Abstract
Treatment with immune checkpoint blockade (ICB) has revolutionized cancer therapy. Until now, predictive biomarkers1,2,3,4,5,6,7,8,9,10 and strategies to augment clinical response have largely focused on the T cell compartment. However, other immune subsets may also contribute to anti-tumour immunity11,12,13,14,15, although these have been less well-studied in ICB treatment16. A previously conducted neoadjuvant ICB trial in patients with melanoma showed via targeted expression profiling17 that B cell signatures were enriched in the tumours of patients who respond to treatment versus non-responding patients. To build on this, here we performed bulk RNA sequencing and found that B cell markers were the most differentially expressed genes in the tumours of responders versus non-responders. Our findings were corroborated using a computational method (MCP-counter18) to estimate the immune and stromal composition in this and two other ICB-treated cohorts (patients with melanoma and renal cell carcinoma). Histological evaluation highlighted the localization of B cells within tertiary lymphoid structures. We assessed the potential functional contributions of B cells via bulk and single-cell RNA sequencing, which demonstrate clonal expansion and unique functional states of B cells in responders. Mass cytometry showed that switched memory B cells were enriched in the tumours of responders. Together, these data provide insights into the potential role of B cells and tertiary lymphoid structures in the response to ICB treatment, with implications for the development of biomarkers and therapeutic targets.
Similar content being viewed by others
Main
Immunotherapy has afforded patients with melanoma and other cancers the potential for long-term survival, and we are beginning to gain insight into the mechanisms of therapeutic responses as well as biomarkers of response and resistance. Considerable progress has been made in this regard, with the identification of several validated biomarkers, particularly for ICB therapy1,2,3,4,5,6,7,8,9,10. It is clear that cytotoxic T cells have a dominant role in responses to ICB and other forms of immunotherapy; however, there is a growing appreciation of other components of the tumour microenvironment that may influence the therapeutic response—including myeloid cells and other subsets of immune cells11.
Tumour-infiltrating B cells have been identified, but their overall functional role in cancer is incompletely understood14,15,19,20,21,22,23,24—some studies suggest that they are tumour-promoting, whereas others show a positive association with improved cancer outcomes, particularly when they are found in association with organized lymphoid aggregates known as tertiary lymphoid structures (TLSs)12,13,16,25,26,27,28.
TLSs have been identified within a wide range of human cancers at all stages of disease, in primary as well as metastatic lesions, but their presence is highly variable between cancer types as well as between patients12,16. Considerable heterogeneity also exists in the cellular constituents of TLSs and their location within tumours, and this may influence the overall effect on anti-tumour immunity and outcome12,13,14,16. These TLS structures are not only a surrogate marker of a brisk immune response; instead, it is thought that they actively modulate anti-tumour immune activity. In this regard, the benefit of a high CD8+ T cell density within a tumour is abrogated in the absence of TLS-associated dendritic cells29. Mature TLSs exhibit evidence for the formation of germinal centres30,31, and oligoclonal B cell responses have previously been identified in cutaneous melanoma and metastases32,33, which suggests an active humoral anti-tumour response within TLSs that is driven by B cells. Notably, although preliminary evidence suggests an association between responses to ICB and the presence of B cells, the precise role of B cells—and in particular TLSs—in response to ICB remains unclear28,34.
A phase 2 clinical trial of neoadjuvant treatment with ICB in patients with high-risk resectable (clinical stage III or oligometastatic stage IV) melanoma was recently conducted to assess the safety and feasibility of this treatment in this patient population (NCT02519322)17. Notably, longitudinal tumour samples were taken in the context of therapy, and molecular and immune profiling was performed to gain insight into the mechanisms of the therapeutic response and resistance. In these studies, known and novel biomarkers of response were identified, and targeted protein expression profiling (via Nanostring Digital Spatial Profiling) revealed significantly higher expression of B cell markers in samples before treatment (baseline) and on-treatment samples of responders to ICB17.
B cells found in the tumours of responders
To gain a deeper understanding of potential mechanisms of therapeutic response to ICB, we performed RNA sequencing (RNA-seq) in longitudinal tumour samples from this patient cohort. In these studies, significantly higher expression of B-cell-related genes such as MZB1, JCHAIN and IGLL5 was observed in patients that respond to ICB treatment versus non-responding patients (‘responders’ and ‘non-responders’, hereafter) at baseline (P < 0.001) with over-representation of these genes compared to T cells and other immune markers (with evaluable tumours from seven responders and nine non-responders) (Fig. 1a, b, Supplementary Tables 1, 2). Other genes that are expected to alter the function of B cells were also significantly enriched in responders versus non-responders, such as FCRL5, IDO1, IFNG and BTLA. Low tumour purity was observed in some samples, particularly in the context of an effective therapeutic response, limiting conventional analysis of RNA-seq data. To address this, we next performed a more focused investigation of the tumour immune microenvironment using the microenvironment cell populations (MCP)-counter method18 on RNA-seq data in baseline and on-treatment tumour samples—focusing more specifically on immune-related genes (Supplementary Table 3), which allowed inclusion of samples with low tumour purity (10 responders and 11 non-responders at baseline, 9 responders and 11 non-responders on-treatment). In these analyses, we again observed enrichment of a B cell signature in responders versus non-responders at baseline and early on-treatment (P = 0.036 and 0.038, respectively). Notably, these analyses included samples from patients with nodal and extra-nodal disease with no obvious contribution based on the site of disease (Fig. 1c, Extended Data Figs. 1a, b, 2a, Supplementary Tables 4, 13), which suggests that B cell signatures were not merely related to the presence of these tumours within lymph nodes. Findings of high B cell lineage scores in responders were replicated in samples from an additional cohort of patients with melanoma treated with neoadjuvant versus adjuvant checkpoint blockade (ClinicalTrials.gov identifier NCT02437279, OpACIN-neo trial) (n = 12 responders, 6 non-responders)35 (Extended Data Figs. 1d, 2c, Supplementary Tables 5, 6, 13). B cell signatures alone were predictive of response in univariable analyses (odds ratio 2.6, P = 0.02 for our trial, and odds ratio 2.9, P = 0.03 for combined melanoma cohorts), but not in multivariable analyses when considering other components of the immune cell infiltrate, which suggests that B cells probably act together with other immune subsets and are not acting in isolation; however, these analyses were limited owing to the low sample size (Supplementary Tables 7, 8). Moreover, these findings were corroborated in translational studies of separate cohorts of patients with melanoma36 and sarcoma37 who were treated with ICB. B cells were not significantly associated with pathological response rates in an analogous trial of neoadjuvant-targeted therapy in patients with BRAF-mutated melanoma38 (Extended Data Fig. 1e, Supplementary Table 9); however, B cells have previously been shown to be positively associated with responses to chemotherapy in other cancer types39,40.
a, Supervised hierarchical clustering of differentially expressed genes (DEG) on RNA-seq analysis by response of melanoma tumour specimens at baseline, with responder defined as having a complete or partial response by RECIST 1.1 and non-responder as having less than partial response (n = 9 non-responders and 7 responders). A cut-off of gene expression fold change of ≥ 2 or ≤ 0.5 and a false discovery rate (FDR) q ≤ 0.05 was applied to select DEGs. Ipi, ipilimumab; nivo, nivolumab. b, Volcano plot depiction of DEG by response from same cohort as in a. R, responders; NR, non-responders. c, Supervised clustering of melanoma tumour specimens by response at baseline (n = 11 non-responders and 10 responders), displaying MCP-counter scores. NK cells, natural killer cells. d, Supervised clustering by clinical response defined as achieving a partial response (PR) according to RECIST 1.1 and non-responders as having progressive disease (PD) of RCC baseline tumour specimens (n = 11 PD and 17 PR) using methodology as in c. P values were determined by two-sided Mann–Whitney U-test. Bev, bevacizumab.
Similar B cell signature observed in RCC
To evaluate the validity of these findings across other cancer types, we next assessed the expression of these immune cell gene expression signatures in a pre-surgical ICB trial for patients with metastatic renal cell carcinoma (RCC) (NCT02210117, PD1 blockade monotherapy versus combined CTLA4 and PD1 blockade versus combined PD1 blockade and bevacizumab) (Supplementary Table 10). Gene expression profiling by microarray and subsequent MCP-counter analysis of baseline tumour samples was performed, demonstrating significantly higher expression of B-cell-related genes in responders versus non-responders (P = 0.0011, n = 17 responders and 11 non-responders) (Fig. 1d, Extended Data Figs. 1c, 2b, 3, Supplementary Tables 11–13). As in the case of melanoma, B cell signatures were predictive of a response in univariable analysis in the RCC cohort (odds ratio 61.2, P = 0.05) but not multivariable analysis, again suggesting cooperative function with other immune subsets; however, sample size was again limited (Supplementary Table 14).
B cells prognostic in TCGA analysis
On the basis of these data and existing data regarding a potential prognostic role for TLSs in melanoma and other cancer types primarily outside the context of ICB treatment18,28,41, we next assessed the expression of these immune-related genes in cutaneous melanoma from The Cancer Genome Atlas (TCGA) platform (TCGA-SKCM, n = 136)42. To this end, we applied the MCP-counter algorithm to available RNA-seq data from a subset of patients with non-recurrent stage III disease (regional lymph node or regional subcutaneous metastases), as these were most comparable to our clinical cohort. In these studies, we identified three distinct melanoma immune classes (MICs), with significantly higher expression of B cells in cluster C than in cluster A (P < 0.0001) or cluster B (P < 0.0001) (Extended Data Fig. 4a, Supplementary Tables 15–17). Importantly, there was no clear association of MICs with known genomic subtypes of melanoma (BRAF, NRAS, NF1 or triple wild type)42 or disease site (nodal or non-nodal) (Extended Data Fig. 4a, Supplementary Table 17). Survival analyses revealed that cases in cluster C had significantly improved overall survival compared with cluster A (P = 0.0068) (Extended Data Fig. 4b). To assess the association with B cell signatures specifically, we next compared overall survival in patients with tumours high for B cell lineage versus low, which demonstrated prolonged survival in patients with B cell-lineage-high tumours (P = 0.053) (Extended Data Fig. 4c). Furthermore, univariable Cox proportional hazards modelling demonstrated that tumours with low infiltration of B cells had significantly increased risk of death (hazard ratio is 1.7 for B-cell-low, P = 0.05) in comparison to the B-cell-high group (Supplementary Table 18). These data are further supported by recent analyses of the TCGA cohort that demonstrate the association of a plasmablast-like B cell signature with survival as well as increased expression of CD8A and infiltration of CD8+ T cells34. Similar analyses were performed to assess the expression of immune-related genes in clear-cell RCC from the TCGA (TCGA-KIRC, n = 526)43. In these analyses, similar immune classes were observed; however, immune infiltration was not associated with survival in these patients (P = 0.24) (Extended Data Fig. 4d–f, Supplementary Tables 19–21), possibly owing to the heterogeneous nature of this disease and other driving mechanisms of patient outcomes.
