Abstract
Current treatments for chronic diarrhea have limited efficacy and several side effects. Probiotics have the potential to alleviate symptoms of diarrhea. This randomized, double-blind, placebo-controlled trial evaluates the effects of administering the probiotic Lactiplantibacillus plantarum P9 (P9) strain in young adults with chronic diarrhea (Clinical Trial Registration Number: ChiCTR2000038410). The intervention period lasts for 28 days, followed by a 14-day post-intervention period. Participants are randomized into the P9 (n = 93) and placebo (n = 96) groups, with 170 individuals completing the double-blind intervention phase (n = 85 per group). The primary endpoint is the diarrhea symptom severity score. Both intention-to-treat (n = 189) and per-protocol (n = 170) analyses reveal a modest yet statistically significant reduction in diarrhea severity compared to the placebo group (20.0%, P = 0.050; 21.4%, P = 0.048, respectively). In conclusion, the results of this study support the use of probiotics in managing chronic diarrhea in young adults. However, the lack of blood parameter assessment and the short intervention period represent limitations of this study.
Similar content being viewed by others
Introduction
Chronic diarrhea is a common functional gastrointestinal disorder affecting 3–20% of the global population1. It is defined as more than three bowel movements or loose stools per day for at least 4 weeks2. Common causes of chronic diarrhea include functional disorders like irritable bowel syndrome with diarrhea (IBS-D) and functional diarrhea, resulting in many clinical and social problems for affected individuals3. Presently, the treatment options for chronic diarrhea include loperamide, diphenoxylate, clonidine, codeine, morphine preparations, and anticholinergic medications4. However, these drugs can cause side effects, such as constipation, nausea, vomiting, bloating, and further episodes of diarrhea5,6. Furthermore, there are concerns regarding antibiotic resistance and persistence associated with their use7. Therefore, managing chronic diarrhea poses significant difficulties. Gut microbial diversity is often reduced in individuals with diarrhea, which increases their susceptibility to pathogens8,9. Many microbe-derived metabolites, including bile acids (BAs), short-chain fatty acids (SCFAs), and amino acids, are involved in gastrointestinal inflammation and colonic homeostasis10,11. Recent studies also focus on the gut phageome, which can modify the gut microbiota and influence gastrointestinal diseases, such as inflammatory bowel disease and colorectal cancer12,13. Therefore, the gut microbiota, metabolome, and phageome are considered targets for managing diarrhea.
Probiotics are defined as “live microorganisms, which when administered in adequate amounts, confer a health benefit on the host”14, such as mitigating disease symptoms and improving gut microbiota balance. They have been used to manage diarrhea, alleviate symptoms through reducing inflammation, restoring gut microbiota balance, and enhancing intestinal mucosal barrier function15. For example, a murine study showed that administering Saccharomyces boulardii relieved diarrhea by modulating the gut microbiota that inhibits gut motility16. Intake of a multispecies probiotic mixture (containing Lactobacillus acidophilus S5, Bacillus subtilis No. Bzg988118, and Saccharomyces cerevisiae SHZ2017) could reduce diarrhea incidence, increase growth performance, and balance the fecal microbiota in pre-weaning calves17. Human clinical trials have yielded controversial results. While some trials involving single- or multi-strain probiotics demonstrated improvements in symptoms and quality of life for patients with chronic diarrhea, particularly in defecation frequency, stool consistency, and health status18,19,20, there have been studies reported no improvement in gastrointestinal and psychological symptoms or quality of life after an eight-week intervention with probiotic fermented milk in individuals with IBS-D21. These discrepancies highlight that the beneficial effects of probiotics on diarrhea are influenced by factors, such as the specific strain used, dosage, sample size, and duration of the clinical trials. Further studies and microbiological analysis are required to delineate their functional role. Probiotics have shown promising effects in improving intestinal homeostasis, which can lead to a range of multi- and reversal functional effects11. For example, in a large randomized trial conducted in India, the probiotic strain, Bifidobacterium animalis subsp. lactis HN019, reduced the incidence of diarrhea and fever during the rainy season when diarrhea incidences were highest22. The same probiotic strain exerted prokinetic effects in constipated subjects23.
