Abstract
Soil organisms are a crucial part of the terrestrial biosphere. Despite their importance for ecosystem functioning, few quantitative, spatially explicit models of the active belowground community currently exist. In particular, nematodes are the most abundant animals on Earth, filling all trophic levels in the soil food web. Here we use 6,759 georeferenced samples to generate a mechanistic understanding of the patterns of the global abundance of nematodes in the soil and the composition of their functional groups. The resulting maps show that 4.4 ± 0.64 × 1020 nematodes (with a total biomass of approximately 0.3 gigatonnes) inhabit surface soils across the world, with higher abundances in sub-Arctic regions (38% of total) than in temperate (24%) or tropical (21%) regions. Regional variations in these global trends also provide insights into local patterns of soil fertility and functioning. These high-resolution models provide the first steps towards representing soil ecological processes in global biogeochemical models and will enable the prediction of elemental cycling under current and future climate scenarios.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Main
As we refine our spatial understanding of the terrestrial biosphere, we improve our capacity to manage natural resources effectively. With ever-growing functional information about the biogeography of aboveground organisms, an unresolved gap in our understanding of the biosphere remains the activity and distribution patterns of soil organisms1,2. The soil biota—including bacteria, fungi, protists and animals—has central roles in every aspect of global biogeochemistry, influencing the fertility of soils and the exchange of CO2 and other gases with the atmosphere3. As such, biogeographical information on the abundance and activity of soil biota is essential for climate modelling and, ultimately, environmental decision-making2,4,5,6. However, the activity of soil organisms is not explicitly reflected in biogeochemical models owing to our limited understanding of their biogeographical patterns at the global scale.
In recent years, pioneering studies in soil biogeography have begun to provide valuable insights into the broad-scale taxonomic diversity patterns of soil bacteria7,8,9,10,11, fungi11,12,13 and nematodes14,15,16,17, and patterns of microbial biomass11,18,19. However, we have been unable to generate a high-resolution, quantitative understanding of the abundance or functional composition of active soil organisms because of two major reasons. First, owing to the methodological challenges in characterizing soil biota, most previous studies have focused on a relatively limited number of spatially distinct sampling sites (fewer than 500) and, therefore, cannot detect high-resolution regional-scale patterns. Second, most global studies have used molecular sequencing approaches, which provide valuable semi-quantitative information on taxonomic diversity but do not provide information on absolute abundances or biomass, which are essential to link biological communities to ecosystem functioning and global biogeochemistry20,21. DNA- and RNA-based approaches cannot unambiguously differentiate between living (that is, either active or dormant) and dead cells, so they cannot be used to quantify the active component of the belowground community22,23. To generate a robust, global perspective of belowground biota and the roles of these soil organisms in biogeochemical cycling, we need a sampling design that provides a thorough global representation of the belowground communities and direct, quantitative abundance data that reflect the active community. Here we use this approach to generate a quantitative understanding of a critical component of the soil food web, for which direct extraction methods enable the quantification of active organisms: nematodes.
Nematodes are a dominant component of the soil community and are by far the most abundant animals on Earth2. They account for an estimated four-fifths of all animals on land24, and feature in all major trophic levels of the soil food web. The functional role of nematodes in soils can be inferred by their trophic position and nematodes are therefore often classified into trophic groups based on feeding guilds (that is, bacterivores, fungivores, herbivores, omnivores and predators). Given their pivotal roles in processing organic nutrients and control of soil microorganism populations25,26,27, they play critical parts in regulating carbon and nutrient dynamics within and across landscapes26 and are a good indicator of biological activity in soils28. However, we lack even a basic understanding of broad-scale biogeographical patterns in nematode abundance and the composition of functional groups. Despite expectations that nematode abundances may peak in warm tropical regions with high plant biomass14,15, previous studies have suggested that the opposite pattern may exist, with high nematode abundances in high-latitude regions with larger standing stocks of soil carbon16,17,29,30,31. Disentangling the effects of these different environmental drivers of soil nematode communities is critical to generate a mechanistic understanding of the global patterns of soil nematodes, and for quantifying their influence on global biogeochemical cycling.
Here we use 6,759 spatially distinct soil samples from all terrestrial biomes and continents to examine the environmental drivers of global nematode communities. By using 73 global layers of climate, soil and vegetation characteristics, we then extrapolate these relationships across the globe to generate spatially explicit, quantitative maps of soil nematode density and functional group composition at a global scale.
Biome-level patterns of soil nematodes
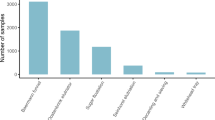
By compiling soil sampling data from all major biomes and continents, we aimed to generate a representative dataset that captured the variation in global nematode densities. Within each sample, we quantified the total abundance of each trophic group using microscopy. To standardize sampling protocols, we focus on the top 15 cm of soil, which is the most biologically active zone of soils6,32. Consistent with previous reports33, nematode abundances are highly variable within and across terrestrial biomes, ranging from dozens to thousands of individuals per 100 g soil (Fig. 1b). This variation highlights the necessity for large datasets in soil biodiversity analyses to reliably predict large-scale patterns, as the accuracy of our mean estimates for any region improves considerably with increasing number of samples (Fig. 2a). Specifically, the confidence in our mean estimates for nematode abundance in any region is relatively low at the scale of individual samples, but high only when calculated with larger sample sizes (that is, more than 400 samples).
a, Sampling sites. A total of 6,759 samples were collected and aggregated into 1,876 1-km2 pixels that were used for geospatial modelling, and abundance data from 39 1-km2 pixels from Antarctica. b, The median and interquartile range of nematode abundances (n = 1,876) per trophic group (top) and per biome (bottom) from all continents. Axes have been truncated for increased readability. Biomes with observations from more than 20 1-km2 pixels are shown.
a, b, The standard error of the observed (a) and predicted (b) mean values of nematode density decrease with increasing sample size. The operation was repeated with 100 and 1,000 random seeds for the observed and predicted mean values, respectively, and the calculated standard errors of the mean are shown. c–h, Heat plots showing the relationships between predicted versus observed nematode abundance values, for total nematode number and each trophic group. Dashed diagonal lines indicate fitted relationships (R2 values are indicated in the bottom right corner), solid diagonal lines indicate a 1:1 relationship between predicted and observed points. i, Bootstrapped (100 iterations) coefficient of variation (standard deviation divided by the mean predicted value) as a measure of prediction accuracy. Sampling was stratified by biome. Overall, our prediction accuracy is lowest in arid regions and in parts of the Amazon and Malay Archipelago.
