain sections is viewed and captured with the Zeiss Axio imager M2m microscope and processed by ImageJ software. For quantifying the expression of Rgs16 and Takusan Gm3500, the counts of the signal dot in each cell within the SCN were measured by ImageJ software. At least 40 neurons were counted for each mouse, and the averaged number from those cells represents the expression intensity for each mouse. The results were calculated as counts per cell. A Student’s two-tailed t-test was used to analyse the quantification data.
Electrophysiology
Whole-cell patch-clamp recordings were performed on tdTomato-labelled GABAergic neurons from the SCN of tdTomato/vgat-cre (wild-type) or Nr1d1loxP/loxPNr1d2loxP/loxP-tdTomato-vgat-cre (knockout) mice. Mice of 8–12 weeks old were deeply anaesthetized with isoflurane and transcardially perfused with a modified ice-cold sucrose-based cutting solution (pH 7.3) containing 10 mM NaCl, 25 mM NaHCO3, 195 mM sucrose, 5 mM glucose, 2.5 mM KCl, 1.25 mM NaH2PO4, 2 mM sodium pyruvate, 0.5 mM CaCl2 and 7 mM MgCl2, bubbled continuously with 95% O2 and 5% CO238. The mice were then decapitated, and the entire brain was removed and immediately submerged in the cutting solution. Slices (250 μm) were cut with a Microm HM 650V vibratome (Thermo Fisher Scientific). Three brain slices were obtained for each mouse, and recordings were made at levels throughout this brain region. The slices were recovered for 1 h at 34 °C and then maintained at room temperature in artificial cerebrospinal fluid (aCSF, pH 7.3) containing 126 mM NaCl, 2.5 mM KCl, 2.4 mM CaCl2, 1.2 mM NaH2PO4, 1.2 mM MgCl2, 11.1 mM glucose and 21.4 mM NaHCO3 saturated with 95% O2 and 5% CO2 before recording. Slices were transferred to a recording chamber and allowed to equilibrate for at least 10 min before recording. The slices were perfused at 34 °C in oxygenated aCSF at a flow rate of 1.8–2 ml min−1. tdTomato-labelled neurons were visualized using epifluorescence and IR-DIC imaging on an upright microscope (Eclipse FN-1, Nikon) equipped with a moveable stage (MP-285, Sutter Instrument). Patch pipettes with resistances of 3–5 MΩ were filled with intracellular solution (pH 7.3) containing 128 mM potassium gluconate, 10 mM KCl, 10 mM HEPES, 0.1 mM EGTA, 2 mM MgCl2, 0.05 mM (Na)2GTP, and 0.05 mM (Mg)ATP. Recordings were made using a MultiClamp 700B amplifier (Axon Instruments), sampled using Digidata 1440A and analysed offline with pClamp 10.3 software (Axon Instruments). Series resistance was monitored during the recording, and the values were generally lower than 10 MΩ and were not compensated. The liquid junction potential was +12.5 mV, and was corrected after the experiment. Data were excluded if the series resistance increased markedly during the experiment or without overshoot for the action potential. Currents were amplified, filtered at 1 kHz and digitized at 20 kHz. The current clamp was engaged to test neural firing frequency and resting membrane potential. The values for resting membrane potential and firing frequency were averaged within 2-min bins. For the mEPSC recordings, the internal recording solution contained 125 mM CsCH3SO3, 10 mM CsCl, 5 mM NaCl, 2 mM MgCl2, 1 mM EGTA, 10 mM HEPES, 5 mM (Mg)ATP, and 0.3 mM (Na)2GTP (pH 7.3 with NaOH)39. The mEPSCs were recorded in whole-cell voltage-clamp mode by holding the membrane potential at Vh = −60 mV in the presence of 1 μM TTX, 50 μM bicuculline. The mIPSC recordings were recorded in whole-cell voltage-clamp mode by holding the membrane potential at Vh = −70 mV. The CsCl-based pipette solution contains 140mM CsCl, 10mM HEPES, 5mM MgCl2, 1mM BAPTA, 5mM (Mg)ATP, and 0.3mM (Na)2GTP (pH 7.30 adjusted with NaOH; 295 mOsm kg−1). The mIPSCs were recorded in the presence of 1μM tetrodotoxin (TTX), 30 μM D-AP5, and 30 μM cyanquixaline (CNQX)40. The mIPSC frequency showed a difference between ZT6 and ZT12 in wild-type mice, which is in line with the sIPSC results of a previous study41. Frequency and peak amplitude were measured using the Mini Analysis program (Synaptosoft). Multiple single neurons were recorded from 3–4 mice in each group and were pooled for statistical analysis.