Lactiplantibacillus (L.) plantarum P9 (P9) is a potential probiotic strain isolated from traditional acidic gruel24. The P9 strain has been found to possess several desirable properties, including high tolerance to gastroenteric fluids, potent immunomodulatory, anti-inflammatory, and intestinal microbial homeostasis modulatory activities25,26. Consuming P9 could alleviate symptoms of chronic constipation, accompanied by increases in functional intestinal microbiota (Lactiplantibacillus plantarum and Ruminiclostridium_E siraeum) and microbial metabolites (L-asparagine, L-pipecolinic acid, valeric acid, and caprylic acid)27. Unlike conventional drugs that typically have a unidirectional action, probiotics work to restore balance in the gastrointestinal tract. Given the multifaceted effects of probiotics, it is intriguing to investigate whether P9 also possesses anti-diarrheal properties.
The aim of this large, randomized, double-blind, placebo-controlled clinical study was to evaluate the efficacy and safety of P9 in the treatment of chronic diarrhea over a 4-week period. The primary endpoint of this study was the diarrhea symptom severity score, while secondary endpoints included stool consistency, frequency, urgency, and gut microbiota and metabolomic changes. This study showed that consuming P9 led to a modest alleviating effect on chronic diarrhea. Importantly, the study also revealed desirable changes in the fecal microbiome and metabolome following P9 treatment. These findings underscore the nuanced role of P9 in modulating gut health and support further exploration of probiotics as a viable strategy for the management of chronic diarrhea.
Results
Demographic data
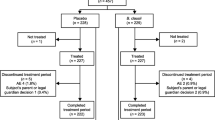
The initial subject recruitment process assessed the eligibility of a total of 215 candidate participants. Of those, 26 were excluded from the intention-to-treat (ITT) population due to not meeting inclusion criteria, declining to participate, or for other reasons (Fig. 1). Thus, the study enrolled a final of 189 patients in the ITT population when the trial was completed, who were randomly assigned to receive either the probiotic P9 (n = 93) or placebo (n = 96). Following a 4-week intervention period, eight and 11 patients were excluded from the P9 and placebo groups, respectively, due to COVID-19 infection, antibiotic intake during the trial period, refusal to participate, or missing diaries (Fig. 1). Consequently, the per-protocol (PP) analysis included 170 patients (85 subjects in each group; Fig. 1).
The baseline characteristics of all 189 patients are summarized in Supplementary Table 1. The mean (SD) age of patients in the P9 and placebo groups was 22.3 (4.0) and 22.0 (4.7) years, respectively. The proportion of male subjects in the P9 and placebo groups was 47.3% and 50.0%, while the proportion of female subjects was 52.7% and 50.0%, respectively. The ratio of Han to other ethnic groups was 90 to three and 91 to five in the P9 and placebo groups, respectively. The mean (SD) body mass index values for the P9 and placebo groups were 22.1 (3.4) and 21.9 (3.3), respectively. The baseline demographic characteristics and medical history of patients, including age, sex distribution, body mass index, ethnicity, history of drug allergies, smoking status, presence of preexisting diseases, and previous drug treatments, exhibited a balanced distribution between the P9 and placebo groups (Supplementary Table 1).