B cells localized in the context of TLSs
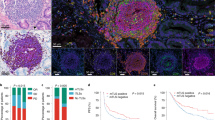
On the basis of the results from gene expression profiling, we next assessed tumour samples histologically to gain insight into the density and distribution of B cells as well as their relationship to TLSs in patients treated with neoadjuvant ICB. The density of CD20+ B cells and TLSs, and the ratio of TLSs to tumour area were higher in responders than in non-responders in our neoadjuvant melanoma cohort, particularly in early on-treatment samples (P = 0.0008, P = 0.001 and P = 0.002, respectively), although statistical significance was not reached for all the markers in the baseline samples (P = 0.132, P = 0.078 and P = 0.037, respectively) (Fig. 2a), which is consistent with previous work that suggested that assessment of early on-treatment immune infiltrate is far more predictive of the response to ICB than assessment of pre-treatment samples1. Findings between gene expression profiling and immunohistochemistry analysis were complementary, and had modest correlation as previously described18 (Extended Data Fig. 5c–e). We also found increased numbers of B-cell-related exosomes (CD20+) in the peripheral blood of responders compared with non-responders at early on-treatment time points (Extended Data Fig. 2d–j).
a, Quantification of CD20 cells by singlet immunohistochemistry and association with response to neoadjuvant ICB in resectable melanoma with responders defined as having complete or partial response by RECIST 1.1 and non-responders as having less than a partial response (n = 11 NR and 10 R at baseline and n = 11 NR and 9 R on treatment). b, c, Density of TLSs (b) and ratio of tumour area occupied by TLSs (c) and correlation by treatment response (n = 7 NR and 7 R at baseline and n = 10 NR and 8 R after treatment). For a–c, bars indicate median values, and errors bars denote interquartile range; individual data points are shown. P values were determined by two-sided Mann–Whitney U-test. d, Representative image of CD20 staining of TLSs in a responder after treatment with ipilimumab and nivolumab. e, Additional staining of boxed area in d showing associated haematoxylin and eosin (H&E) staining and singlet immunostaining of CD20, CD8, CD4, FOXP3 and CD21. f, Multiplex immunofluorescence assay of TLSs as in d for the following markers: CD20, CD21, CD4, CD8, FOXP3 and DAPI. Original magnification, ×20.
Notably, architectural analysis showed that CD20+ B cells were localized in TLSs of tumours of responders, and were colocalized with CD4+, CD8+and FOXP3+ T cells. Colocalization with CD21+ follicular dendritic cells and MECA79+ high endothelial venules was also shown (Fig. 2d–f, Extended Data Figs. 5a, 6a). The vast majority of evaluated TLSs in these patients represented mature secondary-follicle-like TLSs, as indicated by the presence of both CD21+ follicular dendritic cells and CD23+ germinal centre B cells30 (Fig. 2d–f, Extended Data Figs. 5a, 6a). We identify similar mature TLSs in patients with extra-nodal metastases (Extended Data Fig. 5b), which suggests that TLSs may develop in non-nodal sites and are associated with the response to ICB treatment. Analogous immunohistochemical findings were observed in our cohort of patients with RCC treated with pre-surgical ICB, with increased infiltration of CD20+ cells and TLSs density associated with response to treatment (Extended Data Fig. 6b–d); these TLSs are morphologically similar to those found in melanoma (Extended Data Fig. 6e–h). We also assessed the potential functional role of B cells and TLSs in promoting T cell responses in our cohort via additional spatial profiling analyses, and found increased markers of activation on T cells within as compared to those outside these TLSs (Extended Data Fig. 7a–c).
BCR and single-cell RNA-seq offer functional insight
Next, we performed several in-depth analyses to gain insight into the phenotype and function of the infiltrating B cells, and how they might be contributing to responses to ICB. Reasoning that differences in the clonotypes of B cell receptors (BCRs) between responders and non-responders would be indicative of an anti-tumour B cell response, we probed our RNA-seq data for BCR sequences using the modified TRUST algorithm. In these studies, we identified significantly increased clonal counts for both immunoglobulin heavy and light chains (IgH and IgL; P = 0.001 and P = 0.004, respectively) and increased BCR diversity in responders than in non-responders (P = 0.002 and P = 0.0008), which suggests an active role for B cells in anti-tumour immunity (Fig. 3a, Extended Data Fig. 8). To complement these analyses, we analysed single-cell RNA-seq data from baseline and on-treatment samples from an independent cohort of patients with metastatic melanoma treated with ICB (n = 48 tumour samples; 1,760 B cells from 32 patients treated with PD1 blockade monotherapy, CTLA4 blockade monotherapy, or combined blockade of both PD1 and CTLA4, including samples from some patients in our neoadjuvant ICB-treated cohort44) (Supplementary Tables 22, 31). Similar to observations made in our clinical trial cohort, we found that B cells were significantly enriched in tumours from responders versus non-responders and were predictive of a response (odds ratio 1.05, P = 0.02) (Fig. 3b, Extended Data Fig. 9a, Supplementary Table 23). Unbiased analysis for markers of B cells (using all expressed genes in the CD45+CD19+ population only) associated with clinical outcome demonstrated 46 markers were significantly enriched in lesions from responders and 147 markers were significantly enriched in non-responder lesions (Extended Data Fig. 9b, Supplementary Tables 24, 25). Pathways upregulated in responders as compared to non-responders include those consistent with increased immune activity such as CXCR4 signalling, cytokine receptor interaction and chemokine signalling pathways (Supplementary Table 26). Unsupervised clustering of B cells using k-means clustering, after testing for the robustness of each solution, identified four distinct B cell clusters, G1 (B cells, switched, activated IgD− cells), G2 (plasma cells), G3 (B cells unswitched IgD+) and G4 (B cells, switched, activated IgD− cells, with unique markers relative to G1), each of which is associated with different functional states (Fig. 3c, Extended Data Fig. 9c, Supplementary Tables 27, 28). No significant differences were identified when testing for associations of each individual cluster (G1–G4) with the clinical outcome, probably owing to limited sample size. Pathway analysis was also performed on bulk RNA-seq data from our clinical trial cohort, revealing increased immune signalling pathways in responders than in non-responders, including T cell receptor signalling, major histocompatibility complex-mediated antigen presentation and processing, differentiation of T helper 1 and 2 (TH1 and TH2) cells, and costimulatory signalling associated with T cell signalling (Supplementary Tables 29, 30).
a, Normalized clonal counts for BCRs identified in patients with high-risk resectable melanoma treated with neoadjuvant ICB. Both the IgH and IgL are evaluated with responders and non-responders as shown. All samples analysed at baseline. b, Scatter plots demonstrating the percentage of various cell types as indicated between responders (n = 17) and non-responders (n = 31) from a separate cohort of patients with advanced melanoma analysed by single-cell RNA-seq. Samples before and after treatment are combined. B cells are represented by the CD45+CD19+ population. Data are median values with interquartile ranges, and individual data points are shown. P values were determined by two-sided Mann–Whitney U-test. Adjustments for multiple comparisons were not made. c, t-distributed stochastic neighbour embedding (t-SNE) plot of all B cells collected and analysed by single-cell RNA-seq in b. Cells are coloured based on four clusters identified by k-means clustering (G1–G4). Number of cells analysed is 1,760 B cells from 48 tumours arising in 32 patients treated with PD1 blockade monotherapy, CTLA4 blockade monotherapy, or combined PD1 and CTLA4 blockade. d, t-SNE plots demonstrating peripheral blood and intratumoral combined B cell populations from mass cytometric analyses in responders versus non-responders (n = 4 R and 4 NR for peripheral blood and n = 5 R and 3 NR for tumour) from the neoadjuvant ICB trial in patients with advanced melanoma. e, Intratumoral B cell phenotypes included in d grouped by response. f, Quantification of B cell subtypes in e. Plots in d–f represent combined analyses of tumours ran simultaneously with the peripheral blood samples (n = 5 R and 3 NR) and include baseline and on-treatment samples as described in Supplementary Table 31. Statistical analyses including all samples are presented in Extended Data Fig. 10b.
CyTOF shows differential B cell phenotypes
To gain further insight into the potential functional role of B cells in the response to ICB, we performed mass cytometry (CyTOF) in evaluable tumour and peripheral blood samples (seven responders and three non-responders for tumour, and four responders and four non-responders for peripheral blood from our neoadjuvant ICB trial). Sample size was limited owing to the amount of tumour available given prioritization for other studies as well as tumour viability. These analyses included patients with nodal and non-nodal metastases (Extended Data Fig. 10a, Supplementary Tables 31, 32).
We first assessed differences between intratumoral B cells and those in the peripheral blood of patients. In these studies, unique clusters of CD45+CD19+ (B cell) populations including naive (CD19+, CD27−, IgD+), transitional (CD19+, CD24++, CD38++, CD10+, CD27−, IgD+), unswitched and switched memory (CD19+, CD27+, IgD+/−), double-negative (CD19+, CD27−, IgD−), and plasma (-like) cell (CD19+, CD20−, CD22−, CD38++, CD27++) populations were found in peripheral blood and tumour samples, with distinct profiles in the tumour compared with peripheral blood samples (Fig. 3d, Extended Data Figs. 10a, b, 11a, b). Intratumoral B cells had reduced expression of CD21, CD23, CD79b and CXCR5, pointing to distinct functional and migratory profiles compared to similar B cell populations in the peripheral blood (Extended Data Fig. 11b). We next compared the phenotypes of B cells in tumours and peripheral blood from responders and non-responders to ICB treatment. Although B cell subsets (naive, memory and transitional B cells and plasma cells) in the peripheral blood had a similar distribution in responders and non-responders (Fig. 3d, Extended Data Fig. 10b), significant differences were noted in the subsets of B cells in tumours (Fig. 3e, f, Extended Data Fig. 10b). Specifically, tumours from responders had a significantly higher frequency of memory B cells, whereas non-responders had a significantly higher frequency of naive B cells (P = 0.033 for naive and P = 0.033 for memory) (Fig. 3e, f, Extended Data Fig. 10b). Other notable differences included an increase in plasma cells in responders compared with non-responders; however, this did not reach significance and was largely driven by data from one patient (P = 0.3) (Fig. 3e, f, Extended Data Fig. 10b). More granular characterization of the intratumoral B cells reveals an increased percentage of CXCR3+ switched memory B cells (P = 0.0083) in responders than in non-responders; we also note increased CD86+ B cells (P = 0.017) and increased germinal-centre-like (CD19+, CD20++, CD38+, CD27−, IgD−, CD86+, CD95+) B cells (P = 0.24) in responders as compared to non-responders (Extended Data Figs. 10c, d, 11c). Increased proliferation of B cells suggestive of germinal centre formation and activity is observed within TLSs (Extended Data Fig. 7d).
Summary
In summary, we present multiomic data that support a role for B cells within TLSs in the response to ICB in patients with metastatic melanoma and RCC. Although the distinct mechanisms through which B cells contribute are incompletely understood, our data suggest that the same properties of memory B cells and plasma cells desirable for acquired immune responses may also be contributing to an effective T cell response after ICB. Importantly, these B cells are probably acting together with other key immune constituents of the TLS by altering T cell activation and function as well as through other mechanisms. Memory B cells may be acting as antigen-presenting cells, driving the expansion of both memory and naive tumour-associated T cell responses. B cells can also secrete an array of cytokines (including TNF, IL-2, IL-6 and IFNγ), through which they activate and recruit other immune effector cells, including T cells. The observation of switched memory B cells (that can differentiate into plasma cells) in responders suggests that they could be potentially contributing to the anti-tumour response by producing antibodies against the tumours. Although we did not have adequate samples to study this in our cohort, it is an important line of investigation moving forward, and insights could lead to new therapeutic approaches to enhance responses to ICB. Together, findings in these cohorts are provocative and represent important advances in our insight into therapeutic responses to ICB. Further studies are needed in additional (and larger) cohorts across tumour types and stage of disease, as well as with therapeutic regimens. These types of studies along with pre-clinical models will help lend statistical power to the notion that B cells independently contribute to anti-tumour immune function in the context of ICB therapy, and also to better understand the mechanisms through which B cells and TLSs may favourably affect responses. Nonetheless, findings from these unique cohorts provide important insight into the role of B cells and TLSs in therapeutic responses to ICB, and are likely to stimulate further research in this area.