Isolation of sub-hypothalamic brain regions, RT–qPCR, chromatin immunoprecipitation–qPCR and RNA-seq
For isolation of sub-hypothalamic brain regions from tdTomato-vgat-cre (wild-type) or Nr1d1loxP/loxPNr1d2loxP/loxPtdTomato-vgat-cre mice were anaesthetized with CO2 and decapitated. Brains were immediately embedded in OCT and snap-frozen at −80 °C. Coronal brain sections of 100 μm were prepared on the Leica CM1850 cryostat slicer. Coronal slices containing the SCN and other sub-hypothalamic regions were collected. The SCN was recognized by tdTomato fluorescence under a Leica DMi8 inverted microscope and isolated using a 26G needle on a 1-ml syringe. Isolated sub-hypothalamic brain regions were immediately put into a 1.5-ml microcentrifuge tube containing 1 ml TRI reagent (Molecular Research Center), and the tissue was sheared through needles with 30 strokes. RNA extraction and RT–qPCR analyses were processed using the RNeasy Mini Kit (Qiagen), High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems), PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) and QuantStudio 6 instrument (Thermo Fisher Scientific). Standard curves were generated by series dilution of pooled RNA samples. The relative value for each gene was normalized to the 18S RNA of the same sample. Primer pairs flanking or not flanking the floxed regions were used for Nr1d1 and Nr1d2. Given the sequence homology among Takusan members, the primer pair for α7-Takusan is not necessarily specific to α7-Takusan. Therefore, fold-changes can be underestimated. For one chromatin immunoprecipitation (ChIP) reaction, whole hypothalami from five mice were isolated and pulverized together in liquid nitrogen, followed by cross-linking in formaldehyde and sonication in lysis buffer containing 1% SDS. The sample was then diluted to 0.1% SDS and subjected to immunoprecipitation with REV-ERB-α antibody (Abcam ab56754, 4 μg per ChIP reaction). ChIP results were presented as the enrichment percentage after normalizing to the total input. Negative control primers target a gene desert region on mouse chromosome 17 (catalogue 71012, Active Motif). Primer sequences for RT–qPCR and ChIP–qPCR were summarized in Supplementary Table 2. For RNA-seq, an equal amount of SCN RNA from two mice was pooled for one sample. RNA-seq was performed using total RNA extracted from the SCN (n = 3 samples for each genotype) with a total of 50 million reads, PE100, for each sample on the BGISEQ-500 platform. Raw sequence reads from each biological replicate of total RNA were first aligned to the UCSC mm10 genome with TopHat v.2.1.0 using default parameters42. Then, HTSeq was used to obtain read counts from the aligned reads43. Finally, DESeq2 (v.1.8.2) was used to normalized the read counts and perform differential gene analysis44. Significantly differentially expressed genes were identified based on 5% false discovery rate (FDR) threshold.
Recruitment of patients
This study was conducted on a total of 27 patients with type 2 diabetes recruited among hospitalized patients from January 2018 to October 2019. Patients diagnosed with type 2 diabetes at the time of admission into the hospital according to the World Health Organization (WHO) criteria (1999) were considered for this study, whereas patients with other types of diabetes were excluded (Supplementary Table 3). We also excluded patients with a variety of complications, irregular diet or sleep times recently, or who were currently on medications that could confound data interpretation (Supplementary Table 3). The study started four days after the patients were admitted into the hospital. When the study was started, the blood glucose levels were under control using a fixed medication regimen, with fasting glucose 90–180 mg dl−1 and postprandial glucose below 288 mg dl−1. Patients who met the criteria and agreed to participate in the study underwent CGM for 2–3 days. CGM data were compared with fingertip glucometer readings at multiple times throughout the day to assess CGM performance. A predefined exclusion criterion was that patients with fewer than three matched measurements in a day or a correlation coefficient of less than 0.79 would be excluded (Supplementary Table 3). Patients with night-time hypoglycaemia were also excluded to avoid the confounding Somogyi effect (Supplementary Table 3). All procedures involving human participants were reviewed and approved by the Ethics Committee of the Qilu Hospital at Shandong University. Informed consent was obtained from all participants.