P9 administration improved diarrhea symptoms
The primary outcome, the diarrhea symptom severity score, was assessed using the Gastrointestinal Symptom Rating Scale. In the ITT population analysis, the mean (SD) diarrhea symptom severity scores before the intervention were 2.0 (1.0) in the P9 group and 2.1 (1.0) in the placebo group. Following the 28-day intervention, the mean (SD) diarrhea symptom severity score in the P9 group was significantly reduced by 20.0% compared to the placebo group (1.2 [0.8] and 1.5 [0.9] in the P9 and placebo groups, respectively, P = 0.050; Supplementary Table 2). The PP analysis also demonstrated a significantly lower mean (SD) diarrhea symptom severity score in the P9 group compared to the placebo group, reduced by 21.4% (1.1 [0.7] and 1.4 [0.8] in the P9 and placebo groups, respectively, P = 0.048; Fig. 2b, Supplementary Table 3). Similarly, in the PP population, the administration of P9 resulted in a significantly lower mean (SD) stool consistency score in the P9 group compared to the placebo group at day 28 (4.7 [0.7] and 4.9 [0.6] in the P9 and placebo groups, respectively; P = 0.050; Fig. 2b, Supplementary Table 3). However, in the ITT analysis, no significant improvement in stool consistency was observed after P9 intervention (mean [SD] stool consistency score at day 28 = 4.7 [0.7] and 5.0 [0.6] in the P9 and placebo groups, respectively; P = 0.052; Supplementary Table 2). Although these scores also decreased in the placebo group during the intervention period, the magnitude of reduction was significantly greater in the P9 group compared to the placebo group at day 28 (P < 0.01), suggesting that the observed changes in the non-probiotic group could be attributed to a placebo effect. No significant differences were observed between the P9 and placebo groups in bowel frequency, fecal urgency, depression, anxiety, and stress scores during the intervention in both the ITT and PP analyses (P > 0.05; Fig. 2b, Supplementary Fig. 1, Supplementary Tables 2, 3). These results indicate that the P9 intervention modestly but significantly alleviated diarrhea symptoms.
a The workflow of trial design, including the analyses of clinical indicators, fecal metagenomics, and metabolomics. The intervention lasted 28 days (day 0 to day 28), followed by a 14-day post-intervention follow-up period (until day 42). Created with Adobe Illustrator 2021. b Changes in diarrhea symptom severity score, stool consistency score, bowel frequency, and fecal urgency score in the per-protocol population of the P9 (pro; n = 85) and placebo (pla; n = 85) groups, evaluated using the two-sided Wilcoxon rank-sum test. Significant differences are indicated in the plots. “0 d”, “14 d”, “28 d”, and “42 d” represent days 0, 14, 28, and 42, respectively. DASS-21 = depression anxiety stress scales-21; PCA = principal component analysis; PLS-DA = partial least squares-discriminant analysis; SCFAs = short-chain fatty acids; BAs = bile acids. The boxes in the violin plots represent the interquartile range of each group’s distribution of values; the lines inside the boxes represent the median values; the whiskers denote the lowest and highest values within 1.5 times the interquartile range of each group; the dots above/below the upper/lower line are the outliers. Source data can be found in the Source Data file.
Furthermore, to account for any initial variations between treatment groups, we conducted a non-parametric covariance analysis using the sm package in R, incorporating baseline data as a covariate. The results revealed significant differences in the severity of diarrhea between the probiotic and placebo groups at days 14, 28, and 42 (day 14: P = 0.003; day 28: P = 0.000; day 42: P = 0.011). These findings provide strong evidence that the observed diarrhea-alleviating effect was not merely a result of chance but a genuine treatment effect compared to the placebo.
Adverse events reported during the study were recorded. The P9 intervention was well-tolerated by patients, with no serious adverse events reported. Most adverse events following the P9/placebo intervention were mild gastrointestinal complaints, such as diarrhea, constipation, bloating, and nausea, with a few patients experiencing fatigue, insomnia, and dizziness. Although no statistically significant difference was observed in the number of adverse events between the P9 and placebo groups (P > 0.05; Supplementary Table 2), fewer adverse events were reported by patients in the P9 group compared to the placebo group at day 28 (seven versus 15 events) and day 42 (four versus five events). In addition, both the P9 and placebo groups demonstrated high overall compliance with the intervention (mean = 103.7% compliance, range = 53.6% to 107.1% in the placebo group; mean = 102.5%, range = 53.6–107.1% in the P9 group; Supplementary Table 1).
