Abstract
Collagen posttranslational processing is crucial for its proper assembly and function. Disruption of collagen processing leads to tissue development and structure disorders like osteogenesis imperfecta (OI). OI-related collagen processing machinery includes prolyl 3-hydroxylase 1 (P3H1), peptidyl-prolyl cis-trans isomerase B (PPIB), and cartilage-associated protein (CRTAP), with their structural organization and mechanism unclear. We determine cryo-EM structures of the P3H1/CRTAP/PPIB complex. The active sites of P3H1 and PPIB form a face-to-face bifunctional reaction center, indicating a coupled modification mechanism. The structure of the P3H1/CRTAP/PPIB/collagen peptide complex reveals multiple binding sites, suggesting a substrate interacting zone. Unexpectedly, a dual-ternary complex is observed, and the balance between ternary and dual-ternary states can be altered by mutations in the P3H1/PPIB active site and the addition of PPIB inhibitors. These findings provide insights into the structural basis of collagen processing by P3H1/CRTAP/PPIB and the molecular pathology of collagen-related disorders.
Similar content being viewed by others
Introduction
Collagens are a family of ubiquitous proteins in mammals, representing approximately 30% of the total protein content in the human body1. The collagens comprise 29 members to date and play crucial roles in providing biological functions and physical support for organisms. The collagens assemble with various proteins like proteoglycans, laminins, and fibronectins on the exterior surface of cells to form the major organic matter of the extracellular matrix (ECM)2. In general, all 29 subtypes of collagens are homo- or hetero-trimers comprising three α-chain collagens3,4. For instance, type I collagen (Col-I), which represents the major component of the ECM in human tissues such as skin, tendon-bone, and vasculature, consists of two α1 chains and one α2 chain encoded by COL1A1 and COL1A2, respectively. Mutations in COL1A1 and COL1A2 have been linked to various diseases in embryogenesis and adult development, such as osteogenesis imperfecta (OI) and osteoporosis5,6,7.
The biosynthesis of collagens starts from the translocation of the newly translated collagen polypeptide chains (pro-α chains) into the endoplasmic reticulum (ER) lumen, where they are modified and processed to trimerize into a triple helix8. Collagens are proline-rich proteins, and the modifications of proline residues play critical roles in the formation of triple helix and the maintenance of stability. The post-translational modifications (PTMs) on prolines of collagens include prolyl hydroxylation and isomerization. Insufficient prolyl modifications can result in incorrect pro-α chain folding and defects in the intermolecular recognition and fibril assembly9,10. Various PTM enzymes, such as prolyl 4-hydroxylases (P4H), prolyl 3-hydroxylases (P3H), and peptidyl-prolyl cis-trans isomerases, are involved in collagen maturation8,11. Recent studies have shown a link between collagen-related diseases and mutations occurring at genes encoding proteins involved in the collagen I synthesis and assembly pathway, such as P3H1 and peptidyl-prolyl cis-trans isomerase B (PPIB). P3H1 and PPIB were identified as components of an ER-associated ternary complex consisting of P3H1/CRTAP/PPIB12, hereafter designated as PCP complex. In the PCP complex, P3H1 is the core prolyl 3-hydroxylase, specially hydroxylating Pro986 (Pro1164 as numbering in CDS) of the triple-helical domain of collagen type I α1 chain (COL1A1), with the assistance of the cofactors Fe2+ and consuming co-substrates 2-oxoglutarate (2OG) and O213,14. PPIB takes part in the Col-I folding activity by cis/trans isomerization of X-Pro bonds to facilitate the triple helix formation, which could be inhibited by cyclosporin A (CsA), a fungal toxin binding to the active site of PPIB and affecting the maturation of collagen15,16,17,18,19. The third component in the PCP complex is a cartilage-associated protein (CRTAP), a stabilizing factor for PCP complex formation12,13. Pathogenic mutations on the CRTAP gene have also been reported to cause OI in humans20,21,22,23. Taken together, the close associations of osteogenesis imperfecta with all the components of the PCP complex suggest the complex functions as a whole in collagen processing and maturation. Despite the establishment of the PCP complex as a protein complex based on cell-level and co-purification studies8,24, only crystal structures for PPIB were reported16, leaving the overall architecture, complex assembly interactions, and catalytic/regulatory mechanisms unclear.
In this study, we present the cryo-electron microscopic (cryo-EM) structures of the collagen processing machinery, namely the P3H1/CRTAP/PPIB complex, in both the canonical ternary configuration (heterotrimer) and an unexpected dual-ternary configuration (hetero-hexamer). Our findings unveil the hydroxylation-isomerization reaction center at the interface between P3H1 and PPIB. Additionally, through further cryo-EM analysis of the PCP complex supplemented with synthetic peptides that mimic the substrate region of collagen, we observe the existence of multiple collagen recruitment sites within the PCP complex. These findings suggest a coupling mechanism for collagen processing by the PCP complex.
Results
Cryo-electron microscopic analysis on protein complexes of P3H1/CRTAP/PPIB
A stable protein complex comprising all three components was generated through transient co-transfection of Expi293F cells with expression vectors for human P3H1, strep-tagged CRTAP, and flag-tagged PPIB, as estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Supplementary Fig. 1a). Purification of the proteins involved a two-step affinity chromatography approach with the strep tag on CRTAP and the flag tag on PPIB, followed by size-exclusion chromatography (SEC). The protein samples obtained from two eluting peaks, the peak with an elution volume of about 15.1 ml (peak HW) and the peak with an elution volume of about 16.4 ml (peak LW), were pooled separately (Supplementary Fig. 1d). Electron microscopy data were acquired for both protein samples using a Titan Krios transmission electron microscope (FEI) operated at 300 kV. The data were processed using RELION3 and cryoSPARC25,26. Two-dimensional classification of the particles revealed the characteristic features of the ternary complex in peak LW and the features of a dual-ternary complex in peak HW (Supplementary Figs. 2a and 3a). Three-dimensional classification and refinement resulted in an electron microscopy density map with an overall resolution of 3.37 Å and a local resolution ranging from 2.0 Å to 2.6 Å at the core region of the complex for peak LW (Supplementary Fig. 2d) and an overall resolution of 3.62 Å and a local resolution ranging from 2.3 Å to 4.7 Å at the core region of the complex for peak HW (Supplementary Fig. 3). A similar protein production, data collection, and processing protocol was followed for Expi293F cells co-transfected with expression vectors for human P3H1 and CRTAP, resulting in an electron microscopy density map with an overall resolution of 3.65 Å and a local resolution ranging from 2.6 Å to 4.8 Å at the core region of the complex (Supplementary Fig. 4). To gain further insights into the catalytic mechanism of the PCP complex, the co-purified P3H1/CRTAP/PPIB protein samples were supplemented with CsA, 2-oxoglutarate (2OG), and synthetic peptides mimicking the collagen processing substrate. Cryo-EM structures were determined for these samples, achieving resolutions of 3.75 Å, 3.17 Å, and 3.18 Å, respectively (Supplementary Figs. 5–7, Supplementary Table 1).
The architecture of the PCP ternary complex
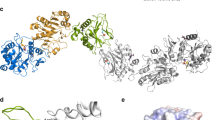
The EM analysis of the protein complex obtained from peak LW showed that human P3H1, CRTAP, and PPIB assemble into a ternary protein complex with 1:1:1 stoichiometry (Fig. 1a, b). The PCP ternary complex has a total molecular weight of approximately 150 kilodaltons and spans over 110 Å along its long axis. The CRTAP and the N-terminal domain (NTD) of P3H1 formed an “X”-shaped base (referred to as the X-base), with PPIB and the C-terminal domain (CTD) of P3H1 positioned on top of the X-base. The P3H1CTD (residues 442-700) occupied one side perpendicular to the plane formed by the X-base, while the corresponding region in CRTAP was absent, creating a binding site for PPIB and resulting in the formation of the P3H1/CRTAP/PPIB ternary complex (Fig. 1c). The X-base of the PCP complex emerges as the primary source of structural stability for the entire complex. A gelatin affinity assay employing P3H1, CRTAP, and PPIB gene knockout cell lines, revealed the essential role of both P3H1 and CRTAP in the collagen recruitment activity of the PCP complex (Fig. 1f). Conversely, the knockout of the PPIB gene showed limited effects on the binding of P3H1 and CRTAP to collagen. These findings emphasize the critical contributions of P3H1 and CRTAP to the collagen-binding function of the PCP complex, underscoring their significance in the overall collagen processing machinery.