Overall, we observed the highest nematode densities in the tundra (median = 2,329 nematodes per 100 g dry soil), boreal forests (median = 2,159) and in temperate broadleaf forests (median = 2,136), and the lowest densities are observed in Mediterranean forests (median = 425), Antarctic sites (median = 96) and hot deserts (median = 81) (Fig. 1b and Supplementary Table 2). To examine the mechanisms that drive the patterns of soil nematode density and functional group composition across biomes, we integrated the nematode abundance data with 73 global datasets of soil physical and chemical properties, and vegetative, climatic, topographic, anthropogenic and spectral reflectance information (Supplementary Table 3). Antarctic sampling points were excluded from the modelling dataset owing to limited coverage of several covariate layers. To match the spatial resolution of our covariates, all samples were aggregated to the 1-km2 pixel level to generate 1,876 unique pixel locations across the world. We analysed a suite of machine-learning models (including random forest, and L1- and L2-regularized linear-regression models) to determine the environmental drivers of the variation in nematode abundance and functional group composition across the globe. We iteratively varied the set of covariates and model hyperparameters across 405 models and evaluated model strength using k-fold cross-validation (with k = 10). This approach allowed us to select the best performing model that had high predictive strength (mean cross-validation R2 = 0.43, overall R2 = 0.86), while taking into account issues that surround multicollinearity, as well as model overparameterization and overfitting. This final model—an iteration of random forests using all 73 covariates—was then used to create per-pixel mean and standard deviation values. Mapping the extent of extrapolation highlighted that our dataset covered most environmental conditions, with the least represented pixels and highest proportion of extrapolation in the Sahara and Arabian Desert (Extended Data Fig. 1b, d). We acknowledge that our models cannot accurately predict nematode abundances at fine spatial scales, as local environmental heterogeneity can cause considerable variation in nematode abundances, even within individual locations. However, the strength of these predictions increases at the larger scales for which our modelled estimates are informed by more data observations (Fig. 2b), ensuring confidence in our estimates. Predicted versus observed plots revealed that, despite the high accuracy in most regions, the models tended to marginally overrepresent the observed numbers at low densities and underrepresent abundances at higher nematode densities (Fig. 2c–h). Moreover, our cross-validation accuracy calculations may be optimistically biased, as we cannot entirely account for the potential effects of overfitting. Our analyses would have ideally included a subset of data that was removed at the beginning of the analyses to enable a fully independent accuracy assessment. However, as the removal of a subset of data would mean the loss of geographical representation, we chose instead to maintain the integrity of the entire dataset and generate spatially explicit maps of model confidence that allow for error propagation throughout the final global calculations (Fig. 2i and Extended Data Fig. 1a, b, d). These maps provide spatial insight into the prediction uncertainties rather than a single accuracy measure for overall model accuracy.
Our statistical models reveal the dominant drivers of nematode abundance across global soils. As with aboveground animals, climatic variables (that is, temperature and precipitation) had important roles in shaping the patterns in total soil nematode abundance. However, soil characteristics (for example, texture, soil organic carbon (SOC) content, pH and cation-exchange capacity) were by far the most important factors driving nematode abundance at a global scale, with effects that largely overwhelmed those of climate (Supplementary Table 3). Linear models enabled us to assess the directionality of these relationships, revealing that both SOC content and cation-exchange capacity had strong positive correlations, whereas pH had a negative effect on total nematode density (Extended Data Fig. 2). These trends support the suggestion that soil resource availability is a dominant factor that structures belowground communities at broad spatial scales, overriding the effect of climate to shape the belowground communities at broad spatial scales2,12,15.
Global biogeography of soil nematodes
The high predictive strength of the top model enabled us to extend the relationships across global soils to construct high-resolution (30 arcsec or approximately 1 km2) quantitative maps of total nematode densities (Fig. 3). These maps reveal notable latitudinal trends in soil nematode abundance, with the highest densities in sub-Arctic regions, a trend that is consistent across all trophic groups (Extended Data Fig. 3a–e). Similar to the regional averages, the highest abundances of soil nematodes are found in boreal forests across North America, Scandinavia and Russia. Whether nematode abundance is expressed as density per gram of soil or per unit area (thus controlling for the differences in soil bulk density), the models reveal a latitudinal gradient in soil nematode abundance (Fig. 3 and Extended Data Figs. 4, 5). Whether soil animals exist at highest abundances in the high or low latitudes has been a contentious issue in the soil ecology literature, with some studies indicating that the highest abundances occur in boreal forests and others suggesting that tropical forests support the greatest abundances14,29,31. Our extensive data from every biogeographical region enabled us to see beyond these contrasting results to reveal a latitudinal pattern of nematode abundance, providing evidence that soil nematodes are present in considerably higher densities in high-latitude Arctic and sub-Arctic regions (Fig. 3).
Along with the latitudinal gradient in nematode abundance, our nematode density map also reveals regional variation that stands out against the global trends. Although nematode abundances were relatively low in tropical regions, our sampling data and models reveal a high nematode abundance in certain tropical peatlands such as the Peruvian Amazon (Figs. 1a, 3). These regions are characterized by high SOC stocks, which support high microbial biomasses that serve as the basic resource for most nematode groups. Similarly, increased SOC stocks at high altitudes compared to lowland regions drive higher nematode abundances in mountainous regions and highlands, such as the Rocky Mountains, Himalayan Plateau and the Alps (Figs. 1a, 3). Although the respective climates of these regions exhibit large differences in mean annual temperature (ranging from less than 0 °C to more than 10 °C), their soils are all characterized by relatively high SOC stocks (that is, more than 50 g kg−1). By contrast, the lowest nematode densities were predicted in hot deserts such as the Sahara, Arabian Desert, Gobi Desert and Kalahari Desert (Fig. 3), regions that are characterized by very low SOC stocks. As such, the spatial variability in nematode abundance is highest in equatorial regions, which exhibit the full range of possible abundances from deserts to biomes characterized by high SOC stocks. This is reflected by the spatial patterns in our model uncertainty, in which low-latitude arid regions with low sampling density and soil nematode abundances are characterized by larger uncertainties (Fig. 2i and Extended Data Fig. 1).
