Abstract
Microbial inoculation involves transplanting microorganisms from their natural habitat to new plants or soils to improve plant performance, and it is being increasingly used in agriculture and ecological restoration. However, microbial inoculants can invade and alter the composition of native microbial communities; thus, a comprehensive analysis is urgently needed to understand the overall impact of microbial inoculants on the biomass, diversity, structure and network complexity of native communities. Here we provide a meta-analysis of 335 studies revealing a positive effect of microbial inoculants on soil microbial biomass. This positive effect was weakened by environmental stress and enhanced by the use of fertilizers and native inoculants. Although microbial inoculants did not alter microbial diversity, they induced major changes in the structure and bacterial composition of soil microbial communities, reducing the complexity of bacterial networks and increasing network stability. Finally, higher initial levels of soil nutrients amplified the positive impact of microbial inoculants on fungal biomass, actinobacterial biomass, microbial biomass carbon and microbial biomass nitrogen. Together, our results highlight the positive effects of microbial inoculants on soil microbial biomass, emphasizing the benefits of native inoculants and the important regulatory roles of soil nutrient levels and environmental stress.





Similar content being viewed by others
Data availability
The sequences used in this study consist of publicly available published data and can be downloaded using the provided accession numbers. All accession numbers, Supplementary Dataset and other relevant data featured in this Article are available via GitHub at https://github.com/aijingjing1314/Microbial-inoculants_Meta-analysis. Source data are provided with this paper.
Code availability
The code used in this study is available via GitHub at https://github.com/aijingjing1314/Microbial-inoculants_Meta-analysis.
References
Kaminsky, L. M., Trexler, R. V., Malik, R. J., Hockett, K. L. & Bell, T. H. The inherent conflicts in developing soil microbial inoculants. Trends Biotechnol. 37, 140–151 (2019).
Jack, C. N., Petipas, R. H., Cheeke, T. E., Rowland, J. L. & Friesen, M. L. Microbial inoculants: silver bullet or microbial Jurassic Park? Trends Microbiol. 29, 299–308 (2021).
Haskett, T. L., Tkacz, A. & Poole, P. S. Engineering rhizobacteria for sustainable agriculture. ISME J. 15, 949–964 (2021).
Mallon, C. A., Van Elsas, J. D. & Salles, J. F. Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol. 23, 719–729 (2015).
Mawarda, P. C., Le Roux, X., Van Elsas, J. D. & Salles, J. F. Deliberate introduction of invisible invaders: a critical appraisal of the impact of microbial inoculants on soil microbial communities. Soil Biol. Biochem. 148, 107874 (2020).
Singh, J. S. & Gupta, V. K. Soil microbial biomass: a key soil driver in management of ecosystem functioning. Sci. Total Environ. 634, 497–500 (2018).
Delgado-Baquerizo, M. et al. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 7, 10541 (2016).
Liu, X., Le Roux, X. & Salles, J. F. The legacy of microbial inoculants in agroecosystems and potential for tackling climate change challenges. iScience 25, 103821 (2022).
Mallon, C. A. et al. The impact of failure: unsuccessful bacterial invasions steer the soil microbial community away from the invader’s niche. ISME J. 12, 728–741 (2018).
Bastida, F. et al. Soil microbial diversity–biomass relationships are driven by soil carbon content across global biomes. ISME J. 15, 2081–2091 (2021).
Zhou, Z., Wang, C. & Luo, Y. Meta-analysis of the impacts of global change factors on soil microbial diversity and functionality. Nat. Commun. 11, 3072 (2020).
Alori, E. T., Dare, M. O. & Babalola, O. O. in Sustainable Agriculture Reviews (ed. Lichtfouse, E.) 281–307 (Springer International Publishing, 2017).
Hernandez, D. J., David, A. S., Menges, E. S., Searcy, C. A. & Afkhami, M. E. Environmental stress destabilizes microbial networks. ISME J. 15, 1722–1734 (2021).
Hartmann, M. & Six, J. Soil structure and microbiome functions in agroecosystems. Nat. Rev. Earth Environ. 4, 4–18 (2023).
Jiang, M. et al. Home‐based microbial solution to boost crop growth in low‐fertility soil. New Phytol. 239, 752–765 (2023).