a The cryo-EM map of the heterotrimeric complex of human P3H1/CRTAP/PPIB (PCP ternary complex) viewed from three angles. b The structural model of the PCP ternary complex depicted as cartoon models, colored according to the corresponding cryo-EM maps shown above. The iron atom is represented as a gray sphere. c Pseudosymmetry observed in the PCP ternary complex. The structural model of the PCP ternary complex is presented as cartoon models. The cartoon models for both P3H1CTD (P3H1442-700) and PPIB were set to transparent for a clear observation of the pseudo-C2-symmetry between P3H1NTD (P3H131-429) and CRTAP. d The structural analysis on CRTAP (left), P3H1 (middle frame), and PPIB (right) in PCP ternary complex. Left: The structure of CRTAP is depicted as a cartoon model, colored in blue, except for the helix pairs of TPR repeats: TPR1 (yellow), TPR2 (orange), TPR3 (green), and TPR4 (cyan). Middle frame: The cartoon representation of P3H1 is viewed from the same relative angle as CRTAP (left in the frame) and the opposite angle (right in the frame). To facilitate clarity, P3H1 CTD and NTD were set to transparent on the left and right of the frame, respectively. Right: The structure of PPIB is depicted as a cartoon model, colored in green, except for the β-strands colored in purple and pink. e Schematic diagram illustrating the organization of secondary structures in CRTAP (upper left), PPIB (lower left), and P3H1 (right). f Collagen recruitment of P3H1, CRTAP, and PPIB. Left: Representative immunoblot results of lysates from 293 T cells with individual knockouts of P3H1, CRTAP, and PPIB genes (two independent KO cell lines for each gene, respectively). Wild-type 293 T cells were used as a control. Right: Representative immunoblot results showing the pulldown of P3H1, CRTAP, and PPIB using Gelatin Sepharose. The representative results from three independent experiments were shown. g 2-OG dependent oxygenase activities of the purified PCP complex. (n = 3 biologically independent samples). Data are presented as mean values +/- SD. Source data are provided as a Source Data file.
Sequence-based prediction indicates that the P3H1CTD belongs to the 2OG-Fe (II) oxygenase superfamily, which catalyzes oxidation in a heme-independent manner with the assistance of bound Fe2+. Structural analysis of the P3H1 CTD revealed a typical “jelly roll” motif characterized by double-stranded β-sheets, namely β-strands a-b-c-l-i-n-e-d and k-j-m-f-g-h (Fig. 1d, e). In addition, the structural superposition of P3H1CTD with other members of the 2OG-Fe(II) oxygenase superfamily, such as hypoxia-inducible factor prolyl hydroxylase PHD1 and PHD227,28, confirmed the assignment of Fe2+ to the non-proteinous electron density hold in the jelly roll motif of P3H1 CTD and coordinated with residues D589, H587, and H659 (Fig. 1d, Supplementary Figs. 10c & 10d). Functional assays examining the 2OG-Fe (II) oxygenase activities demonstrated the oxidation of 2OG by the purified PCP ternary complex, which was inhibited upon depletion of ferrous ions (Fig. 1g).
The P3H1 NTD and CTD are connected by a long N-C linker helix, H19, with an unresolved flexible loop between H19 and H20 (Fig. 1e). Sequence alignment between P3H1 and CRTAP revealed high conservation between P3H1NTD and CRTAP, with an identity of 36.18% between residues 1-420 of P3H1 and CRTAP (Supplementary Fig. 8a). In the cryo-EM structure of the PCP ternary complex, both P3H1NTD and CRTAP exhibit similar protein folding (Fig. 1d). As a result, the X-base shows C-2 pseudosymmetry with a pseudo-symmetric axis at its center between P3H1NTD and CRTAP (Fig. 1c). Both P3H1NTD and CRTAP contain approximately seventeen α-helices, comprising four tetratricopeptide repeats (TPR1-4) in both proteins, respectively (Fig. 1e). TPR is a helix-loop-helix hairpin structural motif and serves as a contact region mediating protein-protein interactions29. In PCP ternary complex, TPR1-4 were formed by helices-pairs H1-2, 6-7, 10-11, and 13-14 in P3H1, while similar helices-loop combinations were observed in the TPR1-4 of CRTAP, except for the H6-7 in P3H1 replaced by H7-8 in CRTAP (Fig. 1d, e).
The interfaces mediating the PCP ternary complex formation
The TPRs of both P3H1NTD and CRTAP are predominantly located at the four “ends” of the X-base and have minimal contribution to the complex formation (Fig. 2a). Instead, the helices 3, 12, and 17 of P3H1 and 4, 12, and 17 of CRTAP constitute the main interface (referred to as the X-interface) at the center of the X-base between P3H1NTD and CRTAP. This interface is characterized by electrostatic interactions such as R76P3H1-D258CRTAP, D106P3H1-R115CRTAP, D263P3H1-K123CRTAP, and F378P3H1-R114CRTAP (cation-π interaction) as well as a series of hydrogen bonds and hydrophobic interactions (Fig. 2d, e). Additionally, a smaller interface could also be identified between P3H1CTD and the helices H4&5 of CRTAP (referred to as the P3H1CTD-CRTAP interface). This interface involves interactions among the residue pairs D692P3H1-K120CRTAP, Q526P3H1-R130/Q131CRTAP, and E527P3H1-S132CRTAP (Fig. 2f). To investigate the significance of these key interacting residues in P3H1, mutagenesis experiments were performed, which revealed a decreased co-precipitation of P3H1 with CRTAP upon mutations at X-interface (R76A/D, R121A/D, D106A/R, and D263A/K of P3H1) (left panel of Fig. 2g). Furthermore, these mutations on P3H1 were found to reduce the binding of PPIB to CRTAP, too, suggesting the importance of the X-base for PPIB recruitment.
a Overall representation of the interactions among the P3H1, CRTAP, and PPIB subunits in the PCP ternary complex. The subunits are depicted as a cartoon model, viewed from two different angles. The secondary structures involved in complex formation are highlighted in red, blue, and green for P3H1, CRTAP, and PPIB, respectively. The interfaces between the components in the complex are indicated by dotted frames. b & c Enlarged views of the PPIB-CRTAP interface (b) and PPIB-P3H1 interface (c). CRTAP and P3H1 are shown as a cartoon model, while PPIB is represented as a transparent surface model. The contacting residues are displayed as a stick model, colored according to the elements. d & e Enlarged views of the X-interface, focusing on two different contacting regions. CRTAP and P3H1 are depicted as a cartoon model, and the contacting residues are shown as a stick model, colored by elements. f Enlarged view of the P3H1CTD-CRTAP interface. CRTAP is represented as a cartoon model, while P3H1 is shown as a transparent surface model. The contacting residues are displayed as a stick model, colored according to the elements. g The interactions among P3H1, CRTAP, and PPIB in the PCP complex. Left: The 293 T cells were co-transfected with CRTAP with strep tag, PPIB, and wildtype P3H1 or P3H1 with mutations at the interfaces of the PCP complex. Shown were the representative immunoblot results from three independent experiments of the pulldown of P3H1 and PPIB by the strep-tagged CRTAP using strep-tactin affinity resin, as well as the representative immunoblot results of lysates. Right: The 293 T cells were co-transfected with flag tagged-PPIB, P3H1, and strep-tagged CRTAP with mutations at the interfaces of the PCP complex. Shown were the representative immunoblot results from three independent experiments of the pulldown of P3H1 and PPIB by the strep-tagged CRTAP using strep-tactin affinity resin, as well as the representative immunoblot results of lysates. Source data are provided as a Source Data file.
The interactions between PPIB and the X-base are relatively limited, with its binding mediated by interactions between the positively charged lysine-rich surface of PPIB and the acidic residues from the C-terminal of CRTAP (Fig. 2b) and the N-terminal of P3H1 (Fig. 2c). These interactions include electrostatic interactions, such as the electrostatic interactions of E41P3H1-K84PPIB and D36P3H1-K71PPIB at the PPIB-P3H1 interface (Fig. 2c) and E386CRTAP-K38PPIB and D255CRTAP-K129PPIB at PPIB-CRTAP interface (Fig. 2b), along with hydrogen bonds and hydrophobic interactions. Mutagenesis experiments were conducted to assess the significance of the PPIB-CRTAP interface. CRTAP with mutations at the interface exhibited decreased binding to PPIB, while their interactions with P3H1 showed minimal changes (right panel of Fig. 2g).
