Abstract
Synthetic membrane nanopores made of DNA are promising systems to sense and control molecular transport in biosensing, sequencing, and synthetic cells. Lumen-tunable nanopore like the natural ion channels and systematically increasing the lumen size have become long-standing desires in developing nanopores. Here, we design a triangular DNA nanopore with a large tunable lumen. It allows in-situ transition from expanded state to contracted state without changing its stable triangular shape, and vice versa, in which specific DNA bindings as stimuli mechanically pinch and release the three corners of the triangular frame. Transmission electron microscopy images and molecular dynamics simulations illustrate the stable architectures and the high shape retention. Single-channel current recordings and fluorescence influx studies demonstrate the low-noise repeatable readouts and the controllable cross-membrane macromolecular transport. We envision that the proposed DNA nanopores could offer powerful tools in molecular sensing, drug delivery, and the creation of synthetic cells.
Similar content being viewed by others
Introduction
Membrane nanopores1, the hollow barrels spanning biological or synthetic membranes, play vital roles in various bodily functions and molecular biology, including signal transduction2, cross-membrane molecular transport3, and label-free molecular sensing4,5. Among the known nanopores in nature, ion channels are the most studied and functionally complex pores with two important properties: highly selective filter and mechanically gated transport6,7. Responding to environmental stimuli, ion channels can dynamically change their inner width of a few nanometers to allow or prevent the passage of cargo. Next-generation nanopores mimicking the dynamic gating behaviors of the natural ion channels have attracted considerable scientific and technological interest8,9, however, existing two challenges are the lumen-tunable pore with a much wider lumen for controllable cross-membrane macromolecular transport and the structurally stable pore for low-noise repeatable electrical readouts in molecular sensing.
The rational route to next-generation synthetic nanopores could be DNA nanotechnology, an ideal tool for building large de novo architectures10. With the strict base-pairing rule and highly predictable precise structures, DNA nanotechnology enables the programmed production and rapid optimization of multifunctional DNA nanostructures11,12,13. Moreover, achieved applications of the DNA nanostructures indicate that the stability14,15 of synthetic nanostructures determines their feasibility in a variety of applications such as precision measurement tools16,17,18 and smart drug carriers19,20,21. Membrane-spanning nanopores entirely engineered by DNA nanotechnology have been previously reported22,23,24,25,26,27,28, but stimuli-responsive DNA nanopores with tunable lumen sizes and low-noise repeatable electrical readouts remain challenges. Nevertheless, the ease of de novo design of nanopores with DNA is apparent.
Existing DNA nanopores are mostly single-layered22 and built based on the parallel alignment of the DNA duplexes; however, the disadvantages are the reported narrow lumen, significant ion leakage29, and difficulty in accommodating the gating mechanism or other functional units. The very recently pioneered DNA nanopores overcome these limitations by arranging the bundled DNA duplexes parallel to the membrane to form multi-layered tube structures9,30. By controlling the length of the nanopore subunits formed by bundled DNA duplexes, the achieved lumen areas range from tens to hundreds of nm2, which is one to two orders of magnitude larger than typical protein pores30. The self-assembled numerous DNA duplexes in the same plane form a broad plate to accommodate wide lumen, conformational transition mechanisms, and stimuli-responsive units. Nevertheless, structural stability was not considered when designing these DNA nanopore designs. In the lumen-tunable static nanopore design30, unstable DNA architecture may lead to lumen size and shape changes, further influencing the precise sensing and controlled transport of protein cargoes. In the proposed stimuli-responsive DNA nanopore9, the conformational transitions are realized by the linkage motion of the quadrilateral frames, while the most known natural ion channels open and close like a diaphragm31. Lumen tuning based on such conformational transition may allow the relatively larger molecules with matched cross-sectional shapes to pass through the constricted quadrilateral nanopores. Moreover, the structural fluctuation of the semi-flexible quadrilateral frames would cause unrepeatable electrical readouts and significant signal noise9. Numerous studies on the DNA nanostructures adopt stable triangular architectures or variants32,33, including triangular subunits, and advanced molecular dynamics simulations are applied to validate the stability of the designed DNA nanostructures34,35. The DNA nanopores with structurally stable triangular shapes can provide repeatable electrical readouts for molecular sensing.
Here, we show a structurally stable triangular DNA nanopore of considerable size and realize mechanical lumen transition. Three pore subunits assembled from side-by-side bundled DNA duplexes are linked to form a triangular frame with a wide lumen. Specific DNA bindings are used to pinch and release the three corners of the triangular frame, thereby mechanically activating the DNA nanopore to switch from the DNA nanopore-expanded state (DNP-E) to the DNA nanopore-contracted state (DNP-C), and vice versa. Rather than that, the very recently pioneered DNA nanopores realize the gated transport by equipping flap-like gates8 or changing the shape of the semi-flexible quadrilateral frames9, the proposed nanopores maintain a triangular configuration in both expanded and contracted states. This property ideally mimics the diaphragm-like lumen transition of natural ion channels and could significantly contribute to structural stability, precise gated transport, and repeatable low-noise signals in electrical sensing. We anticipate that the proposed DNA nanopores could offer valuable insights for developing and applying the next-generation nanopores.
Results
Triangular DNA nanopore
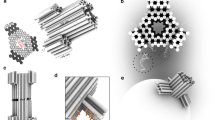
In response to existing challenges in developing next-generation nanopores, we have designed a DNA nanotechnology-engineered triangular nanopore with a tunable lumen for sensing and controlling molecular transport. The triangular DNA nanopore is composed of three subunits interconnected by single-stranded hinges (Fig. 1a). Each subunit adopts a sandwich structure, where the side-by-side bundled long DNA duplexes act as the core layer, and the bundled short DNA duplexes serve as the inner and outer layers. The three-layered middle part of the pore subunit serves as the solid girder, while the two adjacent single-layered ends interconnected by the single-stranded hinge form a v-clip. Triggered by specific DNA bindings (Fig. 1a), the three v-clips can be pinched and released to tune the lumen size of the triangular DNA nanopore. Nine single-stranded DNA (referred to as triggers) are employed to simultaneously pinch the three corners of the large triangular frame during the transition from DNP-E1 to DNP-C2. Conversely, precisely defined twelve single-stranded DNA (referred to as reverse triggers) are utilized to release the three corners of the small triangular frame during the transition from DNP-C1 to DNP-E2. It is even more noteworthy that, during the mechanical transition of the designed nanopore, the lumen retains a triangular configuration in both contracted and expanded states.
a Schematic illustration of characteristic configurations and representative corners of triangular DNA nanopores in expanded and contracted states. b–g Representative TEM images show that the contraction (b) and expansion (e) of DNA nanopores are regulated by triggers and reverse triggers, respectively. Scale bars: 50 nm. Statistical analyses of the cross-sectional area (c, f) and θ angle (d, g) of triangular DNA nanopores in different states. Each set of data is the mean ± SD of 20 nanopores. An unpaired two-tailed t-test was used to evaluate the difference between the two groups. ****p < 0.0001. ns not significant. h 3D views of triangular DNA nanopores in different states equipped with specific staple strands. The Purple and green handles are used to attach cholesterol and Cy3 strands, respectively. i Typical TEM images of DNP-E1 and DNP-C1 bound to SUVs. Three repeats are performed independently. White arrows point to DNA nanopores. Scale bars: 50 nm. j Confocal graphs of Cy3-loaded DNP-E1 (pseudo-colored in green) interacted with GUVs consisting of 90% DOPC and 10% DOPE. Three repeats are performed independently. Scale bars: 10 μm. k Schematics of the insertion of DNA nanopores in different states into lipid bilayers. Source data are provided as a Source Data file.