Liu, X., Mei, S. & Salles, J. F. Inoculated microbial consortia perform better than single strains in living soil: a meta-analysis. Appl. Soil Ecol. 190, 105011 (2023).
Qiu, Z., Egidi, E., Liu, H., Kaur, S. & Singh, B. K. New frontiers in agriculture productivity: optimised microbial inoculants and in situ microbiome engineering. Biotechnol. Adv. 37, 107371 (2019).
Zhang, S., Lehmann, A., Zheng, W., You, Z. & Rillig, M. C. Arbuscular mycorrhizal fungi increase grain yields: a meta‐analysis. New Phytol. 222, 543–555 (2019).
Van Elsas, J. D. et al. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl Acad. Sci. USA 109, 1159–1164 (2012).
Durán, P. et al. Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 175, 973–983.e914 (2018).
Kurkjian, H. M., Akbari, M. J. & Momeni, B. The impact of interactions on invasion and colonization resistance in microbial communities. PLoS Comput. Biol. 17, e1008643 (2021).
Yang, T. et al. Resource availability modulates biodiversity–invasion relationships by altering competitive interactions. Environ. Microbiol. 19, 2984–2991 (2017).
Tecon, R. & Or, D. Biophysical processes supporting the diversity of microbial life in soil. FEMS Microbiol. Rev. 41, 599–623 (2017).
Anthony, M. A., Bender, S. F. & van der Heijden, M. G. Enumerating soil biodiversity. Proc. Natl Acad. Sci. USA 120, e2304663120 (2023).
Montoya, J. M., Pimm, S. L. & Solé, R. V. Ecological networks and their fragility. Nature 442, 259–264 (2006).
Banerjee, S., Schlaeppi, K. & van der Heijden, M. G. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 16, 567–576 (2018).
Herren, C. M. Disruption of cross-feeding interactions by invading taxa can cause invasional meltdown in microbial communities. Proc. R. Soc. B 287, 20192945 (2020).
Shi, S. et al. The interconnected rhizosphere: high network complexity dominates rhizosphere assemblages. Ecol. Lett. 19, 926–936 (2016).
Li, Q. et al. Plant growth‐promoting rhizobacterium Pseudomonas sp. CM11 specifically induces lateral roots. New Phytol. 235, 1575–1588 (2022).
Chen, C., Chen, H. Y., Chen, X. & Huang, Z. Meta-analysis shows positive effects of plant diversity on microbial biomass and respiration. Nat. Commun. 10, 1332 (2019).
Toljander, J. F., Artursson, V., Paul, L. R., Jansson, J. K. & Finlay, R. D. Attachment of different soil bacteria to arbuscular mycorrhizal fungal extraradical hyphae is determined by hyphal vitality and fungal species. FEMS Microbiol. Lett. 254, 34–40 (2006).
Stopnisek, N. et al. Molecular mechanisms underlying the close association between soil Burkholderia and fungi. ISME J. 10, 253–264 (2016).
Batista, B. D. & Singh, B. K. Realities and hopes in the application of microbial tools in agriculture. Microb. Biotechnol. 14, 1258–1268 (2021).
Saad, M. M., Eida, A. A. & Hirt, H. Tailoring plant-associated microbial inoculants in agriculture: a roadmap for successful application. J. Exp. Bot. 71, 3878–3901 (2020).
Schimel, J., Balser, T. C. & Wallenstein, M. Microbial stress‐response physiology and its implications for ecosystem function. Ecology 88, 1386–1394 (2007).
Lori, M., Symnaczik, S., Mäder, P., De Deyn, G. & Gattinger, A. Organic farming enhances soil microbial abundance and activity—a meta-analysis and meta-regression. PLoS ONE 12, e0180442 (2017).
Muhammad, I. et al. Cover cropping enhances soil microbial biomass and affects microbial community structure: a meta-analysis. Geoderma 381, 114696 (2021).
Li, C. et al. Plant and native microorganisms amplify the positive effects of microbial inoculant. Microorganisms 11, 570 (2023).
Tedersoo, L., Bahram, M. & Zobel, M. How mycorrhizal associations drive plant population and community biology. Science 367, eaba1223 (2020).
Jiang, Y. et al. Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356, 1172–1175 (2017).
Bago, B., Pfeffer, P. E. & Shachar-Hill, Y. Carbon metabolism and transport in arbuscular mycorrhizas. Plant Physiol. 124, 949–958 (2000).
