Abstract
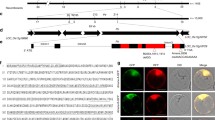
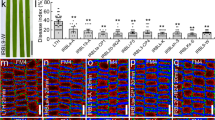
Blast disease caused by the fungus Magnaporthe oryzae is one of the most devastating rice diseases. Disease resistance genes such as Pi-ta or Pi-ta2 are critical in protecting rice production from blast. Published work reports that Pi-ta codes for a nucleotide-binding and leucine-rich repeat domain protein (NLR) that recognizes the fungal protease-like effector AVR-Pita by direct binding. However, this model was challenged by the recent discovery that Pi-ta2 resistance, which also relies on AVR-Pita detection, is conferred by the unconventional resistance gene Ptr, which codes for a membrane protein with a cytoplasmic armadillo repeat domain. Here, using NLR Pi-ta and Ptr RNAi knockdown and CRISPR/Cas9 knockout mutant rice lines, we found that AVR-Pita recognition relies solely on Ptr and that the NLR Pi-ta has no role in it, indicating that it is not the Pi-ta resistance gene. Different alleles of Ptr confer different recognition specificities. The A allele of Ptr (PtrA) detects all natural sequence variants of the effector and confers Pi-ta2 resistance, while the B allele of Ptr (PtrB) recognizes a restricted set of AVR-Pita alleles and, thereby, confers Pi-ta resistance. Analysis of the natural diversity in AVR-Pita and of mutant and transgenic strains identified one specific polymorphism in the effector sequence that controls escape from PtrB-mediated resistance. Taken together, our work establishes that the M. oryzae effector AVR-Pita is detected in an allele-specific manner by the unconventional rice resistance protein Ptr and that the NLR Pi-ta has no function in Pi-ta resistance and the recognition of AVR-Pita.






Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in the paper and its supplementary information files or are available from the corresponding authors on reasonable request.
References
Savary, S. et al. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439 (2019).
Dean, R. et al. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430 (2012).
Baudin, M. et al. Pyricularia oryzae: Lab star and field scourge. Mol. Plant Pathol. 25, e13449 (2024).
Kourelis, J. & van der Hoorn, R. A. L. Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30, 285–299 (2018).
Wang, B., Ebbole, D. J. & Wang, Z. The arms race between Magnaporthe oryzae and rice: diversity and interaction of Avr and R genes. J. Integr. Agric. 16, 2746–2760 (2017).
Cesari, S. Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 219, 17–24 (2018).
Xi, Y., Cesari, S. & Kroj, T. Insight into the structure and molecular mode of action of plant paired NLR immune receptors. Essays Biochem. 66, 513–526 (2022).
Kiyosawa, S. Studies on inheritance of resistance of rice varieties to blast: 3. Inheritance of resistance of a rice variety Pi No. 1 to the blast fungus. Jpn J. Breed. 16, 243–250 (1966).
Orbach, M. J., Farrall, L., Sweigard, J. A., Chumley, F. G. & Valent, B. A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta. Plant Cell 12, 2019–2032 (2000).
Chuma, I. et al. Multiple translocation of the AVR-Pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PLoS Pathog. 7, e1002147 (2011).
Bryan, G. T. et al. tA single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12, 2033–2045 (2000).
Bailey, P. C. et al. Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biol. 19, 23 (2018).
Costanzo, S. & Jia, Y. Alternatively spliced transcripts of Pi-ta blast resistance gene in Oryza sativa. Plant Sci. 177, 468–478 (2009).
Jia, Y., McAdams, S. A., Bryan, G. T., Hershey, H. P. & Valent, B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014 (2000).
Kiyosawa, S. Inheritance of resistance of the rice variety Pi No. 4 to blast. Jpn J. Breed. 17, 165–172 (1967).
Kiyosawa, S. Genetical approach to the biochemical nature of plant disease resistance. Jpn Agric. Res. Q. 6, 72–80 (1971).
Meng, X. et al. The broad-spectrum rice blast resistance (R) gene Pita2 encodes a novel R protein unique from Pita. Rice 13, 19 (2020).
Zhao, H. et al. The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat. Commun. 9, 2039 (2018).
Lee, S., Costanzo, S., Jia, Y., Olsen, K. M. & Caicedo, A. L. Evolutionary dynamics of the genomic region around the blast resistance gene Pi-ta in AA genome Oryza species. Genetics 183, 1315–1325 (2009).
Wang, X., Jia, Y., Shu, Q. Y. & Wu, D. Haplotype diversity at the Pi-ta locus in cultivated rice and its wild relatives. Phytopathology 98, 1305–1311 (2008).
Kiyosawa, S. Establishment of differential varieties for pathogenicity test of rice blast fungus. Rice Genet. Newsl. 1, 95 (1984).
Kiyosawa, S. Genetic studies on host–pathogen relationship in the rice blast disease. In Proc. Symp. Rice Diseases and Their Control by Growing Resistant Varieties and Other Measures 137–153 (Japan International Research Center for Agricultural Sciences, 1967).
Kobayashi, N. et al. Development of new sets of international standard differential varieties for blast resistance in rice (Oryza sativa L.). Jpn Agric. Res. Q. 41, 31–37 (2007).
Huang, C. L., Hwang, S. Y., Chiang, Y. C. & Lin, T. P. Molecular evolution of the Pi-ta gene resistant to rice blast in wild rice (Oryza rufipogon). Genetics 179, 1527–1538 (2008).
Cesari, S. et al. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25, 1463–1481 (2013).
Cesari, S. et al. New recognition specificity in a plant immune receptor by molecular engineering of its integrated domain. Nat. Commun. 13, 1524 (2022).
