Abstract
Fertility requires the faithful proliferation of germ cells and their differentiation into gametes. Controlling these cellular states demands precise timing and expression of gene networks. Nucleic acid binding proteins (NBPs) play critical roles in gene expression networks that influence germ cell development. There has, however, been no functional analysis of the entire NBP repertoire in controlling in vivo germ cell development. Here, we analyzed germ cell states and germline architecture to systematically investigate the function of 364 germline-expressed NBPs in the Caenorhabditis elegans germ line. Using germline-specific knockdown, automated germ cell counting, and high-content analysis of germ cell nuclei and plasma membrane organization, we identify 156 NBPs with discrete autonomous germline functions. By identifying NBPs that control the germ cell cycle, proliferation, differentiation, germline structure and fertility, we have created an atlas for mechanistic dissection of germ cell behavior and gamete production.
Similar content being viewed by others
Introduction
The production of oocytes and sperm (gametes) from germ cells is required for animal reproductive success. To ensure overall fitness, the balance between germ cell proliferation (self-renewal) and differentiation into gametes must be tightly controlled. Model organism research has previously identified regulatory mechanisms that control gene expression and RNA stability to maintain this balance in germ cell behavior. In the Caenorhabditis elegans model, for example, the PUF-domain RNA binding proteins FBF-1/2 promote germline stem cell (GSC) fate, with the mammalian ortholog required for the establishment of female germ cells1,2. Furthermore, the importance of Notch signaling in establishing and maintaining GSC pools was first discovered in C. elegans3. Thus, the C. elegans germ line is a valuable model for discovering conserved mechanisms governing germ cell behavior and fertility. Here, we map the nucleic acid binding protein (NBP) requirements for germ cell and gamete development in C. elegans.
The C. elegans germ line originates from a single embryonic blastomere (P4) that divides into two primordial germ cells (Z2 and Z3)4. These embryonic germ cells are arrested at the G2 stage of the cell cycle and are transcriptionally quiescent to preserve germ cell fate and protect against inappropriate expression of somatic genes5,6. Transcriptional quiescence in these primordial germ cells (PGCs) is maintained by inhibiting RNA Polymerase II (RNA Pol II) and by nucleosome modifications associated with chromatin repression5,7. After embryos hatch and larvae start to feed, germline transcription is activated, and Z2 and Z3 exit G2 to commence mitosis during larval stage 1 (L1)4. Germ cells continue to proliferate throughout larval development and into adulthood, with meiosis commencing from larval stage 3 (L3)8. This initial differentiation program generates sperm, that are deposited in the spermatheca, before the germ line switches to producing oocytes in adults9. The adult germ line is organized in a distal to proximal manner. The distal progenitor zone (PZ) contains stem cells, mitotically dividing germ cells, and meiotic S-phase germ cells, which then enter early meiotic prophase (leptotene and zygotene) in the transition zone (TZ). As germ cells move from the TZ they enter meiotic pachytene prior to gametogenesis. As transcription is activated early during larval development, NBPs likely play important roles in germ cell transitions and cell behavior during larval and adult development. Indeed, previous studies have identified key NBPs that regulate germ cell and gamete development10,11,12,13. However, no systematic in vivo assessment of NBP function in the C. elegans germ line, or in any organism, has been conducted. As a result, we sought to identify the NBPs required for C. elegans germline development.
Based on the presence of a DNA-binding domain, the C. elegans genome is predicted to encode 875 transcription factors (TFs), some of which are experimentally verified14. However, some of these predicted TFs likely have more broad functions in splicing and basal RNA Polymerase II/III regulation. Therefore, we categorize these factors ‘nucleic acid binding proteins’ to embrace other potential functions. Of the 875 NBPs encoded by the C. elegans genome, we identified 364 NBPs that are germline-enriched14,15,16. As most of these NBPs are also expressed in the soma, we dissected their germline functions by exploiting a germline-specific RNA-mediated interference (RNAi) approach16,17,18. Following germline-specific RNAi silencing from the L1 larval stage, we performed semi-automated cell counting and high-content analysis of germline architecture to interrogate a broad spectrum of readouts in adult germ lines. This analysis encompassed the ‘distal germ line’ where mitotic germ cells, Notch signaling, and early meiotic transition were analyzed; and the ‘proximal germ line’ where meiotic progression, oocyte/sperm production, and germline structure were analyzed. We identified 156 NBPs that regulate germ cell development, including 8 NBPs that are essential for fertility. Our research provides a regulatory atlas of germ cell development, which is a valuable resource and experimental platform for understanding how specific germ cell fate decisions and gamete production are controlled.
Results
NBP germline profiling and genetic screen validation
To identify germline-expressed NBPs (GNBPs), we extruded wild-type adult hermaphrodite germ lines and performed RNA-sequencing from extracted RNA (Fig. 1a and Supplementary Data 1). Combining our germline transcriptome dataset with previously published germline transcriptome studies identified 364 GNBPs (Supplementary Data 1 and 2)15,16. These GNBPs were detected in all three of our RNA-sequencing samples, and both germ lines analyzed in Tzur et al., or GNBPs with >10 counts per million in Cao et al. (Supplementary Data 2)15,16. Of the 364 GNBPs, 150 have been ascribed a broad C. elegans reproductive functions (sterility/fertility/brood size) (Supplementary Data 2), however, the precise germline function of these GNBPs is not well understood.
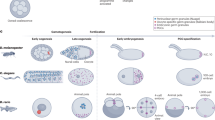
a Schematic of the C. elegans hermaphrodite germ line (left), identification of germline-expressed NBPs by RNA sequencing (center), and representative images and phenotypic readouts for high-content distal and proximal germline analysis (right). The distal germ line is labeled with DAPI (nuclei - blue), SYGL-1 (Notch target - magenta), and pH3 (M-phase chromosomes - green). PZ progenitor zone, TZ transition zone. The proximal germ line is marked with transgenic fluorophores: red = nuclei; green = plasma membrane. Scale bars = 20 μm. b–d Heatmaps showing phenotypes for GNBP silencing in the distal germ line (b ≥20% change compared to control), proximal germ line (c ≥50% of animals with the phenotype) and both distal and proximal germ line (d). b NBP family categories (colored boxes); PZ, TZ, SYGL-1+, pH3+, and mitotic index phenotypes shown as percentage change compared to control (blue = decrease; red = increase); germline expression of each NBP (gray bars = log2 CPM); mammalian ortholog associated with the reproduction GO term (blue boxes = no, black boxes = yes). NBP nucleic acid binding protein family. c Labeling as in b, except purple (≥50% of germ lines with the defect); blue (<50% of germ lines with the defect). Phenotypes shown: small germ line, rachis defect (narrow or wide rachis), meiotic defect (multinucleated cells, abnormal meiotic progression), apoptosis, oocyte defect (delayed expansion at the turn, difference in single-array oocyte number, difference in budded oocyte number, vesiculation), sperm defect (mislocated sperm, incomplete spermatogenesis). d Labeling as in b and c. Note: heatmaps do not include essential GNBPs identified in the screens—this is detailed in Fig. 4 and Supplementary Data 9. e NBP family distribution of the 875 C. elegans NBPs, 106 GNBPs regulating the distal germ line, 60 GNBPs regulating the proximal germ line, and 18 GNBPs regulating both distal and proximal germline development (Supplementary Data 4). NBP families shown: bZIP basic leucine zipper domain, HD homeodomain, HMG high mobility group box domain, MYB myeloblastosis viral oncogene homolog, NHR nuclear hormone receptors, T-box, WH winged helix, ZF zinc finger. Source data are provided as a Source Data file.
By analyzing published NBP expression profiles, we found that most GNBPs are also expressed in the soma (Supplementary Data 2)16,18. Therefore, to circumvent the potential somatic effects of NBP silencing, we utilized a germline-specific RNAi approach to identify germline-specific NBP functions17. Animals lacking the RDE-1 Argonaute are systemically RNAi-deficient19. In our genetic screen, we utilized a rde-1(mkc36) indel mutant in which RDE-1 is re-supplied under the germline-specific sun-1 promoter and 3’ untranslated region (UTR)—thus enabling germline-specific RNAi17. We found that this transgenic rescue strain exhibits wild-type germline development (Supplementary Fig. 2). To test the efficacy of this strain for germline-specific RNAi by feeding, we silenced the RNA Polymerase II encoding gene ama-1 from the first larval stage (L1) for 66 hrs20. sun-1p::rde-1; rde-1(mkc36) animals fed with ama-1 RNAi bacteria grew to adulthood, however, their germ lines did not develop, while ama-1 RNAi wild-type animals arrested as young larvae (Supplementary Fig. 1a). Thus, germline-specific inhibition of transcription prevents germline development without overtly affecting somatic development. As additional proof-of-concept for the germline-specific RNAi approach, we silenced two genes that autonomously control germline development—gld-1 (encodes an RNA binding protein) and mog-7 (encodes a splicing factor). Consistent with previous studies, gld-1 RNAi causes proximal germline tumors (86% of germ lines are tumorous; n = 51) and mog-7 RNAi causes spermatogenic germ lines (32% of germ lines are spermatogenic; n = 66) in sun-1p::rde-1; rde-1(mkc36) animals (Supplementary Fig. 1a, b)21,22,23. Prior to embarking on a large-scale germline-specific screen, we needed to confirm that somatic RNAi was inhibited in sun-1p::rde-1; rde-1(mkc36) animals. We examined somatic phenotypes caused by RNAi silencing of hypodermal (bli-3), intestinal (elt-2), and muscle (unc-112) genes. We found that the knockdown of these genes from the L1 stage caused robust somatic phenotypes in wild-type but not in sun-1p::rde-1; rde-1(mkc36) animals, as shown previously (Supplementary Fig. 1c)17,24.