According to the route designs of the triangular DNA nanopores in different states (Supplementary Figs. 1, 2), DNP-E1 and DNP-C1 were assembled by annealing the scaffold DNA and the multifunctional staple strands (Supplementary Data 1). In the upper region of the representative gel image (Supplementary Fig. 3), homogeneous bands of DNP-E1 and DNP-C1 present different mobilities during gel electrophoresis, providing evidence of the formation of singular assembly products for both DNP-E1 and DNP-C1. After the purification of the assembled products by ultrafiltration units, different morphologies (Fig. 1b, e) of contracted and expanded nanopores were observed by negative-stained transmission electron microscopy (TEM). Based on representative TEM images (Supplementary Figs. 4, 5), it is evident that most of DNP-E1 converted to DNP-C2 upon exposure to the triggers, while the majority of DNP-C1 changed to DNP-E2 with the participation of the reverse triggers. The θ angle and cross-sectional area of each DNA nanopore were determined using a computer vision-assisted method (Supplementary Fig. 6a). Statistical analyses of DNP-E1 and DNP-C2 present a notable decrease of the average cross-sectional area from 787 to 301 nm2 (Fig. 1c and Supplementary Fig. 6b), with no significant change observed in the θ angle (Fig. 1d and Supplementary Fig. 6c). Conversely, the transition of DNP-C1 to DNP-E2 was accompanied by a significant increase of the average cross-sectional area from 303 to 675 nm2 (Fig. 1f and Supplementary Fig. 6b), while the θ angle remained nearly constant (Fig. 1g and Supplementary Fig. 6c).
To facilitate embedding the proposed DNA nanopores into lipid bilayers, we utilized twenty-one handles of staple strands to hybridize the cholesterol strands (Fig. 1h). Cholesterol-modified DNP-E1 and DNP-C1 are shown as upshifted bands in gel electrophoresis analysis (Supplementary Fig. 7), demonstrating that the cholesterol strands can be attached to the DNA nanopores as designed. Small unilamellar vesicles (SUVs) were used to investigate the interaction between cholesterol-tagged DNA nanopores and lipid bilayers (Supplementary Fig. 8a). The signal intensity of the upshifted bands in DNP-C1 samples gradually increased as the lipid concentration of SUVs increased (Supplementary Fig. 8b). In contrast, no upshifted bands were observed for DNP-C1 samples in the absence of SUVs. The signal intensity analysis reveal that the upshifted bands were caused by the interaction between DNP-C1 and SUVs (Supplementary Fig. 8c). In addition to gel electrophoresis, TEM was employed to confirm the binding of synthetic DNA nanopores to SUVs. The shaded areas representing the different lumens of DNP-C1 and DNP-E1 indicate that these DNA nanopores interacted with the SUVs (Fig. 1i). Moreover, to visualize the interaction between cholesterol-tagged DNA nanopores and giant unilamellar vesicles (GUVs), we employed twelve additional handles in DNA nanopores to gather the Cy3 strands (Fig. 1h). Representative confocal images display the efficient binding of abundant Cy3-marked DNA nanopores to GUVs (Fig. 1j).
Electrical characterization
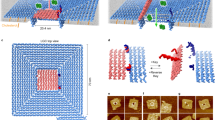
Theoretically, symmetrically distributed cholesterol moieties around the perimeter of triangular DNA nanopores facilitate the insertion of the nanopores into lipid bilayers36, forming well-defined transmembrane channels. To confirm the membrane-spanning nature of both DNA nanopore designs (Fig. 1k), the conductance properties of DNP-C1 and DNP-E1 were tested via single-channel current recordings. In particular, 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC) was used to produce planar lipid bilayers. In every microelectrode cavity of the current recording chip, the electrolyte (1 M KCl, 10 mM HEPES, pH 7.4) was separated by a planar lipid bilayer into two chambers, including a cis side and a trans side. When a potential was applied across the lipid membrane, electrolyte ions were induced, ion flow occurred through the lumen of the embedded DNP-C1 (Fig. 2a), and a constant current of 0.61 nA was observed at an applied voltage of +80 mV. In contrast, when the applied voltage was changed to –80 mV, the recorded current was constant at –0.62 nA. The single-channel current trace (Fig. 2b) indicates the successful insertion of DNP-C1 into the planar lipid bilayer. Statistical analysis of the conductance distribution of DNP-C1 shows the peak conductance with an average of 7.75 ± 0.34 nS (± SD, n = 22) (Fig. 2c).
a–f Current signal studies of DNA nanopores via single-channel current recordings. Schematics of electrolyte ions flow through the DNP-C1 (a) and DNP-E1 (d). Representative current traces of DNP-C1 (b) and DNP-E1 (e) were recorded at +80 mV for the first 0.5 s, after which the applied voltage was switched to −80 mV for the next 0.5 s. Conductance Histograms were obtained from 22 and 20 individual insertions of DNP-C1 (c) and DNP-E1 (f), respectively. The black dashed lines depict Gaussian fits (c, f). g Schematics illustrate that the contraction and expansion of DNA nanopores can be activated by introducing triggers and reverse triggers, respectively. h Representative signal trace demonstrates dynamic lumen changes observed during the transition from DNP-C1 to DNP-E2. i Power spectrum analysis of inserted DNP-C1 and DNP-E2. j Representative single-channel current recordings exhibit dynamic lumen changes observed during the transition from DNP-E1 to DNP-C2. k Power spectrum analysis of inserted DNP-E1 and DNP-C2. Source data are provided as a Source Data file.
DNP-E1 has a larger lumen than DNP-C1. Thus, the inserted DNP-E1 should allow more electrolyte ions to pass through its lumen than the inserted DNP-C1. Consistent with the concept of both schematics (Fig. 2a, d), the constant current of DNP-E1 (1.26 nA) was higher than that of DNP-C1 (0.61 nA) when a voltage of +80 mV was applied across the inserted DNP-E1 and DNP-C1 (Fig. 2b, e). The conductance peak histogram of inserted DNP-E1 appeared at 16.6 ± 0.54 nS (± SD, n = 20) (Fig. 2f), suggesting that the lumen size of DNP-E1 is larger than that of DNP-C1. Notably, the concentrated distribution of the peak conductance of DNP-C1 and DNP-E1 illustrates the repeatable electrical readouts brought by the structurally stable triangular lumen. Moreover, control experiments of DNP-C1 and DNP-E1 without cholesterol modification demonstrate no measurable ionic currents (Supplementary Fig. 9), thus verifying that cholesterol modification is essential for inserting triangular DNA nanopores into planar lipid bilayers. Successful insertion of cholesterol-labeled DNP-C1 and DNP-E1 into lipid membranes was also verified by linear current-voltage relationships with different slopes (Supplementary Fig. 10), as expected for vertically symmetric DNA nanopores.