The strong correlation between temperature and SOC content at a global scale19 makes it challenging to identify the primary driver of the latitudinal gradient in nematode abundances. However, regional deviations from the global biogeographical pattern help to disentangle their relative roles, as they decouple the effects of climate and soil characteristics. For example, low temperatures and high moisture content in high-latitude regions restrict annual decomposition rates, leading to the accumulation of soil organic material19,30. However, the positive effect of SOC in tropical peatland regions (which have not only high soil carbon levels, but also warm temperatures) suggests that it is the content of organic matter, rather than climatic conditions, that ultimately determines the abundance of nematode in soil. These models reinforce the idea of a dominant role of soil characteristics in driving nematode abundances. These trends suggest that the influences of climate on nematode density are not direct, but instead act indirectly by modifying soil characteristics.
We next examined how the structure of nematode communities varied across landscapes by exploring the abundance of each trophic group across our dataset. At the global scale, all trophic groups were positively correlated with one another (Extended Data Fig. 6a), which suggests that biogeographical regions with high nematode abundances are generally hospitable for members of all trophic groups. Despite the distinct feeding habits, the global consistency across trophic groups provides some unity in the biogeography of the soil food web. That is, although different nematodes rely on distinct food sources for their energetic demands, the size of the entire food web is ultimately determined by the availability of soil organic matter. Nevertheless, the relative composition of nematode communities did vary across samples. To characterize the main nematode community types, we clustered the observed relative abundances into four types, on the basis of the relative abundance of each trophic group (Extended Data Fig. 6b). Although there were no clear spatial patterns in these community types, vector analysis revealed that the indices of vegetation cover (for example, the normalized difference vegetation index and enhanced vegetation index) were the best predictors of herbivore-dominated communities, whereas edaphic factors (such as sand content and pH) were strong predictors of communities dominated by bacterivores (Extended Data Fig. 6c).
By summing the nematode density information of each pixel, we can begin to generate a quantitative understanding of the abundances and biomass of soil nematodes at a global scale. We estimate that approximately 4.4 ± 0.64 × 1020 nematodes inhabit the upper layer of soils across the globe (Table 1 and Supplementary Table 5). Of these, 38.7% exist in boreal forests and tundra, 24.5% in temperate regions and 20.5% in tropical and sub-tropical regions (Supplementary Table 6). By combining our estimates of nematode abundance with mean biomass estimates of each functional group (using a database that contains 32,728 nematode samples)34,35, we can approximate that the global biomass of nematodes in the global topsoil is approximately 0.3 gigatonnes (Gt; Table 1). This translates to approximately 0.03 Gt of carbon (Gt C; Table 1 and Supplementary Table 7), which is three times greater than a previous estimate of soil nematode biomass36, and represents 82% of total human biomass on Earth (see Supplementary Methods). Using the same database of nematode metabolic activity34,35, we estimate that nematodes may be responsible for a monthly C turnover of 0.14 Gt C within the global growing season, of which 0.11 Gt C is respired into the atmosphere (Table 1). By comparison, the amount of carbon respired by soil nematodes is equivalent to roughly 15% of carbon emissions from fossil fuel use, or around 2.2% of the total carbon emissions from soils (approximately 9 and 60 Gt C per year, or 0.75 and 5 Gt C per month, respectively)37. As such, our findings indicate that soil nematodes are a major—and, to date, poorly recognized—player in global soil carbon cycling.
Despite high confidence in our estimates of the total abundance and community composition of nematodes, these approximations of the metabolic footprint retain several assumptions that might lead to considerable uncertainty in our estimates. For example, seasonal climatic variation in metabolic activity could influence the values that we present here, and total activity levels might be lower than expected based on these growing season estimates. On the other hand, extraction efficiency can be lower than 50% in some samples, which could lead to underestimation of the actual activity levels. Local variation in land-use types and bias in our sampling data could cause variation in soil nematode abundances at local scales. Furthermore, even though our sampling locations cover the vast majority of environmental conditions on Earth (Extended Data Fig. 1f), certain regions—such as the Sahara and Arabian Desert—are underrepresented in our data, leading to relatively high uncertainties in these regions (Fig. 2i and Extended Data Fig. 1a, b, d). Moreover, as our sampling approach focuses on the top soil layer, we stress that our analysis will underestimate total nematode abundances in, for example, tropical regions where high nematode densities are found in litter layers38. However, the metabolic footprint that we provide enables us to approximate the magnitude of soil nematode contributions to global carbon cycling and highlights their contribution to the total soil carbon budget. Furthermore, our findings emphasize the importance of high-latitude regions, characterized by high soil nematode abundances, for our understanding of soil carbon and feedback effects to ongoing climate change. These regions compose a major reservoir of soil carbon stocks6 and may release much more carbon as a result of increased soil activity of animals and a prolongation of the plant growing season owing to human-induced climate change.