P9 supplementation modulated the diversity and composition of patients’ fecal bacterial microbiota
The fecal microbiota of 169 patients was analyzed by metagenomic sequencing at three time points (days 0, 28, and 42). One patient in the P9 group did not provide stool samples for fecal metagenomic analysis (Fig. 1). No significant differences were observed in alpha diversity (Shannon and Simpson indices, P > 0.05) between the P9 and placebo groups at any time point (Fig. 3a). However, beta diversity analysis (assessed by principal coordinate analysis and adonis test) revealed significant differences in fecal microbiota structure between the P9 and placebo groups at days 28 (R2 = 0.015, P = 0.012) and 42 (R2 = 0.014, P = 0.018; Fig. 3b).
a Shannon and Simpson diversity indices and (b) principal coordinate analysis (PCoA) score plots of the gut microbiota in the placebo (pla; n = 85) and P9 (pro; n = 84) groups at days 0 (0 d), 28 (28 d), and 42 (42 d). The boxplots in (a) show the interquartile range of each group’s distribution of values; the lines inside the boxes represent the median values; the whiskers denote the lowest and highest values within 1.5 times the interquartile range of each group; the dots above/below the upper/lower line are the outliers. In (b), the symbols representing samples of the two groups at different time points are shown in different colors. Results of adonis tests are shown at the lower right corner of the PCoA plots. c Significant differential SGBs between the P9 and placebo groups at different time points. The time points of significant differences are indicated by the color of the taxon name (red, green, and blue correspond to differences on day 28, days 28 and 42, and day 42, respectively). The color scale represents relative abundance, ranging from low abundance (blue) to high abundance (red). Statistically significant differences were evaluated using the two-sided Wilcoxon rank-sum test, with *P < 0.05 considered statistically significant. d Pearson correlation scatter plots show the correlations between differentially abundant SGBs and clinical indicators of diarrhea in the P9 group after 28- and 42-day intervention. The gray error band represents the fitted values, and the shaded area indicates the 95% confidence interval. The blue dots represent the values of the X and Y axes, respectively. Source data can be found in the Source Data file.
We also found that P9 administration was associated with post-intervention changes in species-level fecal microbiota composition. A total of 629 species-level genomic bins (SGBs) were identified within the complete dataset, and 29 of them were significantly differential SGBs during or after the P9 intervention, mainly belonging to Lactobacillaceae, Oscillospiraceae, Ruminococcaceae, and Lachnospira. None of these SGBs showed significant differences between the two groups at baseline (day 0), and they only became differentially abundant at day 28, both days 28 and 42, and day 42 (11, 8, and 10 SGBs, respectively; Fig. 3c; Supplementary Table 4). After the 28-day intervention, the fecal microbiota of the P9 group had significantly more Lactiplantibacillus plantarum and Ruminococcus_A faecicola compared to the placebo group, while the opposite trend was observed for the species, Mediterraneibacter torques, Eubacterium_I ramulus, and Enterocloster sp000431375 (P < 0.05 in all cases; Fig. 3c). At day 42, significantly more Butyricicoccus_A sp002395695 and Streptococcus thermophilus were detected in the P9 group compared to the placebo group, whereas Phascolarctobacterium faecium and Faecalibacterium sp. exhibited the opposite trend (P < 0.05 in all cases; Fig. 3c). Interestingly, eight SGBs showed consistent differences at both days 28 and 42, including significantly more Acutalibacteraceae sp000431775 and Paraprevotella xylaniphila, but significantly fewer Coprococcus sp. and Butyricimonas virosa in the P9 group compared to the placebo group (P < 0.05 in all cases; Fig. 3c). Taken together, these results suggest that P9 supplementation could significantly change the diversity and composition of the gut microbiota in patients with chronic diarrhea.
We further analyzed the correlation between species-level gut microbiota and clinical indicators of diarrhea to investigate whether the improvement in clinical indicators was correlated to changes in specific bacteria after P9 administration (Fig. 3d). The stool consistency score correlated positively with Eubacterium_I ramulus (r = 0.2, P = 0.019); the stress score correlated positively with Enterocloster sp000431375 (r = 0.2, P = 0.008); and bowel frequency showed a significant positive correlation with Mediterraneibacter torques (r = 0.2, P = 0.002). Moreover, the anxiety score showed a significant negative correlation with Ruminococcus_A faecicola (r = −0.2, P = 0.018). These results suggest that the symptom-relief effect was associated with changes in some specific functional gut bacteria.