To explore the dynamic transition of triangular DNA nanopores between expanded and contracted states, we separately added triggers and reverse triggers on the cis sides of membrane-spanning DNP-C1 and DNP-E1 (Fig. 2g). The representative current trace indicates that the current signal of inserted DNP-C1 changed from 0.63 to 1.07 nA with a constant voltage of +80 mV (Fig. 2h), reflecting the lumen expansion induced by reverse triggers. In the power spectrum analysis (Fig. 2i), we found that the noise profile of DNP-E2 exhibited more fluctuations compared to DNP-C1. After treating the membrane-spanning DNP-E1 with triggers, the recorded current signal changed from 1.20 to 0.73 nA (Fig. 2j), validating that the embedded DNA nanopore was contracted. Moreover, signal fluctuations were observed around 1.0 s during the contraction process (Fig. 2j), primarily resulting from transient deformations occurring at specific corners of the embedded DNA nanopore. We attribute the achieved low-noise signal to the stable triangular lumen. Compared to the inserted DNP-E1 without any treatment, the power spectral noise of DNP-C2 was reduced (Fig. 2k), indicating a decrease in lumen size.
To further examine the specificity of the triggers and reverse triggers used to activate conformational transitions of DNA nanopores in different states, random strands (Supplementary Data 1) were separately introduced on the cis sides of membrane-spanning DNP-C1 and DNP-E1. Single-channel current recordings of random strands-incubated DNA nanopores present no apparent fluctuating signals (Supplementary Fig. 11), further emphasizing the importance of triggers and reverse triggers in the expansion and contraction of triangular DNA nanopores. Furthermore, statistical analyses of repetitive current signals demonstrate that the majority of DNP-C1 converted into DNP-E2 in response to reverse triggers, while DNP-E1 transitioned into DNP-C2 in the presence of triggers (Supplementary Fig. 12).
Computational analysis
To assess the structural stability of triangular DNA nanopores in different states, we characterized two different DNA nanopore designs by oxDNA37,38,39 simulations. From a macro-perspective, statistical data of the relative angles in both DNA nanopore designs demonstrate a more pronounced fluctuation in the expanded DNA nanopore compared to the contracted one (Fig. 3a, b). The relative areas of the two DNA nanopore designs did not change significantly during the molecular dynamics simulations, indicating that their distinct lumens can be effectively maintained. In a finer perspective, DNP-E1 exhibited a notably higher root-mean-square deviation (RMSD) as compared to DNP-C1 (Fig. 3c), predominantly due to the elongated core layers positioned at the triangulated nanopore corners. In previous oxDNA simulations, the phenomenon of splaying at the ends of origami structures has been documented40. One plausible explanation for this occurrence could be attributed to end effects. The lack of continuity in nucleotides at the ends weakens the backbone and stacking potentials, thereby facilitating splaying. Also, it is important to note that crossovers in DNA origami-engineered structures are sparse as one approaches the ends, leading to a more loosely bound structure end. Notably, similar behavior of fraying ends of DNA duplexes has been observed in all-atom simulations using CHARMM41 and Amber42 force fields, suggesting a common underlying mechanism across different simulation methodologies. Even though the proportion of broken base pairs remained below 1.5% for both designs throughout the simulations (Fig. 3d), DNP-C1 displayed superior integrity at a finer scale compared to DNP-E1.
a, b Representative morphology and related simulation analyses of DNP-E1 (a) and DNP-C1 (b). All angles and areas of the proposed nanopore at two states are calculated according to the initial model. c, d RMSD (c) and proportion of broken base pairs (d) during coarse-grained oxDNA simulations. Black dashed lines in both panels show the moving average of the plotted observable. e Transverse (top) and side (bottom) conductivity maps of DNP-E1 and DNP-C1 under an applied voltage of 80 mV and salt concentration of 1 M KCl. f Histogram presents the distribution of conductance obtained using SEM and estimated values (arrows) derived using the analytical calculations. Insets represent the cylinder models of DNP-C1 (left) and DNP-E1 (right) nanopore conductance. Source data are provided as a Source Data file.
The conductance of both DNA nanopore designs was initially computed using the simple cylinder model. The conductance ratio of DNP-E1 to DNP-C1 was 2.34, closely aligning with the experimental ratio value (2.14) derived from recorded ionic currents. Notably, membrane nanopores made with DNA inherently possess ionically leaky walls29. To account for this factor, we employed the steric exclusion model (SEM)43,44 to simulate the conductance of both designs based on 3D conductivity maps. Specifically, ten configurations each of DNP-C1 and DNP-E1 were selected (Supplementary Videos 1, 2), mapped into an all-atom representation, and embedded within DPhPC-based lipid bilayers to allow the calculation of 3D conductivity maps (Supplementary Fig. 13a). The conductivity maps were calculated from the distance from lipid or DNA utilizing local conductivity functions specific to the lipid and DNA components (Supplementary Fig. 13b) under an applied voltage of 80 mV and 1 M KCl. Figure 3e depicts the transverse view (top) and side view (bottom) along the center of each system. The peak conductance distributions observed in the SEM data for the two DNA nanopore designs were notably higher than those calculated from the cylinder model because the SEM includes the inherent ion linkage of the DNA structures (Fig. 3f).
While experimental measurements indicate a closely matched conductance ratio of DNP-E1 to DNP-C1 compared to values obtained from SEM simulations or cylinder model calculations, the observed absolute conductance values were significantly smaller than the corresponding theoretical estimates. This discrepancy aligned with findings from prior studies on large DNA nanopores8,30. We hypothesize that the lower experimental conductivities of DNP-C1 and DNP-E1 compared with those of solid-state nanopores of similar internal dimensions45 may be caused by the continuous compression of the surrounding lipid molecules23 and structural distortions29,46 induced by the applied voltages9. Moreover, we note that, for a select few DNA nanopore systems of well-defined transmembrane pore geometry, a good agreement between simulated and experimentally measured conductance values was reported47,48. Future molecular dynamics simulations incorporating interactions between lipids and DNA may reveal factors underlying these disparities. Nevertheless, oxDNA simulations provide evidence for the structural stability of the two DNA nanopore designs, confirming that triangular DNA nanopores in different states possess distinct stable lumens.