Clinical investigations
The study was started an average of four days after the patients were hospitalized. Patients had standard meals provided exclusively by the hospital during the study. All patients had a similar breakfast time (around 07:00) and sleep time (lights off at around 22:00) throughout the hospitalization. Participants underwent CGM for three consecutive days using the iPro2 Professional CGM system (Medtronic). The sensors were inserted by healthcare professionals and fixed properly. The system automatically measured the blood glucose every 10 s and stored an average every 5 min. The mean absolute relative difference (MARD) is the average of the absolute error between all CGM values and matched reference values and is used to assess the performance of CGM systems45. The CGM readings had an average correlation coefficient of 0.95 and MARD of less than 8% with fingertip blood glucose readings (Extended Data Fig. 9). Sleep and activity were monitored using a watch-style wearable activity tracker (Xiaomi Mi Band 2) in the entire study using the built-in software (Supplementary Table 4). Patients recruited after October 2018 were additionally monitored in parallel for sleep and breath by the cardiopulmonary coupling-polysomnography system (CPC-PSG) on the last day of the study (Supplementary Table 5). We define the dawn phenomenon as the absolute increment from the nocturnal glucose nadir to the pre- or post-breakfast values (Δpre-breakfast > 30 mg dl−1 or ∆post-breakfast > 60 mg dl−1) on any day during CGM. Blood samples were collected every 6 h (07:00, 13:00, 19:00 and 01.00) for hormone measurements and gene expression analysis on the second day during CGM. CGM data from the second day during CGM were used to compile the final CGM results for direct correlation with hormone measurements and gene expression data, and to avoid bias due to insertion or removal of the sensors. The serum cortisol, growth hormone and insulin were detected by chemiluminescence analysis (UniCel DxI 800 Access 2 Immunoassay System, Beckman Coulter; ADVIA Centaur XPT, Siemens). Melatonin was detected by high-performance liquid chromatography–tandem mass spectrometry (HPLC–MS/MS, AB SCIEX Triple Quad 4500MD, AB Sciex). The blood monocytes were extracted from fresh blood using Ficoll Histopague-1077 (Sigma), frozen in liquid nitrogen and stored at −80 °C. RNA extraction and RT–qPCR were processed using a RNeasy Mini Kit (Qiagen), PrimeScript RT reagent Kit (Takara), PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) and LightCycler480II instrument (Roche). The RNA quality was assessed by a NanoDrop spectrophotometer (Thermo Fisher Scientific). Blood samples with poor-quality RNA were excluded from the final data analysis. Standard curves were generated by series dilution of pooled RNA samples. The relative value for each gene was normalized to the 18S RNA of the same sample as the housekeeping control. Primer sequences are summarized in Supplementary Table 2.
Statistical analysis
Statistical analyses were performed using SPSS (v.21.0, IBM). No statistical methods were used to pre-determine sample size. Instead, sample sizes were determined based on previous publications46,47,48,49,50,51. Normality was tested by the Shapiro–Wilk test (n < 10) or D’Agostino–Pearson omnibus test (n > 10). For non-normal data or data with nonequivalent variances, the comparisons between two or multiple groups were tested with the Mann–Whitney test or the Kruskal–Wallis test, respectively. All tests were two-sided. All measurements were taken from distinct biological samples (mice or humans). Most comparisons between two groups were analysed using a two-sided unpaired t-test. Food intake and body weight with multiple time points were analysed with a one-way repeated-measures ANOVA with Tukey’s post-hoc test. Multi-group data, such as insulin clamp, electrophysiological data, and gene expression data, were analysed with a two-way ANOVA with Fisher’s least significant difference test or Tukey’s post-hoc test. Kinetic tests, such as GTTs, insulin-tolerance tests (ITTs), PTTs or hormone levels at multiple time points, were analysed with a two-way repeated-measures ANOVA with Holm–Sidak’s multiple comparisons test. CGM data, hormone levels and gene expression in samples from humans were analysed by two-way repeated-measures ANOVA with Holm–Sidak’s test or by the mixed-effects model SAS PROC MIXED (SAS v.9.4) with similar results. The minimum level for significance was set at P < 0.05. Mice of the same genotype were randomized into different groups of treatment or surgery procedures. Most mouse experiments were performed at least twice using independent biological samples or distinct cohorts of mice, except the RNA-seq studies that were performed once. All histology studies were repeated twice with similar results. Experimenters were blinded to the genotype or injection but were not blinded to CNO or doxycycline treatment. Data were excluded using the pre-established criteria. Mice were excluded before metabolic tests if they showed distress, infection, bleeding or anorexia due to surgery. Mice were excluded from final data analyses of metabolic tests if they showed mis-injection at post-mortem examination. Human RNA samples were excluded if the extracted RNA was of poor quality or quantity. Statistical details for results with significant results are summarized in Supplementary Table 1.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
The data that support the findings of this study are freely available from the corresponding authors upon request. RNA-seq data are available at the Gene Expression Omnibus (GEO) with the accession code GSE150840. Source data are provided with this paper.