Methods
Patient cohort(s) and sample collection
For the melanoma neoadjuvant cohort (NCT02519322)17, 23 patients enrolled in a phase II clinical trial of neoadjuvant ICB. Twelve patients received nivolumab monotherapy with 3 mg kg−1 every 2 weeks for up to 4 doses, and 11 patients received ipilimumab 3 mg kg−1 with nivolumab 1 mg kg−1 every 3 weeks for up to 3 doses followed by surgical resection. These patients were treated at the University of Texas MD Anderson Cancer Center and had tumour samples collected and analysed under Institutional Review Board (IRB)-approved protocols (2015-0041, 2012-0846). Of note, these studies were conducted in accordance with the Declaration of Helsinski and approved by the UT MD Anderson Cancer Center IRB. Response was defined as achieving a complete or partial radiographic response by RECIST 1.1 between pre-treatment imaging and post-neoadjuvant treatment imaging before surgical resection. Tumour samples were collected at several time-points for correlative studies including baseline and on-treatment (weeks 3 and 5 for nivolumab monotherapy, weeks 4 and 7 for combination ipilimumab with nivolumab). Tumour samples were obtained as core, punch or excisional biopsies performed by treating clinicians or an interventional radiologist. Samples were immediately formalin-fixed and paraffin-embedded (FFPE), snap-frozen or digested following tissue collection.
Additional patients off-protocol included five patients with widely metastatic melanoma who were treated at the University of Texas MD Anderson Cancer Center and had tumour samples collected and analysed under IRB-approved protocols (LAB00-063 and PA17 – 0261). Samples were immediately FFPE after tissue collection.
For the validation melanoma cohort, we used samples of 18 patients enrolled in the OpACIN-neo trial (NCT02437279). In the phase 1b OpACIN-neo trial, 20 patients with palpable stage III melanoma were randomized 1:1 to receive ipilimumab 3 mg kg−1 and nivolumab 1 mg kg−1, either 4 courses after surgery (adjuvant arm), or 2 courses before surgery and two courses post-surgery (neoadjuvant arm). Coprimary endpoints were safety/feasibility and tumour-specific expansion of T cells. For this current correlative study, response was defined as not having disease relapse. These patients were treated at the Netherlands Cancer Institute (Amsterdam). The study was conducted in accordance with the Declaration of Helsinki and approved by the medical ethics committee of the Netherlands Cancer Institute. All subjects provided informed consent before their participation in the study. Patients underwent a pre-treatment tumour biopsy (1× formalin-fixed and paraffin-embedded (FFPE) and 2× fresh frozen) obtained as a core biopsy performed by a radiologist. RNA was extracted from one frozen biopsy for RNA-seq analysis. We included only 18 patients in our analysis because the tumour purity in the frozen pre-treatment biopsy of 2 patients was too low, therefore no RNA could be isolated and these patients could not be included in this analysis. The clinical responses of this cohort have been previously described35.
The RCC trial was an open-label, randomized, pre-surgical/pre-biopsy trial (NCT02210117) in which adults with metastatic RCC without previous immune checkpoint therapy and anti-VEGF therapy were enrolled and randomized 2:3:2 to receive nivolumab (3 mg kg−1 once every 2 weeks, ×3 doses), nivolumab plus bevacizumab (3 mg kg−1 once every 2 weeks ×3 plus 10 mg kg−1 ×3) or nivolumab plus ipilimumab (3 mg kg−1 once every 2 weeks ×3 1 mg kg−1 ×2), followed by surgery (cytoreductive nephrectomy or metastasectomy), or biopsy at week 8–10, and subsequent nivolumab maintenance therapy for up to 2 years. Response was assessed at 8 weeks and then at ≥12 weeks by RECIST 1.1 criteria. Clinical response data collection is still ongoing. For this current correlative study, clinical response for primary endpoint analysis was defined as achieving a complete or partial response at ≥12 weeks. Blood and tumours before and after treatment were obtained for correlative studies by IRB-approved laboratory protocol PA13-0291. Tumour samples were obtained as core biopsies or surgical resection performed by interventional radiologists or surgeons. Samples were immediately FFPE or snap-frozen after tissue collection.
The single-cell RNA-seq B cell analysis used a dataset from 32 patients with metastatic melanoma (n = 48 samples) treated with anti-PD1 (n = 37), anti-CTLA4 (n = 2), or anti-PD1 and anti-CTLA4 (n = 9)44. Patient response was determined by RECIST criteria: complete response and partial response for responders, or stable disease and progressive disease for non-responders. For the analysis, we focused on individual lesions and classified them into two categories: responder (n = 17) including complete-response and partial-response samples; non-responder (n = 31) including stable-disease and progressive-disease samples, based on radiological tumour evaluations. Samples were collected after patients provided a written consent for research and genomic profiling of collected tissue as approved by the Dana-Farber/Harvard Cancer Center Institutional Review Board (DF/HCC protocol 11-181) and UT MD Anderson Cancer Center (LAB00-063 and 2012-0846).
For the targeted therapy cohort, 13 patients received neoadjuvant and adjuvant dabrafenib and trametinib as part of a single-centre, open-label randomized phase 2 trial for patients with BRAF(V600E) or BRAF(V600K) (that is, Val600Glu or Val600Lys)-mutated melanoma (NCT02231775)—8 weeks of neoadjuvant oral dabrafenib 150 mg twice per day and oral trametinib 2 mg per day followed by surgery, then up to 44 weeks of adjuvant dabrafenib plus trametinib starting 1 week after surgery for a total of 52 weeks of treatment38. Patient radiographic response was determined by RECIST criteria with stable disease (non-responders) and partial response or complete response (responders) noted and coded as indicated; and pathological complete response determined by absence of residual viable malignant cells on H&E staining. These patients were treated at the University of Texas MD Anderson Cancer Center and had tumour samples collected and analysed under IRB-approved protocols. These studies were conducted in accordance with the Declaration of Helsinski.
The authors confirm for all studies involving human research participants we have complied with all relevant ethical regulations.
Gene expression profiling and analysis: RNA extraction for neoadjuvant melanoma ICB-treated cohort
Total RNA was extracted from snap-frozen tumour specimens using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen) following assessment of tumour content by a pathologist, and macrodissection of tumour bed if required. RNA quality was assessed on an Agilent 2100 Bioanalyzer using the Agilent RNA 6000 Nano Chip with smear analysis to determine DV200 and original RNA concentration. On the basis of RNA quality, 40–80 ng of total RNA from each sample then underwent library preparation using the Illumina TruSeq RNA Access Library Prep kit according to the manufacturer’s protocol. Barcoded libraries were pooled to produce final 10–12 plex pools before sequencing on an Illumina NextSeq sequencer using one high-output run per pool of 76-bp paired-end reads, generating 8 fastq files (4 lanes, paired reads) per sample.
RNA-seq data processing and quality check
RNA-seq FASTQ files were first processed through FastQC (v.0.11.5)45, a quality control tool to evaluate the quality of sequencing reads at both the base and read levels. The reads that had ≥ 15 contiguous low-quality bases (phred score < 20) were removed from the FASTQ files. STAR 2-pass alignment (v.2.5.3)46 was then performed on the filtered FASTQ files with default parameters to generate RNA-seq BAM file for each sequencing event. After that, RNA-SeQC (v.1.1.8)47 was run on the aligned BAM files to generate a series of RNA-seq related quality control metrics including read counts, coverage, and correlation. A matrix of Spearman correlation coefficients was subsequently generated by RNA-SeQC among all sequencing events. The correlation matrix was carefully reviewed and the sequencing event generated from one library pool that showed poor correlation with other library pools from the same RNA sample were removed before sample-level merging of BAM files.
Gene expression quantification and normalization
HTSeq-count (v.0.Fig.9.1)48 tool was applied to aligned RNA-seq BAM files to count for each gene how many aligned reads overlap with its exons. The raw read counts generated from HTSeq-count (v.0.9.1)48 were normalized into fragments per kilobase of transcript per million mapped reads (FPKM) using the RNA-seq quantification approach suggested by the bioinformatics team of NCI Genomic Data Commons (GDC; https://gdc.cancer.gov/about-data/data-harmonization-and-generation/genomic-data-harmonization/high-level-data-generation/rna-seq-quantification). In brief, FPKM normalizes read count by dividing it by the gene length and the total number of reads mapped to protein-coding genes using a calculation described below:
in which RCg denotes the number of reads mapped to the gene; RCpc denotes the number of reads mapped to all protein-coding genes; and L denotes the length of the gene in base pairs (calculated as the sum of all exons in a gene). The FPKM values were then log2-transformed for further downstream processes.
RNA-seq analysis for OpACIN-neo trial
RNA-seq and data analysis were performed as previously described35.
Affymetrix microarray for RCC
The Affymetrix microarray data were created using the Affymetrix Clariom D Assay (Human). There are 28 available pre-treatment samples from 3 arms: nivolumab (n = 6), nivolumab plus bevacizumab (n = 14) and nivolumab plus ipilimumab (n = 8). The raw CEL files were normalized using the built-in SST-RMA method of the Affymetrix Transcriptome Analysis Console (TAC, v.4.0) software. The cell lineage scores were calculated using the R package MCP-counter algorithm (v.1.1.0). The Limma R software package49 was used to identify DEGs from normalized microarray data for the RCC cohort.
Identification of DEGs
The HTSeq normalized read count data for all expressed coding transcripts was processed by Deseq2 (v.3.6)50 software to identify DEGs between two response (responders versus non-responders) groups. A cut-off of gene-expression fold change of ≥ 2 or ≤ 0.5 and a FDR q ≤ 0.05 was applied to select the most DEGs. The Limma R software package49 was used to identify DEGs from normalized microarray data for the RCC cohort.
Deconvolution of the cellular composition with MCP-counter
The R package software MCP-counter18 was applied to the normalized log2-transformed FPKM expression matrix to produce the absolute abundance scores for eight major immune cell types (CD3+ T cells, CD8+ T cells, cytotoxic lymphocytes, natural killer cells, B lymphocytes, monocytic lineage cells, myeloid dendritic cells and neutrophils), endothelial cells and fibroblasts. The deconvolution profiles were then hierarchically clustered and compared across response and treatment groups.
Pathway enrichment analyses
The network-based pathway enrichment analysis was performed using DEGs across responder and non-responder groups in the bulk-tissue RNA-seq data from the melanoma neoadjuvant cohort and single-cell RNA-seq data from the metastatic melanoma cohort. In the bulk-tissue, the differentially expressed genes that had a q < 0.05 and log2-transformed fold change >1.5 or < −1.5 were selected as input for network based pathway enrichment analysis using ReactomeFiViz51 application in Cytoscape52,53. In single-cell, the DEGs with q < 0.1 were selected as input for pathway enrichment analysis. Pathway enrichment was calculated using several biological databases (KEGG, NCBI, Reactome, Biocarta and Panther) with hypergeometric test FDR < 0.01.