The bifunctional reaction center with a “face-to-face” configuration of the hydroxylation site and isomerization site
The PCP complex comprises two components that have been reported to possess posttranslational modifying functions: P3H1CTD with the prolyl hydroxylase activity and PPIB with the prolyl isomerase activity. Previous structural and functional studies have elucidated the site of isomerization in PPIB through the binding of cyclosporin A (CsA), an immunosuppressant known for its inhibitory effect on PPIB15,16. In the cryo-EM map of the PCP ternary complex supplemented with CsA (referred to as the PCP-CsA complex), a distinct non-proteinaceous electron density with an annular shape was observed at the corresponding site of PPIB, and the molecular model of CsA fitted well within this density region (Fig. 3a, b). The superposition of the PPIB-CsA complex in the PCP complex with the previously reported crystal structure of PPIB-CsA revealed similar protein conformations and interactions between PPIB and CsA (Supplementary Fig. 9).
a The cryo-EM map of PCP-CsA complex. The density maps for P3H1, CRTAP, and PPIB were colored in red, blue, and green, with the non-proteinous electron density identified near PPIB colored in orange. b The binding of the CsA in the catalytic site of PPIB. The CsA is represented as a stick model colored by elements, while PPIB is displayed as a calculated solvent-accessible electrostatic surface potential map. Residues comprising the PPIB catalytic site are shown as stick models colored by elements (pink-red-blue), and gatekeeper residues are displayed as stick models colored by elements (green-red-blue). c The cryo-EM map of PCP-2OG complex. The density maps for P3H1, CRTAP, and PPIB were colored in transparent gray, blue, and green, respectively. The non-proteinous electron density within is depicted in green, and other non-protein electron densities are shown in yellow. d The binding of the 2-oxoglutarate (2OG) and iron atom in the catalytic site of P3H1. The 2OG was shown as stick model colored by elements, and the iron atom was shown as gray sphere. The P3H1 was shown as a red cartoon model, with the residues interacting with 2OG and iron atom shown as stick models. e The reaction center of the PCP complex. The P3H1CTD and PPIB in the PCP-CsA ternary complex were displayed as surface models, with a sliced view highlighting the “face-to-face” arrangement of the catalytic sites of P3H1 (indicated by a green outline) and PPIB (indicated by a yellow outline). The PCP-2OG complex was superposed with the PCP-CsA complex to illustrate the binding of 2OG in the P3H1 catalytic site. f 2-OG dependent oxygenase activities of the purified PCP complex with mutations at the PH and PI sites (n = 3 biologically independent samples). Data are presented as mean values +/- SD. Source data are provided as a Source Data file.
In the cryo-EM map of PCP ternary complex in the absence of substrate supplementation, the non-proteinous electron densities surrounding the jelly roll motif of P3H1CTD are sparse and weak, except for an apparent electron density corresponding to the Fe2+, which is confirmed by an ICP-MS (Inductively Coupled Plasma Mass Spectrometry) analysis (Supplementary Fig. 10b). To gain further insights into substrate binding in P3H1, we determined the cryo-EM structure of the PCP ternary complex supplemented with 2-oxoglutarate (2OG), the co-substrate involved in prolyl hydroxylation reactions12. In the cryo-EM map of the PCP-2OG complex, a continuous and elongated region of non-proteinous electron density was observed adjacent to the bound Fe2+ (Supplementary Fig. 10a), and the electron density accommodated the 2OG molecule well. Molecular modeling of 2OG was corroborated by superimposing the structure of P3H1CTD with two well-known prolyl hydroxylases, HIF prolyl hydroxylases 1 and 2 (PHD1 and PHD2)27,28. Despite relatively low sequence identities between P3H1CTD and PHD1/PHD2 (approximately 20.8% and 24.6% identities, respectively), the superposition revealed structural conservation in their jelly roll motifs and the arrangement of catalytic residues around the metal ion and 2OG binding (Supplementary Figs. 10c and 10d). The binding pocket of P3H1CTD is located between two sets of β-sheets within the jelly roll motif, and the interaction between the 2OG molecule and P3H1 CTD is mediated by electrostatic interactions involving the 1-carboxyl and 2-keto groups with the Fe2+ ion, as well as the 5-carboxyl group with Arg 569 and Arg 669 (Fig. 3d). While the active mutants P3H1D589A, P3H1H587A, P3H1R669A, and P3H1H659A could form PCP complex with CRTAP and PPIB (Supplementary Fig. 1c), these mutant complexes exhibited significantly decreased 2OG-dependent oxygenase activities (Fig. 3f). Additionally, we identified an OI-related mutation, W675L, located near the active site30, and the PCP complex with mutation W675LP3H1 also showed reduced enzymatic activities (Fig. 3f), suggesting impaired collagen hydroxylation function in affected patients. Remarkably, the proximity of the two crucial catalytic sites, namely the prolyl isomerase site of PPIB (PI-site) and the prolyl hydroxylase site of P3H1 (PH-site), was observed in a face-to-face configuration at the complex level (Fig. 3e). Notably, a narrow slit is formed between the P3H1CTD and PPIB on X-base, where the entrances to the PH- and PI-sites reside on the sides facing the slit. The substrate-binding pockets of P3H1CTD and PPIB are separated by an approximate distance of 20 Å from the geometric centers of their respective pockets. This spatial co-localization of the PI-site and PH-site establishes a bifunctional reaction center, potentially facilitating highly efficient substrate processing.
The collagen binding sites in the PCP complex
The cryo-EM analysis on the PCP complex revealed the presence of non-proteinaceous electron density near CRTAP, a non-enzymatic component of the PCP complex implicated in collagen recruitment, as suggested by Gelatin-Sepharose affinity assay (Fig. 1f). To further investigate collagen recognition and capture by the PCP complex, we designed peptides that mimicked the substrate region in the pro-α peptide of collagen and incubated the PCP complex with these synthetic peptides for electron microscopy analysis. Non-proteinaceous electron densities were observed at multiple positions in the PCP complex supplemented with collagen-derived peptides. Notably, a long continuous blob of electron density was identified at the exact location in the cryo-EM map of the PCP complex supplemented with a 21-amino acid synthetic peptide corresponding to residues 1154-1174 of the collagen α-1(I) chain (COL1A11154-1174), indicating the procollagen recruitment function of CRTAP (Fig. 4a). In isothermal titration calorimetry (ITC) assays, COL1A11154-1174 demonstrates an affinity for the purified PCP complex with an apparent dissociation constant (Kd) of approximately 0.2 mM (Supplementary Figs. 11a and b). Due to the limited size and resolution of the map for the peptide-bound complex, modeling the full-length 21-amino acid peptide is challenging. Nevertheless, the central part of the COL1A11154-1174 (1159-PGPIGPPGPR-1168) fits well in the density.
a Cryo-EM map of PCP complex supplemented with COL1A11154-1174 synthetic peptide (PCP-COL1A1). The non-proteinous electron density is depicted as a green surface model. b The binding of COL1A11154-1174 peptide with CRTAP. CRTAP is shown as a blue cartoon model, while the structural model of residues 1159−1168 of human COL1A1 and its interacting residues on CRTAP are represented as a stick model colored by elements. c The collagen-binding sites (CBS) on the PCP complex. Left: The cryo-EM map of the PCP-COL1A1 complex is sliced to reveal the non-protein electron densities (in green). Right: Two side views of the cryo-EM map of the PCP-COL1A1 complex. d The substrate processing path on the PCP complex. Left: The PCP complex is depicted as a transparent surface model and cartoon model, with the non-protein electron densities represented as a green mesh. The surface model is viewed and sliced as shown in (c) to clearly visualize the reaction center/CBS5. CBS1, 2, and 5, as well as the intervening surfaces between them, are highlighted in yellow. Right: PCP complex is shown as a transparent surface model, with the non-protein electron densities depicted as solid green surfaces. A yellow dotted zone indicates the putative extended substrate processing path connecting CBS1-5. e Representative immunoblot results showing the Gelatin Sepharose-pulldown of PPIB and CBS mutants of P3H1 and CRTAP. The representative results from three independent experiments were shown. Source data are provided as a Source Data file.