To enable functional mapping of the 364 GNBPs, we obtained RNAi feeding clones: 315 from previously generated RNAi libraries and 49 that we cloned (Supplementary Data 3)14,25,26. All RNAi clones were confirmed by sequencing prior to analysis. We profiled GNBP function in the germ line by feeding L1 hermaphrodites with Escherichia coli HT115 bacteria expressing individual NBP RNAi clones and imaged and analyzed one-day adult germ lines (Fig. 1a). We screened GNBP function in hermaphrodite germline development using two parallel approaches for distal and proximal germline analysis (Fig. 1a). Distal germline analysis was performed with confocal imaging and semi-automated cell counting of extruded germ lines of the 3xflag::sygl-1; sun-1p::rde-1; rde-1(mkc36) strain that we confirmed exhibits wild-type germ cell number and brood size (Fig. 1a and Supplementary Fig. 2). Following GNBP RNAi silencing from the L1 stage, we quantified germ cell number in the PZ and TZ by DAPI staining (PZ and TZ size), plus visualized M-phase chromosomes (anti-phospho-histone H3 (pH3) staining) and Notch signaling (SYGL-1 immunofluorescence) in adults (Fig. 1a)27. For proximal germline analysis, we examined gonad architecture, meiotic cell behavior, and gamete development using a transgenic strain co-expressing fluorescent markers targeting chromosomes (mCherry-histone H2B) and plasma membrane (GFP-PH(PLC1δ1)) (Fig. 1a and Supplementary Fig. 3). This approach for analyzing the proximal germ line was previously used for a group of 554 essential genes28, however, their work analyzed <3% of GNBPs and used the RNAi soaking method (not germline-specific) from the L4 stage. Thus, our work is distinct from this previous study with regards to genes investigated, phenotypes analyzed, and methods/timing of gene silencing.
Profiling NBP function in the germ line
To determine the in vivo functional contributions of NBPs in the germ line, we performed two independent reverse genetic screens (as described above) to study the distal and proximal germ line. We selected key germline regulators using the following criteria: distal germline analysis (≥20% significant change in germ cell number of PZ, TZ, SYGL-1+, pH3+, or mitotic index compared to control) and proximal germline analysis (≥4 of 8 germ lines exhibiting a phenotype). This analysis revealed 156 GNBPs that impact germ cell development (Supplementary Data 4 and 9). Silencing of 88 GNBPs caused distal phenotypes, 42 proximal phenotypes, and 18 GNBPs phenotypes in both germline regions. We found that silencing of a further 8 GNBPs in both screens resulted in immature germ lines (Supplementary Data 4), a detailed analysis of which will be discussed later. In addition, the silencing of 10 GNBPs resulted in 20–100% immature germ lines in the proximal screen only, among which 5 also caused distal phenotypes (Supplementary Data 4). We hypothesize that the sterility phenotype of the 10 genes identified only in the proximal screen is either due to increased RNAi sensitivity or a synthetic effect of the RNAi silencing and the transgenes used (mCherry-histone H2B and plasma membrane GFP-PH(PLC1δ1)), as we did not observe sterility in the distal screen (Supplementary Data 4). Finally, of the 156 GNBPs we identified in our screens, 82 have previously reported germline-related phenotypes in C. elegans (Supplementary Data 5).
We classified NBPs that control germline development based on broad DNA-binding domain families and found no obvious over-representation of the NBP family that is functionally required for germline development (Fig. 1e). We wondered whether the NBPs we identified in our primary screen were previously associated with germ cell/gamete development and reproduction. We thus compared our list of NBPs hits to genes associated with the ‘reproduction’ gene ontology (GO) terms (GO:0019952, GO:0050876) in mice, rats and humans (https://www.flyrnai.org/cgi-bin/DRSC_orthologs.pl). We found that 88 GNBPs we identified to regulate C. elegans germline development in our primary screens have mammalian orthologs, of which 43 are associated with germ cell/gamete development and reproduction GO terms (Fig. 1 and Supplementary Data 6).
Regulation of distal germ cell behavior
At the distal PZ, maintenance of the GSC pool is influenced by interaction between Notch ligands (LAG-2/APX-1), expressed on the somatic distal tip cell, and the GLP-1 Notch receptor expressed on the surface of germ cells3,29,30,31,32. This Notch ligand-receptor interaction promotes GSC self-renewal by regulating the two redundantly acting post-transcriptional regulators SYGL-1 and LST-133,34,35,36.
Our primary screen identified 23 GNBPs that, when silenced, have a ≥20% change in PZ size (Fig. 1 and Supplementary Data 4). To identify high-confidence regulators of germline development, we performed a secondary screen of these hits in triplicate (Fig. 2a and Supplementary Fig. 4 and Supplementary Data 10). We found that silencing of 11 GNBPs robustly repeated our initial findings with significant changes in PZ size (Fig. 2a and Supplementary Fig. 4). These factors include two nuclear hormone receptors (NHR-84 and NHR-146), HSF-1/HSF-1, DAF-19/RFX, LET-607/CREB, F54F2.9/DNAJC1, ZK546.5, XND-1, ZTF-23, C02F5.12 and C30G4.4. Of these 11 GNBPs, only HSF-1 and XND-1 have known roles in C. elegans germ cell proliferation11,37.
a Quantification of nuclei number in the PZ of 3xflag::sygl-1; sun-1p::rde-1; rde-1(mkc36) one-day adults. n = 24. Green dashed line = average of the control. P values assessed by two-tailed multiple unpaired t test with no correction. b Quantification of nuclei number of one-day adult germline PZ of sun-1p::rde-1; rde-1(mkc36) treated with control RNAi and glp-1(ar202); sun-1p::rde-1; rde-1(mkc36) treated with control or experimental RNAi. n = 31, 31, 31, 31, 29, 32, 37, 32, 33, 30, 34, 31 and 31. P values assessed by one-way analysis of variance (ANOVA) with no correction for multiple comparisons. c Quantification of nuclei number of one-day adult germline PZ of sun-1p::rde-1; rde-1(mkc36) treated with control RNAi and glp-1(e2141); sun-1p::rde-1; rde-1(mkc36) treated with control or experimental RNAi. n = 30. P values assessed by one-way analysis of variance (ANOVA) with no correction for multiple comparisons. d, e Quantification of SYGL-1+ nuclei number (d) and confocal micrographs of germline PZ (e) of 3xflag::sygl-1; sun-1p::rde-1; rde-1(mkc36) one-day adults. n = 24. Green dashed line = average of the control (d); yellow dashed line = PZ/TZ boundary (e–left) and white line = SYGL-1 boundary (e–right). P values assessed by two-tailed multiple unpaired t test with no correction. f Proliferation rates of the C. elegans germ line after GNBP RNAi. RNAi was performed in triplicate from the L1 stage for 66 hrs before commencing EdU labeling, and germ lines were collected and imaged after 4 and 10 hrs. The 3xflag::sygl-1; sun-1p::rde-1; rde-1(mkc36) strain was used in this experiment. Data for each replicate shown; n = 63, 61, 63, 61, 62, 61, 58, 61, 63, 62, and 61 germ lines for each condition. P values assessed by two-tailed multiple unpaired t test with no correction. g Confocal micrographs showing DAPI and EdU+ nuclei after 4 and 10 hrs of EdU labeling. All RNAi was performed from the L1 stage. All Data was generated from three independent experiments, and results were normalized to respective controls in a–d. Error bars indicate SEM. Scale bars = 20 μm. Source data are provided as a Source Data file.