Molecular sensing
We employed membrane-spanning DNP-C1 and DNP-E1 to explore whether designed DNA nanopores can sense the trypsin molecule (hydrodynamic diameter of 4.0 nm), a model protein with a net positive charge at pH 7.4. In a typical current trace recorded at a constant voltage of +50 mV, the introduction of trypsin regularly blocked the ion flow of inserted DNP-C1. According to these studies8,24,30, we identified an individual blockade as a single event of trypsin translocation, characterized by its dwell time (τoff) and blocking amplitude (A) relative to the open-channel amplitude (Io). As illustrated in Fig. 4a, two distinct types of trypsin translocation events in embedded DNP-C1 were identified by analyzing the relative blocking amplitude (A/Io) and dwell time (Fig. 4b, c). Moreover, to prevent the formation of the dimers and multimers of the trypsin molecule, we tried to shorten the preparation of the trypsin solution used in all protein sensing experiments, and relevant physiological experiments were strictly performed within 30 min to avoid the aggregation of trypsin molecules. More than 50% of trypsin translocation events clustered at a relative blocking amplitude of 30.2 ± 2.74% (± SD, n = 35). The remaining events clustered at a relative blocking amplitude of 53.3 ± 2.26% (± SD, n = 34). Given the current-dependent nature of trypsin translocation on well-developed DNA nanopores8,24, Type I events represent transient interactions of positively charged trypsin with embedded DNP-C1. Contrastingly, Type II events represent that trypsin molecules are fully translocated across the whole lumen of embedded DNP-C1. Intermittent interactions of positively charged trypsin molecules with negatively charged inner walls of DNA nanopores may prolong the dwell time of Type II events. Following this concept, a scatter plot of trypsin translocation events sensed by embedded DNP-C1 indicates that the average dwell time of Type II events (0.69 s) was much longer than that of Type I events (0.04 s).
a Trypsin interacts transiently with inserted DNP-C1 in the Type I event (i). In the Type II event, trypsin passes completely through the inserted DNP-C1 (ii). The schematic diagram portrays the full translocation of trypsin through embedded DNP-E1 (iii). b Single-channel current trace recorded with embedded DNP-C1 at +50 mV in the presence of trypsin in the cis chamber. Insert: a single translocation event is characterized by the dwell time (τoff) and the event amplitude (A) from the open-channel current (Io). c Scatter plot of trypsin translocation events recorded from DNP-C1. d Representative current trace obtained at +50 mV after adding trypsin to the cis chamber of embedded DNP-E1. e Scatter plot of trypsin translocated events recorded with DNP-E1. f Single-channel current trace of trypsin sensing recorded with in situ activated DNP-C2, generated by incubating DNP-E1 inserted in the lipid bilayer with triggers. g Scatter plot of trypsin translocation events recorded from in situ activated DNP-C2. h Single-channel current trace of trypsin sensing recorded with in situ activated DNP-E2, generated by incubating DNP-C1 inserted in the lipid bilayer with reverse triggers. i Scatter plot of trypsin translocation events recorded from in situ activated DNP-E2. Source data are provided as a Source Data file.
Additionally, membrane-spanning DNP-E1 was used to sense the trypsin molecules, which were analyzed by single-channel electrophysiological recordings. Based on the representative current trace of embedded DNP-E1 after the introduction of trypsin (Fig. 4d), trypsin translocation events with diverse relative blocking amplitudes and dwell times were observed. Interestingly, there was only one cluster in the scatter plot (Fig. 4e). Notably, the trypsin translocation events recorded from embedded DNP-E1 exhibited a relative blocking amplitude of 28.1 ± 7.38% (± SD, n = 85), which was narrower than that observed for trypsin translocation events from embedded DNP-C1. Given that DNP-E1 adopted a larger lumen than DNP-C1, we suggest that the majority of trypsin translocated completely through the embedded DNP-E1 rather than interacting transiently with its lumen opening (Fig. 4a). We also carried out trypsin sensing tests using pre-activated DNP-C2 and DNP-E2, which were obtained by treating DNP-E1 and DNP-C1 with triggers and reverse triggers in advance, respectively. These pre-activated nanopores were then separately inserted into the lipid bilayers. The statistical results (Supplementary Fig. 14) obtained from the pre-activated DNA nanopores were consistent with those from the native DNP-E1 and DNP-C1.
Furthermore, the DNP-E1 and DNP-C1 inserted in the lipid bilayers were separately treated with triggers and reverse triggers to generate in situ activated DNP-C2 and DNP-E2 (Fig. 4f, h). The decreased current signal detected 30 min after adding the triggers indicates the successful in situ switch from DNP-E1 to DNP-C2 (Fig. 4f), while the increased current signal shown in Fig. 4h suggests the in situ transition from DNP-C1 to DNP-E2. The statistical data (Fig. 4g, i) of trypsin translocation events recorded from both in situ activated DNP-C2 and DNP-E2 align with those of native DNP-C1 and DNP-E1, as well as pre-activated DNP-C2 and DNP-E2, demonstrating that the proposed stable triangular configurations maintained in both contracted and expanded states contribute to the repeatable electrical readouts in molecular sensing applications. Here, the presence of triggers and reverse triggers did not introduce significant noise in trypsin sensing. We presume that there could be two main reasons: the triggers and reverse triggers are much smaller than the trypsin; and the triggering DNA strands exhibit little interaction with the proposed DNA nanopore or are repelled due to the negative charge of the DNA nanopore as intermittent electrostatic binding of a positively charged trypsin with the negatively charged DNA nanopore is thought to be necessary to resolve translocation events.
Control experiments with embedded DNP-C1 and DNP-E1 demonstrate that adding the same volume of PBS buffer, used to dilute trypsin, to the cis chambers did not lead to apparent signal fluctuations (Supplementary Fig. 15). The noise in the power spectra of both DNP-C1 and DNP-E1 was considerably weaker before the introduction of trypsin treatment than afterward (Supplementary Fig. 16), further verifying that both DNA nanopore designs were capable of sensing trypsin translocation. Statistical analyses of trypsin sensing experiments (Fig. 4 and Supplementary Fig. 14) indicate that contracted nanopores can sense trypsin molecules with low-noise, repeatable readouts compared to expanded nanopores. In more detail, contracted nanopores can distinguish two different types of trypsin translocation events, while expanded nanopores have too large lumens compared with the small trypsin to realize such precise sensing.
Cross-membrane transport
Finally, we investigated whether the different lumens of triangular DNA nanopores allow the transmembrane transport of macromolecules of different sizes. In contrast to SUVs, GUVs provide a better model for simulating physiological cell membranes and can be used to explore the cross-membrane macromolecular transport through the embedded triangular DNA nanopores. We added FITC-dextran (Diameter ≈9.6 nm25) and TRITC-dextran (Diameter ≈22 nm49) to the external solution of GUVs while incubating with cholesterol-free DNA nanopores. No significant increase in fluorescent signal was measured from the interior of GUVs incubated with cholesterol-free DNA nanopores (Supplementary Fig. 17), verifying that cholesterol-free DNA nanopores were unable to insert into lipid membranes, which was consistent with the data obtained from single-channel current recordings (Supplementary Fig. 9).