In the structure of the P3H1/CRTAP/PPIB/COL1A11154-1174 quaternary complex (referred to as the PCP-COL1A1 complex), the binding of collagen peptide to CRTAP is primarily mediated by the hydrophobic interactions. The hydrophobic residues from TPR1 and 2 motifs in CRTAP, e.g., Y55, F150, F153, and F156, stabilize the isoleucine and proline residues in the COL1A11154-1174 (Fig. 4b). Apart from the long blob of electron density, four additional continuous electron densities with smaller sizes were also identified (Fig. 4c and Supplementary Fig. 11c). Although modeling the COL1A11154-1174 peptide into these electron densities was challenging due to size and resolution limitations, the general shape of the four unassigned electron densities, along with the one modeled with COL1A11159-1168, suggests the presence of multiple binding sites for collagens. Therefore, these locations on the PCP complex were designated as Collagen Binding Sites 1-5 (CBS1-5). The CBS1-4 are sitting on the X-base with symmetric distribution, i.e., CBS1 and 2 near the TPR1&2 motifs of CRTAP and P3H1, respectively, and the CBS3 and 4 near the TRP3&4 motifs of CRTAP and P3H1, respectively (Supplementary Fig. 11c). The close proximity of CBS1-CBS5/reaction center-CBS2 on top of the X-base, along with the nearly linear arrangement of the bound electron densities, implies the existence of a substrate interacting zone on the surface of the PCP complex (Fig. 4d). This elongated, belt-like zone is centered around the slit of the bifunctional reaction center formed by P3H1CTD and PPIB, extending symmetrically through the TPR1-2 motifs (CBS1) to the TPR3-4 motifs (CBS3) of CRTAP on one side, and through the TPR1-2 motifs (CBS2) to the TPR3-4 motifs (CBS4) of P3H1 on the other side. Notably, the ITC experiment suggests a stoichiometry closer to 2 (Supplementary 11a), which contrasts with the stoichiometry of 5 CBSs proposed based on the cryo-EM maps. To further assess the collagen-binding capacity of these sites, gelatin-affinity assays were conducted on the PCP complex with mutations at CBS1-4. The results show that mutations at any of these sites impair collagen recruitment by the PCP complex (Fig. 4e), although the overall structure of the ternary complex remains similar to the wild type (Supplementary Fig. 12). Based on the combined EM analysis, ITC results, and gelatin-affinity assays, we suggest that CBSs other than CBS1 and CBS2 may also serve as potential collagen-binding sites to recognize segments flanking the substrate sequence of PCP complex. However, to rule out the potential contribution of biologically irrelevant molecules, such as detergent from sample preparation, further experimental validations are needed. These should include elucidating a native complex structure to confirm the binding between the PCP complex and collagen proteins.
The dual-ternary complex formed by the dimerization of the PCP complex
The purification of the human PCP complex revealed the possibility of a higher-order oligomeric state, as indicated by size-exclusion chromatography (Supplementary Fig. 1d). Further Native-PAGE analysis on cell lysates confirmed the existence of both the dual-ternary complex upon overexpression and endogenous dual-ternary complex (Supplementary Fig. 13). The cryo-EM analysis of the sample from HW fractions resulted in an electron density map larger than that of the PCP ternary complex (Supplementary Fig. 3). Molecular modeling studies demonstrated that this complex consists of two copies of the PCP ternary complex, hence referred to as the PCP dual-ternary complex or dual-PCP (Fig. 5a). The two copies of PCPs in the dual-PCP complex, subcomplex I and II (PCPI and PCPII), exhibited an overall architecture similar to the PCP ternary complex but interestingly assembled in an asymmetric manner (Fig. 5a and Supplementary Fig. 3d). The inter-subcomplex contacts were primarily mediated by interactions between two P3H1 protomers at the P3H1I-P3H1II interface, as well as between CRTAP of PCPI and PPIB of PCPII at the CRTAPI-PPIBII interface. Notably, compared to the PCP ternary complex (Fig. 5b), one of the major conformational changes observed in the dual-PCP complex was the tilting of helix 19 in P3H1I (N-C linkerI) which extends to the vicinity of TPR1 and 2 of P3H1II and established close interactions with helices 2 and 7 through electrostatic interactions involving P3H1I E420 - P3H1II R60 and P3H1I E427 - P3H1II R63 (Fig. 5d). Another notable conformational change involved the displacement of the C-terminal domains (CTDs) of both P3H1 protomers. Due to the assembly manner of the two sub-complexes, spatial conflicts arose between both PPIBs and the CTD of the P3H1 protomer from the other subcomplex, respectively (Fig. 5b). In the structural model of the dual-PCP complex, both P3H1 CTDs were “forced” out of their original positions. However, the cryo-EM map for the dislocated P3H1CTD could not be resolved, likely due to the loss of stabilizing effects from its interaction with PPIB in the PCP ternary complex.
a The cryo-EM map of the hexameric PCP complex viewed at different angles. The electron density corresponding to the different components of the dual-PCP complex was colored as the scheme shown below. b The spatial clashes of the location of P3H1CTD (shown as gray surface) in the PCP ternary complex with the neighboring subcomplex in the dual-PCP ternary complex. The PCP ternary complex was superposed with the PCPII and PCPI of the dual-PCP ternary complex in the upper and lower panel, respectively. The dual-PCP ternary complex was shown as cartoon model colored to the scheme in (a) and the PCP ternary complex was shown as cartoon model in gray, with the P3H1CTD shown as surface model in gray. c The assembly of two subcomplexes in the dual-PCP ternary complex. The dual-PCP ternary complex was shown as a cartoon model and viewed from two angles, and the interfaces between PCPI and PCPII were highlighted with dotted frames. d An enlarged view of the interfaces shown in (c). The contacting residues in the CRTAPI-PPIBII interface and P3H1I-P3H1II interface were shown as stick model and colored by elements.
The identification of a hetero-hexameric form of the PCP complex represents an unexpected conformation not previously reported. Its biological relevance remains to be elucidated. Size exclusion chromatography (SEC) experiments demonstrated that the oligomeric states could vary depending on the enzymatic activities of the PCP complex. The addition of the PPIB inhibitor CsA resulted in a single elution peak in SEC analysis, corresponding to the trimer, indicating the crucial role of PPIB activity in hexameric complex formation (Supplementary Fig. 1h). Introduction of an enzymatic mutation at the PI-site, R95A, produced a PCP complex with a similar trimer-only SEC profile, further supporting the role of PPIB activity in hexamer formation (Supplementary Fig. 1h). Interestingly, when PPIB was excluded from co-transfection in PCP complex overexpression, a purified P3H1/CRTAP binary complex was obtained (Supplementary Figs. 1b and 1e), and cryo-EM analysis revealed its 3.65-Å structure (Supplementary Fig. 4).
The P3H1/CRTAP binary complex displayed an overall architecture and component conformation similar to the PCP ternary complex, except for the absence of PPIB (Supplementary Fig. 14). Neither SEC nor cryo-EM analyses suggested a higher oligomeric state in the case of P3H1/CRTAP co-expression, suggesting PPIB is essential for hexamer formation but not for P3H1-CRTAP association. Meanwhile, we observed elevated P3H1 activities in PPIB-free (binary complex) or -inhibited (PPIBR95A) state (Fig. 3f), and a slightly lower P3H1 activity could be observed in HW fractions compared with that in LW fractions from SEC-FPLC (Supplementary Fig. 1f), implying the ternary complex might represent a conformation more active than the dual-ternary complex. However, this hydroxylation enhancement was absent in the PCP complex supplemented with PPIB inhibitor CsA. We speculate that this could result from steric hindrance due to CsA binding in the reaction center. The dynamic balance of the PCP complex between its ternary and dual-ternary states makes it difficult to distinguish their respective processing activity levels precisely. However, SEC analysis revealed that mutations in the PH-site favor the higher oligomeric state (Supplementary Fig. 1g), while PI-site mutation R95A stabilizes the ternary state while enhancing P3H1 activities (Fig. 3f and Supplementary Fig. 1h). These findings, coupled with the displacement of catalytic P3H1CTD from its original position in the “face-to-face” reaction center, implied that the dual-ternary state might function as a low-activity reservoir for the enzymatic complex due to the disassembled “face-to-face” configuration in PCP hexamer.
Although these results suggest a potential relationship between the balance of ternary and dual-ternary states and the collagen processing functions of the PCP complex, we did not observe a significant shift in this balance upon the addition of synthetic peptides in SEC analysis (Supplementary Fig. 1i). However, it is important to note that this observation does not preclude the possibility of collagen binding regulating the ternary/dual-ternary balance, as the synthetic peptides used in our study may not fully mimic the native procollagen peptide chain. Therefore, further investigations are warranted to study the shift and balance of PCP oligomeric states in a context closer to the native ER environment, as well as the specific enzymatic activity of different oligomeric states.
Discussion
The P3H1/CRTAP/PPIB complex represents multi-functional collagen-processing machinery, its roles encompassing prolyl residue isomerization, hydroxylation, and molecular chaperoning during collagen maturation. Although the physical interaction among these crucial collagen-processing factors has been documented, the lack of structural information hampers our understanding of the coordination and mechanistic linkage within this molecular machinery, as well as the molecular pathology of related OI conditions. Our findings reveal a collagen processing machinery characterized by a bifunctional reaction center situated atop an X-shaped base. Within this architecture, the prolyl isomerase and hydroxylase sites are closely localized, with a distance of approximately 20 Å between the centers of the two sites. Although the conformation of the PCP complex with a substrate peptide bound in the reaction center is yet to be revealed for elucidation of the catalytic mechanism, this close spatial configuration, along with the density of the non-PCP components identified in the bifunctional reaction center, suggests a coupled modification mechanism for collagen processing. In this proposed mechanism, the substrate sequence covering the prolyl to be modified, along with the flanking residues, would enter the bifunctional reaction center for efficient hydroxylation and isomerization in the ER lumen. While this bi-functional reaction center might not be necessary for P3H1 for its hydroxylase activity, as suggested by its normal catalytic activities upon the inhibition of PPIB or in the PPIB-free state (PC binary complex), it could be critical for PPIB’s function. This is indicated by the significantly lower activity towards the hydroxylated substrate observed in the free active PPIB, suggesting the role of substrate recruitment of the PCP complex for prolyl/hydroxyprolyl isomerization by PPIB. Certainly, the coupled processing mechanism would not exclude the possibility of PPIB independently catalyzing the prolyl isomerization without hydroxylation. An extensive survey of the alternative substrates of the PCP complex and their structural characterization would be of research interest in the future.