Xiao, Y., Zhao, Z., Chen, L. & Li, Y. Arbuscular mycorrhizal fungi and organic manure have synergistic effects on Trifolium repens in Cd-contaminated sterilized soil but not in natural soil. Appl. Soil Ecol. 149, 103485 (2020).
Gu, Y. et al. Invader–resident community similarity contribute to the invasion process and regulate biofertilizer effectiveness. J. Clean. Prod. 241, 118278 (2019).
Hu, L. et al. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 9, 2738 (2018).
Zuluaga, M. Y. A. et al. Inoculation with plant growth-promoting bacteria alters the rhizosphere functioning of tomato plants. Appl. Soil Ecol. 158, 103784 (2021).
Kong, Z. & Liu, H. Modification of rhizosphere microbial communities: a possible mechanism of plant growth promoting rhizobacteria enhancing plant growth and fitness. Front. Plant Sci. 13, 920813 (2022).
Orwin, K. H. et al. Linkages of plant traits to soil properties and the functioning of temperate grassland. J. Ecol. 98, 1074–1083 (2010).
Dong, L. et al. Biofertilizers regulate the soil microbial community and enhance Panax ginseng yields. Chin. Med. 14, 20 (2019).
Neuenkamp, L., Prober, S. M., Price, J. N., Zobel, M. & Standish, R. J. Benefits of mycorrhizal inoculation to ecological restoration depend on plant functional type, restoration context and time. Fungal Ecol. 40, 140–149 (2019).
Scheffer, R. A. & Aerts, R. Root decomposition and soil nutrient and carbon cycling in two temperate fen ecosystems. Oikos 91, 541–549 (2000).
Davis, M. A., Grime, J. P. & Thompson, K. Fluctuating resources in plant communities: a general theory of invasibility. J. Ecol. 88, 528–534 (2000).
Nazaries, L. et al. The response of soil multi-functionality to agricultural management practices can be predicted by key soil abiotic and biotic properties. Agric. Ecosyst. Environ. 307, 107206 (2021).
Li, H. et al. Effects of PGPR microbial inoculants on the growth and soil properties of Avena sativa, Medicago sativa, and Cucumis sativus seedlings. Soil Tillage Res. 199, 104577 (2020).
Bai, X. et al. Extracellular enzyme activity and stoichiometry: the effect of soil microbial element limitation during leaf litter decomposition. Ecol. Indic. 121, 107200 (2021).
Trabelsi, D. & Mhamdi, R. Microbial inoculants and their impact on soil microbial communities: a review. Biomed. Res. Int. 2013, 863240 (2013).
Ma, Z. & Chen, H. Y. Positive species mixture effects on fine root turnover and mortality in natural boreal forests. Soil Biol. Biochem. 121, 130–137 (2018).
Zhang, T. A., Chen, H. Y. & Ruan, H. Global negative effects of nitrogen deposition on soil microbes. ISME J. 12, 1817–1825 (2018).
Anderson, T.-H. Microbial eco-physiological indicators to asses soil quality. Agric. Ecosyst. Environ. 98, 285–293 (2003).
Moscatelli, M. C., Lagomarsino, A., Marinari, S., De Angelis, P. & Grego, S. Soil microbial indices as bioindicators of environmental changes in a poplar plantation. Ecol. Indic. 5, 171–179 (2005).
Sadegh Kasmaei, L. et al. Influence of plant growth promoting rhizobacteria, compost, and biochar of Azolla on rosemary (Rosmarinus officinalis L.) growth and some soil quality indicators in a calcareous soil. Commun. Soil Sci. Plant Anal. 50, 119–131 (2019).
Allison, S. D. & Martiny, J. B. Resistance, resilience, and redundancy in microbial communities. Proc. Natl Acad. Sci. USA 105, 11512–11519 (2008).
Jiao, S. et al. Core phylotypes enhance the resistance of soil microbiome to environmental changes to maintain multifunctionality in agricultural ecosystems. Glob. Change Biol. 28, 6653–6664 (2022).
Fan, K. et al. Biodiversity of key-stone phylotypes determines crop production in a 4-decade fertilization experiment. ISME J. 15, 550–561 (2021).
Bastida, F. et al. The active microbial diversity drives ecosystem multifunctionality and is physiologically related to carbon availability in Mediterranean semi‐arid soils. Mol. Ecol. 25, 4660–4673 (2016).