The PZ houses self-renewing stem cells, germ cells undergoing the final round of the mitotic cell cycle, and germ cells in meiotic S-phase38. To better understand the cellular and molecular effects underpinning the changes in PZ size, we analyzed the roles of these 11 GNBPs in Notch signaling and the mitotic cell cycle (Fig. 2a–g and Supplementary Fig. 4 and 5). First, we used a glp-1(ar202) temperature-sensitive gain-of-function mutant that at the semi-permissive temperature of 15 °C generates ~25% more PZ germ cells than wild-type animals (Fig. 2b and Supplementary Fig. 4e). We found that RNAi silencing of hsf-1 and xnd-1 decreased the number of PZ germ cells in glp-1(ar202gf) animals at the semi-permissive temperature (Fig. 2b). Next, we used a glp-1(e2141) temperature-sensitive loss-of-function mutant that at the semi-permissive temperature of 15 °C generates ~25% fewer PZ germ cells than wild-type animals (Fig. 2c and Supplementary Fig. 4e). We found that only RNAi silencing of xnd-1 decreased the number of PZ germ cells in glp-1(e2141lf) animals at the semi-permissive temperature (Fig. 2c). Together, these data suggest that xnd-1 regulates PZ germ cell number independently of glp-1. In contrast, RNAi silencing of C02F5.12, C30G4.4, daf-19, F54F2.9, and ZK546.5 increases PZ size in wild-type animals but not in glp-1 loss or gain-of-function animals at the semi-permissive temperature. This suggests that to regulate PZ cell number these GNBPs require wild-type GLP-1 function (Fig. 2a–c).
Maintenance of the GSC pool in the PZ is regulated by two direct GLP-1 target genes SYGL-1 and LST-133,34. SYGL-1 and LST-1 proteins are present in partially overlapping gradients through the GSC pool33. Previous studies showed that overexpression of SYGL-1 or LST-1 can expand the GSC pool33 and sygl-1 RNAi knockdown can reduce the PZ length39. Therefore, we quantified endogenous 3xFLAG::SYGL-1 and 3xFLAG::LST-1 after silencing the 11 GNBPs that control PZ size to determine their potential roles in regulating Notch signaling (Fig. 2d, e and Supplementary Fig. 4b–d). We found that RNAi silencing of C02F5.12 and ZK546.5 increased the number of SYGL-1+ germ cells, consistent with their knockdown leading to an increase in PZ size (Fig. 2a, d, e). This implies that C02F5.12 and ZK546.5 inhibit Notch signaling. Given that RNAi silencing of C02F5.12 and ZK546.5 did not affect the glp-1(gf) increase of PZ size, they may function upstream of GLP-1. We also found that RNAi silencing of hsf-1 and xnd-1 decreased the number of SYGL-1+ germ cells, consistent with their knockdown leading to a decrease in PZ size (Fig. 2a, d, e). This suggests that HSF-1 and XND-1 promote Notch signaling. Given that HSF-1 and XND-1 are required for glp-1(gf) induced increase in PZ size, they likely function downstream of GLP-1. Interestingly, both HSF-1 and XND-1 have ChIP peaks in the promoter regions of sygl-1 (ENCSR512EIF, ENCSR327NRA), suggesting a direct regulatory relationship with this Notch target gene40,41. In contrast to the SYGL-1+ analysis, we did not detect changes in LST-1+ in the PZ after silencing these 11 GNBPs (Supplementary Fig. 4c, d), even though we could detect small changes in LST-1+ cells in glp-1 mutants (Supplementary Fig. 4e, f). These data suggest that the change in PZ size following silencing of C02F5.12, ZK546.5, hsf-1 and xnd-1 may be due to altered Notch-dependent SYGL-1 regulation.
PZ size is also impacted by the mitotic cell cycle that is genetically separable to Notch signaling38. Within the PZ, a cohort of germ cells act as stem cells that self-renew by mitosis or differentiate into gametes42,43. To examine whether any of the 11 GNBPs impact progression through the cell cycle, we quantified cell proliferation using 5-ethynyl-2′-deoxyuridine (EdU) staining (Fig. 2f, g)38,44. We silenced each GNBP from the L1 stage, fed the resultant adults with EdU-labeled bacteria for 4 hrs or 10 hrs, and then counted EdU+ germ cells (Fig. 2f, g). Using these data, we calculated the rate of proliferation as the number of germ cells incorporating EdU per hour (Fig. 2f, g). As previously reported, control animals had a proliferation rate of ~23 cells/hour (Fig. 2f)38. We found that RNAi silencing of C02F5.12, daf-19, and ZK546.5 increased the germ cell proliferation rate, which may contribute to increases in PZ size observed by increasing the number of mitotic cells (Fig. 2f, g). Taken together, these results reveal key GNBPs that control PZ size, likely through fine-tuning Notch signaling, cell cycle dynamics, and possibly other mechanisms.
Antibody staining to detect phosphorylation of histone H3 (pH3) at serine 10 is used to mark mitotic M-phase germ cells45,46. No pH3+ cells are observed after proliferative zone cells have entered meiosis38. Therefore, changes in the number of pH3+ germ cells could indicate changes in the total number of mitotic gem cells, or changes in M-phase length with respect to the entire cell cycle. We found that of the 11 GNBPs analyzed, only hsf-1 silencing increased the number of pH3+ germ cells (Supplementary Fig. 5a, b). This suggests that hsf-1 silencing increases mitotic germ cell number or M-phase length, or potentially accelerates other cell cycle phases. However, further investigation of hsf-1 functions in mitosis and early meiosis is required to explain the contradictory results of decreased PZ size, decreased SYGL-1+ cells, and unchanged proliferation rate following hsf-1 silencing. A previous study found that germline-specific HSF-1 protein depletion from hatching causes a severe reduction in germ cells and depletion from the mid-L3 stage causes a decrease in EdU labeled germ cells11. These contradictory findings suggest the effect of HSF-1 knockdown on the germ line is dosage or developmental stage dependent. Another method frequently used to assess germline proliferation is the mitotic index (MI), which is the percentage of PZ cells that are M-phase47. In addition to hsf-1, we found silencing of F54F2.9 and nhr-146 caused changes in MI without changing the absolute number of pH3+ cells (Supplementary Fig. 5a–c). Although silencing of F54F2.9 and nhr-146 caused changes in PZ size, there were no changes in the number of SYGL-1+ cells or proliferation rate (Fig. 2d, f). Thus, it is possible that these genes are not involved in mitosis, and the change in MI is a result of changes in overall PZ size, which also includes meiotic M-phase cells.
Proximal to the PZ, the germline TZ houses a cohort of crescent-shaped nuclei that is characteristic of early meiotic prophase (leptotene and zygotene) germ cells48. We found that some GNBPs are also required for normal TZ size (ZK546.5, HSF-1/HSF1, XND-1, ZTF-23, and NHR-146), suggesting that meiotic prophase progression and/or chromosome pairing may be perturbed following silencing of these GNBPs (Supplementary Fig. 5d, e). Interestingly, the changes in TZ size paralleled changes in PZ size—ZK546.5 silencing increased PZ and TZ size; hsf-1, xnd-1, ztf-23, and nhr-146 silencing decreased PZ and TZ size (Fig. 2h). This suggests that when these genes are silenced, germ cell output from the PZ impacts TZ size. Finally, we found that 5 out of the 11 GNBPs that are important for distal germ cell behavior also impact the proximal germ line. ZK546.5 silencing caused incomplete spermatogenesis (7/8 germ lines), ztf-23 and C02F5.12 silencing caused changes in the number of single array or budded oocytes, and C30G4.4 and nhr-146 silencing caused germ cell multinucleation (Supplementary Data 4). Thus, these GNBPs likely play roles in both mitotic and meiotic germ cell behavior. Taken together, our distal germline screen discovered multiple GNBPs that control germ cell behavior.
Regulation of meiotic cell behavior and gamete development
As germ cells progress proximally, they mature into sperm during late larval development before committing to the oogenic program. To examine the function of GNBPs in these processes, we utilized a transgenic strain in which fluorescent markers targeting chromosomes (mCherry-histone H2B) and plasma membrane (GFP-PH(PLC1δ1)) enable gonad architecture to be visualized (Fig. 1a and Supplementary Fig. 3)28,49. We identified GNBP functions in the proximal germ line including germline structure (small germ line, narrow rachis), meiosis (multinucleation, meiotic progression, and apoptosis), sperm production and behavior (spermatogenesis, sperm localization), and oocyte production (germ cell expansion, number of oocytes, oocyte budding and vesiculation) (Supplementary Fig. 3 and Supplementary Data 4). The most prominent phenotype we observed was multinucleation of germ cells, and thus, we focused our subsequent analysis on this phenotype.