In conclusion, our maps provide spatially explicit, quantitative information on belowground biota at the global scale. In addition to providing baseline information about soil nematodes as a fundamental component of terrestrial ecosystems, it also alters some of our most basic assumptions about the terrestrial biosphere by highlighting that soil animal abundances peak in high-latitude zones. The high numbers of nematodes that are present across all global soils highlights their functional importance in the dynamics of global soil food webs, nutrient cycling and terrestrial ecosystem functioning. This quantitative understanding of these belowground animals enables us to begin to comprehend the order of magnitude of their influence on the global carbon cycle and the spatial patterns in these processes. By providing quantitative information about the variation in biological activity in soils around the world, our models can provide the information necessary to explicitly represent soil biotic activity levels in spatially explicit biogeochemical models. That is, this information can now be used to parameterize, scale or benchmark spatially explicit model predictions of organic matter turnover under current or future climate change scenarios. We highlight that this global nematode study can and should be supplemented with similar future efforts to understand the biogeography of other important soil organisms, including fungi, bacteria and protists. Our soil nematode abundance and biomass data can serve as a stepping stone to facilitate future modelling efforts that add additional layers of soil biodiversity information to build a thorough understanding of the overwhelming abundance of life belowground and its influence on global ecosystem functioning.
Methods
Data reporting
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Data acquisition
We collected data on soil nematode abundances from previously published studies and unpublished data collections that morphologically quantified nematodes and determined taxa to the level of trophic groups or taxonomic groups. Rather than taxonomic diversity, we decided to focus on trophic groups as this gives more functional information. Trophic groups were assigned according to a previously published study39. We retained only samples that contained the following metadata: longitude and latitude, season or date sampled, sampling depth, information on land use (agriculture or natural sites) and whether samples were collected from soils or litter. We then standardized our efforts by focusing on all samples that were derived from soils and in which samples were representative of the nematode functional group composition in the top 15 cm of soils. This resulted in a final subset of 6,759 samples that were used for further analyses. Of these, 32.8% originate from agricultural or managed sites and 67.2% from natural sites. All data points that fell within the same 30 arcsec (approximately 1-km2) pixels were aggregated as an average, resulting in a total of 1,915 unique pixels across the globe as inputs into the models (Supplementary Table 1). The 39 pixels located in Antarctica were removed from the dataset as the covariate layers have limited coverage in these regions. This resulted in a total of 1,876 unique pixels that were used for geospatial modelling.
Acquisition of global covariate layers
To create spatial predictive models of nematode abundance, we first sampled our prepared stack of 73 ecologically relevant, global map layers at each of the point locations within the dataset. These layers included climatic, soil nutrient, soil chemical, soil physical and vegetative indices, radiation and topographic variables and one anthropogenic covariate (Supplementary Table 3). All covariate map layers were resampled and reprojected to a unified pixel grid in EPSG:4326 (WGS84) at 30 arcsec resolution (approximately 1 km2 at the equator). Layers with a higher original pixel resolution were downsampled using a mean aggregation method; layers with a lower original resolution were resampled using simple upsampling (that is, without interpolation) to align with the higher resolution grid.
Geospatial modelling
Using the ClustOfVar package40 in R, we reduced the covariates of interest to the most representative and least collinear few. As we did not have a specific number of variables defined a priori to use as a parameter for the clustering procedure, we put a range of cluster numbers (that is, 5, 10, 15 and 20) into the ClustOfVar functions to compute multiple covariate groups to test machine learning models. Using these selections of variables, we used a ‘grid search’ procedure to iteratively explore the results of a suite of machine-learning models trained on each group of covariates computed from the ClustOfVar function. Moreover, following recent advancements in machine learning for spatial prediction41, we tested models using all covariates with and without latitude/longitude data as well as a specific selection of covariates that represented principal ecosystem components plus satellite-based spectral reflectance. In addition to grid searching through models trained on different groupings of the covariates, we also explored the parameter space of multiple machine-learning algorithms (including random forests and regularized linear regression with both L1 and L2 regularization) and optional post hoc image convolution using kernels of various pixel sizes. During the grid search procedure, we assessed each model using k-fold cross-validation, to test the performance and overfitting across each of the 405 models. For each fold, a 10% subset of the data was extracted and held back for validation. Then, the model was trained on the remaining data and tested on the validation data. To test each model on the entire dataset, this process was performed 10 times for each model (that is, k = 10). The coefficient of determination values for each fold were then used to compute mean and standard deviation values for the cross-validated model. These mean and standard deviation values were the basis for choosing the best model of all 405 explored models using the grid search procedure, which was an iteration of random forests using all 73 non-spatial covariates. The grid search procedure was performed using the total nematode abundance data, and this final model was then used to model the sub-functional group abundance. The final R2 value for the ensembled total nematode abundance model (also assessed using tenfold cross-validation) was 0.43.
Model uncertainty
To create a per-pixel mean and standard deviation, we created an ensemble model using multiple versions of the best model; as the best model was an iteration of random forests using all 73 non-spatial covariates, the ensemble procedure was to rerun this model 10 times (each with different random seed values) and then averaging the model results. Using these values, we calculated the coefficient of variation (standard deviation divided by the mean predicted value) as a measure of the prediction accuracy of our model (Extended Data Fig. 1a).
To create statistically valid per-pixel confidence intervals, we performed a stratified bootstrapping procedure with the ‘total number’ collection of nematode point data. The stratification category was the sampled biomes of each point feature (to avoid biases), and the number of bootstrap iterations was 100. Each of the bootstrap iterations required the classification of the composite raster data—that is, 209,000,000 pixels classified 100 times. Doing so allows us to generate per pixel, statistically robust 95% confidence intervals (Fig. 2i).
Next, we tested the extent of extrapolation in our models by examining how many of the Earth’s pixels existed outside the range of our sampled data for each of the 73 global covariate layers. To evaluate the sampled range, we extracted the minimum and maximum values of each covariate layer of the pixels in which our sampling sites were located. Then, using the final model, we evaluated the number of variables that fell outside the sampled range, across all terrestrial pixels. Next, we created a per-pixel representation of the relative proportion of interpolation and extrapolation (Extended Data Fig. 1b). This revealed that our samples covered the vast majority of environmental conditions on Earth, with 84% of Earth’s pixels values falling within the sampled range of at least 90% of all bands (Extended Data Fig. 1c). Across all environmental layers, the percentage of pixels with values within the sampled range is generally above 85% (Extended Data Fig. 1f).