TCGA SKCM and KIRC data downloading and patient selection
The normalized RNA-seq expression data of TCGA skin cutaneous melanoma (TCGA-SKCM) and Kidney Renal Clear Cell Carcinoma (TCGA-KIRC) was downloaded from NCI Genomic Data Commons (GDC; https://portal.gdc.cancer.gov) and the relevant clinical data were downloaded from recent TCGA PanCancer clinical data study54. The information of SKCM genomic subtypes was obtained from the TCGA-SKCM study42.To achieve a uniform cohort of patients with stage III (non-recurrent) melanoma for analysis, we applied an appropriate set of sequential filters: the TCGA-SKCM cohort was filtered to include patients with biospecimen tissue sites that included regional lymph node or regional subcutaneous metastases. We excluded patients presenting with stage IV disease. Then, to exclude patients with recurrent stage III disease, we excluded all patients for whom the number of days from the diagnosis of the primary to the accession date was more than 90 days. In addition, for a patient to be included, their tumour must also have had a defined melanoma driver type. Finally, we eliminated those lacking sufficient gene expression data, yielding a final stage III TCGA-SKCM cohort of n = 136. Survival data were missing for 9 of 136 samples, so n = 127 samples were available for overall survival analyses. For TCGA-KIRC, the cases without available expression data were excluded and a total of 526 cases were taken into subsequent analysis.
Survival analyses
In TCGA cohort, survival data were not available for nine samples and these were excluded from survival analysis. As previously described42, the survival time for each patient for the SKCM melanoma cohort was ‘curated TCGA survival’ (that is, from time of TCGA biospecimen procurement). The time to event was defined as the time interval from date of accession for each sample to date of death or censoring from any cause (curated value CURATED_TCGA_days_to_death_or_last_follow-up; aka TCGA post-accession survival). The survival analysis was performed using Cox proportional hazards model and survival curves were plotted using Kaplan–Meier method. The statistical comparison of the survival curves was done using the log-rank test. The analysis was done using R package survival (https://cran.r-project.org/web/packages/survival/index.html).
Statistical analyses
The statistical comparison between responder and non-responder groups for a given continuous variable was performed using two-sided Mann–Whitney U-test. The association between two continuous variables was assessed using Spearman’s rank correlation coefficient. To control for multiple comparisons, we applied the Benjamini–Hochberg method55 and calculated adjusted P values. Univariable and multivariable analysis predicting response to ICB was performed using logistic regression modelling. Biological replicates are indicated in the individual figure legends. Technical replicates were constrained to n = 1 per time point, owing to limited tissue availability in patient-derived samples as well as prioritization for multiple studies. No statistical methods were used to predetermine sample size. The experiments were not randomized, and investigators were not blinded to allocation during experiments and outcome assessment unless stated otherwise.
Single immunohistochemistry
H&E and immunohistochemistry staining were performed on FFPE tumour tissue sections. The tumour tissues were fixed in 10% formalin, embedded in paraffin, and serially sectioned. Four-micrometre sections were used for the histopathological study.
Sections were stained with mouse or rabbit anti-human monoclonal antibodies against CD20 (Dako, M0755, 1:1,400), CD21 (Novocastra, NCL-L-CD21-2G9, 1:10 or Leica, CD21-2G9; 1:20), CD23 (Leica, CD23-1B12, 1:15), CD4 (Novocastra, CD4-368-L-A, 1:80) CD8 (Thermo Scientific, MS-457-S, 1:25), FOXP3 (Biolegend, 320102, 1:50). All sections were counterstained with haematoxylin, dehydrated and mounted. All sections were processed with peroxidase-conjugated avidin/biotin and 3′-3-diaminobenzidine (DAB) substrate (Leica Microsystem) and slides were scanned and digitalized using the scanscope system from Scanscope XT, Aperio/Leica Technologies.
Quantitative analysis of immunohistochemistry staining was conducted using the image analysis software ImageScope-Aperio/Leica. Five random areas (1 mm2 each) were selected using a customized algorithm for each marker in order to determine the number of positive cells at high power field. The data are expressed as a density (total number of positive cells per mm2 area). Immunohistochemistry staining was interpreted in conjunction with H&E stained sections.
TLS quantification
TLSs were qualified and quantified using both H&E and CD20 immunohistochemistry staining. Structures were identified as aggregates of lymphocytes having histological features with analogous structures to that of lymphoid tissue with germinal centres (including B cells (CD19/20), T cells (CD3), follicular dendritic cells (CD21) and high endothelial venules (MECA79), appearing in the tumour area13,56,57,58. For the current study, criteria used for the quantification of TLS includes: (1) the total number of structures identified either within the tumoral area or in direct contact with the tumoral cells on the margin of the tumours (numbers of TLS per mm2 area); and (2) a normalization of the total area occupied by the TLNs in relation of the total area of the tumour analysed (ratio: area of TLS/area tumour + TLNs).
Multiplex immunofluorescence assay and analysis
For images shown in Fig. 2 and Extended Data Fig. 6, for immunofluorescence multiplex staining, we followed the staining method for the following markers: CD20 (Dako, M0755, 1:500) with subsequent visualization using fluorescein Cy3 (1:50); CD21 (Novocastra, NCL-L-CD21-2G9, 1:10) with subsequent visualization using fluorescein Cy5 (1:50); CD4 (CM153BK, Biocare, 1:25) with subsequent visualization using fluorescein Cy5.5 (1:50); CD8 (1:200, M7103, Dako) with subsequent visualization using fluorescein Cy3.5 (1:50); FOXP3 (Biolegend, 320102, 1:50) with subsequent visualization using fluorescein FITC (1:50) and nuclei visualized with DAPI (1:2,000). All of the sections were coverslipped using Vectashield Hardset 895 mounting medium.
The slides were scanned using the Vectra slide scanner (PerkinElmer). For each marker, the mean fluorescent intensity per case was then determined as a base point from which positive calls could be established. For multispectral analysis, each of the individually stained sections was used to establish the spectral library of the fluorophores. Five random areas on each sample were analysed blindly by a pathologist at 20× magnification.
For additional multiplex images shown in Extended Data Fig. 5, for additional multiplex staining, we followed similar methods to the above for the following markers: MECA79-Dy550 (Novus, MECA-79, 1:100); CD20-Dy594 (Novus, IGEL/773; 1:100); CD4-AF647 (abcam, ERP6855, 1:100); and nuclei visualized with Syto13 at 500 nM. The slides were scanned with the GeoMx DSP machine as described below.
GeoMx Digital Spatial Profiling: microscope and fluidics system overview
For immune profiling of T cells located within and outside TLS structures in patient samples, the GeoMx Digital Spatial Profiler (NanoString), a custom-built high-speed automated system and integrated instrument software, was used. A multiplexed cocktail of primary antibodies with UV photocleavable indexing oligonucleotides (GeoMx Immune Profile Core; 22 targets, including 3 isotype controls and 4 additional modules; IO Drug Target, Immune Activation Status, Immune Cell Typing, and Pan Tumour) and 4 fluorescent markers was applied to a slide-mounted FFPE tissue section. For the fluorescent markers, we used Syto13 at 500 μM for nuclei visualization; CD20-Dy594 (Novus, IGEL/773; 1:100); CD3-AF647 (Novus, C3e/1308; 1:100); and PMEL-Dy550 (Novus, HMB45; 1:100) with S100B-Dy550 (Novus, 15F4NB; 1:100). Images at ×20 magnification were assembled to yield a high-resolution image of the tissue area of interest. The specific regions of interest (ROIs) for molecular profiling were then selected based on location (TLS or non-TLS areas of tumour) and CD3-positive staining and sequentially processed by the microscope automation. ROIs were selectively illuminated with UV light to release the indexing oligos by coupling UV LED light with a double digital mirror device (DDMD) module. Following each UV illumination cycle, the eluent was collected from the local region via microcapillary aspiration and transferred to an individual well of a microtiter plate. Once all ROIs were processed, pools of released indexing oligos were hybridized to NanoString optical barcodes for digital counting and subsequently analysed with an nCounter Analysis System.
nCounter hybridization assay for photocleaved oligo counting
Hybridization of cleaved indexing oligonucleotides to fluorescent barcodes was performed using the nCounter Protein PlexSet reagents based on manufacturer’s directions. Hybridizations were performed at 65 °C overnight in a thermocycler. After hybridization, samples were processed using the nCounter Prep Station and Digital Analyzer as per manufacturer instructions. Data were normalized to technical controls and area. Data were calculated against isotype controls to generate signal-to-noise ratios. Protein targets with a signal to noise ratio less than 2 were removed from downstream analysis.
B cell clonotype analyses
The modified TRUST algorithm59 was applied to extract the B cell immunoglobin hypervariable regions from the bulk RNA-seq data and assemble the complementarity-determining region 3 (CDR3) sequences of the B cell heavy chain (IgH) and light chain (IgL). BCR clonotypes were identified and the clonal fraction was automatically calculated by TRUST. The output of TRUST was parsed by the R package tcR (v.3.4.1)60 for downstream analyses. Only in-frame productive clonotypes were taken into subsequent analysis. The total number of BCR clonotypes detected per sample was normalized by the corresponding sequencing depth of each individual sample and calculated as per 100 million mapped reads. The top five clonotypes were selected by their clonal expression abundance. The BCR repertoire diversity was calculated by entropy from the tcR package60.
Single-cell sequencing and analysis of CD45+ B cells
Fresh isolated tumour samples were dissociated using the human tumour dissociation kit (Miltenyi Biotec; 130-095-929), sorted into 96-well plates containing 10 µl of TCL buffer (Qiagen) with 1% β-mercaptoethanol, using the following anti-human antibodies: FcX (Biolegend, 422302), CD45-PE (Biolegend, 304008), CD3-APC (Biolegend, 300412), CD235a-APC/Cy7 (Biolegend, 349116) and HLA-A,B,C-FITC (Biolegend, 311426). Sorting of viable cells was performed using the live/dead dye Zombie Violet (Biolegend, 77477). Single-cell libraries were generated using a modified version of the full-length Smart-seq2 protocol as previously described61, and were sequenced on a NextSeq 500 sequencer (Illumina), resulting in a median of approximately 1.4 million paired-end reads and a median of 2,588 genes detected per cell. A cutoff of log2(transcripts per million (TPM) + 1) ≥ 2 was used to define a gene as expressed in each single cell. For each sample, we computed the fraction of B cells using pre-defined markers (CD19 and/or MS4A1). Notably, this is a plate-based protocol; thus, for each patient, we collected and sequenced the same number of cells (n = 384 CD45+ cells per plate). Thus, the number of cells per patient is equal, and the frequency reflects patients with either high or low B cell infiltrate.
Unsupervised clustering of immune cells
To cluster all cells that passed quality control, we applied the k-means algorithm with a correlation distance metric, testing \(k=3,\ldots ,15\). The algorithm was applied using all genes with variance >6, yielding approximately 4,000 genes. This value was selected based on the relation between the variance and the fraction of cells expressing each gene. To determine the optimal number of clusters we applied the following steps: (1) we first examined how much of the complexity each cluster captures by applying the elbow method. This was done by computing the Pearson correlation matrix R and the distance matrix D as (1 − R). We then computed the sum of pairwise distances between all cells in different clusters, \({{\rm{Dis}}}_{b}={\sum }_{l=1}^{k}({\sum }_{i\in {C}_{l},j\notin {C}_{l}}D(i,j))\), and the total distance, \({{\rm{Dis}}}_{t}={\sum }_{i,j}D(i,j)\), in which i and j stand for each pair of single cells. The ratio between these two measures, \(V\,={{\rm{Dis}}}_{b}/{{\rm{Dis}}}_{t}\,\) was used to estimate the variance explained by a given solution, such that in the extreme case in which all cells are clustered together or the case in which each cell is a single cluster, this ratio would be 0 and 1, respectively. Exploring this ratio, we then select the solutions that are near plateau (\(k=10,\ldots ,15\)). (2) We then performed differential expression analysis (see ‘Differential expression analysis’) to search for gene markers that are significantly more highly expressed in a specific cluster as compared to all other clusters. Then, to avoid complex solutions, we excluded solutions with clusters that have too few marker genes (<20) distinguishing between them and the rest of the cells. (3) Finally, we performed a robustness analysis and selected the clustering solution with the highest median robustness score. Specifically, to determine the robustness of each clustering solution, we performed 100 iterations in which we randomly removed 10% of the cells, and re-ran the k-means algorithm and checked the stability of the clustering solution. We quantified the agreement of a given solution with the original one as the number of pairs of cells that were either clustered together, or not clustered together, in both solutions, divided by the total number pairs shared between the runs. This process yielded a median robustness measure of 0.96 for the selected k=11.