The C. elegans hermaphrodite gonad is a syncytium with germ cell nuclei partially enclosed by a plasma membrane. Multinucleated germ cells (MNCs) can occur in both wild-type and mutant animals at varying frequency50. Persistent MNCs would generate multinuclear oocytes and infertile embryos and are therefore cleared by apoptosis prior to generating gametes. Earlier studies showed that failed germ cell division in the PZ and an unstable rachis can cause multinucleation, while in other cases, including in wild-type animals, MNCs form at specific germline structures called folds50,51. In our primary screen, we identified 26 GNBPs that have >25% increase in germ lines with MNCs compared to control animals (Supplementary Data 4). To confirm this result, we selected 10 GNBPs with the strongest penetrant phenotypes (DMD-7/DMRTC1, BAZ-2/BAZ2A, JUN-1/JUN, MTER-4, LIN-26, MADF-5, NHR-40/HNF4, NHR-114/NR1H3, NHR-161/PPAR-alpha, and UNC-62/MEIS) and repeated RNAi silencing in triplicate (Fig. 3). We confirmed that silencing of all 10 GNBPs increased MNCs in both one-day and two-day adults (Fig. 3a, b and Supplementary Fig. 6a, b). Of these GNBPs, only NHR-114 had previously been associated with germ cell multinucleation in C. elegans52. We used confocal microscopy to analyze the PZ of one-day adults following GNBP silencing and detected no MNCs, suggesting that MNC formation is not due to aberrant mitotic division, as shown previously51.
a Quantification of multinucleated germ cells of pie-1p::mCherry::his-58; pie-1p::GFP::PH(PLC1δ1); sun-1p::rde-1; rde-1(mkc36) two-day adults following RNAi from the L1 stage. n = 30, 30, 31, 30, 31, 33, 30, 30, 30, 30 and 30. P values assessed by one-way ANOVA with no correction for multiple comparison. b Fluorescence micrographs of wild-type or ced-3(rp190) two-day adult germ lines in the pie-1p::mCherry::his-58; pie-1p::GFP::PH(PLC1δ1); sun-1p::rde-1; rde-1(mkc36) strain. Control RNAi and RNAi of dmd-7 and baz-2 were applied from the L1 stage in wild-type animals. Dash lines = border between the TZ (above) and pachytene region (below) of the germ line. Asterisks = multinucleated cells; triangles = apoptotic cells. c, d Quantification of apoptotic germ cells (c) and multinucleated germ cells (d) in two-day adults of pie-1p::mCherry::his-58; pie-1p::GFP::PH(PLC1δ1); sun-1p::rde-1; rde-1(mkc36) strain (wild-type and ced-3(rp190) animals). n = 30 (c), 30 and 33 (d). P values assessed by two-tailed unpaired t test. e Quantification of multinucleated germ cells in ced-3(rp190), pie-1p::mCherry::his-58; pie-1p::GFP::PH(PLC1δ1); sun-1p::rde-1; rde-1(mkc36) two-day adults following RNAi from the L1 stage. n = 30. P values assessed by one-way ANOVA with no correction for multiple comparison. f, g Confocal micrographs of DAPI (white) and phalloidin (pink) staining (f) and quantification of germline folding events in the pachytene region (g) of sun-1p::rde-1; rde-1(mkc36) one-day adults. Images for control RNAi (L4440) and lin-26 RNAi are shown in f (note—only one plane is shown). Yellow dashed line = border between the TZ (right) and pachytene region (left) of the germ line. n = 36, 29, 31, 24, 29, 32, 29, 24, 23, 28 and 26. P values assessed by one-way ANOVA with no correction for multiple comparison. All RNAi was performed from the L1 stage. Data was generated from three (a, c–e) or four independent experiments (g). Error bars indicate SEM. Scale bars = 20 μm. Source data are provided as a Source Data file.
As part of normal germline development, multinucleate cells are removed by physiological apoptosis to prevent them from potentially generating multiploid oocytes50,53. The CED-3 executioner caspase is essential for physiological germline apoptosis53, and thus germ lines lacking ced-3 contain many MNCs50. To determine whether the regulation of germ cell multinucleation by GNBPs is dependent on CED-3, we silenced the 10 GNBPs that inhibit MNC formation in ced-3 mutant animals. As the ced-3 gene is linked to the pie-1p::mCherry::his-58 transgene that is used to count multinucleation events, we used CRISPR/Cas9 genome editing to generate a 3951 bp ced-3(rp190) deletion allele (Supplementary Fig. 7). These animals exhibit overt persistent apoptotic corpses confirming defective apoptosis (Supplementary Fig. 7b, c). Further, we did not detect apoptosis in the germ lines of ced-3(rp190) animals, and loss of CED-3 caused an increase in multinucleated germ cells (Fig. 3b–d), confirming that apoptosis is involved in the clearance of multinucleated germ cells50.
We next counted germline MNCs in ced-3(rp190) two-day adults after GNBP RNAi silencing from the L1 stage (Fig. 3e). We found that RNAi silencing of jun-1, mter-4, lin-26, nhr-40, nhr-114, nhr-161, and unc-62 caused no change in MNC number in the ced-3(rp190) background, suggesting that silencing these GNBPs lead to an increase in multinucleated cells by interfering with apoptosis. In contrast, silencing of dmd-7, baz-2, and madf-5 increased MNCs independent of loss of ced-3, suggesting that these GNBPs promote mono-nucleation through mechanisms that are independent of apoptosis. However, caspase-3 can play roles outside apoptosis54,55, therefore it is possible that CED-3 is involved in multinucleation formation through other mechanisms. In addition, we observed that MNCs are primarily located in the distal half of the pachytene region in wild-type and ced-3(rp190) animals (Fig. 3b), suggesting that mechanisms independent of physiological apoptosis can remove/resolve multinucleation.
A previous study revealed that meiotic germline structure impacts the location and occurrence of multinucleation50. During proliferation, folds in the germline syncytium are formed and multinucleate cells occur during fold eversion50. As the size and complexity of germline folds is correlated with multinucleation, we assessed whether the 10 GNBPs with high numbers of MNCs regulate germline folding. We used phalloidin to stain the germline actin cytoskeleton after RNAi silencing of these GNBPs and measured germline folding by counting the rows and numbers of cells protruding inwards from the single layer of cells that form the tube-like structure in the pachytene region (Fig. 3f, g and Supplementary Fig. 6c). We found that RNAi silencing of dmd-7, baz-2, mter-4, lin-26, nhr-114, and nhr-161 caused excess germline folding that may result in the increased multinucleation observed (Fig. 3f, g and Supplementary Fig. 6c). Taken together, these data reveal 10 GNBPs that act in the germ line to maintain uninuclear germ cells. Cell behavior is likely controlled by these GNBPs through gene networks connected to the apoptotic pathway and germline morphogenesis.
Essential regulators of germline development
Our initial distal germline screen identified eight GNBPs (CDC-5L, ZNF-622, VEP-1, F23B12.7, F33H1.4, F58G1.2, REPO-1, and TBP-1) that when silenced from the L1 stage result in small germ lines that do not generate embryos in one-day adults (Supplementary Data 4). These GNBPs are mostly uncharacterized but have predicted general functions in controlling gene expression (Supplementary Data 5). Analysis of broods after silencing these GNBPs from the L1 stage showed almost complete sterility (Fig. 4a). An exception was repo-1 RNAi, which generated ~230(±30.47) progeny, however, most resultant embryos did not hatch (Fig. 4a). To investigate when these GNBPs are required during development, we performed temporal germline analysis by silencing each GNBP from the L1 stage and examining germline size and morphology at each larval stage through to adult (Fig. 4b and Supplementary Fig. 10).
a Quantification of hatched larvae and dead eggs (brood size) following germline-specific RNAi from the L1 stage. P values were assessed by two-tailed unpaired t test comparing the brood size (hatched larvae + dead eggs) of experimental RNAi with control RNAi. n = 16, 18, 18, 15, 17, 17, 18, 17, and 18. b Timeline of temporal germline analysis showing the time and respective life stages when germline imaging and analysis were performed following egg-laying on control and RNAi plates (RNAi from the L1 stage). c Quantification of germline length of pie-1p::mCherry::his-58; sun-1p::rde-1; rde-1(mkc36) animals at the L4 stage following RNAi from the L1 stage. Data was generated from three independent experiments, and results normalized to respective controls. n = 30, 28, 30, 27, 30, 29, 27, 26, and 27. P values assessed by one-way ANOVA with no correction for multiple comparison. d DIC (left) and fluorescent micrographs (right) of pie-1p::mCherry::his-58; sun-1p::rde-1; rde-1(mkc36) germ lines at the L4 stage following control, cdc-5L and znf-622 RNAi. Blue arrow = vulva. Scale bar = 20 μm. e Heatmap showing the percentage of germ lines producing sperm, oocytes and embryos in day 1, day 2 and day 3 of adulthood following germline-specific RNAi from the L1 stage. For day 2 and day 3 adult analysis, worms were selected from day 1 sterile animals, except for the control group, and incubated for another 48 hrs or 72 hrs prior to analysis. n = 30. f Quantification of hatched larvae and dead eggs (brood size) following germline-specific RNAi from the L4 stage. n = 18, 18, 17, 18, 18 17, 18, 18 and 18. P values assessed by two-tailed unpaired t test comparing the brood size (hatched larvae + dead eggs) of experimental RNAi with control RNAi. g–j Quantification of nuclei number of PZ (g), TZ (h), pH3+ (i), and SYGL-1+ (j) of 3xflag::sygl-1; sun-1p::rde-1; rde-1(mkc36) one-day adult germ lines following RNAi treatment from the L4 stage. n = 22, 24, 23, 24, 22, 24, 24, 24 and 24. P values were assessed by one-way ANOVA with no correction for multiple comparisons. All data generated from three independent experiments. Error bars indicate SEM. Source data are provided as a Source Data file.