With the assistance of cholesterol moieties, membrane-spanning DNP-C1 allowed the influx of smaller FITC-dextran into GUVs while preventing the influx of larger TRITC-dextran (Fig. 5a, b and Supplementary Video 3). Representative confocal images and associated fluorescence traces demonstrate that both macromolecules can enter the interior of the corresponding GUVs through the wide lumen of embedded DNP-E1 (Fig. 5c, d and Supplementary Video 4). After 180 min of incubation, more than 40% of GUVs exhibited FITC influx in the presence of DNP-C1 treatment, with only a few GUVs were filled with TRITC-dextran (Fig. 5e and Supplementary Fig. 18). In the case of DNP-E1 treatment, over 30% of GUVs displayed influx of both macromolecules, indicating that the expanded DNA nanopores allowed the transmembrane transport of FITC-dextran and TRITC-dextran (Fig. 5f and Supplementary Fig. 18). Collectively, these results confirm that cholesterol-modified triangular DNA nanopores with different lumens can spontaneously insert into the lipid membranes of GUVs. These findings demonstrate that both states of triangular DNA nanopores can function as size-selective gateways to control the transmembrane transport of various molecules.
a Time series of corresponding confocal images of GUVs (pseudo-colored in magenta) incubated with DNP-C1 and different macromolecules (FITC-dextran and TRITC-dextran). Three repeats are performed independently. Scale bars: 10 μm. b Fluorescence traces recorded from the interior of DNP-C1-incubated GUVs. The influx of FITC-dextran follows exponential kinetics. c Confocal images at different time points illustrate that embedded DNP-E1 allowed FITC-dextran and TRITC-dextran to be transported across lipid membranes. Three repeats are performed independently. Scale bars: 10 μm. d Fluorescence traces recorded from the internal areas of GUVs treated with DNP-E1. The influx of both TRITC-dextran and FITC-dextran follows exponential kinetics. e, f Bar graphs show the percentage of GUVs after 15, 60, 120, and 180 min, respectively. Data (mean ± SD) for DNP-C1 (e) and DNP-E1 (f) are from three independent experiments with n = 45 GUVs and n = 57 GUVs, respectively. Source data are provided as a Source Data file.
Discussion
In this work, we propose a triangular DNA nanopore with a tunable lumen. The main advantages of the developed nanopore are the large enough lumen size to accommodate complex protein, stable triangular structure made with DNA, and lumen tuning by the diaphragm-like configuration transition mimicking natural ion channels. In contrast, the majority of existing DNA nanopores utilized for molecular sensing24 and transmembrane transport25 have limited lumen sizes and lack lumen tunability. Dey and Yan et al. 8 proposed a DNA nanopore that allows stimulus-controlled transport of functional proteins across bilayer membranes. It features a large lumen and a nanomechanical lid, which can be controllably closed and reopened via a lock-and-key mechanism. Xing and Howorka et al. 30 innovatively created various nanopores by arranging the bundled DNA duplexes parallel to the membrane. In their follow-up research9, they realized the lumen tuning by forming the DNA duplexes at the corners to stabilize the semi-flexible quadrilateral frames. TEM images and electrical characterizations indicated that this nanopore had an unstable configuration in the closed state, leading to inconsistent conductance and electrical readouts in protein sensing. Compared with other works, the proposed DNA nanopores maintain stable triangular configurations before and after switching between contracted and expanded states, and feature more concentrated conductance distributions and more repeatable relative blocking amplitudes in trypsin sensing.
Single-channel current recordings demonstrate successful in situ switching of the lumen-tunable DNA nanopore; however, current signal fluctuations were observed during the contraction process (Fig. 2j). The main reason could be transient deformations occurring at specific corners of the embedded DNA nanopore, as the transition from DNP-E1 to DNP-C2 requires nine single-stranded DNA molecules to pinch the three corners of the nanopore. This multi-step chemical process, with its varied configuration changes, continuously influenced the electrical properties of the nanopore over a short duration, resulting in signal fluctuations. Moreover, two distinct clusters were observed in the statistical analyses recorded from native DNP-C1 (Fig. 4c), in situ activated DNP-C2 (Fig. 4g), and pre-activated DNP-C2 (Supplementary Fig. 14b), consistent with previous findings that utilized a nanopore equipped with a lid8. In contrast, no apparent two clusters were found in expanded DNA nanopores (Fig. 4e, i and Supplementary Fig. 14d). We infer that expanded nanopores have too large lumens compared with the small trypsin to realize such precise sensing. This suggests that maintaining a closer match between the lumen and the molecule contributes to more precise sensing. It is also a primary motivation for developing a DNA nanopore with a large tunable lumen.
Proposed nanopores entirely engineered by DNA nanotechnology allow rational de novo design of complex structures. The end-to-end linked numerous DNA duplexes are arranged parallel to the membrane to form a triangular frame capable of accommodating a wide lumen and stimuli-responsive units. This design route offers unprecedented freedom in tuning the sizes and shapes of DNA nanopores to match different analytes. For instance, more pore subunits could be linked to form quadrilateral, hexagonal frames, and even subcircular nanopores. Here, we tend to choose the triangular frame because it could provide relatively higher structural stability. The lengths of the subunits can be changed to tune the pore sizes, and they do not need to be equal. Thus, there is nearly no limit to the selection of pore size and shape. Moreover, by adjusting the relative locations and lengths of the outer and inner layers to the core layers of the pore subunits, we could customize the mechanical contraction and expansion of the nanopore, enabling the mechanically gated transport of specific proteins. In the future, the highly programable sizes, shapes, and mechanical transitions provided by our unique nanopore design could deliver a substantial change to the traditional approach of using static nanopores in molecular sensing and cross-membrane transport.
In conclusion, by mimicking the diaphragm-like transition of natural ion channels and adopting a stable triangular configuration, we present a lumen-tunable DNA nanopore for high-precision molecular sensing and controllable cross-membrane transport. It offers solutions to the two main challenges in developing next-generation synthetic nanopores: achieving controllable transmembrane cargo exchange similar to natural ion channels, and obtaining low-noise, repeatable readouts for molecular sensing. We envision that these proposed DNA nanopores could become powerful tools in biosensing and synthetic biology.
Methods
Materials
M13mp18 single-stranded DNA (N4040S) was obtained from New England Biolabs. All oligonucleotides were purchased from Sangon Biotech. All lipid products involved, such as 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, 850375), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE, 850725), and DPhPC (850356) were procured from Avanti Polar Lipids. CellMaskTM Deep Red Plasma Membrane Stain (C10046) was obtained from Invitrogen. Unless otherwise stated, all other reagents and solvents were purchased from Sigma-Aldrich.
Preparation of DNA nanopores
DNA nanopores in both states were designed by using the honeycomb lattice version of caDNAno250. Detailed route designs for both structures are shown in Supplementary Fig. 1. The nucleotide sequences of all used staple strands are shown in Supplementary Data 1. In a typical reaction, scaffold DNA (10 nM) was mixed with fundamental staple strands (50 nM) and specific staple strands (100 nM) in TE-Mg2+ buffer (10 mM Tris-HCl, 1 mM EDTA, and 10 mM MgCl2, pH 8.0). The mixture was then annealed with a LifeTouch thermal cycler (Bioer Technology, China). The DNA nanopores were assembled using a 24 h annealing procedure: initially, the mixture was cooled from 95 to 60 °C at a rate of 1 °C per 10 min, then to 25 °C at a rate of 1 °C per 30 min, and finally kept at 4 °C until ultrafiltration. After the annealing reaction, the synthetic DNA nanopores were filtered with centrifugal filters (Amicon Ultra-0.5 ml 100 kDa) to remove excess staple strands. The ultrafiltration was conducted at 2500×g for 3 min and repeated three times. In addition, cholesterol strands (5’-GATGCATAGAAGGAA-3’-Chol) were mixed with purified DNA nanopores at a ratio of two times relative to the total number of cholesterol attachment sites on the DNA nanopores and incubated at 37 °C for 12 h to produce cholesterol-tagged DNA nanopores. Optionally, Cy3 fluorophores can be attached to DNA nanopores by treating purified DNA nanopores with a tenfold excess concentration of Cy3 strands (5’-GCTAGCATGCTG-3’-Cy3) at room temperature for 12 h. After additional modifications with cholesterol moieties and Cy3 fluorophores, the desired DNA nanopores were ultimately obtained either through three rounds of ultrafiltration or by using a DNA extraction kit (D1200, Solarbio, China).