Amor, D. R., Ratzke, C. & Gore, J. Transient invaders can induce shifts between alternative stable states of microbial communities. Sci. Adv. 6, eaay8676 (2020).
Hang, X. et al. Trichoderma-amended biofertilizer stimulates soil resident Aspergillus population for joint plant growth promotion. NPJ Biofilms Microbiomes 8, 57 (2022).
Tao, C. et al. Bio-organic fertilizers stimulate indigenous soil Pseudomonas populations to enhance plant disease suppression. Microbiome 8, 137 (2020).
Deng, X. et al. Bio‐organic soil amendment promotes the suppression of Ralstonia solanacearum by inducing changes in the functionality and composition of rhizosphere bacterial communities. New Phytol. 235, 1558–1574 (2022).
Wen, T. et al. Deciphering the mechanism of fungal pathogen‐induced disease‐suppressive soil. New Phytol. 238, 2634–2650 (2023).
Zhang, L., Zhou, J., George, T. S., Limpens, E. & Feng, G. Arbuscular mycorrhizal fungi conducting the hyphosphere bacterial orchestra. Trends Plant Sci. 27, 402–411 (2022).
Li, C. et al. Mineral-solubilizing microbial inoculant positively affects the multifunctionality of anthropogenic soils in abandoned mining areas. J. Environ. Manage. 344, 118553 (2023).
Bashan, Y., de-Bashan, L. E., Prabhu, S. & Hernandez, J.-P. Advances in plant growth-promoting bacterial inoculant technology: formulations and practical perspectives (1998–2013). Plant Soil 378, 1–33 (2014).
Qiu, L. et al. Erosion reduces soil microbial diversity, network complexity and multifunctionality. ISME J. 15, 2474–2489 (2021).
Fierer, N., Bradford, M. A. & Jackson, R. B. Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364 (2007).
Janssen, P. H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72, 1719–1728 (2006).
Zhao, J. et al. Pyrosequencing reveals contrasting soil bacterial diversity and community structure of two main winter wheat cropping systems in China. Microb. Ecol. 67, 443–453 (2014).
Stokstad, E. The nitrogen fix. Science 353, 1225–1227 (2016).
Yang, J. et al. Mechanisms underlying legume–rhizobium symbioses. J. Integr. Plant Biol. 64, 244–267 (2022).
Yan, J., Han, X., Lu, X., Chen, X. & Zou, W. Land use indirectly affects the cycling of multiple nutrients by altering the diazotrophic community in black soil. J. Sci. Food Agric. 102, 3788–3795 (2022).
Mo, Y. et al. Low shifts in salinity determined assembly processes and network stability of microeukaryotic plankton communities in a subtropical urban reservoir. Microbiome 9, 128 (2021).
Ju, F. & Zhang, T. Bacterial assembly and temporal dynamics in activated sludge of a full-scale municipal wastewater treatment plant. ISME J. 9, 683–695 (2015).
Liu, W. et al. Dynamic microbial assembly processes correspond to soil fertility in sustainable paddy agroecosystems. Funct. Ecol. 34, 1244–1256 (2020).
Sun, C. et al. Seasonal dynamics of the microbial community in two full-scale wastewater treatment plants: diversity, composition, phylogenetic group based assembly and co-occurrence pattern. Water Res. 200, 117295 (2021).
Zhou, J. et al. Functional molecular ecological networks. mBio 1, e00169–00110 (2010).
Ling, N., Wang, T. & Kuzyakov, Y. Rhizosphere bacteriome structure and functions. Nat. Commun. 13, 836 (2022).
Moher, D., Liberati, A., Tetzlaff, J., Altman, D. G. & PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann. Intern. Med. 151, 264–269 (2009).
McGrath, S. et al. Estimating the sample mean and standard deviation from commonly reported quantiles in meta-analysis. Stat. Methods Med. Res. 29, 2520–2537 (2020).
Ruehlmann, J. & Körschens, M. Calculating the effect of soil organic matter concentration on soil bulk density. Soil Sci. Soc. Am. J. 73, 876–885 (2009).
Nel, T., Hardie, A. G. & Clarke, C. E. Simple and multivariate linear regression models for pH conversion between measurement techniques. Commun. Soil Sci. Plant Anal. 53, 1797–1808 (2022).