We found that germline size and morphology were mostly intact during early larval development (L1 through L3) following GNBP silencing (Supplementary Fig. 8). However, from the L4 stage, GNBP silencing began to reveal germline defects—silencing of all eight essential GNBPs caused a significant decrease in germline length (Fig. 4c). Of note, cdc-5L and znf-622 silencing resulted in small germ lines (~50% the length of controls) that contained large nuclei suggesting arrest in the G2 stage of the cell cycle (Fig. 4c, d and Supplementary Fig. 8). We continued to monitor germline development for all 8 GNBP RNAi experiments during the first three days of adulthood to determine the effect on sperm, oocyte, and embryo development (Fig. 4e). We found that silencing of some genes prevented or delayed sperm generation (cdc-5L, znf-622, vep-1 and F23B12.7), with all genes defective or delayed in oocyte production and embryogenesis (Fig. 4e). In addition, silencing of cdc-5L, znf-622, vep-1 and tbp-1 caused sterility in three-day adults (Fig. 4e). We wondered whether these eight GNBPs are continuously required for germline development or perform functions during larval development to enable fertility in adults. We explored this by performing germline-specific RNAi silencing from the mid-L4 stage at which gonad development and spermatogenesis are largely complete (Fig. 4f). We found that silencing of repo-1, F33H1.4, F23B12.7, F58G1.2, tbp-1, and vep-1 had similar broods to control animals, albeit with different degrees of embryonic lethality (Fig. 4f). In contrast, cdc-5L and znf-622 silencing caused approximately >50% reduction in brood size (with cdc-5L RNAi causing 100% embryonic lethality), suggesting post-developmental roles in germline maintenance (Fig. 4f).
To examine the function of the essential GNBPs in the oogenic germ line, we quantified distal germ cell numbers in one-day adults following RNAi from the L4 stage (Fig. 4g–j). We identified subtle changes in PZ/TZ size, pH3+ cells, and SYGL-1+ staining (Fig. 4g–j). However, these changes are negligible compared to the detrimental effect of silencing these GNBPs from the L1 stage (Fig. 4a), suggesting that the germ cell cycle and transition to meiosis are largely intact. Further, the minor changes in distal germ line cell numbers suggest that the brood size decrease we observed following cdc-5L and znf-622 silencing are likely caused by defects in gametogenesis. Taken together, we have identified multiple NBPs that control the timing and function of gamete generation and fertility in C. elegans.
Discussion
In this study, we profiled the function of germline-expressed NBPs in C. elegans to gain a comprehensive understanding of germline regulation. To identify autonomous regulatory functions, we first determined that 364 GNBPs are germline expressed and then depleted their expression specifically in the germ line. We used two independent screening approaches to investigate GNBP functions from the proliferative germ cell stage to gamete production. Using high-content imaging, semi-automated germ cell counting, and cell behavior analysis of >6000 germ lines and >3 million germ cells, we identified 156 GNBPs that influence germline development, >75% of which are undescribed. Furthermore, we identified 8 factors that are essential for fertility. As a result, this study provides a rich resource for mechanistic dissection of NBP function in germ cells and potentially new opportunities for manipulating cell fates.
Previous C. elegans forward and reverse genetic screening approaches identified genes that cause sterility or reduced brood sizes56,57,58. Further, a large-scale screen of essential genes used the RNAi soaking method to examine proximal germline development28; however, <3% of these genes overlapped with our screen. Whether phenotypes observed in these previous studies result from autonomous germline functions or somatic deficits is, however, unclear. Thus, our use of a germline-specific RNAi approach to survey autonomous NBP functions, allows for a more refined and robust evaluation of critical germline regulators.
Our primary screen identified 8 factors that are essential for fertility: CDC-5L, ZNF-622, VEP-1, F23B12.7, F33H1.4, F58G1.2, REPO-1, and TBP-1. Loss of these factors had previously produced sterile/fertility phenotypes in genetic mutants and/or large-scale RNAi screens21,26,59,60,61,62. Our analysis shows that these factors act autonomously and predominantly during larval development to enable germline function as we observed robust sterility when these factors were silenced from hatching, but not when silenced late in development. Major exceptions were silencing of cdc-5L, repo-1, and znf-622, which caused reduced brood sizes and high proportions of dead eggs even when they were silenced late in development. Previous studies have identified conserved germline functions in mammals for some of these factors. Porcine CDC-5L is required for fertility and oocyte maturation63, which is consistent with the oogenesis defect induced by C. elegans cdc-5L silencing. In addition, murine BDP1 is required for spermatogonial stem cell gene expression64, which may be the cause of sterility and delayed sperm development when the BDP1 C. elegans ortholog VEP-1 is silenced.
We also identified 128 GNBPs that control germ cell behavior in the distal germ line, including multiple NBPs previously reported. For example, our screen confirmed previous studies identifying a zinc finger TF LSL-1 that limits TZ size, and the heat shock factor HSF-1 and chromatin factor XND-1 that promote germ cell proliferation in the PZ10,11,37. Our secondary screen focused on the GNBPs important for controlling PZ size to define whether these factors potentially control Notch signaling and/or mitotic cell cycle—two critical determinants of PZ germ cell behavior. Our analysis identified 5 GNBPs that limit and 6 NBPs that promote PZ size. Key findings from this work suggest that HSF-1 and XND-1 are required for excess germ cell proliferation caused by GLP-1/Notch overexpression. Further, loss of the zinc finger NBPs C02F5.12 and ZK546.5 enhances expression of the GLP-1 target gene SYGL-1 and germ cell proliferation rate, suggesting that these genes act in the Notch signaling pathway. Interestingly, C02F5.12 is a paralog of ZIM-1, ZIM-2, ZIM-3, and HIM-8 that are required for chromosome pairing65. However, as with C02F5.12, we found that loss of ZIM-2 additionally limits PZ size suggesting a pre-meiotic function for these proteins. Further, ZK546.5 also limits the size of the TZ—a phenotype it shares with its paralog LSL-1. Finally, we found that the RFX-type TF DAF-19 limits the germ cell proliferation in C. elegans. Interaction analysis predicts that DAF-19 physically interacts with the serine-threonine kinase ATL-1 and checkpoint kinase CHKR-1, two kinases that are required for fertility, suggesting a potential mechanism for DAF-19 function in the germ line66. A mouse ortholog of DAF-19, RFX2, is expressed in the testis and required for spermatogenesis67. Thus, future analysis of DAF-19 functions in sperm development in C. elegans hermaphrodites and males is warranted.
Meiotic development in the C. elegans germ line is predominantly regulated by post-transcriptional networks governed by the redundant GLD-1, GLD-2, and SCFPROM-1 proteins68,69,70,71. However, the pachytene region is transcriptionally active prior to the transcriptional silencing that occurs in oocytes72,73. Our analysis of NBP function in the proximal germ line identified functions in spermatogenesis, oogenesis, gonad structure, and meiotic progression. However, the most common phenotype we observed was increased multinucleation of meiotic cells where silencing of 26 GNBPs caused a robust phenotype. We focused on the 10 GNBPs with the most penetrant multinucleation phenotype for secondary analysis. Previous studies revealed that folding of the germline structure and apoptosis impacts multinucleation50. In consensus with this, we found that silencing of 6/10 of these GNBPs (DMD-7, BAZ-2, MTER-4, LIN-26, NHR-114, and NHR-161) caused excess folding in the germline syncytium that may contribute to the multinucleation phenotype. Furthermore, we found that the increase in multinucleation caused by silencing of 7/10 of these GNBPs is dependent on the presence of the apoptotic executioner caspase, CED-3. Caspase-3 is a well-known essential apoptosis inducer, but it can also regulate non-apoptotic processes, including axonal differentiation, growth, routing, regeneration, and degeneration55. Considering that the molecular mechanism of multinucleation formation and removal is unknown, it is challenging to speculate if CED-3 only controls this event by inducing apoptosis, or if it has additional functions.