Differential expression analysis
In all cases, differential expression analysis was applied to all genes that had an average expression level log2(TPM+ 1 ) > 2 in either tested groups, G1 and G2. Then, for each gene i, we count the number of cells in G1 and G2 that express it with an expression level log2(TPM + 1) > 2 or ≤ 2. We then apply Fisher’s exact test for the corresponding 2 × 2 table. To identify significant differences, we considered genes with a Bonferroni-corrected q ≤ 0.05 and log2-transformed fold change > 0.5.
CyTOF antibody conjugation
In-depth characterization of B cells from responders and non-responders was performed using metal-tagged antibodies. Metal conjugated antibodies were purchased from Fluidigm or conjugated to unlabelled antibodies in-house. All unlabelled antibodies were purchased in carrier-free form and conjugated with the corresponding metal tag using Maxpar X8 polymer per manufacturer’s instructions (Fluidigm). Metal isotopes were acquired from Fluidigm and indium (III) chloride was acquired from Sigma-Aldrich. Antibody concentration was determined by measuring the amount of A280 protein using Nanodrop 2000 (Thermo Fisher Scientific). Conjugated antibodies were diluted using PBS-based antibody stabilizer supplemented with 0.05% sodium azide (Sigma-Aldrich) to a final concentration of 0.5 mg ml−1. Antibodies used with the corresponding metal tag isotopes: CD45 (Fluidigm, HI30, 89Y), CD80 (Biolegend, 2D10, 115In), CD138 (BD Biosciences, MI15, 141Pr), CD19 (Fluidigm, HIB19, 142Nd), CD5 (Fluidigm, UCHT2, 143Nd), HLA-ABC (BD Biosciences, EMR8-5, 144Nd), CD178 (Biolegend, NOK1, 145Nd), IgD (Biolegend, IA6-2, 146Nd), CD20 (Fluidigm, 2H7, 147Sm), PDL1 (Fluidigm, 29E.2A3, 148Nd), HLA-DR (Biolegend, L243, 149Sm), CD25 (BD Biosciences, 2A3, 150Nd), IGM (Biolegend, MHM-88, 151Eu), CD95 (BD Biosciences, DX2, 152Sm), CXCR5 (Fluidigm, RF8B2, 153Eu), CD86 (BD Biosciences, IT2.2, 154Sm), CD27 (Fluidigm, L128, 155Gd), CXCR3 (Biolegend, G025H7, 156Gd), CD10 (Fluidigm, HI10a, 158Gd), PDL-2 (Biolegend, 24F.10C12, 159Tb), CD39 (Fluidigm, A1, 160Gd), BAFF-R (Biolegend, 11C1, 161Dy), CD79b (Fluidigm, CB3.1, 162Dy), CD1d (Biolegend, 51.1, 163Dy), CD23 (Fluidigm, EBVCS-5, 164Dy), CD40 (Biolegend, 5C3, 165Ho), CD24 (BD Biosciences, ML5, 166Er), CD38 (BD Bioscience, HIT2, 167Er), CD21 (Biolegend, Bu32, 168Er), ICOS (Biolegend, C398.4A, 169Tb), CTLA4 (Fluidigm, 14D3, 170Er), CD9 (Biolegend, HI9a, 171Yb), CD11c (Biolegend, Bu15, 172Yb), CD14 (Biolegend, HCD14, 173Yb), PD1 (Miltenyi, PD1.3.1.3, 174Yb), CXCR4 (Biolegend, 12G5, 175Lu), CD22 (Biolegend, HIB22, 176Yb), CD3 (Biolegend, UCHT-1, 194Pt), cisplatin (Fluidigm, 198Pt) and CD16 (Fluidigm, 3G8, 209Bi).
Sample preparation and acquisition
Peripheral blood mononuclear cells and tumour cells were collected and washed twice with wash buffer (0.5% bovine serum albumin (BSA) in PBS). For tumour, this included 9 responders and 9 non-responders, and for peripheral blood mononuclear cells, 8 responders and 8 non-responders. To determine the live population, cells were stained with 1 µM cisplatin for 3 min. The reaction was stopped with FACS buffer (2% fetal bovine serum (FBS) in PBS), and the cells were washed once with wash buffer. Cells were then incubated with 5 µl of Fc receptor blocking buffer reagent (Miltenyi) for 10 min at room temperature. Cells were incubated with surface antibodies at room temperature for 60 min, washed twice with wash buffer and stored overnight in 1 ml of 1.6% paraformaldehyde (EMD Biosciences) in PBS with 125 nM iridium nucleic acid intercalator (Fluidigm). The next day, samples were washed twice with cell staining buffer, re-suspended in 1 ml of MilliQ dH2O, filtered through a 35-µm nylon mesh (cell strainer cap tubes, BD) and counted. Before analysis, samples were resuspended in MilliQ dH2O supplemented with EQ four element calibration beads at a concentration of 0.5 × 105 per ml. Samples were acquired at 300 events per second on a Helios instrument (Fluidigm) using the Helios 6.5.358 acquisition software (Fluidigm).
Data analysis
Mass cytometry data were normalized based on EQ four element signal shift over time using Fluidigm normalization software 2. Initial data processing was performed using Flowjo version 10.2. Mass cytometry data were normalized based on EQTM four element signal shift over time using Fluidigm normalization software 2. Initially, all responder and non-responder normalized FCS files were either concatenated or separately exported for downstream analyses. Data were processed and analysed using Cytobank; CD19+ sample ‘clean-up’ was performed by gating on intact (191Ir+ DNA stain), no beads (140Ce−), live (198Pt−), no T-cells CD3− (194Pt), no monocytes CD14− (173Yb) and CD45+ (89Y), no natural killer cells CD16− (209Bi), CD19+ B cells. Mass cytometry complex data were analysed using viSNE, in combination with heat map, to identify distinct subpopulations using the following parameters: CD19 (142Nd), CD20 (147Sm), CD5 (143Nd), HLA-ABC (144Nd), IgD (146Nd), PDL1 (148Nd), HLA-DR (149Sm), CD25 (150Nd), IgM (151Eu), CD95 (152Sm), CXCR5 (153Eu), CD86 (154Sm), CD27 (155Gd), CXCR3 (156Gd), CD10 (158Gd), CD39 (160Gd), BAFFR (161Dy), CD79b (162Dy), CD1d (163Dy), CD23 (164Dy), CD40 (165Ho), CD24 (166Er), CD38(167Er), CD9(171Yb), CD11c (172Yb), CXCR4 (175Lu), and CD22 (176Yb). Samples with fewer than 200 CD45+CD19+ B cells were not used for downstream analyses. Percentages of different subpopulations of B cells were measured in aggregated responder and non-responder peripheral blood cells and tumour samples for each run; statistical analyses performed via unpaired Student’s t-test.
Analysis of peripheral blood exosomes from human plasma
Approximately 1 ml of plasma per patient sample contained in a cryovial was thawed rapidly in a 37 °C water bath. The plasma was transferred into a 1.5-ml Eppendorf tube and centrifuged at room temperature for 5 min at 800g and 10 min at 2,000g. The supernatant was filtered with a 0.22-µm filter (6789-1302) directly into an ultracentrifuge tube (Z80615SCA, 331372). A distinct filter was used for each 500 µl of plasma filtered, and each filter was subsequently cleared with 2 × 1 ml PBS), all of which was collected into the ultracentrifuge tube. Additional PBS was added to the ultracentrifuge tube to reach 11 ml. The tubes were the ultracentrifuged at 4 °C for 15–16 h at 100,000g using a Beckman Optima XE-90 ultracentrifuge. The pellet was resuspended in 200–300 µl of PBS by pipetting up and down. The exosomes contained in this resuspension were stored at −80 °C until further use.
Flow cytometric analyses of exosomes
Exosomes were thawed on ice. Concentration was determined using the NanoSight NS300 nanoparticle tracking analyser according to the manufacturer’s directions, and 15 µl of exosomes (which was equivalent to approximately 4 × 109 particles on average) were mixed with 30 µl of pre-washed anti-human CD63-coated Dynabeads (Invitrogen, 10606D). For one sample, the Nanosight measurement was erroneous and excluded. All samples were included in the flow cytometric analyses. Round-bottom 2-ml tubes were used. All pre-wash and washes thereafter were performed using 0.22 µm filtered 0.1% BSA in PBS (0.1% BSA/PBS) and the samples were mixed well by pipetting up and down at each wash steps. One-hundred microlitres of 0.1% BSA/PBS was added to beads + exosomes mixture for a final volume of 145 µl (15 µl of exosomes + 30 µl of Dynabeads + 100 µl of 0.1% BSA/PBS). The samples were mixed by pipetting up and down and allowed to incubate for 4–16 h at room temperature on a benchtop rotator. Three-hundred microlitres of 0.1% BSA/PBS was added to the samples and the samples were placed on a magnet (1 min incubation minimum). The supernatant was discarded and the beads (and bound exosomes) were washed once with 400 µl 0.1% BSA/PBS.
The beads (with bound exosomes) were resuspended in 400 µl of 0.1% BSA/PBS and subsequently split into four distinct round-bottom 2-ml tubes, each containing 100 µl. To each of these tubes, either antibodies or isotype control were added. These include: PE/Cy7 anti-human CD20 (Biolegend, 302312, clone 2H7) or isotype control PE/Cy7 mouse IgG2b (Biolegend, 400326, clone MCP-11); APC/Cy7 anti-human CD27 (Biolegend, 356424, clone M-T271) or isotype control APC/Cy7 mouse IgG1 (Biolegend, 400128, clone MOPC-21); PE/Cy7 anti-human CD9 (Biolegend, 312116, clone HI9a) or isotype control PE/Cy7 mouse IgG1 (Biolegend, 400126, clone MOPC-21); and Alexa Fluor 647 anti-human CD63 (Biolegend, 353016, clone H5C6) or isotype control Alexa Fluor 647 mouse IgG1 (Biolegend, 400130, clone MOPC-21). For each antibody or isotype control, 0.4 µg was added to each tube. The samples were allowed to incubate at room temperature for 1–3 h, in the dark. Three-hundred microlitres of 0.1% BSA/PBS was added to the samples and the samples were placed on a magnet (1 min incubation). The supernatant was discarded and the beads (and bound exosomes) were washed once with 400 µl 0.1% BSA/PBS. The beads were visible on the magnet at each step of the procedure described above. The supernatant was discarded and the beads were resuspended in 200 µl of 0.1% BSA/PBS and transferred into flow cytometry tubes for flow cytometry analysis. The flow cytometry data were captured within 24 h of completing the staining of the beads-exosomes samples. If not read immediately after completing the staining, the flow cytometry tubes were stored at 4 °C in the dark. The data were subsequently analysed using FlowJo. Responder versus non-responder status was blinded until flow cytometry data capture and FlowJo analyses were completed.