Paliy, O. & Shankar, V. Application of multivariate statistical techniques in microbial ecology. Mol. Ecol. 25, 1032–1057 (2016).
Zhou, Z., Zheng, M., Xia, J. & Wang, C. Nitrogen addition promotes soil microbial beta diversity and the stochastic assembly. Sci. Total Environ. 806, 150569 (2022).
Pittelkow, C. M. et al. Productivity limits and potentials of the principles of conservation agriculture. Nature 517, 365–368 (2015).
Rosenberg, M. S. MetaWin: statistical software for meta-analysis: version 2 (Sinauer, 2000).
Butler, O. M., Elser, J. J., Lewis, T., Mackey, B. & Chen, C. The phosphorus‐rich signature of fire in the soil–plant system: a global meta‐analysis. Ecol. Lett. 21, 335–344 (2018).
Jin, Z. C., Zhou, X. H. & He, J. Statistical methods for dealing with publication bias in meta‐analysis. Stat. Med. 34, 343–360 (2015).
Rosenberg, M. S. The file‐drawer problem revisited: a general weighted method for calculating fail‐safe numbers in meta‐analysis. Evolution 59, 464–468 (2005).
Rosenthal, R. The file drawer problem and tolerance for null results. Psychol. Bull. 86, 638–641 (1979).
Ji, X., Liu, M., Yang, J. & Feng, F. Meta-analysis of the impact of freeze–thaw cycles on soil microbial diversity and C and N dynamics. Soil Biol. Biochem. 168, 108608 (2022).
Flora of China Editorial Committee Beijing (China): Flora of China (Science Press, 1999).
Bates, D., Maechler, M., Bolker, B. & Walker, S. lme4: linear mixed-effects models using ‘Eigen’ and S4. R version 1.1–27.1 https://cran.r-project.org/package=lme4 (2021).
Oksanen, J. Vegan: community ecology package. R version 1.8–5 https://www.cran.r-project.org (2007).
Leinonen, R., Sugawara, H., Shumway, M. & Collaboration, I. N. S. D. The Sequence Read Archive. Nucleic Acids Res. 39, D19–D21 (2010).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Liu, Y. X. et al. EasyAmplicon: an easy‐to‐use, open‐source, reproducible, and community‐based pipeline for amplicon data analysis in microbiome research. iMeta 2, e83 (2023).
Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584 (2016).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Cole, J. R. et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642 (2014).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Yuan, M. M. et al. Climate warming enhances microbial network complexity and stability. Nat. Clim. Change 11, 343–348 (2021).
Deng, Y. et al. Molecular ecological network analyses. BMC Bioinformatics 13, 113 (2012).
Bastian, M., Heymann, S. & Jacomy, M. Gephi: an open source software for exploring and manipulating networks. ICWSM 8, 361–362 (2009).
Guimera, R. & Nunes Amaral, L. A. Functional cartography of complex metabolic networks. Nature 433, 895–900 (2005).
Olesen, J. M., Bascompte, J., Dupont, Y. L. & Jordano, P. The modularity of pollination networks. Proc. Natl Acad. Sci. USA 104, 19891–19896 (2007).
Peng, G.-s & Wu, J. Optimal network topology for structural robustness based on natural connectivity. Phys. A 443, 212–220 (2016).
Herren, C. M. & McMahon, K. D. Cohesion: a method for quantifying the connectivity of microbial communities. ISME J. 11, 2426–2438 (2017).
Bashan, Y., Prabhu, S., de-Bashan, L. E. & Kloepper, J. W. Disclosure of exact protocols of fermentation, identity of microorganisms within consortia, formation of advanced consortia with microbe-based products. Biol. Fertil. Soils 56, 443–445 (2020).