Preparation of lipid vesicles
For SUVs, the DOPC solution (10 mg/mL in chloroform, 50 μL) was dried by argon flow in a glass vial (2 mL) to yield thin films. The obtained films were suspended by HEPES-Na+ buffer (50 mM HEPES, 500 mM NaCl, pH 7.4) and then sonicated for 30 min at room temperature using a water bath sonicator (power: 80 W, frequency: 40 kHz) to generate homogeneous SUVs. Dynamic light scattering was performed using a Zetasizer Nano analyzer (Malvern, UK) to determine the size distribution of the acquired SUVs. For GUVs, the lipid solution (DOPC/DOPE = 9.9:0.1, 10 mg/mL in chloroform, 10 μL) was loaded onto an indium tin oxide (ITO)-coated glass slide. After 5 min of evaporation, dried lipid films were formed. The glass slide carrying lipid films was first placed into a Vesicle-Pre-Pro device (Nanion Technologies, Germany). An O-ring was placed around the lipid films, and then sorbitol (1 M, 300 μL) was added to the lipid films to fill the entire O-ring. Subsequently, another ITO glass slide was mounted on top of the O-ring to ensure that a sealed chamber was formed between the two ITO glass slides. Finally, an alternating electric field was applied for 2 h to produce GUVs. Notably, all obtained lipid vesicles (SUVs and GUVs) were stored at 4 °C and used within 48 h.
Agarose gel electrophoresis
For samples containing only DNA nanopores, the mobility of various samples was analyzed using agarose gel electrophoresis (1.0%, TE-Mg2+ buffer) at 4 °C. For samples of cholesterol-tagged DNA nanopores, they were subjected to agarose gel electrophoresis (1.0%, PBS buffer). To study the interaction of cholesterol-tagged DNA nanopores and SUVs, equal amounts of DNP-C1 (5 nM) were incubated with a range of lipid concentrations of SUVs (0, 5, 10, 20, and 40 nM), respectively. After 1 h of incubation, the mixed samples were subjected to agarose gel electrophoresis (1.5%, HEPES-Na+ buffer). All agarose gels were premixed with 0.01% SYBRTM Safe (Invitrogen, USA) and run at 100 V for 60 min. All gel images were captured with a Tanon 4600SF Multifunctional Imaging System (Tanon, China).
TEM characterization
The configurations of triangular DNA nanopores and the bindings of SUVs with cholesterol-tagged DNA nanopores were separately characterized using negative-stain TEM. Briefly, diverse samples (2 μL) were adsorbed on glow-discharged (30 s, PDC-32G-2, Harrick Plasma), carbon-coated TEM copper grids (300 meshes, BZ11143A, EMCN) for 5 min, respectively. After that, the remaining solution on these TEM grids was carefully removed with filter paper. These grids were then treated with a staining solution (1.0% uranyl acetate containing NaOH, pH 7) for 30 s. Immediately, the additional staining solution was absorbed with filter paper. Finally, TEM images of all samples were taken with a JEM 1400 Plus transmission electron microscope (JEOL, Japan) at 100 kV.
Single-channel current recordings
An integrated chip-based parallel bilayer recording device (Orbit Mini, Nanion Technologies, Germany) was equipped with a multielectrode-cavity-array (MECA) chip (IONERA, Germany) to perform single-channel current recordings. The electrolytic buffer (150 μL) consisting of 1 M KCl and 10 mM HEPES at pH 7.4 should be initially added to the MECA chip. Next, an appropriate volume (1–2 μL) of DPhPC solution (10 mg/mL in octane) should be smeared on every cavity of the MECA chip to form a planar lipid bilayer. The successful formation of planar lipid bilayers can be verified by monitoring the resistance and capacitance of the corresponding cavities. Meanwhile, each cavity of the MECA chip was divided into two sides: cis side and trans side. Cholesterol-tagged DNA nanopores (1 nM, 2 μL) were then added to the cis side for spontaneous insertion into planar lipid bilayers. The insertion of triangular DNA nanopores can be accelerated by adding 1 μL of 0.5% n-octyl-oligo-oxyethylene (40530) to the cis side. In molecular sensing experiments, equal volumes (10 μM, 1.5 μL) of trypsin solution (T4549) were added to the cis sides of embedded DNP-C1 and DNP-E1, respectively. Trypsin translocation events with different relative blocking amplitudes were recorded at +50 mV. The Orbit Mini device was grounded at the trans side. Single-channel current traces were acquired at 10 kHz using Element Data Reader 3.8.21 (Elements, Italy). All obtained current traces were analyzed with Clampfit 11 (Molecular Devices, USA).
In situ switching in molecular sensing
Triggers (10 nM, 2 μL) and reverse triggers (10 nM, 2 μL) were separately introduced to the cis sides of embedded DNA nanopores to trigger in situ switching between contracted and expanded states. Notably, the applied transmembrane voltage should be disconnected before performing this operation. After an incubation period of at least 30 min, a transmembrane voltage of +50 mV was applied to monitor changes in the current signal. A significant change in the current signal indicated successful in situ switching of the DNA nanopores. Subsequently, trypsin solution was added to the cis chambers of the in situ activated DNP-E2 and DNP-C2, respectively. Trypsin translocation events were then recorded from multiple independently activated DNP-E2 and DNP-C2.
OxDNA simulations
The dynamics of DNP-E1 and DNP-C1 are captured using oxDNA237,38,39. The interactions between the nucleotides include a backbone potential, excluded volume, stacking, coaxial stacking, cross stacking, electrostatic repulsions, and hydrogen-bonding between complementary base pairs, that have been parameterized to reproduce the structural, mechanical, and thermodynamic properties of double-stranded and single-stranded DNA. Molecular dynamics simulations of DNP-E1 and DNP-C1 were performed using GPU-implemented sequence-dependent oxDNA2 in the canonical NVT (constant number of particles N, volume V, and temperature T) ensemble at 300 K and 1 M NaCl. Initial geometries of DNP-E1 and DNP-C1 were obtained using oxView’s rigid-body dynamics51. Standard oxDNA relaxation procedures52 were then used to remove any nucleotide overlaps and overly long bonds that would otherwise cause the molecular dynamics to fail due to excessively large forces. OxDNA simulations were performed with the Anderson-like John thermostat using a 15-fs time step. Both nanopores were energy minimized for 10,000 steps and relaxed for 30.3 μs prior to production simulations that spanned 8 ms for DNP-C1 and DNP-E1.