P9 supplementation modulated the diversity and structure of patients’ fecal phageome
Since phages play a part in the remodeling of the gut microbiota, we investigated intervention-associated changes in the diversity and composition of the patient’s fecal phageome. A total of 94,384 non-redundant virus operational taxonomic units (vOTUs) were annotated by comparing our dataset against the Metagenomic Gut Virus catalog. Among these, 41,059 vOTUs were assigned to 13 bacteriophage families, including 7,587 prophages and 33,472 non-prophages (Fig. 4a). Taxonomic annotation of these sequences revealed a high prevalence of Siphoviridae (33.2%), Myoviridae (9.3%), Microviridae (5.1%), Podoviridae (1.9%), and crAss-phage (1.0%; Fig. 4a), with the majority belonging to the order Caudovirales (87.7%; Fig. 4a).
a The circle diagram shows whether an annotated virus operational taxonomic unit (vOTU) is classified as a “prophage” or “non-prophage”, its quality level and completeness (the three innermost circles), and its order- and family-level taxonomic distribution according to the International Committee on Taxonomy of Viruses (ICTV) classification (the two outermost circles). b Principal coordinates analysis (PCoA) and (c) Simpson’s diversity index of the gut microbiota in the two groups at days 0 (0 d), 28 (28 d), and 42 (42 d). The boxplots represent the interquartile range of each group’s distribution of values; the lines inside the boxes represent the median values; the whiskers denote the lowest and highest values within 1.5 times the interquartile range of each group; the dots above/below the upper/lower line are the outliers. P-values shown in the plot were calculated using the two-sided Wilcoxon rank-sum test. d Pearson correlation between the Shannon diversity index of the gut bacteria and phage microbiota, revealing a strong positive correlation (R = 0.928; P = 9.21e-21). e Procrustes analysis performed on the gut species-level genome bins (SGB) and bacteriophages of the two groups at different time points confirmed a positive cooperativity between the gut bacterial microbiota and phageome (correlation = 0.818; P = 0.001). f Family-level classification of phages and their bacterial hosts at the genus and phylum levels. g Pearson correlation scatter plots show the correlation between family-level bacteriophages and clinical indicators of diarrhea in the P9 group after 28 and 42 days of intervention. The gray error band represents the fitted values, and the shaded area indicates the 95% confidence interval. The blue dots represent the values on the X and Y axes, respectively. Source data can be found in the Source Data file.
Beta diversity analysis demonstrated no significant differences in the overall phageome between the P9 and placebo groups at days 0, 28, and 42 (Fig. 4b). Consistently, there were no significant differences in the alpha diversity of the phageome between the two groups at days 0 and 42 (P > 0.05; Fig. 4c), except for day 28, where the Simpson’s diversity index of the patients’ fecal phageome in the P9 group was numerically lower but did not reach statistical significance compared to the placebo group (P = 0.056; Fig. 4c). Interestingly, we observed a significant positive correlation between the Shannon index of the gut bacterial microbiota and the phageome (R = 0.928, P = 9.21e-21; Fig. 4d), which was consistent with the results of the Procrustes analysis (R = 0.818, P = 0.001; Fig. 4e). These findings suggest a strong cooperativeness between the gut phageome and its bacterial hosts.
Phages are obligate intracellular parasites that reside in bacterial hosts, so specific genomic associations between phages and bacteria reflect the phage infection history. To further explore the interplay between gut bacteriophages and bacteria, we investigated the distribution of phage sequences within bacterial host genomes (Fig. 4f). A total of 21,103 vOTUs annotated to 12 known bacteriophage families were analyzed; 94.2% of these vOTUs (corresponding to 19,874 vOTUs) were predicted to be associated with specific bacteria hosts, and 12,870 of them were connected to known host bacterial genera. Siphoviridae, the most prevalent enteric phage family, were mainly associated with Firmicutes and Bacteroidota hosts (including the genera Ruminococcus, Bacteroides, Faecalibacterium, Eubacterium, and Lachnospira). Myoviridae and Microviridae are two widespread and abundant human intestinal phage families that commonly infect Firmicutes and Bacteroidota hosts (including the genera Faecalibacterium, Lachnospira, and Bacteroides). In addition, crAss-phage primarily infected Bacteroides hosts (Bacteroides and Prevotella), and Herelleviridae mainly infected Firmicutes hosts (Flavonifractor). Surprisingly, the levels of most of these infected bacterial hosts, including Faecalibacterium, Eubacterium, and Lachnospira, changed significantly after P9 intervention.