Change history
15 June 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41586-021-03654-5
References
Shi, S. Q., Ansari, T. S., McGuinness, O. P., Wasserman, D. H. & Johnson, C. H. Circadian disruption leads to insulin resistance and obesity. Curr. Biol. 23, 372–381 (2013).
Coomans, C. P. et al. Detrimental effects of constant light exposure and high-fat diet on circadian energy metabolism and insulin sensitivity. FASEB J. 27, 1721–1732 (2013).
O’Neal, T. B. & Luther, E. E. Dawn phenomenon. https://www.statpearls.com/articlelibrary/viewarticle/20266/ (StatPearls Publishing, 2020).
Monnier, L., Colette, C., Dejager, S. & Owens, D. Magnitude of the dawn phenomenon and its impact on the overall glucose exposure in type 2 diabetes: is this of concern? Diabetes Care 36, 4057–4062 (2013).
Hastings, M. H., Maywood, E. S. & Brancaccio, M. Generation of circadian rhythms in the suprachiasmatic nucleus. Nat. Rev. Neurosci. 19, 453–469 (2018).
Zhang, R., Lahens, N. F., Ballance, H. I., Hughes, M. E. & Hogenesch, J. B. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl Acad. Sci. USA 111, 16219–16224 (2014).
Cho, H. et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485, 123–127 (2012).
Zhang, Y. et al. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science 348, 1488–1492 (2015).
Doi, M. et al. Circadian regulation of intracellular G-protein signalling mediates intercellular synchrony and rhythmicity in the suprachiasmatic nucleus. Nat. Commun. 2, 327 (2011).
Tu, S. et al. Takusan: a large gene family that regulates synaptic activity. Neuron 55, 69–85 (2007).
Panda, S. et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320 (2002).
Adelmant, G., Bègue, A., Stéhelin, D. & Laudet, V. A functional Rev-erbα responsive element located in the human Rev-erbα promoter mediates a repressing activity. Proc. Natl Acad. Sci. USA 93, 3553–3558 (1996).
Carroll, M. F. & Schade, D. S. The dawn phenomenon revisited: implications for diabetes therapy. Endocr. Pract. 11, 55–64 (2005).
Porcellati, F., Lucidi, P., Bolli, G. B. & Fanelli, C. G. Thirty years of research on the dawn phenomenon: lessons to optimize blood glucose control in diabetes. Diabetes Care 36, 3860–3862 (2013).
Cuesta, M., Boudreau, P., Cermakian, N. & Boivin, D. B. Rapid resetting of human peripheral clocks by phototherapy during simulated night shift work. Sci. Rep. 7, 16310 (2017).
Akashi, M. et al. Noninvasive method for assessing the human circadian clock using hair follicle cells. Proc. Natl Acad. Sci. USA 107, 15643–15648 (2010).
la Fleur, S. E., Kalsbeek, A., Wortel, J., Fekkes, M. L. & Buijs, R. M. A daily rhythm in glucose tolerance: a role for the suprachiasmatic nucleus. Diabetes 50, 1237–1243 (2001).
Coomans, C. P. et al. The suprachiasmatic nucleus controls circadian energy metabolism and hepatic insulin sensitivity. Diabetes 62, 1102–1108 (2013).
Foppen, E., Tan, A. A., Ackermans, M. T., Fliers, E. & Kalsbeek, A. Suprachiasmatic nucleus neuropeptides and their control of endogenous glucose production. J. Neuroendocrinol. 28, https://doi.org/10.1111/jne.12365 (2016).
Kalsbeek, A., Yi, C.-X., La Fleur, S. E. & Fliers, E. The hypothalamic clock and its control of glucose homeostasis. Trends Endocrinol. Metab. 21, 402–410 (2010).
Bolli, G. B. et al. Demonstration of a dawn phenomenon in normal human volunteers. Diabetes 33, 1150–1153 (1984).