Ionic current calculations
From each of the simulated DNP-E1 and DNP-C1 oxDNA2 trajectories, we randomly selected ten configurations for the calculation of the ionic current, ensuring a minimum separation of at least 20 μs between the chosen configurations. Subsequently, these chosen configurations were transformed into an all-atom representation via a custom script using the mrDNA package53. Each of these all-atom configurations was embedded at the center of a DPhPC lipid bilayer patch (130 × 130 nm2) created by tiling a pre-equilibrated 13 × 13 nm2 patch using VMD 1.9.4. Lipids having atoms within 15 Å of DNA atoms and those within the central cavity were both removed. Employing the volmap tool of VMD54, we computed the three-dimensional map of the number density of lipid atoms with 1 Å resolution, marking voxels with a non-zero density as “occupied” by lipid. Equivalent maps were obtained for the DNA using the density of N1 purine atoms. For each occupancy map, the distance to the nearest occupied voxel was computed and used to look up the conductivity from a linear ramp for lipids, ranging from zero to the bulk conductivity value (11.18 S/m) between 3 and 10 Å, and a previously determined55 tabulated dependence of the conductivity on distance from DNA. For each system, the two conductivity maps (lipid and DNA) were combined by using the minimum of the two values at each location. The resulting combined conductivity maps were used as inputs for the continuum SEM43, yielding simulated ionic currents. To validate the conductance obtained via SEM, we considered an analytical model of the conductance of a cylinder that fits within the hollow cavities of the DNP-C1 and DNP-E1. The cylinder model provides a rough estimate of the conductance of the DNA nanopore designs. Two internally tangent cylinders of the DNP-C and DNP-E were utilized to represent the nanopore at two states. The two cylinders have radii of 9.5 and 14.5 nm, and a height of 10 nm. The conductance of the cylinder is given by the bulk conductivity times the cylinder area over its height. The bulk conductivity at 1 M KCl is 11.12 nS/nm.
Confocal microscopy
Cholesterol-tagged DNP-C1 (10 nM, 10 μL) and DNP-E1 (10 nM, 10 μL) were incubated with CellMaskTM-labeled GUVs solutions (890 μL) for 30 min, respectively, to ensure the spontaneous insertion of DNA nanopores into lipid membranes. These mixtures were then added separately to 35 mm μ-Dishes (Ibidi, Germany). Subsequently, each μ-Dish was placed on the objective stage of the microscope and left for another 10 min to allow the GUVs to settle to the bottom of the corresponding μ-Dish. In a typical macromolecular influx assay, equal volumes (20 μM, 100 μL) of FITC-dextran (FD40S) and TRITC-dextran (T1287) were then added to the μ-Dishes to investigate the transmembrane transport capacity of DNP-C1 and DNP-E1. The treated GUVs were then dynamically monitored for 3 h at room temperature with a Nikon C2 confocal microscope (Nikon, Japan) at 20× or 40× magnification. All confocal images were acquired using NIS-Elements AR 5.11. Fluorescent intensity changes of the obtained confocal images were measured and analyzed using ImageJ 1.52a. When the mean interior intensity of an individual GUV was stronger than half of the mean exterior intensity, the corresponding GUV was considered as a FITC-dextran/TRITC-dextran influx GUV. Otherwise, it was classified as a no-influx GUV.
Statistical analysis
Current traces and images are shown as representative of all independent experiments. All data were presented as mean ± SD unless otherwise stated. Data analysis was performed using GraphPad Prism 9.5 (GraphPad, USA). An unpaired two-tailed t-test was applied to evaluate the differences between the two groups. P values less than 0.05 were considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that the source data supporting the findings of this study are available within the paper and its Supplementary information files. Source data are available for Figs. 1–5 and Supplementary Figs. 3, 6–16, 18 in the associated source data file. Source data are provided with this paper.
Code availability
Scripts used to convert oxDNA configurations to all-atom models and calculate the ionic current have been deposited in the Illinois Data Bank under accession code IDB-6464580 https://doi.org/10.13012/B2IDB-6464580_V1.
References
Howorka, S. Building membrane nanopores. Nat. Nanotechnol. 12, 619–630 (2017).
Ballesteros, A. & Swartz, K. J. Regulation of membrane homeostasis by TMC1 mechanoelectrical transduction channels is essential for hearing. Sci. Adv. 8, eabm5550 (2022).
Wang, J., Terrasse, R., Bafna, J. A., Benier, L. & Winterhalter, M. Electrophysiological characterization of transport across outer‐membrane channels from Gram‐negative bacteria in presence of lipopolysaccharides. Angew. Chem. Int. Ed. 59, 8517–8521 (2020).
Manrao, E. A. et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat. Biotechnol. 30, 349–353 (2012).
Wloka, C. et al. Label-free and real-time detection of protein ubiquitination with a biological nanopore. ACS Nano 11, 4387–4394 (2017).
Sun, Y. et al. A highly selective and recyclable NO-responsive nanochannel based on a spiroring opening−closing reaction strategy. Nat. Commun. 10, 1323 (2019).
Zhang, M. et al. Designing biomimic two-dimensional ionic transport channels for efficient ion sieving. ACS Nano 15, 5209–5220 (2021).
Dey, S. et al. A reversibly gated protein-transporting membrane channel made of DNA. Nat. Commun. 13, 2271 (2022).
Xing, Y., Dorey, A. & Howorka, S. Multi-stimuli-responsive and mechano-actuated biomimetic membrane nanopores self-assembled from DNA. Adv. Mater. 35, 2300589 (2023).
Xing, Y., Rottensteiner, A., Ciccone, J. & Howorka, S. Functional nanopores enabled with DNA. Angew. Chem. Int. Ed. 62, e202303103 (2023).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Seeman, N. C. & Sleiman, H. F. DNA nanotechnology. Nat. Rev. Mater. 3, 17068 (2017).
Chen, Y., Wang, F., Feng, J. & Fan, C. Empowering single-molecule analysis with self-assembled DNA nanostructures. Matter 4, 3121–3145 (2021).
Keller, A. & Linko, V. Challenges and perspectives of DNA nanostructures in biomedicine. Angew. Chem. Int. Ed. 59, 15818–15833 (2020).
Linko, V. & Keller, A. Stability of DNA origami nanostructures in physiological media: the role of molecular interactions. Small 19, 2301935 (2023).
Nickels, P. C. et al. Molecular force spectroscopy with a DNA origami–based nanoscopic force clamp. Science 354, 305–307 (2016).
Kosuri, P., Altheimer, B. D., Dai, M., Yin, P. & Zhuang, X. Rotation tracking of genome-processing enzymes using DNA origami rotors. Nature 572, 136–140 (2019).
Nummelin, S. et al. Robotic DNA nanostructures. ACS Synth. Biol. 9, 1923–1940 (2020).
Li, S. et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 36, 258–264 (2018).