To evaluate how well our data spread throughout the full multivariate environmental covariate space, we performed a principal component analysis (PCA)-based approach. After performing a PCA on the sampled data, we used the centring values, scaling values and eigenvectors to transform the composite image into the same PCA spaces. Then, we created convex hulls for each of the bivariate combinations from the first 11 principal components (which collectively covered more than 80% of the sample space variation). Using the coordinates of these convex hulls, we classified whether each pixel falls within or outside each of these convex hulls. In total, 62% of the world’s pixels fell within the entire set of 55 PCA convex hull spaces computed from our sampled data, with most of the outliers existing in arid regions (Extended Data Fig. 1d).
Geospatial analyses and extrapolation were performed in Google Earth Engine42. Additional model results have been supplied as raw source code (see ‘Code availability’).
Nematode density values
To compute the original nematode density values (which were calculated as numbers of nematodes per 100 g of soil), we performed the following calculations at a per-pixel level. First, we multiplied the value by 10 to compute the number of nematodes per 1 kg of soil; the new units, per pixel, became the ‘number of nematodes per 1 kg of soil’. Then, we multiplied this value by the per-pixel bulk density values as produced by SoilGrids43; bulk density values were then produced in ‘kg of soil per 1 m3’. Finally, the new units after multiplication are the ‘number of nematodes per 1 m3 of soil’. Next, we multiplied this value by 0.15 m to compute the ‘number of nematodes per 1 m2 of soil (in the top 15 cm)’. For pixels that had a soil layer shallower than 15 cm, the pixel value was multiplied by the depth to bedrock values as produced by SoilGrids43. These respective pixel values were then multiplied by the area of each pixel presumed to have soil (that is, we exclude areas of ‘permanent snow/ice’ and ‘open water’ from the calculations, following the Consensus Land Cover classes found at https://www.earthenv.org/landcover); the units at this point, per pixel, are the total number of nematodes (in the first 15 cm of soil). Finally, all pixel values were summed to compute the final nematode abundance values across all pixels (that is, across the entire globe).
Clustering
To delineate main nematode ‘community types’ (that is, the relative frequency of each trophic group in a given sample), we first defined the number of clusters for the analysis. On the basis of pairwise distances and partitioning around medoids (k-medoids) clustering, we chose to select four clusters. Each of the four community types was then plotted (Extended Data Fig. 6b) to reveal their composition. To examine which environmental variables best explained each of the community types, we plotted each of the samples using non-metric multidimensional scaling (stress = 0.0691) and fitted environmental variables as vectors (Extended Data Fig. 6c).
Biomass estimates
Using publicly available data34,35, a database with taxon-specific body-size values (that is, length and width) of 32,728 nematode taxa (including 9,497 observations of adult nematodes and 23,231 observations of juveniles) was created to calculate the biomass, and respiration and assimilation rates for each trophic group. A nematode community typically contains numerous juveniles35; we assume the presence of 70% juveniles and 30% adults. For all calculations described in this section, we calculated per-trophic group means using per-taxon observations. To produce the final values, we multiplied the mean calculated values per trophic group with the predicted number of individuals per trophic group and per biome. The biomass of an assemblage of nematodes can be calculated as the sum of the weights of the number of individuals of each species that is present. According to a previous study44, the fresh weight of individual nematodes is calculated by
in which Wfresh is the fresh weight (μg) per individual, L is the length (μm) of the nematode and D is the greatest body diameter (μm)44. Assuming a dry weight of nematodes as 20% of fresh weight and the proportion of carbon in the body as 52% of dry weight45,46, the dry weight (Wdry) of an individual nematode can be calculated as
Daily carbon used in production
To calculate the total carbon used per nematode per day, we assumed that life-cycle length in days can be approximated as 12 times the colonizer–persister (cp) scale47,48 and that the accumulation of fresh weight is linear. Then, the daily increase in fresh weight is
in which Wt and cpt are the adult weight and cp value for a nematode of trophic group t, respectively. Then, we calculate the normalized daily carbon used in production (PC) as
in which cpt is the mean cp value of the respective trophic group. For a nematode assemblage, the daily carbon used in production can be calculated as
for nt individuals of each trophic group present in the assemblage.
Carbon respiration
To estimate the carbon respiration rates of an assemblage of nematodes, we assume relationships between respiration rates and body weights for poikilothermic organisms, so that
in which R is the respiration rate, W is the fresh weight (μg) per individual, and a and b are regression parameters49,50. Following literature51,52, we assume that b is equal to 0.75. The parameter a varies with temperature and the time interval on which the rate is based. For example, an average a value of approximately 1.40 nl O2 h−1 for 68 nematode species has previously been determined53. This converts to an a value of 2.43 ng CO2 h−1 at 15 °C. To estimate CO2 respiration in μg per day, we make the assumption of an a value of 2.43 × 24/1,000 (= 0.058) for our calculations. Using the relative molecular weights of carbon and oxygen in CO2 (12/44 = 0.273), we can calculate the total rate of carbon respiration for all nematodes in the system as
or
in which nt is the number of individuals and Wt the median body weight of each of the trophic groups summed over t trophic groups.
Total daily carbon budget
The total carbon budget (in μg per day) for each trophic group is the sum of the amounts that are respired and used for production, that is:
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
All raw data, sampled covariate layer data, models and maps are available at https://gitlab.ethz.ch/devinrouth/crowther_lab_nematodes. The total nematode abundance map is accessible online at https://www.crowtherlab.com.
Code availability
All source code and models are available at https://gitlab.ethz.ch/devinrouth/crowther_lab_nematodes.
References
Cameron, E. K. et al. Global gaps in soil biodiversity data. Nat. Ecol. Evol. 2, 1042–1043 (2018).
Bardgett, R. D. & van der Putten, W. H. Belowground biodiversity and ecosystem functioning. Nature 515, 505–511 (2014).
Paul, E. A. Soil Microbiology, Ecology, and Biochemistry 4th edn (Elsevier, 2015).
Wieder, W. R., Bonan, G. B. & Allison, S. D. Global soil carbon projections are improved by modelling microbial processes. Nat. Clim. Change 3, 909–912 (2013).