Overall, our detailed map of GNBP function throughout the C. elegans germ line is a valuable resource for mechanistic studies of germ cell and gamete development. Our discovery of multiple NBPs with specific functions in the developing germ line suggests that many distinct pathways act to fine-tune gene networks and cell behavior. We anticipate that specific GNBP will act downstream of key signaling pathways (e.g., TGF, mTOR, AMPK, and insulin signaling) to integrate environmental and genetic cues for optimizing germ cell proliferation, meiotic transition, and progeny production. Together, these regulatory outputs, when combined, likely provide robustness to the process of delivering the next generation.
We wish to highlight three limitations of our study. First, the categorization of NBPs was based on the presence of DNA-binding domains that may regulate gene expression at the transcriptional and post-transcriptional level14. As such, these factors may directly control transcription through their DNA-binding domain or may control other aspects of gene expression, such as RNA splicing (CDC-5L) and translation (ZNF-622), thus care must be taken when planning mechanistic dissection of their function. Second, we found that the transgenic strain we used to analyze the proximal germ line (OD95) identified more GNBPs that cause a sterility phenotype than the strain used for distal analysis. This OD95 strain expresses two independently integrated transgenes to mark germ cell nuclei and membranes that may cause sensitivity to certain genetic perturbations. Third, in our analysis of essential NBPs, gene knockdown from the L4 stage may not be sufficient to engender robust germline phenotypes.
Methods
Caenorhabditis elegans strains and maintenance
C. elegans strains were maintained on Nematode Growth Medium (NGM) plates and fed with OP50 Escherichia coli bacteria at 20 °C, unless otherwise stated. All strains used in this study are listed in Supplementary Data 7.
RNA sequencing
RNA sequencing was performed in triplicate. For each replicate, at least 15 germ lines were extracted by dissecting one-day adult hermaphrodites. Germ lines were isolated from the rest of the worms and embryos by dissecting at the middle of the spermatheca. mRNA was isolated using RNAeasy Micro Kit (Qiagen). A minimum of 17 ng mRNA was used to perform low-input RNA sequencing. RNA sequencing was performed at Micromon Genomics (Monash University). RNA samples were quantified using Invitrogen Qubit and associated chemistry (Invitrogen, Carlsbad CA., USA), and RNA integrity was measured using Agilent Bioanalyzer 2100 microfluidic device, in conjunction with the associated hardware and chemistry (Agilent Technologies, Waldbronn, Germany). Illumina sequencing libraries were constructed using 2.5 ng DNA and Clontech SMART-Seq® v4 Ultra Low-Input RNA Kit. RNA samples were sequenced on an Illumina NextSeq500 and 150c High-Output SBS chemistry. RNA-sequencing data were converted to gene read counts by the Monash Bioinformatics Platform. Raw fastq files were analyzed using the RNAsik pipeline using STAR aligner to the Caenorhabditis elegans reference genome WBcel23574. Reads were quantified using featureCounts75 producing the raw genes count matrix and various quality control metrics. Raw counts were then analyzed with Degust, a web tool that performs normalization using trimmed mean of M values (TMM)76. The gene expression count was converted to log2-counts per million (logCPM) using R-package Mean-variance modeling at the observational level (voom)/Linear Models for Microarray Data (limma)77. NBPs were identified by gene ontology analysis. Note that while dissecting the adult germ line, some of the somatic gonad is likely also isolated in the preparation, and thus the derived dataset is likely germline-enriched.
RNAi experiments
HT115(DE3) E. coli bacteria containing control (L4440 plasmid), or experimental RNAi plasmids were cultured overnight in LB containing 50 μg/ml ampicillin. RNAi bacteria were seeded uniformly over plates containing IPTG (Isopropyl β-d-1-thiogalactopyranoside), and dried for at least 24 hours. For RNAi feeding from the L1 stage, two-day hermaphrodite adults were picked to RNAi plates to lay eggs for 1 hour, and the resultant progeny analyzed. For RNAi feeding from the L4 stage, mid-L4 (Christmas Tree vulva) animals were picked to RNAi plates. For long-term RNAi experiments, worms were added to freshly seeded RNAi plates every 3 days. All RNAi plasmids used in this study are listed in Supplementary Data 3.
RNAi plasmid cloning
400–750 bp genomic sequence for each gene was amplified using primers listed in Supplementary Data 8 and HindIII or XbaI sites were introduced at both ends of the amplicon. The empty vector (L4440) and amplified product were digested using HindIII or XbaI enzyme. The amplified sequence was ligated into the vector using T4 DNA ligase. After verifying the inserted sequences using Sanger sequencing, plasmids were transformed into HT115(DE3) E. coli bacteria for RNAi experiments. Note that the genes ztf-25 and F10E7.11 are targeted by the RNAi clones against lsy-27 and T07F8.4, respectively, and thus were not specifically targeted (Supplementary Data 3).
Fluorescence microscopy for proximal germline phenotypic analysis
For live imaging, worms were anesthetised in 0.01% tetramisole on a 5% agarose pad. Images were taken using a Zeiss AXIO Imager M2 upright fluorescence microscope and ZEN 2.0 software at ×63 objective, unless otherwise specified. For the initial screen and validation of multinucleation phenotype, oocytes and the adjacent pachytene region were imaged in two layers (upper and mid layer of pachytene cells). Eight individual germ lines were imaged for each RNAi treatment. For analyzing multinucleation in ced-3 mutants, the mid and upper layers of the entire pachytene region were imaged.
Proximal germline phenotypic analysis
For screening the proximal germ line, the pie-1p::mCherry::his-58; pie-1p::GFP::PH(PLC1δ1); sun-1p::rde-1; rde-1(mkc36) strain was used. This strain expresses mCherry tagged Histone 2B (nucleus) and GFP fusion binding PI4, 5P2 (plasma membrane), which allows observation of nuclei and plasma membrane in live animals28,49. Live imaging was performed as described in the previous section. Phenotypic analysis was performed by two people independently, blinded to RNAi knockdown. The images were analyzed by a second experimenter and the original experimenter was consulted when differences in the two analyses were observed. All phenotypes analyzed are listed in Supplementary Fig. 3 and the results shown in Supplementary Data 4 are after deducting phenotypes observed in the respective control.
Germline staining
Dissected germ lines were fixed on poly-L-lysine coated slides by immersing in methanol (precooled at −20 °C) for 1 min followed by 4% paraformaldehyde for 30 min. After fixation, samples were washed twice in phosphate-buffered saline (pH 7.4) containing 0.5% Tween 20 (PBST) and blocked using 30% normal goat serum. Subsequently, the samples were incubated in primary antibodies for at least 16 hours at 4 °C. After washing twice with PBST, samples were incubated in secondary antibodies and DAPI for 1.5 hours. Samples were mounted using Fluoroshield mounting media (Sigma) after washing twice with PBST. Primary antibodies/probes used: anti-phospho-Histone H3 (Ser10) (06-570, Merk, 1:500), monoclonal ANTI-FLAG® M2(F1804, Sigma-Aldrich, 1:500), Fluor™ 568 Phalloidin (Invitrogen). Secondary antibodies used: goat anti-rabbit 647 (A21245, Invitrogen, 1:1000), goat anti-mouse 555 (A21422, Invitrogen, 1:1000).
EdU (5-Ethynyl-2’deoxyuridine) labeling
EdU labeling was performed by feeding worms with EdU bacteria as in previous studies45,78. EdU feeding plates were prepared as follows: NGM agar plates (-peptone, 60 μg/ml carbenicillin) were seeded with concentrated E. coli MG1693 that was grown in M9 minimal media (3 g/L KH2PO4, 6 g/L Na2HPO4, 0.5 g/L NaCl, 1 g/L NH4Cl, 2 mM MgSO4, 0.1 mM CaCl2, 0.4% glucose, 1.25 μg/ml thiamin) supplemented with 0.5 μM thymidine and 20 μM EdU for 24 hours. Worms were fed with RNAi bacteria for 66 hrs from hatching, then transferred to EdU feeding plates. Dissection of germ lines was performed 4 hours and 10 hours after commencing EdU feeding. Sample fixation was performed as described above. The Click reaction was performed according to the manufacturer’s instructions (Invitrogen™, C10337), and DNA was stained with DAPI. Z-stack images were taken using confocal microscopes Leica SP5 and Leica Stellaris5 (×63 objectives). Automated nuclei counting was performed using Imaris software as described below. Proliferation rate (the number of germ cells per hour) = (EdU+ germ cells at 10 hours—EdU+ germ cells at 4 hours)/6 hours.