Wu, X., Wu, T., Liu, J. & Ding, B. Gene therapy based on nucleic acid nanostructure. Adv. Healthc. Mater. 9, 2001046 (2020).
Jiang, S. et al. nanostructures for therapeutics. Chem 7, 1156–1179 (2021).
Burns, J. R., Seifert, A., Fertig, N. & Howorka, S. A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nat. Nanotech. 11, 152–156 (2016).
Krishnan, S. et al. Molecular transport through large-diameter DNA nanopores. Nat. Commun. 7, 12787 (2016).
Diederichs, T. et al. Synthetic protein-conductive membrane nanopores built with DNA. Nat. Commun. 10, 5018 (2019).
Thomsen, R. P. et al. A large size-selective DNA nanopore with sensing applications. Nat. Commun. 10, 5655 (2019).
Arnott, P. M. & Howorka, S. A temperature-gated nanovalve self-assembled from DNA to control molecular transport across membranes. ACS Nano 13, 3334–3340 (2019).
Lanphere, C., Arnott, P. M., Jones, S. F., Korlova, K. & Howorka, S. A biomimetic DNA‐based membrane gate for protein‐controlled transport of cytotoxic drugs. Angew. Chem. 133, 1931–1936 (2021).
Offenbartl‐Stiegert, D., Rottensteiner, A., Dorey, A. & Howorka, S. A light‐triggered synthetic nanopore for controlling molecular transport across biological membranes. Angew. Chem. Int. Ed. 61, e202210886 (2022).
Plesa, C. et al. Ionic permeability and mechanical properties of DNA origami nanoplates on solid-state nanopores. ACS Nano 8, 35–43 (2014).
Xing, Y., Dorey, A., Jayasinghe, L. & Howorka, S. Highly shape- and size-tunable membrane nanopores made with DNA. Nat. Nanotechnol. 17, 708–713 (2022).
Perozo, E., Marien, D., Cortes & Cuello, L. G. Structural rearrangements underlying K + -channel activation gating. Science 285, 73–78 (1999).
Simmel, S. S., Nickels, P. C. & Liedl, T. Wireframe and tensegrity DNA nanostructures. Acc. Chem. Res. 47, 1691–1699 (2014).
Sala, L. et al. Cisplatin-cross-linked DNA origami nanostructures for drug delivery applications. ACS Appl. Nano Mater. 5, 13267–13275 (2022).
Liu, F. et al. A tetrahedral DNA nanorobot with conformational change in response to molecular trigger. Nanoscale 13, 15552–15559 (2021).
Ahmad, K. et al. Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations. Nat. Commun. 14, 3630 (2023).
Lanphere, C. et al. Design, assembly, and characterization of membrane-spanning DNA nanopores. Nat. Protoc. 16, 86–130 (2021).
Ouldridge, T. E., Louis, A. A. & Doye, J. P. K. Structural, mechanical, and thermodynamic properties of a coarse-grained DNA model. J. Chem. Phys. 134, 085101 (2011).
Šulc, P. et al. Sequence-dependent thermodynamics of a coarse-grained DNA model. J. Chem. Phys. 137, 135101 (2012).
Snodin, B. E. K. et al. Introducing improved structural properties and salt dependence into a coarse-grained model of DNA. J. Chem. Phys. 142, 234901 (2015).
Chhabra, H. et al. Computing the elastic mechanical properties of rodlike DNA nanostructures. J. Chem. Theory Comput. 16, 7748–7763 (2020).
Maffeo, C., Luan, B. & Aksimentiev, A. End-to-end attraction of duplex DNA. Nucleic Acids Res. 40, 3812–3821 (2012).
Choi, T., Li, Z., Song, G. & Chen, H.-F. Comprehensive comparison and critical assessment of RNA-specific force fields. J. Chem. Theory Comput. 20, 2676–2688 (2024).
Wilson, J., Sarthak, K., Si, W., Gao, L. & Aksimentiev, A. Rapid and accurate determination of nanopore ionic current using a steric exclusion model. ACS Sens. 4, 634–644 (2019).
Choudhary, A., Maffeo, C. & Aksimentiev, A. Multi-resolution simulation of DNA transport through large synthetic nanostructures. Phys. Chem. Chem. Phys. 24, 2706–2716 (2022).
Kowalczyk, S. W., Grosberg, A. Y., Rabin, Y. & Dekker, C. Modeling the conductance and DNA blockade of solid-state nanopores. Nanotechnology 22, 315101 (2011).
Hemmig, E. A. et al. Optical voltage sensing using DNA origami. Nano Lett. 18, 1962–1971 (2018).
Göpfrich, K. et al. Large-conductance transmembrane porin made from DNA origami. ACS Nano 10, 8207–8214 (2016).
Göpfrich, K. et al. Ion channels made from a single membrane-spanning DNA duplex. Nano Lett. 16, 4665–4669 (2016).
Fragasso, A. et al. Reconstitution of ultrawide DNA origami pores in liposomes for transmembrane transport of macromolecules. ACS Nano 15, 12768–12779 (2021).
Douglas, S. M. et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009).
Bohlin, J. et al. Design and simulation of DNA, RNA and hybrid protein–nucleic acid nanostructures with oxView. Nat. Protoc. 17, 1762–1788 (2022).
Doye, J. P. K. et al. The oxDNA coarse-grained model as a tool to simulate DNA origami. Methods Mol. Biol. 2639, 93–112 (2023).
Maffeo, C. & Aksimentiev, A. MrDNA: a multi-resolution model for predicting the structure and dynamics of DNA systems. Nucleic Acids Res. 48, 5135–5146 (2020).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Belkin, M. & Aksimentiev, A. Molecular dynamics simulation of DNA capture and transport in heated nanopores. ACS Appl. Mater. Interfaces 8, 12599–12608 (2016).
Acknowledgements
This research was supported in part by the National Natural Science Foundation of China (Grant 62273052 to X.L.), the Beijing Natural Science Foundation (IS23062 to T.A.), and in part by the Grant-in-Aid for Scientific Research (23K22712 to T.A.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. H.C., C.M., and A.A. acknowledge support from the Human Frontier Science Project (RGP0047/2020), and the supercomputer time provided through the ACCESS allocation grant (MCA05S028).
Author information
Authors and Affiliations
Contributions
X.L. and F.L. conceived the idea of the project. T.A., Q.H., and X.L. supervised the project. F.L. constructed and characterized the DNA nanopores. F.L. and X.L. performed all electrophysiological experiments. H.C., C.M., and A.A. performed all computational characterizations. F.L. conducted the gel electrophoresis and confocal microscopy. Z.C. analyzed the TEM images using computer vision-assisted methods. F.L., H.C., and Z.C. drew the schematic illustrations. F.L., X.L., and H.C. collected and interpreted data. X.L. and F.L. wrote the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Veikko Linko and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, X., Liu, F., Chhabra, H. et al. A lumen-tunable triangular DNA nanopore for molecular sensing and cross-membrane transport. Nat Commun 15, 7210 (2024). https://doi.org/10.1038/s41467-024-51630-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51630-0
- Springer Nature Limited