For GPC1 staining, three tubes of beads with exosomes were processed in parallel. One tube did not receive any antibody (exosomes alone), one tube received primary antibody (1 h) followed by secondary antibody (1 h), and one tube received secondary antibody only (1 h). All three tubes were processed similarly, including for a wash step after one hour (post primary antibody incubation, 300 µl 0.1% BSA/PBS was added to the samples and the samples were placed on a magnet for 1 min incubation, and then resuspended into 100 µl of 0.1% BSA/PBS), and again another hour later (after the secondary antibody incubation), before transferred into a FC tube. All incubations were carried out at RT and covered from light, and beads were visible at each step when placed on the magnet. Rabbit anti-human GPC1 antibody was used (Sigma, SAB2700282, 3 µl per tube), and Alexa Fluor 488 conjugated goat anti-rabbit IgG (Invitrogen, A-11008, 2 µl per tube) were used. The samples were analysed by flow cytometry.
Nanoimager analyses
Beads with exosomes stained for flow cytometry analysis for CD63 (Alexa Fluor 647 anti-human CD63) or isotype control as described above (see ‘Flow cytometric analyses of exosomes’) were evaluated by using the Nanoimager S Mark I from ONI (Oxford Nanoimaging) with the lasers 405 nm/150 mW, 488 nm/200 mW, 561 nm/300 mW, 640 nm/1 W and dual emission channels split at 560 nm. Data were processed on NimOS (v.1.25) from ONI. In brief, 25 µl of sample was spotted onto a slide (Fisher Scientific, 12-550-15), covered with a 1.5H coverslip (Zeiss, 474030-9000), and immediately placed on the stage. All images were captured using HILO mode (highly inclined and laminated optical sheet) at an illumination angle of 35.0° with a 10.0-ms exposure setting for 200 frames. To minimize photobleaching, the focal plane of the beads was found under the 405 nm laser at 37% power, then switched to the 640 nm laser at 25% power for image acquisition.
Electron microscopy analyses
Bead only and beads with exosomes were prepared as described above (‘Flow cytometric analyses of exosomes’). The samples were magnetized and resuspended in 50 µl of 1% glutaraldehyde in PBS at 4 °C, or in 30µl of 0.1% BSA/PBS, and mixed with 30 µl of warm (60 °C) 1% agarose in distilled water. The agarose-bead mixture was allowed to cool on ice, and the gels were cut into approximately 1-mm3 pieces and placed in 1% glutaraldehyde in PBS at 4 °C. Fixed samples were washed in 0.1 M sodium cacodylate buffer and treated with 0.1% Millipore-filtered cacodylate buffered tannic acid, postfixed with 1% buffered osmium, and stained en bloc with 1% Millipore-filtered uranyl acetate. The samples were dehydrated in increasing concentrations of ethanol, infiltrated, and embedded in LX-112 medium. The samples were polymerized in a 60 °C oven for approximately 3 days. Ultrathin sections were cut in a Leica Ultracut microtome (Leica), stained with uranyl acetate and lead citrate in a Leica EM Stainer, and examined in a JEM 1010 transmission electron microscope (JEOL) at an accelerating voltage of 80 kV. Digital images were obtained using AMT Imaging System (Advanced Microscopy Techniques). Two-sided Mann–Whitney U-test was used to determine significance.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
The additional datasets generated during and/or analysed during the current study for Clinical Trial NCT02519322 are now available in the European Genome-phenome Archive repository (EGAS00001003178). Other datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Code availability
The authors declare that the code for reproducibility of data are publicly available or will be available upon request.
References
Chen, P. L. et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 6, 827–837 (2016).
Taube, J. M. et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin. Cancer Res. 20, 5064–5074 (2014).
Cottrell, T. R. & Taube, J. M. PD-L1 and emerging biomarkers in immune checkpoint blockade therapy. Cancer J. 24, 41–46 (2018).
Yarchoan, M., Hopkins, A. & Jaffee, E. M. Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 377, 2500–2501 (2017).
Ayers, M. et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 127, 2930–2940 (2017).
Subudhi, S. K. et al. Clonal expansion of CD8 T cells in the systemic circulation precedes development of ipilimumab-induced toxicities. Proc. Natl Acad. Sci. USA 113, 11919–11924 (2016).
Jacquelot, N. et al. Predictors of responses to immune checkpoint blockade in advanced melanoma. Nat. Commun. 8, 592 (2017).
Gopalakrishnan, V. et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018).
Matson, V. et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108 (2018).
Routy, B. et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018).
Fridman, W. H., Zitvogel, L., Sautès-Fridman, C. & Kroemer, G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 14, 717–734 (2017).
Colbeck, E. J., Ager, A., Gallimore, A. & Jones, G. W. Tertiary lymphoid structures in cancer: drivers of antitumor immunity, immunosuppression, or bystander sentinels in disease? Front. Immunol. 8, 1830 (2017).
Dieu-Nosjean, M. C., Goc, J., Giraldo, N. A., Sautès-Fridman, C. & Fridman, W. H. Tertiary lymphoid structures in cancer and beyond. Trends Immunol. 35, 571–580 (2014).
Sarvaria, A., Madrigal, J. A. & Saudemont, A. B cell regulation in cancer and anti-tumor immunity. Cell. Mol. Immunol. 14, 662–674 (2017).
Tsou, P., Katayama, H., Ostrin, E. J. & Hanash, S. M. The emerging role of B cells in tumor immunity. Cancer Res. 76, 5597–5601 (2016).
Sautès-Fridman, C., Petitprez, F., Calderaro, J. & Fridman, W. H. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat. Rev. Cancer 19, 307–325 (2019).
Amaria, R. N. et al. Neoadjuvant immune checkpoint blockade in high-risk resectable melanoma. Nat. Med. 24, 1649–1654 (2018).
Becht, E. et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 17, 218 (2016).
Yuen, G. J., Demissie, E. & Pillai, S. B lymphocytes and cancer: a love-hate relationship. Trends Cancer 2, 747–757 (2016).
Chiaruttini, G. et al. B cells and the humoral response in melanoma: the overlooked players of the tumor microenvironment. OncoImmunology 6, e1294296 (2017).
Erdag, G. et al. Immunotype and immunohistologic characteristics of tumor-infiltrating immune cells are associated with clinical outcome in metastatic melanoma. Cancer Res. 72, 1070–1080 (2012).
Iglesia, M. D. et al. Genomic analysis of immune cell infiltrates across 11 tumor types. J. Natl. Cancer Inst. 108, (2016).
Ladányi, A. et al. Prognostic impact of B-cell density in cutaneous melanoma. Cancer Immunol. Immunother. 60, 1729–1738 (2011).
Garg, K. et al. Tumor-associated B cells in cutaneous primary melanoma and improved clinical outcome. Hum. Pathol. 54, 157–164 (2016).
Ladányi, A. et al. Density of DC-LAMP+ mature dendritic cells in combination with activated T lymphocytes infiltrating primary cutaneous melanoma is a strong independent prognostic factor. Cancer Immunol. Immunother. 56, 1459–1469 (2007).
Martinet, L. et al. High endothelial venules (HEVs) in human melanoma lesions: Major gateways for tumor-infiltrating lymphocytes. OncoImmunology 1, 829–839 (2012).
Avram, G. et al. The density and type of MECA-79-positive high endothelial venules correlate with lymphocytic infiltration and tumour regression in primary cutaneous melanoma. Histopathology 63, 852–861 (2013).
Messina, J. L. et al. 12-Chemokine gene signature identifies lymph node-like structures in melanoma: potential for patient selection for immunotherapy? Sci. Rep. 2, 765 (2012).
Goc, J. et al. Dendritic cells in tumor-associated tertiary lymphoid structures signal a Th1 cytotoxic immune contexture and license the positive prognostic value of infiltrating CD8+ T cells. Cancer Res. 74, 705–715 (2014).
Posch, F. et al. Maturation of tertiary lymphoid structures and recurrence of stage II and III colorectal cancer. OncoImmunology 7, e1378844 (2017).
Siliņa, K. et al. Germinal centers determine the prognostic relevance of tertiary lymphoid structures and are impaired by corticosteroids in lung squamous cell carcinoma. Cancer Res. 78, 1308–1320 (2018).
Cipponi, A. et al. Neogenesis of lymphoid structures and antibody responses occur in human melanoma metastases. Cancer Res. 72, 3997–4007 (2012).
Selitsky, S. R. et al. Prognostic value of B cells in cutaneous melanoma. Genome Med. 11, 36 (2019).
Griss, J. et al. B cells sustain inflammation and predict response to immune checkpoint blockade in human melanoma. Nat. Commun. 10, 4186 (2019).
Blank, C. U. et al. Neoadjuvant versus adjuvant ipilimumab plus nivolumab in macroscopic stage III melanoma. Nat. Med. 24, 1655–1661 (2018).
Cabrita, R. L. et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature https://doi.org/10.1038/s41586-019-1914-8 (2020).
Petitprez, F. et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature https://doi.org/10.1038/s41586-019-1906-8 (2020).
Amaria, R. N. et al. Neoadjuvant plus adjuvant dabrafenib and trametinib versus standard of care in patients with high-risk, surgically resectable melanoma: a single-centre, open-label, randomised, phase 2 trial. Lancet Oncol. 19, 181–193 (2018).
Reddy, S. M. et al. Poor response to neoadjuvant chemotherapy correlates with mast cell infiltration in inflammatory breast cancer. Cancer Immunol. Res. 7, 1025–1035 (2019).
Song, I. H. et al. Predictive value of tertiary lymphoid structures assessed by high endothelial venule counts in the neoadjuvant setting of triple-negative breast cancer. Cancer Res. Treat. 49, 399–407 (2017).
Germain, C. et al. Presence of B cells in tertiary lymphoid structures is associated with a protective immunity in patients with lung cancer. Am. J. Respir. Crit. Care Med. 189, 832–844 (2014).
Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell 161, 1681–1696 (2015).
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49 (2013).
Sade-Feldman, M. et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175, 998–1013 (2018).
FastQC. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
DeLuca, D. S. et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28, 1530–1532 (2012).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Love, M. I., Anders, S., Kim, V. & Huber, W. RNA-Seq workflow: gene-level exploratory analysis and differential expression. F1000 Res. 4, 1070 (2015).
Wu, G., Dawson, E., Duong, A., Haw, R. & Stein, L. ReactomeFIViz: a Cytoscape app for pathway and network-based data analysis. F1000 Res. 3, 146 (2014).
Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P. L. & Ideker, T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 (2011).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Liu, J. et al. An integrated TCGA Pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173, 400–416 (2018).
Benjamini, Y., Drai, D., Elmer, G., Kafkafi, N. & Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125, 279–284 (2001).
Pimenta, E. M. & Barnes, B. J. Role of tertiary lymphoid structures (TLS) in anti-tumor immunity: potential tumor-induced cytokines/chemokines that regulate TLS formation in epithelial-derived cancers. Cancers (Basel) 6, 969–997 (2014).
Hiraoka, N. et al. Intratumoral tertiary lymphoid organ is a favourable prognosticator in patients with pancreatic cancer. Br. J. Cancer 112, 1782–1790 (2015).
Ruddle, N. H. Lymphatic vessels and tertiary lymphoid organs. J. Clin. Invest. 124, 953–959 (2014).
Hu, X. et al. Landscape of B cell immunity and related immune evasion in human cancers. Nat. Genet. 51, 560–567 (2019).