Van Cauter, E., Polonsky, K. S. & Scheen, A. J. Roles of circadian rhythmicity and sleep in human glucose regulation. Endocr. Rev. 18, 716–738 (1997).
Boden, G., Chen, X. & Urbain, J. L. Evidence for a circadian rhythm of insulin sensitivity in patients with NIDDM caused by cyclic changes in hepatic glucose production. Diabetes 45, 1044–1050 (1996).
Radziuk, J. & Pye, S. Diurnal rhythm in endogenous glucose production is a major contributor to fasting hyperglycaemia in type 2 diabetes. Suprachiasmatic deficit or limit cycle behaviour? Diabetologia 49, 1619–1628 (2006).
Albus, H., Vansteensel, M. J., Michel, S., Block, G. D. & Meijer, J. H. A GABAergic mechanism is necessary for coupling dissociable ventral and dorsal regional oscillators within the circadian clock. Curr. Biol. 15, 886–893 (2005).
Choi, H. J. et al. Excitatory actions of GABA in the suprachiasmatic nucleus. J. Neurosci. 28, 5450–5459 (2008).
Freeman, G. M., Jr, Krock, R. M., Aton, S. J., Thaben, P. & Herzog, E. D. GABA networks destabilize genetic oscillations in the circadian pacemaker. Neuron 78, 799–806 (2013).
Vong, L. et al. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron 71, 142–154 (2011).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Siepka, S. M. & Takahashi, J. S. Methods to record circadian rhythm wheel running activity in mice. Methods Enzymol. 393, 230–239 (2005).
Atasoy, D., Aponte, Y., Su, H. H. & Sternson, S. M. A FLEX switch targets channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J. Neurosci. 28, 7025–7030 (2008).
Sprengel, R. & Hasan, M. T. Tetracycline-controlled genetic switches. Handb. Exp. Pharmacol. 178, 49–72 (2007).
Ochoa, C. D., Alexeyev, M., Pastukh, V., Balczon, R. & Stevens, T. Pseudomonas aeruginosa exotoxin Y is a promiscuous cyclase that increases endothelial tau phosphorylation and permeability. J. Biol. Chem. 287, 25407–25418 (2012).
Hockemeyer, D. et al. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell 3, 346–353 (2008).
Roth, B. L. DREADDs for neuroscientists. Neuron 89, 683–694 (2016).
Krashes, M. J. et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J. Clin. Invest. 121, 1424–1428 (2011).
Armbruster, B. N., Li, X., Pausch, M. H., Herlitze, S. & Roth, B. L. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl Acad. Sci. USA 104, 5163–5168 (2007).
Ren, H. et al. FoxO1 target Gpr17 activates AgRP neurons to regulate food intake. Cell 149, 1314–1326 (2012).
Liu, T. et al. Fasting activation of AgRP neurons requires NMDA receptors and involves spinogenesis and increased excitatory tone. Neuron 73, 511–522 (2012).
Fenselau, H. et al. A rapidly acting glutamatergic ARC→PVH satiety circuit postsynaptically regulated by α-MSH. Nat. Neurosci. 20, 42–51 (2017).
Itri, J., Michel, S., Waschek, J. A. & Colwell, C. S. Circadian rhythm in inhibitory synaptic transmission in the mouse suprachiasmatic nucleus. J. Neurophysiol. 92, 311–319 (2004).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Danne, T. et al. International consensus on use of continuous glucose monitoring. Diabetes Care 40, 1631–1640 (2017).
Faraco, G. et al. Dietary salt promotes neurovascular and cognitive dysfunction through a gut-initiated TH17 response. Nat. Neurosci. 21, 240–249 (2018).
Peixoto, R. T., Wang, W., Croney, D. M., Kozorovitskiy, Y. & Sabatini, B. L. Early hyperactivity and precocious maturation of corticostriatal circuits in Shank3B−/− mice. Nat. Neurosci. 19, 716–724 (2016).
Witton, J. et al. Hippocampal circuit dysfunction in the Tc1 mouse model of Down syndrome. Nat. Neurosci. 18, 1291–1298 (2015).
Xu, P. et al. Estrogen receptor-α in medial amygdala neurons regulates body weight. J. Clin. Invest. 125, 2861–2876 (2015).
Perusini, J. N. et al. Optogenetic stimulation of dentate gyrus engrams restores memory in Alzheimer’s disease mice. Hippocampus 27, 1110–1122 (2017).