Germline imaging and analysis
The number of germ cells was assessed by counting the number of DAPI-stained nuclei. The PZ is defined as germ cells between the distal tip cell and the TZ. The TZ is predominantly composed of cells in early meiotic prophase, with DAPI-stained nuclei in this region having a distinctive crescent shape. In this study, the TZ is defined as a continuous region where germ cell rows have ≥60% crescent-shaped nuclei. Nuclei numbers in the progenitor and TZs were analyzed by immunostaining and 3D modeling27. Z-stack images were taken using confocal microscopes Leica SP5 and Leica Stellaris5 (×63 objectives). Semi-automatic nuclei counting was performed using Imaris software27. Briefly, the surface tool was used to define the germline regions of interest, and the spot tool was used to count the number of DAPI-stained nuclei in each selected region. Nuclear diameters were defined according to our previous publication27, and accuracy was confirmed by confirming that the number of nuclei in the PZ of control samples is within the previously reported range27.
Generation of a ced-3 null mutant using CRISPR-Cas9 genome editing
The ced-3 gene and the transgenic reporter ltIs37 [pie-1p::mCherry::his-58 + unc-119(+)] are both on chromosome IV. To facilitate analysis of multinucleation in live animals lacking CED-3 caspase, a ced-3 null mutant strain was generated using CRISPR-Cas9 to remove the ced-3 genomic sequence from strain ltIs37 IV; ltIs38. Two crRNAs used to target ced-3 were designed and ordered using the online tool provided by https://sg.idtdna.com (Supplementary Data 8). The following mix was then injected into ltIs37 IV; ltIs38 animals: 0.5 µg Cas9 protein, 2 µg universal tracrRNA, 1.1 µg each crRNA, myo-2::mCherry plasmid (4 ng/µl). Individual F1 progeny of injected worms were isolated, and F2 progeny were screened for ced-3 deletion by PCR. After confirmation of deletion by Sanger sequencing, the ced-3 null mutant was outcrossed three times prior to analysis.
Brood size analysis
L4 hermaphrodites were picked onto individual NGM plates seeded with OP50 or RNAi bacteria. Worms were allowed to lay eggs for 24 hours and then mothers were moved to new plates. After a further 24 hours, the number of larvae and embryos was counted. This process was repeated for six days. The sum of the number of larvae and embryos is the total brood size.
Bioinformatic analysis
Heatmaps were generated using the ComplexHeatmap package in R79. Published RNAi and allele phenotypes were collected using the Simplemine online tool from wormbase (https://wormbase.org//tools/mine/simplemine.cgi). We used Amigo 2 gene ontology database to identify mouse, rat, and human orthologs involved in reproduction80. The mouse, rat, and human orthologous genes encoding NBPs in C. elegans were identified using the online tool DIOPT Ortholog Finder81. The orthologous genes with high or moderate scores were selected for further analysis.
Quantification and statistical analysis
Statistical analyses were performed in GraphPad Prism 9 using one-way analysis of variance, multiple unpaired t test or unpaired t tests. We did not apply corrections to our data prior to statistical analysis as each GNBP RNAi knockdown was independent of each other, and thus the group variance was not considered in our experiments. Specific statistical tests used for each experiment are detailed in the Figure legends. Values are expressed as mean ± S.E.M. Differences with a P value < 0.05 were considered significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data are available in the main text, supplementary materials. Source data are provided in Supplementary Data 9 and 10. The germline-enriched RNA-sequencing data generated in this study have been deposited in the GEO database under accession code GSE254738. No unique identifiers, or weblinks are in our study and there are no restrictions on data availability. Materials are available upon request from Roger Pocock.
References
Crittenden, S. L. et al. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417, 660–663 (2002).
Mak, W., Fang, C., Holden, T., Dratver, M. B. & Lin, H. An important role of pumilio 1 in regulating the development of the mammalian female germline. Biol. Reprod. 94, 134 (2016).
Austin, J. & Kimble, J. Glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C-Elegans. Cell 51, 589–599 (1987).
Fukuyama, M., Rougvie, A. E. & Rothman, J. H. C. elegans DAF-18/PTEN mediates nutrient-dependent arrest of cell cycle and growth in the germline. Curr. Biol. 16, 773–779 (2006).
Furuhashi, H. et al. Trans-generational epigenetic regulation of C. elegans primordial germ cells. Epigenet. Chromatin 3, 15 (2010).
Seydoux, G. et al. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature 382, 713–716 (1996).
Batchelder, C. et al. Transcriptional repression by the Caenorhabditis elegans germ-line protein PIE-1. Genes Dev. 13, 202–212 (1999).
Hansen, D., Hubbard, E. J. & Schedl, T. Multi-pathway control of the proliferation versus meiotic development decision in the Caenorhabditis elegans germline. Dev. Biol. 268, 342–357 (2004).
Ellis R., Schedl T. Sex determination in the germ line. WormBook, 1–13 (2007).
Rodriguez-Crespo, D., Nanchen, M., Rajopadhye, S. & Wicky, C. The zinc-finger transcription factor LSL-1 is a major regulator of the germline transcriptional program in Caenorhabditis elegans. Genetics 221, iyac039 (2022).
Edwards, S. L. et al. Insulin/IGF-1 signaling and heat stress differentially regulate HSF1 activities in germline development. Cell Rep. 36, 109623 (2021).
Curran, S. P., Wu, X., Riedel, C. G. & Ruvkun, G. A soma-to-germline transformation in long-lived Caenorhabditis elegans mutants. Nature 459, 1079–1084 (2009).
Chi, W. & Reinke, V. Promotion of oogenesis and embryogenesis in the C. elegans gonad by EFL-1/DPL-1 (E2F) does not require LIN-35 (pRB). Development 133, 3147–3157 (2006).
Jimeno-Martin, A. et al. Joint actions of diverse transcription factor families establish neuron-type identities and promote enhancer selectivity. Genome Res. 32, 459–473 (2022).
Tzur, Y. B. et al. Spatiotemporal gene expression analysis of the caenorhabditis elegans germline uncovers a syncytial expression switch. Genetics 210, 587–605 (2018).
Cao, J. et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017).
Zou, L. et al. Construction of a germline-specific RNAi tool in C. elegans. Sci. Rep. 9, 2354 (2019).
Serizay, J. et al. Distinctive regulatory architectures of germline-active and somatic genes in C. elegans. Genome Res. 30, 1752–1765 (2020).
Tabara, H. et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123–132 (1999).
Timmons, L. & Fire, A. Specific interference by ingested dsRNA [letter]. Nature 395, 854 (1998).
Kerins, J. A., Hanazawa, M., Dorsett, M. & Schedl, T. PRP-17 and the pre-mRNA splicing pathway are preferentially required for the proliferation versus meiotic development decision and germline sex determination in Caenorhabditis elegans. Dev. Dyn. 239, 1555–1572 (2010).
Cao W., Tran C., Archer S. K., Gopal S., Pocock R. Functional recovery of the germ line following splicing collapse. Cell Death Differ. 29, 772–787 (2021).
Nayak, S., Goree, J. & Schedl, T. fog-2 and the evolution of self-fertile hermaphroditism in Caenorhabditis. PLoS Biol. 3, e6 (2005).
Kumsta, C. & Hansen, M. C. elegans rrf-1 mutations maintain RNAi efficiency in the soma in addition to the germline. PLoS One 7, e35428 (2012).
Rual, J. F. et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14, 2162–2168 (2004).
Kamath, R. S. et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237 (2003).
Gopal, S., Boag, P. & Pocock, R. Automated three-dimensional reconstruction of the Caenorhabditis elegans germline. Dev. Biol. 432, 222–228 (2017).
Green, R. A. et al. A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell 145, 470–482 (2011).
Austin, J. & Kimble, J. Transcript analysis of glp-1 and lin-12, homologous genes required for cell interactions during development of C. elegans. Cell 58, 565–571 (1989).
Henderson, S. T., Gao, D., Lambie, E. J. & Kimble, J. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120, 2913–2924 (1994).
Gao, D. L. & Kimble, J. Apx-1 can substitute for its homolog lag-2 to direct cell-interactions throughout caenorhabditis-elegans development. Proc. Natl. Acad. Sci. USA 92, 9839–9842 (1995).
Tax, F. E., Yeargers, J. J. & Thomas, J. H. Sequence of C. elegans lag-2 reveals a cell-signalling domain shared with Delta and Serrate of Drosophila. Nature 368, 150–154 (1994).