A surprising revelation from our structural analysis is the existence of the PCP dual-ternary complex. Through electron microscopy and biochemical investigations, we demonstrate that the ternary and dual-ternary states can coexist and undergo interconversion. The equilibrium between these states is dependent on the enzymatic activities of the PCP complex. For instance, specific mutations at the P3H1 active site, such as H659A and D589A, induce a ternary-free state, wherein only the dual-ternary complex or even higher oligomeric complexes are present (Supplementary Fig. 1g). Furthermore, we observed that the OI-related mutation W675L30, located near the active site, results in significantly reduced 2OG-dependent oxygenase activities and a ternary-free state, as confirmed by SEC analysis (Supplementary Fig. 1g). Considering the crucial role of P3H1CTD displacement in hexamer formation, as suggested by structural comparisons between the PCP complex in the ternary and dual-ternary states (Fig. 5b), it is plausible that the active site mutations promote hexamer formation by destabilizing P3H1CTD in the trimeric PCP, thereby favoring the transition of the PCP complex from the ternary to the dual-ternary state. In addition to the impact of P3H1 mutations on the ternary/dual-ternary balance, we also observed an intriguing inverse effect of PPIB activity on the PCP complex. Both the addition of a PPIB inhibitor and the active site mutation of PPIB led to a ternary-only state, in which the majority of the PCP complex adopts a trimeric configuration. The functional significance of this transition between the trimeric and hexameric forms of the PCP complex remains unclear. However, our findings suggest that the activities of P3H1 and PPIB act as reciprocal regulatory factors that drive the cycle of trimer-hexamer transition during substrate processing of the PCP complex (Fig. 6), and the PCP dual-ternary complex might serve as a low-activity pool where the P3H1 is in a non-catalytic state. Further investigations are warranted to elucidate the precise functional interplay between P3H1 and PPIB in the regulation of the PCP complex and its role in collagen processing.
The assembly of the PCP complex initiates with the association between P3H1 and CRTAP, resulting in the formation of the binary complex. Subsequently, PPIB joins the binary complex to form the PCP ternary complex. A dynamic equilibrium between the ternary and dual-ternary conformations may exist, regulated by the enzymatic activities of P3H1 and PPIB. This suggests the potential role of an inactive pool for the PCP dual-ternary complex.
While CRTAP shares a protein sequence homology of 36% with P3H1, it lacks the C-terminal oxygenase catalytic domain (Supplementary Fig. 8). Consequently, CRTAP has not been associated with any enzymatic activity thus far. However, CRTAP plays a crucial role in stabilizing the PCP complex and facilitating the 3-prolyl hydroxylation activities of P3H122,31. The deficiency in 3-hydroxylation at Pro986 and bone development defects observed in patients carrying mutations in CRTAP and Crtap-/- mice further support the importance of CRTAP in collagen processing. Our structural findings clearly demonstrate how CRTAP implements its two significant functions within the PCP complex: complex maintenance and substrate recruitment. As a major component of the X-shaped base of the PCP complex, both CRTAP and P3H1 are indispensable for the formation of a functional collagen processing enzyme complex. Mutations at the X-interface, such as R121A/DP3H1, D106A/DP3H1, and R76DP3H1, significantly decrease the formation of the PCP complex, while the mutations at CRTAP-PPIB interface, such as D255A/KCRTAP and E386A/KCRTAP, disrupt the recruitment of PPIB onto the P3H1/CRTAP complex (Fig. 2g). Additionally, our PCP-COL1A1 complex structure reveals that the TPR1-2 domain of CRTAP serves as the primary site for recruiting collagen peptides. OI-related mutations identified in CBSs, such as L67P, L151P, and K157E20,23,32 in CBS1, and G219S in CBS333, highlight the critical role of CBS1/3 of CRTAP in collagen processing. In contrast, to date, no disease-related mutation has been found in CBS2/4 of P3H1. All four mutations could lead to low collagen-binding of the PCP complex, as shown in Supplementary Fig. 15. These observations underscore the essential contributions of CRTAP to the proper functioning of the PCP complex, both in terms of maintaining the complex structure and facilitating collagen substrate recruitment.
Methods
Protein expression and purification
Recombinant human PPIB was successfully produced from Escherichia coli, whereas full-length human CRTAP and P3H1 could not be overexpressed with appropriate thermostability in either E. coli or the baculovirus/insect cell system. Therefore, recombinant expression of all genes of interest was carried out to conduct structural and functional studies in a HEK293-based system. The coding cassette for Human P3H1 (P3H1a isoform, UniProt ID Q32P28-1) was synthesized and subsequently cloned into the pcDNA3.4 vector (Thermo Fisher) without any tag. The cDNAs of human CRTAP and PPIB were obtained from CCSB-Broad Lentiviral Expression Library. CRTAP was then cloned into a modified pcDNA3.4 vector with an HRV 3 C Protease cleavage site and a carboxy-terminal twin strep tag at the C-terminal of CRTAP. PPIB was cloned into a modified pcDNA3.4 vector, incorporating a Tobacco Etch Virus (TEV) protease cleavage site, followed by a 3×FLAG tag and an octameric poly-histidine tag at the C-terminal of PPIB. The QuikChange II Site-directed mutagenesis kit was utilized to introduce point mutations, insertions, or deletions in the constructs.
For recombinant expression, P3H1, CRTAP, and PPIB were co-expressed in Expi293F cells (Thermo Fisher, A14527) cultured in chemically defined Union-293 medium (Union-Biotech, UP0050) at 37 °C with 5% CO2 in an incubator shaker. Typically, when the cell density reached about 2.5 × 106 cells ml-1 with 95% viability, plasmids for overexpression of P3H1, CRTAP, and PPIB in 1:1:1 molar ratio (wild-type or the variants) were transiently co-transfected into the cells using polyethylenimine (PEI, Polysciences). For 1 liter of cell culture, 1 mg of plasmids were gently mixed with 3 mg PEI in 100 ml Opti-MEM reduced serum medium for 20 min before transfection. For the P3H1/CRTAP binary complex, plasmids for overexpression of P3H1 and CRTAP were transiently co-transfected into the cells. Following transfection, the cells were cultured for 72 hours before harvesting. The transfected Expi293F cells were collected and then resuspended in lysis buffer (150 mM NaCl, 20 mM HEPES pH7.4, 10% glycerol) supplemented with 4 mM MgCl2, 0.2 mg mL-1 Dnase I, and 1× protease inhibitor (MCE).
To purify the PCP complex or P3H1/CRTAP binary complex, a two-step affinity purification method was employed at 4 °C or under ice bath conditions. Initially, the cell suspension was sonicated on ice, followed by centrifugation at 46,000 g for 45 minutes at 4 °C to remove cellular debris. The resulting supernatant was collected and subjected to Strep-Tactin resin affinity chromatography. The protein complex was eluted using lysis buffer supplemented with 50 mM D-biotin (B105433, Aladdin Biochemical). The eluents were then subjected to a second round of affinity chromatography using anti-Flag beads (GeneScript), and the proteins were eluted with lysis buffer supplemented with 250 μg mL-1 Flag peptide. In the case of the P3H1/CRTAP complex, a single-step affinity purification was performed. Subsequently, the protein was concentrated and further purified using size-exclusion chromatography with a Superose 6 Increase 10/300 GL column (Cytiva), pre-equilibrated with a buffer containing 150 mM NaCl and 20 mM HEPES pH 7.4. For the purification of the PCP-CsA and PCP-2OG complexes, the mobile phases of the size-exclusion chromatography were supplemented with 20 μM Cyclosporin A (HY-B0579, MCE) and 2 mM 2-oxoglutarate (K1875, Sigma), respectively. Peak fractions were collected, pooled, and concentrated to approximately 6 mg mL-1 for the peak with an elution volume of about 15 mL (peak HW) and 15 mg mL-1 for the peak with an elution volume of about 16 mL (peak LW) for cryo-EM sample preparation. In the case of the P3H1/CRTAP binary complex, the peak fractions were pooled and concentrated to approximately 10 mg mL-1 for cryo-EM sample preparation.