Guo, L. et al. Specific recognition of two MAX effectors by integrated HMA domains in plant immune receptors involves distinct binding surfaces. Proc. Natl Acad. Sci. USA 115, 11637–11642 (2018).
Ortiz, D. et al. Recognition of the Magnaporthe oryzae effector AVR-Pia by the decoy domain of the rice NLR immune receptor RGA5. Plant Cell 29, 156–168 (2017).
Xi, Y., Chalvon, V., Padilla, A., Cesari, S. & Kroj, T. The activity of the RGA5 sensor NLR from rice requires binding of its integrated HMA domain to effectors but not HMA domain self-interaction. Mol. Plant Pathol. 23, 1320–1330 (2022).
Stein, J. C. et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat. Genet. 50, 285–296 (2018).
He, N. et al. Analysis of a rice blast resistance gene Pita-Fuhui2663 and development of selection marker. Sci. Rep. 12, 14917 (2022).
Coates, J. Armadillo repeat proteins: beyond the animal kingdom. Trends Cell Biol. 13, 463–471 (2003).
Tewari, R., Bailes, E., Bunting, K. A. & Coates, J. C. Armadillo-repeat protein functions: questions for little creatures. Trends Cell Biol. 20, 470–481 (2010).
Athiyannan, N., Aouini, L., Wang, Y. & Krattinger, S. G. Unconventional R proteins in the botanical tribe Triticeae. Essays Biochem. 66, 561–569 (2022).
Le Naour-Vernet, M. et al. Adaptive evolution in virulence effectors of the rice blast fungus Pyricularia oryzae. PLoS Pathog. 19, e1011294 (2023).
Han, J. et al. The fungal effector Avr-Pita suppresses innate immunity by increasing COX activity in rice mitochondria. Rice 14, 12 (2021).
Veillet, F., Durand, M., Kroj, T., Cesari, S. & Gallois, J.-L. Precision breeding made real with CRISPR: illustration through genetic resistance to pathogens. Plant Commun. 1, 100102 (2020).
Faivre-Rampant, O. et al. Characterization of the model system rice–Magnaporthe for the study of nonhost resistance in cereals. New Phytol. 180, 899–910 (2008).
Leung, H., Borromeo, E. S., Bernardo, M. A. & Notteghem, J. L. Genetic analysis of virulence in the rice blast fungus Magnaporthe grisea. Phytopathology 78, 1227–1233 (1988).
Rao, Z. M. et al. Genetic dissection of blast resistances at different growth stages in rice (Oryza sativa L.). Rice Genet. Newsl. 19, 39 (2004).
Berruyer, R. et al. Identification and fine mapping of Pi33, the rice resistance gene corresponding to the Magnaporthe grisea avirulence gene ACE1. Theor. Appl. Genet. 107, 1139–1147 (2003).
Silue, D., Tharreau, D. & Notteghem, J. L. Identification of Magnaporthe grísea avirulence genes to seven rice cultivars. Phytopathology 82, 1462–1467 (1992).
Sweigard, J. A., Chumley, F., Carroll, A., Farrall, L. & Valent, B. A series of vectors for fungal transformation. Fungal Genet. Rep. 44, 52–53 (1997).
Miki, D. & Shimamoto, K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45, 490–495 (2004).
Lei, Y. et al. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 7, 1494–1496 (2014).
Miao, J. et al. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23, 1233–1236 (2013).
Bohnert, H. U. A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 16, 2499–2513 (2004).
Foster, A. J. et al. CRISPR-Cas9 ribonucleoprotein-mediated co-editing and counterselection in the rice blast fungus. Sci. Rep. 8, 14355 (2018).
Toki, S. et al. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 47, 969–976 (2006).
Jalilian, A. et al. The RLCK subfamily VII-4 controls pattern-triggered immunity and basal resistance to bacterial and fungal pathogens in rice. Plant J. 115, 1345–1356 (2023).
Acknowledgements
Work in the lab of T.K. was funded by ANR projects Resistance Proteins (ANR-15-CE20-0007) and RePairs (ANR-21-CE20-0017). Work in the lab of B.Z. at IRRI was funded by Swiss National Science Foundation. M.B. was funded by an E.U. Marie Skłodowska-Curie individual fellowship (No. 896153). N.L. was funded by a PhD grant from the French Ambassy in Thailand and by Campus France. A.J. was supported by a visiting scholarship from the Iranian Ministry of Science, Research and Technology (28-11-2018). G.X. and J.W. were supported by the National Natural Science Foundation of China (32372505, U20A2021 and 32172422), the Major Science and Technology Project of Hunan Province (2021NK1001) and the Natural Science Foundation of Hunan Province of China (2021JJ30486).
Author information
Authors and Affiliations
Contributions
G.X., S.C., D.T., B.Z., J.W. and T.K. designed research. G.X., N.L., S.C., K.L., M.B., A.J., M.J.T.-Y., V.C. and I.M. performed research. G.X., N.L., S.C., K.L., M.B., A.J., E.F., D.T. and T.K. analysed data. G.X., S.C., B.Z., J.W. and T.K. acquired funding. G.X., N.L., S.C., M.B., D.T., B.Z. and T.K. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–7, legends of supplementary figures, and source data for Supplementary Figs. 2b and 3.
Supplementary Data 1
Source data for Supplementary Fig. 6a–c and for their statistical analysis in Supplementary Table 3.
Supplementary Tables
Supplementary Tables 1–6 containing supplementary information.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xiao, G., Laksanavilat, N., Cesari, S. et al. The unconventional resistance protein PTR recognizes the Magnaporthe oryzae effector AVR-Pita in an allele-specific manner. Nat. Plants 10, 994–1004 (2024). https://doi.org/10.1038/s41477-024-01694-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-024-01694-z
- Springer Nature Limited