Shin, H. et al. SYGL-1 and LST-1 link niche signaling to PUF RNA repression for stem cell maintenance in Caenorhabditis elegans. Plos Genet. 13, e1007121 (2017).
Chen, J. et al. GLP-1 Notch-LAG-1 CSL control of the germline stem cell fate is mediated by transcriptional targets lst-1 and sygl-1. PLoS Genet. 16, e1008650 (2020).
Ferdous, A. S. et al. LST-1 is a bifunctional regulator that feeds back on Notch-dependent transcription to regulate C. elegans germline stem cells. Proc. Natl Acad. Sci. USA 120, e2309964120 (2023).
Haupt, K. A. et al. The molecular basis of LST-1 self-renewal activity and its control of stem cell pool size. Development 146, dev181644 (2019).
Mainpal, R., Nance, J. & Yanowitz, J. L. A germ cell determinant reveals parallel pathways for germ line development in Caenorhabditis elegans. Development 142, 3571–3582 (2015).
Fox, P. M. & Schedl, T. Analysis of germline stem cell differentiation following loss of GLP-1 notch activity in Caenorhabditis elegans. Genetics 201, 167–184 (2015).
Kershner, A. M., Shin, H., Hansen, T. J. & Kimble, J. Discovery of two GLP-1/Notch target genes that account for the role of GLP-1/Notch signaling in stem cell maintenance. Proc. Natl Acad. Sci. USA 111, 3739–3744 (2014).
Luo, Y. et al. New developments on the Encyclopedia of DNA Elements (ENCODE) data portal. Nucleic Acids Res. 48, D882–D889 (2020).
Consortium, E. P. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Crittenden, S. L., Troemel, E. R., Evans, T. C. & Kimble, J. GLP-1 is localized to the mitotic region of the C. elegans germ line. Development 120, 2901–2911 (1994).
Crittenden, S. L., Leonhard, K. A., Byrd, D. T. & Kimble, J. Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol. Biol. Cell 17, 3051–3061 (2006).
Seidel, H. S. & Kimble, J. Cell-cycle quiescence maintains Caenorhabditis elegans germline stem cells independent of GLP-1/Notch. Elife 4, e10832 (2015).
Crittenden S. L., Seidel H. S., Kimble J. Analysis of the C. elegans germline stem cell pool germine stem cell pool. Methods Mol. Biol. 1463, 1–33 (2017).
Hsu, J. Y. et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102, 279–291 (2000).
Crittenden, S. L., Seidel, H. S. & Kimble, J. Analysis of the C. elegans germline stem cell pool. Methods Mol. Biol. 2677, 1–36 (2023).
Hubbard, E. J. Caenorhabditis elegans germ line: a model for stem cell biology. Dev. Dyn. 236, 3343–3357 (2007).
Essex, A., Dammermann, A., Lewellyn, L., Oegema, K. & Desai, A. Systematic analysis in Caenorhabditis elegans reveals that the spindle checkpoint is composed of two largely independent branches. Mol. Biol. Cell 20, 1252–1267 (2009).
Raiders, S. A., Eastwood, M. D., Bacher, M. & Priess, J. R. Binucleate germ cells in Caenorhabditis elegans are removed by physiological apoptosis. PLoS Genet. 14, e1007417 (2018).
Lan, H. et al. An extracellular matrix protein promotes anillin-dependent processes in the Caenorhabditis elegans germline. Life Sci. Alliance 2, e201800152 (2019).
Gracida, X. & Eckmann, C. R. Fertility and germline stem cell maintenance under different diets requires nhr-114/HNF4 in C. elegans. Curr. Biol. 23, 607–613 (2013).
Gumienny, T. L., Lambie, E., Hartwieg, E., Horvitz, H. R. & Hengartner, M. O. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126, 1011–1022 (1999).
Eskandari, E. & Eaves, C. J. Paradoxical roles of caspase-3 in regulating cell survival, proliferation, and tumorigenesis. J. Cell Biol. 221, e202201159 (2022).
Dehkordi, M. H., Munn, R. G. K. & Fearnhead, H. O. Non-canonical roles of apoptotic caspases in the nervous system. Front. Cell Dev. Biol. 10, 840023 (2022).
Hirsh, D. & Vanderslice, R. Temperature-sensitive developmental mutants of Caenorhabditis elegans. Dev. Biol. 49, 220–235 (1976).
Argon, Y. & Ward, S. Caenorhabditis elegans fertilization-defective mutants with abnormal sperm. Genetics 96, 413–433 (1980).
Maeda, I., Kohara, Y., Yamamoto, M. & Sugimoto, A. Large-scale analysis of gene function in Caenorhabditis elegans by high- throughput RNAi. Curr. Biol. 11, 171–176 (2001).
Simmer, F. et al. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1, E12 (2003).
Fraser, A. G. et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330 (2000).
Sun, Y. et al. A genome-wide RNAi screen identifies genes regulating the formation of P bodies in C. elegans and their functions in NMD and RNAi. Protein Cell 2, 918–939 (2011).
Sonnichsen, B. et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434, 462–469 (2005).
Liu, X. M. et al. Single-cell transcriptome sequencing reveals that cell division cycle 5-like protein is essential for porcine oocyte maturation. J. Biol. Chem. 293, 1767–1780 (2018).
Sisakhtnezhad, S. & Heshmati, P. Comparative analysis of single-cell RNA sequencing data from mouse spermatogonial and mesenchymal stem cells to identify differentially expressed genes and transcriptional regulators of germline cells. J. Cell Physiol. 233, 5231–5242 (2018).
Phillips, C. M. & Dernburg, A. F. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev. Cell 11, 817–829 (2006).
Garcia-Muse, T. & Boulton, S. J. Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. EMBO J. 24, 4345–4355 (2005).
Wu, Y. et al. Transcription factor RFX2 is a key regulator of mouse spermiogenesis. Sci. Rep. 6, 20435 (2016).
Francis, R., Maine, E. & Schedl, T. Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics 139, 607–630 (1995).
Kadyk, L. C. & Kimble, J. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 125, 1803–1813 (1998).
Jantsch, V. et al. Caenorhabditis elegans prom-1 is required for meiotic prophase progression and homologous chromosome pairing. Mol. Biol. Cell 18, 4911–4920 (2007).
Nayak, S. et al. The Caenorhabditis elegans Skp1-related gene family: diverse functions in cell proliferation, morphogenesis, and meiosis. Curr. Biol. 12, 277–287 (2002).
Gibert, M. A., Starck, J. & Beguet, B. Role of the gonad cytoplasmic core during oogenesis of the nematode Caenorhabditis elegans. Biol. Cell 50, 77–85 (1984).
Starck, J. Radioautographic study of RNA synthesis in Caenorhabditis elegans (Bergerac variety) oogenesis. Biol. Cell 30, 181–182 (1977).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
Kocsisova, Z., Kornfeld, K. & Schedl, T. Rapid population-wide declines in stem cell number and activity during reproductive aging in C. elegans. Development 146, dev173195 (2019).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Carbon, S. et al. AmiGO: online access to ontology and annotation data. Bioinformatics 25, 288–289 (2009).
Hu, Y. et al. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinform. 12, 357 (2011).
Acknowledgements
We thank members of the Pocock and Gopal laboratories for their advice and comments on the manuscript. RNA sequencing and Bioinformatics performed at Monash Micromon Genomics and Monash Bioinformatics Platform. Imaging performed at Monash Microimaging. We thank Nuria Flames for their kind gift of NBP RNAi clones. Some strains were provided by the Caenorhabditis Genetics Center (University of Minnesota), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by the following grants: Australian Research Council DE190100174 (S.G.) and DP200103293 (R.P.); National Health and Medical Research Council GNT1161439 (S.G.), GNT1105374 (R.P.), GNT1137645 (R.P.) and GNT2018825 (R.P., W.C., and Q.F.).
Author information
Authors and Affiliations
Contributions
Conceptualization: R.P., S.G. Methodology: R.P., S.G., W.C., Q.F., R.G. Investigation: R.P., S.G., W.C., Q.F., R.G., G.A., D.B. Visualization: R.P., S.G., W.C., Q.F., R.G., G.A., D.B. Funding acquisition: S.G., R.P. Project administration: R.P., S.G., W.C. Supervision: R.P., S.G., W.C. Writing—original draft: R.P. Writing—review & editing: R.P., S.G., W.C., Q.F., R.G., G.A., D.B.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Peter Okkema and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cao, W., Fan, Q., Amparado, G. et al. A nucleic acid binding protein map of germline regulation in Caenorhabditis elegans. Nat Commun 15, 6884 (2024). https://doi.org/10.1038/s41467-024-51212-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51212-0
- Springer Nature Limited