Before cryo-EM sample preparation, the HW PCP complex was supplemented with 0.05% (w/v) CHAPS (Anatrace, 75621-03-3) as a protective agent, while the LW-PCP complex was supplemented with 3 mM fluorinated Fos-choline 8, (Anatrace, 313997-22-7) as a protective agent. For the cryo-EM sample preparation of the PCP-COL1A1 complex, 1 mM synthetic peptides (GL Biochem) were added to the LW-PCP complex and incubated for 30 minutes before the addition of fluorinated Fos-choline 8.
Cryo-EM sample preparation and data collection
Three-microliter aliquots of the samples were applied to glow-discharged holey carbon grids (Quantifoil R1.2/1.3 Au, 300 mesh), and after 20 s of incubation, the grids were blotted for 1 s and rapidly plunged into liquid ethane cooled by liquid nitrogen, using a Vitrobot Mark IV (FEI) operated at 8 °C and 100% humidity. The grids were imaged using a Titan Krios transmission electron microscope (FEI) operated at 300 kV, with the specimen maintained at liquid nitrogen temperatures. Images were automatically collected with EPU software (FEI) on a K3 camera (Gatan) operated in super-resolution counting mode, placed at the end of a GIF Quantum energy filter (Gatan), functioning in zero-energy-loss mode with a slit width of 15 eV. Data was typically collected at a nominal magnification of 81,000 (corresponding to a physical pixel size of 1.1 Å), with a defocus range between -1.5 and -2.6 μm. The dose rate was set to ~15.7 electrons/Å2/s, and the total exposure time was 3.85 s, resulting in a total dose of 50 electrons/Å2, fractionated into 32 frames.
Cryo-EM data processing
For the PCP ternary complex dataset, a total of 3,754 cryo-EM images were collected, and motion correction was performed on the dose-fractioned image stacks using MotionCor2 with dose-weighting34,35. The contrast-transfer function (CTF) parameters of each image were determined with Gctf36, and automatic particle-picking was carried out using Gautomatch-v0.56 (https://www2.mrc-lmb.cam.ac.uk/download/gctf_gautomatch_cu10-1-tar-gz/) on the basis of templates generated from a few hundred manually picked particles. Subsequent image processing steps were performed with RELION-3.137 and cryoSPARC25. An overview of the data processing procedure is shown in Supplementary Fig. 2a. The particles were first extracted with 4× binning (4.4 Å/pixel), and then junk particles were removed by two rounds of 2D classifications. The remaining particles were re-extracted without binning (1.1 Å/pixel) and subjected to heterogeneous refinement using six initial references generated by cryoSPARC. The particles corresponding to the best class are further processed with non-uniform refinement and 3D auto-refinement with C1 symmetry using cryoSPARC and RELION-3.1, respectively. After the first-round 3D auto-refinement, CTF refinement, and Bayesian polishing with RELION-3.1, a map with an overall resolution of 3.42Å was generated. A further 3D classification and the second-round 3D auto-refinement, CTF refinement with RELION-3.1, were performed. Finally, a total of 1,047,611 good particles were subjected to 3D auto-refinement, which generated a map with a high resolution.
For the PCP dual-ternary dataset, a total of 5423 cryo-EM movie stacks were collected. After motion correction and CTF estimation in RELION, automatic particle-picking with Gautomatch was performed. An overview of the data processing corresponding to this dataset is shown in Supplementary Fig. 3a. Briefly, the particles were first extracted with 4× binning, and junk particles were removed by two rounds of 2D classifications. The remaining particles were re-extracted without binning and then subjected to heterogeneous refinement in cryoSPARC. The particles corresponding to the best class were further processed with several rounds of heterogeneous refinement and non-uniform refinement, which generated a map with an overall resolution of 3.96Å. After 3D auto-refinement, CTF refinement, and Bayesian polishing were performed with RELION, the polished particles were re-subjected to cryoSPARC, and then non-uniform refinement with C1 symmetry was performed, thereby yielding a high-resolution map.
For the PCP binary complex dataset, a total of 3255 cryo-EM movie stacks were collected. After motion correction and CTF estimation in RELION, automatic particle-picking with Gautomatch was performed. An overview of the data processing corresponding to this dataset is shown in Supplementary Fig. 4a. Briefly, the particles were first extracted with 4× binning, and junk particles were removed by two rounds of 2D classifications. The remaining particles were re-extracted without binning and then subjected to heterogeneous refinement in cryoSPARC. The particles corresponding to the best class were further processed with multi-rounds of heterogeneous refinement and non-uniform refinement using cryoSPARC. The good particles were re-subjected to RELION for further 3D classification to exclude the particles of the ternary complex for the contaminated endogenous PPIB. After 3D auto-refinement and Bayesian polishing with RELION, a total of 162,963 good particles were re-subjected to cryoSPARC, and then non-uniform refinement with C1 symmetry was performed, thereby yielding a high-resolution map.
For the PCP-CsA complex dataset, a total of 4868 cryo-EM movie stacks were collected. After motion correction and CTF estimation in RELION, automatic particle-picking with Gautomatch was performed. An overview of the data processing corresponding to this dataset is shown in Supplementary Fig. 5a. Briefly, the particles were first extracted with 4× binning, and junk particles were removed by two rounds of 2D classifications. The remaining particles were re-extracted without binning and then subjected to heterogeneous refinement in cryoSPARC. The particles corresponding to the best class were further processed with heterogeneous refinement and non-uniform refinement using cryoSPARC to yield a map with 3.96 Å. The good particles were re-subjected to RELION for further 3D auto-refinement, CTF refinement, and Bayesian polishing. Again, the particles were re-subjected to cryoSPARC for heterogeneous refinement and non-uniform refinement. Finally, with a total of 277,801 good particles, a map with an overall resolution of 3.53Å was generated. At the same time, the particles yielding the map with 3.96Å were performed with skip alignment 3D classifications to get a total of 138,386 particles with good CsA density, which generated a map with an overall resolution of 3.75 Å used for model building after further 3D auto-refinement, Bayesian polishing and non-uniform refinement with C1 symmetry imposed.
For the PCP-2OG complex dataset, a total of 6191 cryo-EM movie stacks were collected. After motion correction and CTF estimation in RELION, automatic particle-picking with Gautomatch was performed. An overview of the data processing corresponding to this dataset is shown in Supplementary Fig. 6a. Briefly, the particles were first extracted with 4× binning, and junk particles were removed by 2D classifications. The remaining particles were re-extracted without binning and then subjected to heterogeneous refinement in cryoSPARC. The particles corresponding to the best class were further processed with rounds of heterogeneous refinement and non-uniform refinement to yield a total of 745,296 good particles. The particles were re-subjected to RELION for 3D auto-refinement and Bayesian polishing. Again, the particles were re-subjected to cryoSPARC for multi-rounds of heterogeneous refinement and non-uniform refinement. Finally, with a total of 699,771 good particles, a map with an overall resolution of 3.17 Å was generated.
For the PCP-COL1A1 complex dataset, a total of 9507 cryo-EM movie stacks were collected. After motion correction and CTF estimation in RELION, automatic particle-picking with Gautomatch was performed. An overview of the data processing corresponding to this dataset is shown in Supplementary Fig. 7a. Briefly, the particles were first extracted with 4× binning, and junk particles were removed by two rounds of 2D classifications. The remaining particles were re-extracted without binning and then subjected to heterogeneous refinement in cryoSPARC. The particles corresponding to the best class were further processed with non-uniform refinement. The good particles were re-subjected to RELION-3.1. By performing subset selection to exclude the particles from the bad-quality micrographs, a total of 1,779,834 good particles were selected to process with 3D auto-refinement and Bayesian polishing. Finally, a map with an overall resolution of 3.18Å was generated.
Local resolution estimation was performed by cryoSPARC and RELION-3.1, and all the resolutions were estimated with the gold-standard Fourier shell correlation 0.143 criterion with high-resolution noise substitution26,38,39.
Model building and refinement
The AlphaFold2-predicted structures of human P3H1 (https://alphafold.ebi.ac.uk/entry/Q32P28) and the CRTAP (https://alphafold.ebi.ac.uk/entry/O75718) and the crystal structure model of human PPIB (PDB ID: 1CYN) were used as references for initial model building using Phenix40. The initial models were then docked into the electron density map using Chimera41, followed by iterative manual adjustment in COOT42 and real space refinement using Phenix40.