Nazarov, V. I. et al. tcR: an R package for T cell receptor repertoire advanced data analysis. BMC Bioinformatics 16, 175 (2015).
Villani, A. C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017).
Acknowledgements
The correlative research was funded by philanthropic support from the MD Anderson Melanoma Moon Shot Program, the Parker Institute for Cancer Immunotherapy, the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, and the AIM at Melanoma Foundation. It was supported by The Immunotherapy Platform and the Core grant CA016672 (SMF) to support the Sequencing and Microarray Facility at UT-MDACC as well as the the Office of the Assistant Secretary for Defense for Health Affairs grant (W81XWH-16-1-0120 and W81XWH-16-1-0121). S.M.R. received support from National Institutes of Health T32 CA 009666 and Cancer Prevention Research Institute of Texas RR190020. B.A.H. received support from National Institutes of Health T32 CA 009599 and the MD Anderson Cancer Center support grant P30 CA016672. A.P.C. is supported by the CPRIT Research Training Program (RP170067) and the United States Department of State’s Bureau of Educational and Cultural Affairs. E.Z.K. is supported by National Institutes of Health grant T32 CA009599. F.G.K. is supported with funding from the Odyssey Program at the MD Anderson Cancer Center with support from the Theodore N. Law Endowment for Scientific Achievement. Research in the Kalluri laboratory is supported by the Cancer Prevention and Research Institute of Texas and National Cancer Institute grants CA213233, P01CA117969, and CA195733. K.M.M. and M.A.Z. are supported by CPRIT RP190507. The High Resolution Electron Microscopy Facility at UT-MDASS is supported by CCSG grant NIGH P30CA016672. We thank to O. Contrares for technical support on the multiplex immunofluorescence and for M. Andrews for technical support on RNA-seq library preparation and K. McAndrews, D. P. Dowlatshahi, L. Snowden, J. Leveille and S. Yang for support with analyses of exosomes.
Author information
Authors and Affiliations
Contributions
B.A.H., S.M.R., J.G., S.Z. and R.B. contributed equally. Jointly supervising authors include P.S., K.R., M.T.T., L.W. and J.A.W. Corresponding authors are B.A.H. and J.A.W. J.A.W. designed the study. J.E.G., R.N.A., P.H., M.A.D., H.A.T., S.P., A.L., S. Woodman, C.U.B., T.N.S., P.S. and J.A.W. collected samples for correlative analyses. B.A.H., S.M.R., J.G., S.Z., R.B., R.T., K.Y., M.S.-F., J.B., G.H., V.G., Y.X., H.Z., A.P.C., W.L., V.S.L., F.G.K., A.L., R.A., S. Woodman, E.Z.K., P.-O.G., A.R., C.N.S., E.M.B., L.E.H., A.J.L., R.Z., C.W.H., D.A.L., S.O., M.B., S. Warren, D.R., O.K., E.A.R., D.P., C.U.B., T.N.S., L.H.B., M.A.Z., K.M.M., R.K., J.A., F.P., W.H.F., C.S.-F. and N.H. analysed and interpreted data. Specifically, K.Y., M.S.F. and N.H. assisted with single-cell RNA-seq analyses. S.O., M.B. and S. Warren assisted with Nanostring Digital Spatial Profiling analyses. R.B. and K.R. assisted with CyTOF. S.Z., R.T., L.W., M.A.Z. and K.M.M. assisted with BCR analyses. J.B., S.O., M.B., S. Warren, C.H., D.L. and M.T.T. assisted with singlet and multifocal immunohistochemistry. S.Z., L.W., J.E.G. and A.J.L. assisted with analyses of the TCGA patient cohorts. S.Z., R.T. and L.W. assisted with all bioinformatics and statistical analyses. R.T. performed pathway analyses. V.S.L., F.G.K. and R.K. assisted with analyses of exosomes. D.R., O.K., E.A.R., D.P., C.U.B. and T.N.S. provided assistance with analyses of patients from the OpACIN-neo trial. J.G., J.B. and P.S. assisted with analyses of the RCC patient cohort. All authors developed and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
J.A.W. is an inventor on a US patent application (PCT/US17/53.717) submitted by the University of Texas MD Anderson Cancer Center that covers methods to enhance immune checkpoint blockade responses by modulating the microbiome. J.A.W. reports compensation for speaker’s bureau and honoraria from Imedex, Dava Oncology, Omniprex, Illumina, Gilead, PeerView, Physician Education Resource, MedImmune and Bristol-Myers Squibb. J.A.W. serves as a consultant/advisory board member for Roche/Genentech, Novartis, AstraZeneca, GlaxoSmithKline, Bristol-Myers Squibb, Merck, Biothera Pharmaceuticals and Microbiome DX. J.A.W. also receives research support from GlaxoSmithKline, Roche/Genentech, Bristol-Myers Squibb, and Novartis. J.A.W., S.M.R. and B.A.H. are co-inventors on an unpublished patent application related to methods of targeting B cells to enhance response to immune checkpoint blockade. M.T.T. reports advisory board participation and speaker paid honorarium from Nanostring and Myriad Genetics. M.A.D. serves as a paid consultant for BMS, Novartis, and Roche/Genentech. M.A.D. also reports to be a principal investigator of a research grant from Roche/Genentech and an unpaid consultant to Nanostring. C.U.B. reports an advisory role in BMS, MSD, Roche, Novartis, GSK, AZ, Pfizer, GenMab and Pierre Fabre. C.U.B. receives research funding from BMS, Novartis and Nanostring. C.U.B. reports stock ownership from Uniti Cars and Neon Therapeutics. N.H. is a founder, stockholder and SAB member of Neon Therapeutics. W.H.F. serves as a consultant for AstraZeneca, Ipsen, Adaptimmune, OxfordBiotherapeutics, and Catalym. W.H.F. reports participation in data transparency committee for Servier and data management committee for Novartis. O.K. receives grant support from BMS. J.E.G. is a contributor of UpToDate-melanoma staging and prognosis. J.E.G. reports to be an unpaid member of Melanoma Research Foundation and Melanoma Research Alliance. J.E.G. reports to be on advisory board of Merck. R.K. reports to be scientific founder, stockholder and consultant of Codiak Biosciences. A.J.L. reports consultancies and research support from BMS, Genentech/Roche and MedImmune/Astra-Zeneca. P.S. reports a patent licensed to Jounce Therapeutics. P.S. serves as a consultant for Constellation, Jounce Therapeutics, Neon, BioAtla, Pieris, Oncolytics, Forty-Seven, Polaris, Apricity, Marker, Codiak, ImaginAb, Hummingbird, Dragonfly, Lytix and Bristol-Myers Squibb (BMS). P.S. has ownership interests in Jounce Therapeutics, Neon, Constellation, Oncolytics, BioAtla, Forty-Seven, Apricity, Polaris, Marker, Codiak, ImaginAb, Hummingbird, Dragonfly and Lytix. L.H.B. reports to be on StemImmune/Calidi Scientific and Medical Advisory, Scientific Advisory Board of BoardNextCure, Replimmune, Western Oncolytics, Torque Therapeutics Khloris, Pyxis, Cytomix. L.H.B. reports to be Chair of Food and Drug Administration Cellular, Tissues and Gene Therapies Advisory Committee. R.Z. reports a patent application related to work on GITR, PD1 and CTLA4. R.Z. is a consultant for Leap Therapeutics. P.H. is on advisory board for Dragonfly, GlaxoSmithKline, Immatics and Sanofi. S.W., S.O. and M.B. are employees and stockholders of NanoString Technologies. All other authors report no competing interests directly relevant to this work.
Additional information
Peer review information Nature thanks James Mulé, Caroline Robert and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 MCP-counter results in patients with melanoma and RCC treated with pre-surgical ICB or targeted therapy.
a, Supervised clustering by response of MCP-counter scores in on-treatment samples from a cohort of high-risk patients with resectable melanoma treated with neoadjuvant ICB, with responders defined as achieving a complete or partial response by RECIST 1.1 (n = 11 NR and 9 R). b, Analysis shown by unsupervised hierarchical clustering of baseline (n = 11 NR and 10 R) and on-treatment samples (n = 11 NR and 9R) from the neoadjuvant melanoma cohort. Unique clusters identified are indicated by shaded boxes on top row. c, Unsupervised hierarchical analysis shown for metastatic RCC patients (same cohort as Fig. 1d; n = 11 PD and 17 PR). Response (PR, partial response) or non-response (PD, progressive disease) as measured by RECIST 1.1. Unique clusters identified are indicated by shaded boxes on top row. d, Supervised clustering by response of MCP-counter scores from OpACIN-neo clinical trial (NCT02437279) of neoadjuvant versus adjuvant ICB in high-risk resectable melanoma (n = 6 NR and 12 R). Responders were defined as patients who did not have a relapse. e, Supervised clustering by response of MCP-counter scores in combined pre-treatment and on-treatment biopsies from a cohort of high-risk resectable melanoma patients treated with neoadjuvant targeted therapy (dabrafenib and trametinib) as part of NCT02231775 (n = 7 patients for baseline and n = 8 patients for on-treatment samples) with responder defined as achieving a complete or partial response by RECIST 1.1 and non-responder defined as having stable or progressive disease. Pathological response is defined by the presence or absence of viable tumour at time of surgical resection. P values were made using two-sided Mann–Whitney U-test.
Extended Data Fig. 2 Representation of MCP-counter scores for all patient cohorts and analyses of peripheral blood exosomes.
a–c, Box plot representation of heat maps for patients with: high-risk resectable melanoma treated with neoadjuvant ICB (n = 11 NR and 10 R for baseline and n = 11 NR and 9 R on treatment) as presented in Fig. 1c and Extended Data Fig. 1a, b (a); metastatic RCC treated with pre-surgical ICB as presented in Fig. 1d and Extended Data Fig. 1c (n = 11 PD and 17 PR) (b); and high-risk resectable melanoma treated with ICB as part of OpACIN-neo trial as presented in Extended Data Fig. 1d (n = 6 NR and 12 R) (c). For a–c, medians with interquartile range are shown. P values were determined by two-sided Mann–Whitney U-test. d, Schematic for exosomal analyses of serum samples from patients with melanoma on neoadjuvant ICB trial. e, Representative transmission electron micrographs showing Dynabead with exosomes present after immunocapture. f, Nanoimager-captured images of the beads coated with CD63+ exosomes as compared with isotype control. g, h, Exosomal concentration (g) and mean exosomal size (h) for serum samples for responders and non-responders at the time point indicated. i, Ratio of mean fluorescent intensity (MFI) of beads stained with anti-CD63 as compared to isotype control. j, Ratio of mean fluorescent intensity of beads stained with anti-CD9, -CD20, -CD27 and -GPC1 antibodies as compared to isotype control (or secondary antibody only for GPC1). For e–j, bars indicate median values and individual data points representing 8 R and 5 NR (unless indicated in the Methods) in addition to interquartile ranges. P values were determined using two-sided Mann–Whitney U-test.
Extended Data Fig. 3 Transcriptional analysis of tumour specimens from patients with metastatic RCC treated with pre-surgical ICB.
a, Supervised hierarchical clustering by response of RCC tumour specimens at baseline of most DEGs by microarray analysis, with response defined as having a partial response by RECIST 1.1 and non-response as having progressive disease (n = 11 PD and 17 PR). Fold change and P values are calculated by the limma package as described in the Methods. A cut-off of gene expression fold change of ≥ 2 or ≤ 0.5 and a FDR q ≤ 0.05 was applied to select DEGs. b, Volcano plot depiction of DEGs by response from same cohort.