Bradford, M. A. et al. A test of the hierarchical model of litter decomposition. Nat. Ecol. Evol. 1, 1836–1845 (2017).
Crowther, T. W. et al. Quantifying global soil carbon losses in response to warming. Nature 540, 104–108 (2016).
Delgado-Baquerizo, M. et al. A global atlas of the dominant bacteria found in soil. Science 359, 320–325 (2018).
Thompson, L. R. et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463 (2017).
Lozupone, C. A. & Knight, R. Global patterns in bacterial diversity. Proc. Natl Acad. Sci. USA 104, 11436–11440 (2007).
Fierer, N. & Jackson, R. B. The diversity and biogeography of soil bacterial communities. Proc. Natl Acad. Sci. USA 103, 626–631 (2006).
Bahram, M. et al. Structure and function of the global topsoil microbiome. Nature 560, 233–237 (2018).
Tedersoo, L. et al. Global diversity and geography of soil fungi. Science 346, 1256688 (2014).
Davison, J. et al. Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism. Science 349, 970–973 (2015).
Nielsen, U. N. et al. Global-scale patterns of assemblage structure of soil nematodes in relation to climate and ecosystem properties. Glob. Ecol. Biogeogr. 23, 968–978 (2014).
Wu, T., Ayres, E., Bardgett, R. D., Wall, D. H. & Garey, J. R. Molecular study of worldwide distribution and diversity of soil animals. Proc. Natl Acad. Sci. USA 108, 17720–17725 (2011).
Boag, B. & Yeates, G. W. Soil nematode biodiversity in terrestrial ecosystems. Biodivers. Conserv. 7, 617–630 (1998).
Song, D. et al. Large-scale patterns of distribution and diversity of terrestrial nematodes. Appl. Soil Ecol. 114, 161–169 (2017).
Xu, X., Thornton, P. E. & Post, W. M. A global analysis of soil microbial biomass carbon, nitrogen and phosphorus in terrestrial ecosystems. Glob. Ecol. Biogeogr. 22, 737–749 (2013).
Serna-Chavez, H. M., Fierer, N. & van Bodegom, P. M. Global drivers and patterns of microbial abundance in soil. Glob. Ecol. Biogeogr. 22, 1162–1172 (2013).
Geisen, S. et al. Integrating quantitative morphological and qualitative molecular methods to analyse soil nematode community responses to plant range expansion. Methods Ecol. Evol. 9, 1366–1378 (2018).
Darby, B. J., Todd, T. C. & Herman, M. A. High-throughput amplicon sequencing of rRNA genes requires a copy number correction to accurately reflect the effects of management practices on soil nematode community structure. Mol. Ecol. 22, 5456–5471 (2013).
Carini, P. et al. Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat. Microbiol. 2, 16242 (2016).
Blazewicz, S. J., Barnard, R. L., Daly, R. A. & Firestone, M. K. Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J. 7, 2061–2068 (2013).
Platt, H. M. in The Phylogenetic Systematics of Freeliving Nematodes (ed. Lorenzen, S.) i–ii (The Ray Society, 1994).
Ingham, R. E., Trofymow, J. A., Ingham, E. R. & Coleman, D. C. Interactions of bacteria, fungi, and their nematode grazers: effects on nutrient cycling and plant growth. Ecol. Monogr. 55, 119–140 (1985).
Ferris, H. Contribution of nematodes to the structure and function of the soil food web. J. Nematol. 42, 63–67 (2010).
Crowther, T. W., Boddy, L. & Jones, T. H. Species-specific effects of soil fauna on fungal foraging and decomposition. Oecologia 167, 535–545 (2011).
Neher, D. A. Role of nematodes in soil health and their use as indicators. J. Nematol. 33, 161–168 (2001).
Procter, D. L. Global overview of the functional roles of soil-living nematodes in terrestrial communities and ecosystems. J. Nematol. 22, 1–7 (1990).
Fierer, N., Strickland, M. S., Liptzin, D., Bradford, M. A. & Cleveland, C. C. Global patterns in belowground communities. Ecol. Lett. 12, 1238–1249 (2009).
Sohlenius, B. Abundance, biomass and contribution to energy flow by soil nematodes in terrestrial ecosystems. Oikos 34, 186–194 (1980).
Jobbágy, E. G. & Jackson, R. B. The vertical distribution of soil organic carbon and its relation to climate and vegetation. Ecol. Appl. 10, 423–436 (2000).
Ettema, C. H. Soil nematode diversity: species coexistence and ecosystem function. J. Nematol. 30, 159–169 (1998).
Ferris, H. Nematode Ecophysiological Parameters. http://nemaplex.ucdavis.edu/Ecology/EcophysiologyParms/EcoParameterMenu.html (2018).
Mulder, C. & Vonk, J. A. Nematode traits and environmental constraints in 200 soil systems: scaling within the 60–6000 μm body size range. Ecology 92, 2004 (2011).
Bar-On, Y. M., Phillips, R. & Milo, R. The biomass distribution on Earth. Proc. Natl Acad. Sci. USA 115, 6506–6511 (2018).
Schlesinger, W. H. & Bernhardt, E. S. in Biogeochemistry 419–444 (2013).
Powers, T. O. et al. Tropical nematode diversity: vertical stratification of nematode communities in a Costa Rican humid lowland rainforest. Mol. Ecol. 18, 985–996 (2009).
Yeates, G. W., Bongers, T., De Goede, R. G., Freckman, D. W. & Georgieva, S. S. Feeding habits in soil nematode families and genera—an outline for soil ecologists. J. Nematol. 25, 315–331 (1993).
Chavent, M., Kuentz-Simonet, V., Liquet, B. & Saracco, J. ClustOfVar: an R package for the clustering of variables. J. Stat. Softw. 50, 1–16 (2012).
Hengl, T., Nussbaum, M., Wright, M. N., Heuvelink, G. B. M. & Gräler, B. Random forest as a generic framework for predictive modeling of spatial and spatio-temporal variables. PeerJ 6, e5518 (2018).