Cell culture, transfections, affinity capture, and immunoblotting
HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Sigma-Aldrich) supplemented with 10% fetal bovine serum (Wisent Corporation, catalog # 080–150) at 37 °C with 5% CO2. To study the interactions among P3H1, CRTAP, and PPIB, cells were co-transfected with a total of 8 μg plasmids (as the combinations of various expression vectors as indicated below) per 6-cm dish using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. For P3H1 and CRTAP interaction, the expression vector for wild-type P3H1 or mutants was co-transfected with the wild-type CRTAP in a 1:1 molar ratio. For CRTAP and PPIB interaction, the expression vector for wild-type CRTAP or mutants was co-transfected with the PPIB expression vector in the presence or absence of P3H1 in 1:1 or 1:1:1 molar ratio, respectively. After 48 hours of incubation, cells were harvested, washed with Dulbecco’s phosphate-buffered saline (DPBS), and resuspended in a lysis buffer containing 150 mM NaCl, 20 mM HEPES pH7.4, 10% glycerol supplemented with 1.5% (w/v) DDM, 4 mM MgCl2, 0.2 mg mL-1 Dnase I, and 1×protease inhibitor (MCE). All experiments were conducted at 4 °C or under ice bath conditions. Each sample was solubilized at 4 °C for 3 h, followed by centrifugation at 16,900 g for 20 min at 4 °C, and the supernatants were collected as the whole cell lysates.
For all the interaction assays, cleared supernatants were subjected to affinity chromatography using Strep-Tactin XT superflow resin (IBA). The resin was then washed three times with the lysis buffer containing 150 mM NaCl, 20 mM HEPES pH 7.4, and 10% glycerol and resuspended in the loading buffer of electrophoresis. The samples were subjected to SDS-PAGE and then transferred to PVDF membranes for immunoblotting.
For immunoblotting, the P3H1 antibody (mouse monoclonal, sc-393003, Santa Cruz, 1:1000) was used to detect the expression of the wild-type P3H1 and variants. Anti-CRTAP antibody (rabbit, 15724-1-AP, Proteintech, 1:2000) was used to detect the expression of the wild-type CRTAP and variants. The PPIB antibody (mouse monoclonal, sc-390193, Santa Cruz, 1:1000) was used to detect the expression of PPIB and variants. The anti-rabbit GAPDH antibody (A19056, ABclonal, 1:10000) was used to detect the expression of GAPDH to evaluate the cell quantity. The secondary antibody is anti-mouse IgG, HRP-linked antibody (7076S, Cell Signaling, 1:1000) and anti-rabbit IgG, HRP-linked antibody (HS101-01, Transgen, 1:10000). All the antibodies were diluted by 3%(w/v) BSA in TBST in prior to use. Images were captured with an Amersham Imager 600.
CRISPR-Cas9 Knockout and Collagen Binding Analysis
The generation of P3H1, CRTAP, and PPIB knockout 293 T cell lines was achieved using the CRISPR-Cas9 technique. Briefly, gRNA sequences targeting P3H1, CRTAP, and PPIB were cloned into lentiCRISPRv2GFP (Addgene, plasmid #82416), which facilitated the co-expression of GFP, Cas9, and the respective gRNA. Two gRNAs were designed for each gene: human P3H1 gRNA1 (CTCAGGATTGCCCACGAAGA) and gRNA2 (GACTATTACCAAACCATGTC); the human CRTAP gRNA1 (TGCCGGTAGGCCGACTCGAG) and gRNA2 (GCGGCATCAGCTCGTCCCGT); the human PPIB gRNA1 (TTCTTCTCATCGGCCGCAGA) and (TTGCCGCCGCCCTCATCGCG). HEK293T cells were transfected with the plasmid using Lipofectamine 2000, as described above. After 48 hours, cells expressing GFP were collected and isolated as single cells into 96-well plates through FACS sorting. The disruption of the target gene in each monoclonal cell line was confirmed through immunoblot analysis using specific antibodies.
For the collagen binding assay, cells lacking P3H1, CRTAP, or PPIB in 6-cm dishes were harvested and washed with DPBS. Subsequently, the cells were lysed using 1.5% (w/v) DDM in a lysis buffer supplemented with 4 mM MgCl2, 0.2 mg mL−1 Dnase I, and 1× protease inhibitor. After solubilization at 4 °C for 3 hours, the lysates underwent centrifugation at 16,900 g for 20 minutes at 4 °C. The resulting supernatants were collected and utilized as whole-cell lysates for the subsequent collagen binding assay.
These lysates were combined with Gelatin-Sepharose (cytiva) and incubated at 4 °C for 3 hours. Following incubation, the Sepharose resin was subjected to three washes using the lysis buffer. The resin, coupled with Gelatin, along with the proteins interacting with the denatured collagen (Gelatin), were resuspended in the electrophoresis loading buffer. Subsequently, the samples were subjected to SDS-PAGE and immunoblotting to detect the specific proteins of interest.
Biochemical assessment of 2-OG dependent oxygenase activities
The 2-OG-dependent oxygenase activity, catalyzed by P3H1 within the PCP complex, was quantified through a two-step procedure involving succinate measurement. Initially, reactions (5 μL total volume) were conducted in an assay buffer (10 mM NaCl, 20 mM HEPES pH 7.4). These reactions comprised 0.4 mg mL−1 enzymatic complex proteins, 1 mM peptide (GLNGLPGPIGPPGPRGRTGDA, synthesized by GL Biochem), 100 μM 2-OG (Sigma, K1875), and 100 μM FeCl2 (Sigma, 44939). To ensure purity, all samples underwent purification through size-exclusion chromatography with a Superose 6 Increase 10/300 GL column. The reactions were incubated at 37 °C for 1 hour and subsequently terminated by heating the samples at 95 °C for 2 minutes. The cooled samples were then transferred to a white 384-well plate (Corning, 3572) for the second part of the procedure, which involved succinate detection. For this, reactions were conducted using the Succinate-GloTM JmjC Demethylase/Hydroxylase Assay kit (Promega, V7990). In detail, 5 μL of Succinate-Glo reagent I was introduced to the sample and co-incubated for 1 hour at 25 °C. Subsequently, 10 μL of Succinate-Glo reagent II was added to the reactions. After a 10-minute incubation, luminescence was measured using a luminometer. All experiments were conducted in triplicate simultaneously, and each sample underwent at least three repetitions.
Isothermal titration calorimetry (ITC) analysis
Isothermal titration calorimetry (ITC) was employed to assess the interaction between the PCP complex and synthetic COL1A11154-1174 peptides (GL Biochem). First, the PCP complex was purified and concentrated to 37 μM in a buffer containing 150 mM NaCl and 20 mM HEPES pH 7.4. Similarly, the synthetic COL1A11154-1174 peptides were dissolved in the same buffer and diluted to a final concentration of 1.5 mM. Binding isotherms were generated by titrating the peptides into the solution containing the PCP complex. The ITC measurements were conducted using a MicroCal PEAQ ITC instrument, with a sample volume of 280 μL and 13 ligand injections of 3 μL each. The cell temperature was maintained at 16 °C, and stirring was performed at 750 rpm. The reference power was set to 5 DP with a spacing of 150 s between injections. A blank control containing the same buffer without the PCP complex was subjected to the same procedure.
Inductively-Coupled Plasma-Mass Spectrometry (ICP-MS) analysis
The PCP complex was purified and then diluted to 13.5 μg/ml or 27 μg/ml (theoretical [Fe] = 5 and 10 ppb, as calculated with a PCP:Fe molar ratio of 1:1). ICP-MS analysis was performed using a NexION 2000 ICP Mass Spectrometer (PerkinElmer) with a protocol that combined external calibration with internal standardization. To generate a calibration curve for the instrument response, solutions containing different concentrations of FeCl2 (corresponding to 4–32 ppb net weight of Fe) were used as standards of known analyte concentrations. The element scandium (Sc) was designated as the internal standardization element. Prior to analysis, the PCP complex samples were digested with 1 ml of nitric acid (HNO3) for 50 minutes at 98 °C using the Sample Preparation Block 50-24 (PerkinElmer). The digested samples were then centrifuged at 3200 g for 10 minutes to remove any debris. Data analysis was performed using Origin (version 9.8.0.200), with the standard curve treated using linear regression through zero.