Acknowledgements
We sincerely thank all the researchers whose valuable data were included in this global synthesis. We would like to express our gratitude to Y. Wu, who contributed to the project development through valuable discussions. C.L. is grateful for the partial financial support from the Postgraduate Research and Practice Innovation Program of Jiangsu Province (KYCX21_0915) and the China Scholarship Council (202108320300). J.Z. acknowledges the funding support from Jiangsu Science and Technology Plan Project (BE2022420), the Innovation and Promotion of Forestry Science and Technology Program of Jiangsu Province (LYKJ[2021]30), the Scientific Research Project of Baishanzu National Park (2021ZDLY01) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Contributions
C.L., Z.J., B.Z., X.L. and J.Z. conceived the study. C.L., S.M. and J.Q. collected and organized the data. C.L., X.C. and L.Z. analysed the data. C.L. wrote the first draft of the paper. X.C., Z.J., L.Z., B.Z., U.G., X.L., J.Z. and C.M. reviewed the paper before submission. The authors have approved the final paper for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Ecology & Evolution thanks Elly Morriën, Xavier Roux and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The effects of microbial inoculants on soil microbial biomass.
a, bacterial biomass; b, fungal biomass; c, actinobacterial biomass; d, diazotrophic biomass. Bars around the means denote 95% confidence intervals (CIs). Mean values < 0 indicate a higher value in control treatment (yellow dots), while mean values > 0 indicate a higher value in microbial inoculant treatment (blue dots). Bacteria, fungi, and AMF in front of each subgroup name represent bacterial inoculants, fungal inoculants, and arbuscular mycorrhizal fungi (AMF) inoculants, respectively. The number of observations is beside each attribute. The between-group heterogeneity (Qbetween) statistic is computed using the one-sided chi-square test. A significance level is set at Prandom < 0.05 to determine the significance of Qbetween. Differences among subgroups are deemed significant when their CIs do not overlap. Source data are provided as a Source Data file.
Extended Data Fig. 2 Effects of microbial inoculant on soil microbial biomass associated with soil background physicochemical properties.
Points signify the values predicted by the partial regressions of soil background physicochemical properties. Black lines represent the average responses with their 95% confidence intervals (CIs) shaded in grey. n represents the number of observations. The one-sided F-test is used to calculate P values.
Extended Data Fig. 3 The effects of microbial inoculants on soil microbial alpha diversity.
a, Bacterial Shannon diversity; b, bacterial richness diversity; c, fungal Shannon diversity; d, fungal richness diversity. Bars around the means denote 95% confidence intervals (CIs). Bacteria, fungi, and AMF in front of each subgroup name represent bacterial inoculants, fungal inoculants, and arbuscular mycorrhizal fungi (AMF) inoculants, respectively. The number of observations is beside each attribute. The between-group heterogeneity (Qbetween) statistic is computed using the one-sided chi-square test. A significance level is set at Prandom < 0.05 to determine the significance of Qbetween. Differences among subgroups are deemed significant when their CIs do not overlap. Source data are provided as a Source Data file.
Extended Data Fig. 4 The effects of microbial inoculants on soil bacterial alpha diversity and community structure based on reanalysis of amplicon data.
a, bacterial alpha diversity, including Shannon, Pielou, ACE, Chao, and Richness diversity. Error bars on the columns represent standard errors (SD). Statistical comparisons are assessed through two-tailed Wilcoxon’s rank sum tests. P values are adjusted using the Benjamini–Hochberg false discovery rate (FDR) correction. All dots signify the change in response ratio between the control and microbial inoculant bacterial diversity at 95% confidence intervals (CIs). The number of observations is provided beside each attribute. b, Principal Coordinate Analysis (PCoA) plots depict the Bray-Curtis distance of bacterial communities in CK and microbial inoculant treatments (CK n = 453 vs. microbial inoculant n = 1076) c, Three non-parametric multivariate analyses, including non-parametric multivariate analysis of variance (Adonis), analysis of similarity (ANOSIM), and multi-response permutation procedure (MRPP), consistently support the significant alteration of bacterial community structure by microbial inoculants. P values are adjusted using the Benjamini–Hochberg method with sequentially modified Bonferroni correction. Source data are provided as a Source Data file.
Extended Data Fig. 5 The effects of microbial inoculants on soil microbial community.
a, bacterial community structure; b, bacterial beta diversity; c, fungal community structure; d, fungal beta diversity. Bars around the means denote 95% confidence intervals (CIs). Mean values < 0 indicate a higher value in control treatment (yellow dots), while mean values > 0 indicate a higher value in microbial inoculant treatment (blue dots). Bacteria, fungi, and AMF in front of each subgroup name represent bacterial inoculants, fungal inoculants, and arbuscular mycorrhizal fungi (AMF) inoculants, respectively. RRStructure < 0 indicates that microbial inoculant has no effect on microbial community structure, and a greater positive value of RRStructure indicates a greater magnitude of change in the community structure. The number of observations is beside each attribute. The between-group heterogeneity (Qbetween) statistic is computed using the one-sided chi-square test. A significance level is set at Prandom < 0.05 to determine the significance of Qbetween. Differences among subgroups are deemed significant when their CIs do not overlap. Source data are provided as a Source Data file.