Extended Data Fig. 4 Immune infiltrate is prognostic of improved disease-specific survival in TCGA cutaneous melanoma cohort but not the clear-cell RCC cohort.
a, Unsupervised hierarchical analysis of TCGA SKCM RNA-seq data using MCP-counter scores identifies three MICs with differential presence of individual cell types as indicated. Numbers of patients in each class is shown on top of the plot. P value determined by two-sided Kruskal–Wallis rank-sum test and q value calculated by FDR. b, Kaplan–Meier estimates of overall survival of MIC groups. c, Kaplan–Meier estimates of overall survival by B cell lineage scores shown by high and low groups dichotomized by median values. Overall survival was defined as the time interval from date of accession for each sample to date of death or censoring from any cause (Methods). d, Unsupervised hierarchical analysis of TCGA KIRC RNA-seq data using MCP-counter scores identifies three immune classes with differential presence of individual cell types as indicated. Numbers of patients in each class are shown at top of plot. P values determined by two-sided Kruskal–Wallis rank-sum test q value calculated by FDR. e, Kaplan–Meier estimates of overall survival probability of immune class groups. f, Kaplan–Meier estimates of overall survival probability by B cell lineage scores shown by high and low groups dichotomized by median values. For both, overall survival was defined as the time interval from date of accession for each sample to date of death or censoring from any cause. For b, c, e, f, patient numbers are included in the table below the graph and P values were calculated by log-rank test.
Extended Data Fig. 5 TLSs found in nodal and non-nodal metastases are consistent with mature secondary follicular-like TLSs with modest correlation with gene expression data.
a, Representative TLSs in tumours from patients with melanoma treated with neoadjuvant ICB demonstrating maturation status as indicated by the presence of follicular dendritic cells (CD21) and germinal centre B cells (CD23). We also include multiplex immunohistochemistry for SYTO13, MECA79, CD20 and CD4 (with magnified view of individual TLSs indicated by white box on the right). Circles denote defined TLSs based on multiplex immunohistochemistry. Black line approximates tumour border. b, Representative TLSs from non-lymph node metastases on additional patients with metastatic melanoma indicated by H&E staining, as well as singlet staining for CD20, CD21 and CD23. Black line on H&E image denotes tumour border. c, Comparison of baseline and on-treatment gene expression with MCP-counter analyses for B cell lineage as well as MS4A1 expression by RNA-seq for patients with high-risk resectable melanoma treated with ICB as part of clinical trial (n = 11 NR and 10 R for baseline and n = 11 NR and 9 R for on-treatment). Response and treatment arm as indicated. Medians with interquartile range are shown. P values were determined by two-sided Mann–Whitney U-test. d, Linear regression modelling of MCP-counter values for B cell lineage with regards to CD20 counts (n = 10 NR and 7 R) and TLS density (n = 10 NR and 6 R) as indicated. e, Linear regression modelling of MS4A1 gene expression with regards to CD20 counts (n = 10 NR and 7 R) and TLS density (n = 10 NR and 6 R) as indicated. These represent on-treatment time points. For d, e, r, values calculated by linear regression and P values for non-zero slope as calculated by Prism v.8.0.0.
Extended Data Fig. 6 TLSs are associated with response in RCC similar to those observed in melanoma.
a, Multiplex immunohistochemistry images from three additional patients with advanced melanoma treated with neoadjuvant ICB. Staining as indicated and similar to Fig. 2. b, Quantification of CD20 cells by singlet immunohistochemistry and association with response to neoadjuvant ICB in metastatic RCC, with responders defined as having partial response and non-responders as having progressive disease by RECIST 1.1 (n = 10 PD and 8 PR at baseline and n = 5 PD and 11 PR on treatment). c, d, Density of TLSs (n = 10 PD and 9 PR at baseline and n = 5 PD and 9 PR on treatment) (c) and ratio of tumour area occupied by TLSs (n = 10 PD and 7 PR at baseline and n = 5 PD and 11 PR on treatment) (d) and correlation by treatment response. Bars indicate median values and interquartile ranges are shown. P values were determined by two-sided Mann–Whitney U-test. e–g, Representative image of CD20 staining in responder with TLSs, associated H&E slide, singlet stains and characterization by multiplex immunofluorescence of TLSs. h, Multiplex immunohistochemistry images from three additional patients with RCC treated with pre-surgical ICB. Staining as indicated and similar to g.
Extended Data Fig. 7 TLSs are associated with markers of T cell activation and response and B cell proliferation.
NanoString GeoMx Digital Spatial Profiling was used to perform high-plex proteomic analysis with spatial resolution. a, Example of selection of ROIs (200 μm × 200 μm) from representative patients with melanoma treated with neoadjuvant ICB including ROI containing TLSs and ROIs outside the context of a TLS (non-TLS). ROI selection was completed using H&E staining and confirmed with immunofluorescence as shown using S100B and PMEL, SYTO13, CD3 and CD20. Masking for B cells and T cells as indicated based on CD3 and CD20 staining. b, Fold change (log2-transformation) in expression of various markers of T cell activation and response in TLS-associated T cells as compared to T cells found outside the TLS per individual slide. Data show individual TLS ROI values divided by the average non-TLS value of that slide. Increased expression in the context of TLSs is represented by shaded pink box (>0). c, Average log2-transformed fold change of expression for TLS-associated T cells as compared to non-associated T cells. Individual dots represent individual patients/slides as indicated. Data show the average log2-transformed count per TLS ROI value minus the average log2-transformed count per non-TLS ROI value per slide for each protein queried. For b and c, increased expression in the context of TLSs is represented by shaded pink box (>0). Median and interquartile range are indicated. Error bars indicate 95% confidence intervals. d, Levels of Ki67 protein expression in B cell masks of non-TLSs and TLS ROIs by individual patient as indicated. Counts are represented as signal-to-noise ratios of Ki67 compared to geometric means of isotype controls. Median and interquartile range are indicated. Error bars indicated 95% confidence ratios, and P values were determined by Student’s t-test. For a–d, the number of ROIs analysed for each patient are 11 for patient 1, 12 for patient 2, 12 for patient 10, 7 for patient 17 and 7 for patient 19.
Extended Data Fig. 8 BCR analyses of intratumoral B cells in patients with advanced melanoma before and after treatment with neoadjuvant ICB.
a, Clonal counts for all identified clonotypes for both IgH and IgL after treatment with ICB. Patients are grouped as responders and non-responders and identified as indicated in which each bar represents individual patient. b, Clonal proportion for all identified clonotypes for both IgH and IgL for baseline samples further evaluated in Fig. 3a and on-treatment samples in a. Patients are grouped as responders and non-responders and identified as indicated in which each bar represents individual patient. c, d, Summed expression of top five clonotypes in normalized read counts (c) and BCR repertoire diversity (d) for responders and non-responders for both IgH and IgL at baseline (n = 11 NR and 10 R for IgH and IgL) and on-treatment (n = 10 NR and 9 R for IgH and n = 11 NR and 9 R for IgL). Box plot shows median and interquartile range. P values determined by two-sided Mann–Whitney U-test.
Extended Data Fig. 9 Single-cell RNA-seq analysis reveals unique clusters of B cells associated with response to ICB.
a, Scatter plots comparing the percentage of CD45+ cells staining positive for CD19+ (B cells) as indicated between responder (n = 17) and non-responder (n = 31) samples with all time points combined or stratified by pre- and post-treatment as indicated. Data are median and interquartile range. P values were determined by two-sided Mann–Whitney U-test. b, Heat map displaying scaled expression values (log2(TPM + 1) of discriminative genes from all B cells between responder (blue) and non-responder (red) samples. Top marker genes are shown for each group. c, Heat map showing scaled expression values (log2(TPM + 1)) of discriminative gene sets per cluster as defined in Fig. 3c. A list of representative genes is shown per cluster next to the left margin. For both heat maps, colour scheme is based on z-scores from −2.5 (blue) to 2.5 (yellow).
Extended Data Fig. 10 Mass cytometry reveals significant differences in Bcell populations between responder and non-responder tumours.
a, Pie charts representing composition of individual tumour and peripheral blood samples for patients with melanoma treated with ICB used in all analyses for mass cytometry. Matched patient samples are located directly beneath one another. Samples from patients with lymph node or non-lymph-node metastases as indicated. Cell types as indicated. Asterisk indicates samples included in t-SNE plots and pie charts in c, Fig. 3d–f, and phenographs in Extended Data Fig. 11. b, Scatter plots demonstrating quantification of different peripheral blood and intratumoral B cell phenotypes. Median and interquartile range are shown. All samples are represented in b (for tumour, n = 7 R and 3 NR and, for peripheral blood, n = 4 R and 4 NR). P values were determined by one-sided Mann–Whitney U-test. c, t-SNE plots demonstrating intratumoral B cell phenotypes from the neoadjuvant ICB trial in patients with advanced melanoma grouped by response and including further breakdown of memory cell subtypes and germinal centre B cells. Plots represent combined analyses of tumours ran simultaneously with the peripheral blood samples (n = 5 R and 3 NR) and include baseline and on-treatment samples as detailed in Supplementary 31. d, Quantification of B cell subtypes in tumour from mass cytometric analyses in responders and non-responder from all tumours (n = 7 R and 3 NR). Median and interquartile range are shown. P values were determined by one-sided Mann–Whitney U-test.
Extended Data Fig. 11 Surface expression of markers analysed by mass cytometry.
a, Individual phenographs for surface expression of each marker analysed as indicated. These data represent combined tumour and peripheral blood samples from patients with melanoma treated with ICB ran together (8 tumour n = 5 R and n = 3 NR and 8 peripheral blood samples n = 4 R and n = 4 NR), thus eliminating batch effect. b, Percentage of CD45+CD19+ cells by tissue type—peripheral blood versus tumour—that are positive for each of the surface markers indicated. c, Percentage of CD45+CD19+ cells in tumour by response—responder versus non-responder—that are positive for each of the surface markers indicated. For b and c, all samples are represented (for tumour, n = 7 R and 3 NR and, for peripheral blood, n = 4 R and n = 4 NR). Error bars indicate median and interquartile range. P values were determined by two-sided Mann–Whitney U-test.
Supplementary information
Supplementary Tables
This file contains Supplementary Tables 1, 5, 7-8, 10, 14, 18, 22-23, 26, 28-32.
Supplementary Tables
This file contains Supplementary Tables 2-4, 6, 9, 11-13, 15-17, 19-21, 24-25, and 27.
Supplementary Information
This file contains a full Supplementary Tables Guide for tables 1-32.
Rights and permissions
About this article
Cite this article
Helmink, B.A., Reddy, S.M., Gao, J. et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 577, 549–555 (2020). https://doi.org/10.1038/s41586-019-1922-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1922-8
- Springer Nature Limited
This article is cited by
-
Single-cell sequencing reveals the heterogeneity of B cells and tertiary lymphoid structures in muscle-invasive bladder cancer
Journal of Translational Medicine (2024)
-
Phenotypic and spatial heterogeneity of CD8+ tumour infiltrating lymphocytes
Molecular Cancer (2024)
-
Genomic and transcriptomic analysis of breast cancer identifies novel signatures associated with response to neoadjuvant chemotherapy
Genome Medicine (2024)
-
CD19-immunoPET for noninvasive visualization of CD19 expression in B-cell lymphoma patients
Biomarker Research (2024)
-
B cells in head and neck squamous cell carcinoma: current opinion and novel therapy
Cancer Cell International (2024)