Native-PAGE analysis coupled with immunoblotting
Expi293F cells were either left untreated (untreated group) or co-transfected with P3H1, CRTAP, and PPIB (PCP overexpression group). The cells were harvested, and the cell membranes were disrupted using the detergent LMNG (Anatrace) for 2 hours at 4 °C. The lysates were then centrifuged at 16800 g for 30 minutes at 4 °C, and the supernatants containing the lysates were collected. The lysates were mixed with 2× native loading buffer and loaded onto a native gel along with a protein marker (DM2631, Coolaber). The gel was run for 2 hours at 120 V at 4 °C. Following electrophoresis, the proteins were transferred from the gel to a membrane, and immunoblotting was performed using standard procedures.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The coordinates have been deposited in the PDB with the accession codes 8K0F (PCP ternary complex), 8K0E (P3H1/CRTAP binary complex), 8K0I (P3H1/CRTAP/PPIB dual-ternary complex), 8K0M (PCP-2OG complex), 8K17 (PCP/COL1A11154-1174 quaternary complex), and 8KC9 (PCP-CsA complex). The cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes EMDB-36763 (PCP ternary complex), EMDB-36762 (P3H1/CRTAP binary complex), EMDB-36765 (P3H1/CRTAP dual-ternary complex), EMDB-36774 (PCP-2OG complex), EMDB-36787 (PCP/COL1A11154-1174 quaternary complex) and EMDB-37097 (PCP-CsA complex,). All the other data are available from the corresponding authors. Source data are provided in this paper.
References
Ricard-Blum, S. The collagen family. Cold Spring Harb. Perspect. Biol. 3, a004978 (2011).
Walker, C., Mojares, E. & Del Rio Hernandez, A. Role of extracellular matrix in development and cancer progression. Int. J. Mol. Sci. 19, 3028 (2018).
Kadler, K. E., Holmes, D. F., Trotter, J. A. & Chapman, J. A. Collagen fibril formation. Biochem J. 316, 1–11 (1996).
Myllyharju, J. & Kivirikko, K. I. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet 20, 33–43 (2004).
Uitterlinden, A. G. et al. Relation of alleles of the collagen type Ialpha1 gene to bone density and the risk of osteoporotic fractures in postmenopausal women. N. Engl. J. Med 338, 1016–1021 (1998).
Prockop, D. J. & Kivirikko, K. I. Collagens: molecular biology, diseases, and potentials for therapy. Annu Rev. Biochem 64, 403–434 (1995).
Grant, S. F. et al. Reduced bone density and osteoporosis associated with a polymorphic Sp1 binding site in the collagen type I alpha 1 gene. Nat. Genet 14, 203–205 (1996).
Wu, J. et al. Characterization of PPIB interaction in the P3H1 ternary complex and implications for its pathological mutations. Cell Mol. Life Sci. 76, 3899–3914 (2019).
Pokidysheva, E. et al. Biological role of prolyl 3-hydroxylation in type IV collagen. Proc. Natl Acad. Sci. USA 111, 161–166 (2014).
Agarwal, G. et al. DDRs and Collagen Fibrillogenesis. in Discoidin Domain Receptors in Health and Disease 23–56 (2016).
Zeng, B. et al. Chicken FK506-binding protein, FKBP65, a member of the FKBP family of peptidylprolyl cis-trans isomerases, is only partially inhibited by FK506. Biochem J. 330, 109–114 (1998).
Vranka, J. A., Sakai, L. Y. & Bachinger, H. P. Prolyl 3-hydroxylase 1, enzyme characterization and identification of a novel family of enzymes. J. Biol. Chem. 279, 23615–23621 (2004).
Marini, J. C., Cabral, W. A., Barnes, A. M. & Chang, W. Components of the collagen prolyl 3-hydroxylation complex are crucial for normal bone development. Cell Cycle 6, 1675–1681 (2007).
Cabral, W. A. et al. Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nat. Genet 39, 359–365 (2007).
Kozlov, G. et al. Structural basis of cyclophilin B binding by the calnexin/calreticulin P-domain. J. Biol. Chem. 285, 35551–35557 (2010).
Mikol, V., Kallen, J. & Walkinshaw, M. D. X-ray structure of a cyclophilin B/cyclosporin complex: comparison with cyclophilin A and delineation of its calcineurin-binding domain. Proc. Natl Acad. Sci. USA 91, 5183–5186 (1994).
Price, E. R. et al. Human cyclophilin B: a second cyclophilin gene encodes a peptidyl-prolyl isomerase with a signal sequence. Proc. Natl Acad. Sci. USA 88, 1903–1907 (1991).
Steinmann, B., Bruckner, P. & Superti-Furga, A. Cyclosporin A slows collagen triple-helix formation in vivo: indirect evidence for a physiologic role of peptidyl-prolyl cis-trans-isomerase. J. Biol. Chem. 266, 1299–1303 (1991).
van Dijk, F. S. et al. PPIB mutations cause severe osteogenesis imperfecta. Am. J. Hum. Genet 85, 521–527 (2009).
Baldridge, D. et al. CRTAP and LEPRE1 mutations in recessive osteogenesis imperfecta. Hum. Mutat. 29, 1435–1442 (2008).
Barnes, A. M. et al. Deficiency of cartilage-associated protein in recessive lethal osteogenesis imperfecta. N. Engl. J. Med 355, 2757–2764 (2006).
Morello, R. et al. CRTAP is required for prolyl 3- hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell 127, 291–304 (2006).
Van Dijk, F. S. et al. CRTAP mutations in lethal and severe osteogenesis imperfecta: the importance of combining biochemical and molecular genetic analysis. Eur. J. Hum. Genet 17, 1560–1569 (2009).
Ishikawa, Y., Wirz, J., Vranka, J. A., Nagata, K. & Bachinger, H. P. Biochemical characterization of the prolyl 3-hydroxylase 1.cartilage-associated protein.cyclophilin B complex. J. Biol. Chem. 284, 17641–17647 (2009).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Chowdhury, R. et al. Structural basis for oxygen degradation domain selectivity of the HIF prolyl hydroxylases. Nat. Commun. 7, 12673 (2016).
Ahmed, S. et al. 1,2,4-Triazolo-[1,5-a]pyridine HIF prolylhydroxylase domain-1 (PHD-1) inhibitors with a novel monodentate binding interaction. J. Med Chem. 60, 5663–5672 (2017).
Blatch, G. L. & Lassle, M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21, 932–939 (1999).
Barbirato, C. et al. Mutational characterization of the P3H1/CRTAP/CypB complex in recessive osteogenesis imperfecta. Genet Mol. Res 14, 15848–15858 (2015).
Chang, W., Barnes, A. M., Cabral, W. A., Bodurtha, J. N. & Marini, J. C. Prolyl 3-hydroxylase 1 and CRTAP are mutually stabilizing in the endoplasmic reticulum collagen prolyl 3-hydroxylation complex. Hum. Mol. Genet 19, 223–234 (2010).
Caparros-Martin, J. A. et al. Molecular spectrum and differential diagnosis in patients referred with sporadic or autosomal recessive osteogenesis imperfecta. Mol. Genet Genom. Med 5, 28–39 (2017).
Arvai, K. et al. Next-generation sequencing of common osteogenesis imperfecta-related genes in clinical practice. Sci. Rep. 6, 28417 (2016).
Grant, T. & Grigorieff, N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 A reconstruction of rotavirus VP6. Elife 4, e06980 (2015).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166 (2018).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D. Struct. Biol. 75, 861–877 (2019).
Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr D. Biol. Crystallogr 66, 486–501 (2010).
Acknowledgements
The authors thank Drs. Lu Zhou, Ming Lei, and Lijun Wang, and Ms. Ying Cui (for scientific discussion). We thank the staff members of the Protein Technology Platform, the Electron Microimaging Center and Bioimaging Facility, and Proteomics Platform at Shanghai Institute of Precision Medicine, Shanghai Jiao Tong University School of Medicine, and the Mass Spectrometry facility of the School of Pharmacy, Fudan University, for providing technical support and assistance in data collection. This work is supported by the National Natural Science Foundation of China (82072468 and 82272519, Y. C.; 82130073, J. Z.; 82372430, A. Q.), the Shanghai Science and Technology Committee (20S11902000, Y.C.), and the Class IV Peak Disciplines (Shanghai Institute of Precision Medicine) from the Shanghai Municipal Education Commission. This work was also supported by Innovative Research Team of High-level Local Universities from the Shanghai Municipal Education Commission (SHSMU-ZLCX20211700, Y.C.), Shanghai Frontiers Science Center of Degeneration and Regeneration in Skeletal System, Shanghai Jiao Tong University School of Medicine, and Shanghai Key Laboratory of Orthopedic Implants (KFKT202207), Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine.
Author information
Authors and Affiliations
Contributions
Y.C. and J.Z. conceived the study. Y.C., W.L., J.P., and D.Y. designed the experiments. W.L. and J.P. performed biochemistry and cell-based experiments. Y.C., W.L., B.R., Y.S., Y.X., Q.W., and M.C. performed structural biology experiments. Y.C., W.L., B.R., S.L., and D.Y. built and refined structural models. All authors analyzed the results. Y.C., W.L., J.P., P.M., R.L., A.Q., and J.Z. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, W., Peng, J., Yao, D. et al. The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex. Nat Commun 15, 7844 (2024). https://doi.org/10.1038/s41467-024-52321-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52321-6
- Springer Nature Limited