Extended Data Fig. 6 Effects of microbial inoculant on soil microbial diversity associated with soil background physicochemical properties.
Points signify the values predicted by the partial regressions of soil background physicochemical properties. n represents the number of observations. The one-sided F-test is used to calculate P values.
Extended Data Fig. 7 Phylogenetic tree showing the top 200 bacterial amplicon sequences variants (ASVs) with the highest cumulative relative abundance.
The color of the inner ring represents the taxonomy at the phylum level, while the name of the inner ring corresponds to the genus. Medium ring 1 illustrates the relative abundance of 200 ASVs in the CK treatment, and Medium ring 2 depicts the relative abundance of 200 ASVs in the microbial inoculant treatment. Medium ring 3 displays the relative abundance of ASVs across different treatments, with yellow indicating higher enrichment in the CK treatment and blue indicating higher enrichment in the microbial inoculant treatment. The symbol * represents a significance level of P < 0.05, determined through two-tailed Wilcoxon’s rank sum tests. P values are adjusted using the Benjamini–Hochberg false discovery rate (FDR) correction. The outer ring represents taxonomy at the class level.
Extended Data Fig. 8 The composition of bacterial communities under CK and microbial inoculant treatments at the phylum level.
a, the composition of bacterial communities under CK and microbial inoculant treatments at the phylum level; b, differences at the phylum level caused by microbial inoculants; c, differences at the phylum level caused by bacterial inoculants; d, differences at the phylum level caused by AMF inoculants; e, differences at the phylum level caused by fungal inoculants; f, differences at the phylum level caused by Mix inoculants. ▲ represents the increase of taxa under microbial inoculant treatments; ▼represents the decrease of taxa under microbial inoculant treatments. Statistical comparisons are assessed through two-tailed Wilcoxon’s rank sum tests. P values are adjusted using the Benjamini–Hochberg false discovery rate (FDR) correction.
Extended Data Fig. 9 The composition of bacterial communities under CK and microbial inoculant treatments at the class level.
a, the composition of bacterial communities under CK and microbial inoculant treatments at the class level; b, differences at the class level caused by microbial inoculants; c, differences at the class level caused by bacterial inoculants; d, differences at the class level caused by AMF inoculants; e, differences at the class level caused by fungal inoculants; f, differences at the class level caused by Mix inoculants. ▲ represents the increase of taxa under microbial inoculant treatments; ▼represents the decrease of taxa under microbial inoculant treatments. Statistical comparisons are assessed through two-tailed Wilcoxon’s rank sum tests. P values are adjusted using the Benjamini–Hochberg false discovery rate (FDR) correction.
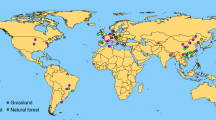
Extended Data Fig. 10 The experimental locations in this meta-analysis.
a, the experimental locations of 335 publications included in this meta-analysis (microbial attributes); b, the experimental locations of 48 publications included in this meta-analysis (reanalysis of amplicon data based on upper microbial attributes database).
Supplementary information
Source data
Source Data Fig. 1
Source data.
Source Data Fig. 2
Source data.
Source Data Fig. 3
Source data.
Source Data Fig. 4
Source data.
Source Data Fig. 5
Source data.
Source Data Extended Data Fig. 1
Source data.
Source Data Extended Data Fig. 3
Source data.
Source Data Extended Data Fig. 4
Source data.
Source Data Extended Data Fig. 5
Source data.
Source Data Extended Data Fig. 7
Source data.
Source Data Extended Data Fig. 8
Source data.
Source Data Extended Data Fig. 9
Source data.
Source Data Extended Data Fig. 10
Source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, C., Chen, X., Jia, Z. et al. Meta-analysis reveals the effects of microbial inoculants on the biomass and diversity of soil microbial communities. Nat Ecol Evol 8, 1270–1284 (2024). https://doi.org/10.1038/s41559-024-02437-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-024-02437-1
- Springer Nature Limited