Wang, W. et al. Chemogenetic activation of prefrontal cortex rescues synaptic and behavioral deficits in a mouse model of 16p11.2 deletion syndrome. J. Neurosci. 38, 5939–5948 (2018).
Acknowledgements
We thank M. Lazar for the Nr1d1loxP and Nr1d2loxP mice; Q. Tong and Y. Xu for critical reading of the manuscript and technical guidance; H. Liu for technical consultation; X. Wang for statistics consultation; C. Yu and S. S. B. Lee for technical assistance; K. Oka and B. Arenkiel for viral vector production; and C. Ljungberg (U54HD083092 and 1S10OD016167) for some of the histology studies. The Mouse Metabolism and Phenotyping Core at Baylor College of Medicine was supported by R01DK114356 and UM1HG006348. The authors were supported by grants from the National Natural Science Foundation of China (81971458 and 31671222 to G.D.); the American Diabetes Association (ADA1-17-PDF-138 to Y. He, ADA1-19-PDF-012 to W.Z. and NIH P20 GM135002 to Y. He); the US Department of Agriculture (Cris51000-064-01S to Y.X.); the National Key R&D Program of China (2016YFC0901204 and 2018YFC1311801 to L.C. and 2017YFC1001300 to G.D.); and the NIH (R01DK111436, R01HL153320, RF1AG069966 and R01ES027544 to Z.S.). We also thank the John S. Dunn Foundation, the Mrs. Clifford Elder White Graham Endowed Research Fund, the Cardiovascular Research Institute at Baylor College of Medicine, the Dan L Duncan Comprehensive Cancer Center (P30CA125123), the Texas Medical Center Digestive Diseases Center (P30DK056338), the SPORE program in lymphoma at Baylor College of Medicine (P50 CA126752) and the Gulf Coast Center for Precision Environmental Health (P30ES030285).
Author information
Authors and Affiliations
Contributions
Z.S. conceived the study. G.D. identified the mouse phenotype and oversaw the human study. X.L. performed gene expression analysis, chemogenetic studies, and stereotaxic injections in mice. X.H. recruited study participants and supervised the human study. W.Z. performed histological studies and ChIP. Y.G. coordinated the insulin clamp. W.L. made DNA constructs. S.Q. performed some of the mouse metabolic tests. J.S. performed gene expression analyses in human samples. J.W., F.L., J.W. and C.C. collected human blood samples, made clinical measurements and coordinated clinical studies. Y. He performed patch clamp recording. P.B. performed the initial mouse crossbreeding. G.D., X.L. and Y.G. maintained the mouse lines. P.S. performed CLAMS and insulin clamp analyses. G.D., X.L., W.Z., Y.G., J.S., J.W., Y. He, P.B., T.Y. and Z.S. analysed the data. K.Z., Y. Han and C.I.A. performed statistical analyses. Y.X., X.H. and Z.S. interpreted the data. L.C. and Z.S. obtained funding. G.D. and Z.S. wrote the manuscript with input from other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no financial or non-financial conflict of interest. No patent was involved in the study.
Additional information
Peer review information Nature thanks Erik D. Herzog, Satchidananda Panda and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Behavioural characterization of knockout mice.
a, RNAscope analysis of Nr1d1 (REV-ERB-α) gene expression at ZT6–9 and ZT18–21 in three-month-old wild-type mice. Scale bars, 200 μm b, Representative wheel-running actogram for five-month-old mice in light–dark conditions (LD) or constant darkness (DD). c, Phase angle of the light entrainment in the last day of the light–dark conditions (n = 7 mice). d–i, Representative chi-square periodograms (d, e, h, i) and period length (f, g) for five-month-old mice in light–dark conditions or constant darkness (n = 7 mice). j, Average wheel-running activity in constant darkness after normalization to the intrinsic period (tau) (n = 7 mice). Data are mean ± s.e.m. *P <0.05 by two-sided t-test. Statistical details are in Supplementary Table 1.