Gorelick, N. et al. Google Earth Engine: planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 202, 18–27 (2017).
Hengl, T. et al. SoilGrids250m: global gridded soil information based on machine learning. PLoS ONE 12, e0169748 (2017).
Andrassy, I. Die Rauminhalts und Gewichtsbestimmung der Fadenwürmer (Nematoden). Acta Zool. Hung. 2, 1–15 (1956).
Mulder, C., Cohen, J. E., Setälä, H., Bloem, J. & Breure, A. M. Bacterial traits, organism mass, and numerical abundance in the detrital soil food web of Dutch agricultural grasslands. Ecol. Lett. 8, 80–90 (2005).
Persson, T. Influence of soil organisms on nitrogen mineralization in a northern Scots pine forest. In Proc. VIII Int. Colloq. Soil Zool. (eds Lebrun, P. et al.) 117–126 (1983).
Bongers, T. The maturity index: an ecological measure of environmental disturbance based on nematode species composition. Oecologia 83, 14–19 (1990).
Bongers, T. The Maturity Index, the evolution of nematode life history traits, adaptive radiation and cp-scaling. Plant Soil 212, 13–22 (1999).
Kleiber, M. Body size and metabolism. Hilgardia 6, 315–353 (1932).
West, G. B., Brown, J. H. & Enquist, B. J. A general model for the origin of allometric scaling laws in biology. Science 276, 122–126 (1997).
Atkinson, H. J. in Nematodes as Biological Models vol. 2 (ed Zuckerman, B. M.) 122–126 (Academic, 1980).
Klekowski, R. Z., Wasilewska, L. & Paplinska, E. Oxygen consumption in the developmental stages of Panagrolaimus rigidus. Nematologica 20, 61–68 (1974).
Klekowski, R. Z., Wasilewska, L. & Paplinska, E. Oxygen consumption by soil-inhabiting nematodes. Nematologica 18, 391–403 (1972).
Acknowledgements
This research was supported by a grant from DOB Ecology to T.W.C., a grant from the Netherlands Organization for Scientific Research (grant 016.Veni.181.078) to S.G., grants from NSF (OPP 1115245, 1341736, 0840979) to B.J.A., by a Ramon y Cajal fellow award (RYC-2016-19939) to R.C.H., a grant from UNEP & Global Environment Facility to J.E.C., a grant from NERC (NE/M017036/1) to T.C., a grant from FAPEMIG/FAPESP/VALE S.A.(CRA-RDP-00136-10) to L.B.C., through the strategic programme UID/BIA/04050/2013 (POCI-01-0145-FEDER-007569) awarded to S.R.C., a grant from CNPq PROTAX (562346/2010-4) to J.M.d.C.C., a grant from DFG (CRC990) to V.K. and S.S., a grant from the MSHE of Russia (AAAA-A17-117112850234-5) to A.A.K., grants from the Chinese Academy of Sciences (XDB15010402) and the National Natural Science Foundation of China (41877047) to Q.L., grants from the National Natural Science Foundation of China (31330011, 31170484) to W.L., grants from NERC (NE/M017036/1) to M.M., grants from the Spanish Ministry of Innovation (CGL2009-14686-C02-01/ 02, CGL2013-43675-P) to J.A.R.M., grants from NSF (DEB-0450537, DEB-1145440) to P.M., T.O.P. and K. Powers, grants from the German Academic Exchange Service (PKZ 91540366) and NAFOSTED (106.05 – 2017.330) to T.A.D.N., by an ARC Discovery project (DP150104199) to U.N.N., by the National Key Research and Development Program of China (2016YFC0502101) and the National Natural Science Foundation of China (31370632) to K. Pan, a grant from the Natural Environment Research Council (NERC) to D.G.W., a grant from BAPHIQ (106AS-9.5.1-BQ-B3) J.-i.Y. The James Hutton Institute receives financial support from the Scottish Government Rural and Environment Science and Analytical Services (RESAS) division. Investigations in northwest Russia were carried out under state order for IB KarRC RAS and are partially supported by the Russian Foundation for Basic Research (18-34-00849). We thank E. Clark and A. Orgiazzi for review of the manuscript; and R. Bouharroud, Z. Ferji, L. Jackson and E. Mzough for providing data.
Author information
Authors and Affiliations
Contributions
J.v.d.H., S.G., D.R. and T.W.C. designed and performed the data analyses. J.v.d.H., D.R. and T.W.C. designed and performed geospatial analyses. J.v.d.H., S.G., H.F., R.G.M.d.G. and C.M. designed and performed biomass calculations. S.G., H.F., W.T., D.A.W., R.G.M.d.G., B.J.A., W.A., W.S.A., R.D.B., M.B., R.C.-H., J.E.C., T.C., X.C., S.R.C., R.C., J.M.d.C.C., M.D., L.d.B.C., D.D., M.E., B.S.G., C.G., K.H., D.K., P.K., A.K., G.K., V.K., A.A.K., Q.L., W.L., M. Magilton, M. Marais, J.A.R.M., E.M., E.H.M., C.M., P.M., R.N., T.A.D.N., U.N.N., H.O., J.E.P.R., K. Pan, V.P., L.P., J.C.P.d.S., C.P., T.O.P., K. Powers, C.W.Q., S.R., S.S.M., S.S., H.S., A.S., A.V.T., J.T., W.H.v.d.P., M.V., C.V., L.W., D.H.W., R.W., D.G.W. and J.-i.Y. contributed data. J.v.d.H., S.G. and T.W.C. wrote the first draft of the manuscript with input from D.A.W. All authors contributed to editing of the paper.