Extended Data Fig. 2 Metabolic characterization of knockout mice on a normal chow diet.
a, Daily food intake in three-month-old mice in home cages (n = 4 cages across 20 days). b, c, Food intake measured by the comprehensive laboratory animal monitoring system (CLAMS) in the 6 h (b) or 12 h (c) before GTT analyses at four months old (n = 5 mice). Box plot centre lines, box limits and whiskers represent the median, quartiles and minimum and maximum values, respectively. d, Blood glucose levels in four-month-old mice (n = 14 wild-type mice; n = 10 knockout mice). e, Serum insulin levels (n = 10 mice per group). f, Blood glucagon levels (n = 12 mice per group). g, Blood corticosterone levels (n = 11 mice per group). h, Blood GLP-1 levels (n = 12 wild-type mice; n = 10 knockout mice). i, Blood growth hormone (GH) levels (n = 11 wild-type mice; n = 12 knockout mice). j, k, GTTs in five-month-old mice at ZT6–8 (j) or ZT12–14 (k), with vgat-cre mice serving as the wild-type control (n = 7 mice). l, Body weight for clamp analyses at five months old (n = 4 mice). m, n, Blood glucose levels (m) and glucose infusion rate (n) during clamp analyses (n = 4 mice). o, Hyperinsulinaemia-mediated suppression of hepatic glucose production in the clamp analyses (n = 4 mice). Data are mean ± s.e.m. *P < 0.05 by two-way ANOVA or two-sided t-test. Statistical details are in Supplementary Table 1.
Extended Data Fig. 3 Metabolic characterization of knockout mice on a high-fat diet.
a, Body weight on HFD. HFD started at 10 months (n = 12 mice). b, GTTs at ZT6–8 after two weeks on HFD (n = 12 mice). c, GTTs at ZT12–14 after three weeks on HFD (n = 12 mice). d, ITTs at ZT6–8 after four weeks on HFD (n = 12 mice). e, ITTs at ZT12–14 after five weeks on HFD (n = 12 mice). f, Injection of streptozotocin (STZ) six weeks after HFD. g, h, Body weight (g) and blood glucose levels (h) at ZT10 after STZ injection (n = 12 mice). i, Blood glucose levels at the indicated ZTs two weeks after STZ injection, n = 12 mice. Data are mean ± s.e.m. *P < 0.05 by two-way ANOVA or two-sided t-test. Statistical details are in Supplementary Table 1.
Extended Data Fig. 4 Gene expression analysis of different brain regions.
a–e, RT–qPCR analysis of the indicated brain-region-specific marker genes (Vip (a), Pmch (b), Crf (c), Rfrp (also known as Npvf) (d) and Pomc1 (also known as Pomc) (e)) for brain regions isolated from both wild-type and knockout mice at ZT6 at the age of three months (n = 12 mice). Box plot centre lines, box limits and whiskers represent the median, quartiles and minimum and maximum values, respectively. f–i, RT–qPCR analysis comparing mRNA expression of Nr1d1 (f), Nr1d2 (g), Bmal1 (h) and Npas2 (i) in wild-type and knockout mice at ZT6 at the age of three months (n = 6 mice). Data are mean ± s.e.m. *P < 0.05 by two-way ANOVA or two-sided t-test. Statistical details are in Supplementary Table 1.
Extended Data Fig. 5 Electrophysiological and molecular characterization of knockout mice.
a–c, Brain slice patch-clamp representative traces for spontaneous firing (a), mEPSCs (b) and mIPSCs (c) at ZT12–14. d–g, Temporal pattern of expression of Rgs16 (d), α7-Takusan (Gm10406) (e), Nr1d1 (f) and Nr1d2 (g) in the hypothalamus in light–dark conditions from CircaDB (http://circadb.hogeneschlab.org). h–k, RT–qPCR analysis of the mRNA levels of Nr1d1 (h), Nr1d2 (i), Bmal1 (j) and Npas2 (k) in the SCN of three-month-old mice (n = 6 mice). Primers for Nr1d1 and Nr1d2 did not span the floxed exons. l, RNAscope of Rgs16 at the SCN in wild-type and knockout mice at the indicated ZTs. Scale bars, 100 μm. m, Quantification of Rgs16 staining (n = 5 wild-type mice at ZT4; n = 3 knockout mice at ZT4; n = 4 wild-type or knockout mice at ZT16). n, In situ hybridization analysis of Takusan Gm3500 staining. Scale bars, 25 μm. o, Quantification of in situ hybridization analysis of Takusan Gm3500 (n = 4 wild-type mice at ZT4; n = 6 wild-type mice at ZT16; n = 3 knockout mice at ZT4 or ZT16). p, Genome browser views of transcription start sites (green arrows) and nearby AGGTCA elements (red arrows) for the indicated genes on GRCm38. q, ChIP–qPCR analysis of Nr1d1 in the hypothalamus of three-month-old wild-type mice at ZT9 and ZT21—the peak and the trough of REV-ERB-α expression, respectively (n = 4 samples). Hypothalami from five mice were pooled as one sample. The negative control primers target a gene desert region on chromosome 17. The primer sequences of ChIP–qPCR assays are in Supplementary Table 6. Data are mean ± s.e.m. *P < 0.05 by two-way ANOVA or two-sided t-test. Statistical details are in Supplementary Table 1.