Corresponding authors
Ethics declarations
Competing interests
W.S.A. is an employee of Nature Communications, a sister journal from the same publisher; he did not have any access to or involvement with the editorial process at Springer Nature. All other authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks Nico Eisenhauer, Deborah Neher and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Fig. 1 Model accuracy assessment and extent of interpolation and extrapolation across all terrestrial pixels in 73 global covariate layers.
a, Coefficient of variation (standard deviation as a fraction of the mean predicted value) as a measure of the prediction accuracy of our model. b, Proportional extent of interpolation (purple) versus extrapolation (red) in the univariate space. c, Percentage of pixels that fall within the convex hulls of the first 11 principal component spaces (collectively covering >80% of the sample space variation). d, Percentage of pixels interpolated as a function of the percentage of global environmental conditions covered by the sample set. On the global scale, 86% of the Earth’s pixels have at least 90% of the covariate bands falling within the sampled range of environmental conditions. e, Percentage of pixels falling within the 55 convex hull spaces of the first 11 principal components (collectively explaining >80% of the variation). On the global scale, 62% of the Earth’s pixels fell within 100% of 55 convex hull spaces. f, Percentage of terrestrial pixels falling within the sampled range, per covariate band.
Extended Data Fig. 2 Linear regression models of the most important variables from the final random forest model and annual mean temperature.
SOC and cation-exchange capacity have a positive correlation with total nematode abundance, whereas pH is negatively correlated. These linear regression models (n = 1,809) were not used to create global perspectives of nematode distribution patterns. The grey area represents the 95% confidence interval of the mean.
Extended Data Fig. 3 Global maps of nematode trophic group abundance.
a, Bacterivores. b, Fungivores. c, Herbivores. d, Omnivores. e, Predators. Scales differ per map. Most trophic groups show similar patterns, but predators (e) are predicted to be present in particularly high abundances in some arid soils—for example, in the Sahara and Arabian Desert. Pixel values were binned into seven quantiles to create the colour palette.
Extended Data Fig. 4 Global map of total nematode abundance per unit area (m2).
Correcting for the lower bulk density in soils that are high in organic matter, this map shows the same global patterns of nematode abundance as in Fig. 3. Hence, it is not the low bulk density of soils in boreal regions that result in the observed patterns, but rather the high nematode abundances. Pixel values were binned into seven quantiles to create the colour palette.
Extended Data Fig. 5 Global maps of nematode trophic group abundance per unit area (m2).
a, Bacterivores. b, Fungivores. c, Herbivores. d, Omnivores. e, Predators. Scales differ per map. Correcting for the lower bulk density in soils that are high in organic matter, these maps show the same global patterns of nematode trophic group abundance as in Extended Data Fig. 3a–e. Pixel values were binned into seven quantiles to create the colour palette.
Extended Data Fig. 6 Community types and driving variables of community type composition.
a, Correlations between trophic groups. Overall, correlations of predators with other trophic groups are the least positive. b, On the basis of the relative abundance of each trophic group, soil nematode communities can be classified in four distinct types. We find that these soil nematode communities are dominated by herbivores (type 1), herbivores and bacterivores (type 2), bacterivores (type 3) or have a mixed composition (type 4). c, Non-metric multidimensional scaling to highlight environmental conditions that drive the composition of each of the four main community types. Vegetation-type indices, such as the normalized difference vegetation index (NDVI) and enhanced vegetation index (EVI), drive the dominance of herbivores in nematode communities (type 1), whereas edaphic characteristics are correlated with communities dominated by microbivores (types 3 and 4). The names of the environmental variables are listed in Supplementary Table 3.
Supplementary information
41586_2019_1418_MOESM2_ESM.csv
Supplementary Table Supplementary Table 1 | Nematode abundance data and corresponding metadata values. Abundance data for each trophic group and associated metadata from 1,876 1-km2 pixels that were used for geospatial modelling and abundance data from 39 1-km2 pixels from Antarctica. (.csv file).
41586_2019_1418_MOESM3_ESM.csv
Supplementary Table Supplementary Table 2 | Summary of mean, median and sample size values per biome. The number of sites corresponds to the number of 1-km2 pixels into which the samples were aggregated. (.csv file).
41586_2019_1418_MOESM4_ESM.xlsx
Supplementary Table Supplementary Table 3 | Global covariate layers used for geospatial modelling. A total of 73 global covariate layers was used in our modelling approach. The 7 Nadir Reflectance Band layers (i.e., MCD43A4.005 BRDF-Adjusted Reflectance 16-Day Global 500m) are summarised as one entry in the table. (.xlsx file).
41586_2019_1418_MOESM5_ESM.xlsx
Supplementary Table Supplementary Table 4 | Variable importance metrics. Edaphic characteristics emerged as the most important variables. As the full dataset includes collinear variables leading to a false representation of the variable importance metrics, analysis was performed on a selection of main variables. (.xlsx file).
41586_2019_1418_MOESM6_ESM.csv
Supplementary Table Supplementary Table 5 | Number of soil nematodes per trophic group, per biome. Summing the predicted number of nematodes per 1 km2 pixel across biomes we estimate a total of 4.4 × 1020 nematodes are present in the top 15 cm of soil across the globe. (.csv file).
41586_2019_1418_MOESM7_ESM.csv
Supplementary Table Supplementary Table 6 | Relative abundance of soil nematodes per trophic group, per biome. (.csv file).
41586_2019_1418_MOESM8_ESM.csv
Supplementary Table Supplementary Table 7 | Nematode biomass per trophic group, per biome. Note that values are presented in megatons (106 tons) carbon. (.csv file).
Rights and permissions
About this article
Cite this article
van den Hoogen, J., Geisen, S., Routh, D. et al. Soil nematode abundance and functional group composition at a global scale. Nature 572, 194–198 (2019). https://doi.org/10.1038/s41586-019-1418-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1418-6
- Springer Nature Limited
This article is cited by
-
Increasing the number of stressors reduces soil ecosystem services worldwide
Nature Climate Change (2023)
-
Land and deep-sea mining: the challenges of comparing biodiversity impacts
Biodiversity and Conservation (2023)
-
Native diversity buffers against severity of non-native tree invasions
Nature (2023)
-
Global relationships in tree functional traits
Nature Communications (2022)
-
Phytoparasitic nematodes of organic vegetables in the Argan Biosphere of Souss-Massa (Southern Morocco)
Environmental Science and Pollution Research (2021)