Extended Data Fig. 6 Metabolic characterization of mice overexpressing RGS16 or α7-Takusan in SCNGABA neurons.
a, Validation of the injection with GFP fluorescence signals. Scale bar, 200 μm. b–e, GTTs (b, c) and ITTs (d, e) at the indicated ZTs in four-month-old vgat-cre mice injected with AAV-FLEX vectors encoding GFP, RGS16 or α7-Takusan (n = 7 mice). f, Body weight of three-month-old vgat-cre mice at three weeks after AAV injection (n = 14 mice). Data are mean ± s.e.m. *P < 0.05 for RGS16 or α7-Takusan versus the GFP control by two-way ANOVA followed by Holm–Sidak’s test. Statistical details are in Supplementary Table 1.
Extended Data Fig. 7 Rhythmicity of SCNGABA neuron firing in glucose metabolism.
a, Experimental design for chemogenetic activation of the SCNGABA neurons in wild-type mice with hM3Dq. b, Body weight of vgat-cre mice injected with AAV expressing hM3Dq or control mCherry (n = 11 mice). Mice were injected at the age of two months. c, Experimental design for chemogenetic repression of the SCNGABA neurons in wild-type and knockout mice with hM4Di. d, Body weight of wild-type and knockout mice injected with AAV expressing hM4Di (n = 12 wild-type mice; n = 14 knockout mice). Mice were injected at the age of two months. e, f, GTTs in wild-type or knockout mice injected with AAV expressing hM4Di at the indicated ZTs in the presence of saline (e) or CNO (f) (n = 12 wild-type mice; n = 14 knockout mice). Data are mean ± s.e.m. *P < 0.05 by two-sided t-test.
Extended Data Fig. 8 Rhythmicity of REV-ERB expression in SCNGABA neurons in glucose metabolism.
a, Experimental design for inducible re-expression of REV-ERB-α in the SCNGABA neurons of knockout mice. Mice were injected virus at the age of 2.5 months. b, Body weight at the time of euthanasia (n = 9 mice). c, d, GTTs in 4–4.5-month-old mice at ZT6–8 after injection of doxycycline at ZT0 (c) or ZT18 (d) (n = 9 mice). e, RT–qPCR analysis of the SCN from knockout mice with inducible re-expression of REV-ERB-α. Doxycycline was injected at ZT0 and the brain was collected at ZT12–14 (n = 4 mice). Data are mean ± s.e.m. *P < 0.05 by two-sided t-test. Statistical details are in Supplementary Table 1.
Extended Data Fig. 9 Assessment of CGM performance.
a, A representative comparison between fingertip glucometer reading and CGM reading for an individual at different times of the day. b, Pearson correlation coefficient between CGM and fingertip readings (n = 16 DP−; n = 11 DP+). c, MARD, the average of the absolute error between all CGM values and matched reference values (n = 16 DP−; n = 11 DP+). Data are mean ± s.e.m.
Supplementary information
Supplementary Tables
This file contains Supplementary Tables 1-6. Supplementary Table 1. Statistical tests. Statistical details were shown for results with significant differences, including the names of the statistical methods, p values, and sample sizes for those not included in the main figure legends due to limited space. Supplementary Table 2. Primer sequences for RT-qPCR and ChIP-qPCR. Nucleic acid sequences (from the 5’ end to the 3’ end) were shown for RT-qPCR primers, ChIP-qPCR primers, and the ISH probe. Supplementary Table 3. Inclusion and exclusion criteria for patient recruitment. Supplementary Table 4. Characteristics of human subjects. Supplementary Table 5. Cardiopulmonary Coupling-Polysomnography (CPC-PSG) of human subjects. Supplementary Table 6. Medication usage in human subjects.
Source data
Rights and permissions
About this article
Cite this article
Ding, G., Li, X., Hou, X. et al. REV-ERB in GABAergic neurons controls diurnal hepatic insulin sensitivity. Nature 592, 763–767 (2021). https://doi.org/10.1038/s41586-021-03358-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03358-w
- Springer Nature Limited