Significantly fewer Podoviridae and Myoviridae sequences were detected in the P9 group compared to the placebo group at day 42, and the level of Podoviridae was also significantly reduced at day 42 compared to baseline (P < 0.05; Supplementary Fig. 2a). Correlation analysis was then performed between clinical indicators and 12 known bacteriophage families at days 28 and 42 (Fig. 4g). Only three bacteriophage families showed weak correlations with fecal urgency score and bowel frequency. Specifically, both Microviridae (r = 0.2, P = 0.012) and crAss-phage (r = 0.2, P = 0.003) exhibited a weak positive correlation with fecal urgency score; and Herelleviridae showed a weak positive correlation with bowel frequency (r = 0.2, P = 0.019). We further investigated the correlation between specific vOTUs and clinical parameters. Surprisingly, some vOTUs showed a stronger correlation with diarrhea symptom scores. For example, certain Microviridae vOTUs (vOTU-39925 and vOTU-59687) showed a significant positive correlation with fecal urgency; and several crAss-phage vOTUs (vOTU-02356, vOTU-73019, vOTU-49715, vOTU-55801, vOTU-32262, and vOTU-44203) displayed a significant positive correlation with fecal urgency (r ≥ 0.3 and P < 0.05 in all cases; Supplementary Fig. 2b). These results suggest that specific phage sequences may be associated with the improvement of diarrhea after P9 supplementation. However, owing to the unavailability of high-resolution viral genome assembly tools and databases, the role of the intestinal phageome in chronic diarrhea and its remission is still inconclusive.
P9 supplementation modulated patients’ predicted gut bioactive metabolites and carbohydrate-active enzymes (CAZymes)
A genome-centric metabolic reconstruction was established to identify intervention-associated changes in gut metabolic modules (GMMs) encoded within the entire dataset of 629 SGBs using the MetaCyc and the Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. A total of 72 GMMs belonging to 11 metabolite categories were identified, including SCFAs, amino acids, tryptophan, and its derivatives, unsaturated fatty acids, monosaccharides, disaccharides, polysaccharides, neurotransmitters, vitamins, BAs, and other metabolic modules (Fig. 5a). These identified modules were encoded by 34 bacterial orders. The modules of acetate synthesis, quinolinic acid degradation, and S-adenosylmethionine synthesis were common to most orders (Fig. 5a). However, some orders exhibited a higher degree of metabolic diversity than others. In particular, the Bacteroidales order exhibited the highest metabolic diversity, followed by Lachnospirales and Oscillospirales (both belonging to the Firmicutes phylum). These orders encoded a wide array of metabolic modules related to SCFA, amino acid, vitamin, and BA metabolism. Interestingly, several bacteria belonging to these three orders (including Lactiplantibacillus plantarum, Paraprevotella xylaniphila, and Acutalibacteraceae sp000431775) showed significant differential abundance after the P9 intervention. The modulation of these metabolically diverse taxa potentially impacts the overall biochemical functions of the gut microbiota.
a Distribution of 72 identified GMMs belonging to 11 metabolic modules, including short-chain fatty acids (SCFAs), amino acids, tryptophan and its derivatives, unsaturated fatty acids, monosaccharides, disaccharides, polysaccharides, neurotransmitters, vitamins, bile acids, and other metabolic modules across 34 bacterial orders (corresponding to 629 species-level genome bins, SGBs). Each metabolic module is shown in a different color. The size of the circle on the heatmap represents the proportion of SGBs encoding that specific module. b Shannon diversity index and (c) principal coordinates analysis (PCoA) score plots of the gut microbiota in the two groups at days 0 (0 d), 28 (28 d), and 42 (42 d). Adonis test results are displayed in the lower right corner of the PCoA plots. P-values shown in (b) were calculated using the two-sided Wilcoxon rank-sum test. d Boxplots comparing the abundances of predicted significantly differential bioactive metabolites that were responsive to P9 treatment at day 28 of the intervention. P-values < 0.05 in all cases, evaluated by the two-sided Wilcoxon rank-sum test. e Heatmap showing the enrichment of specific families of CAZymes (carbohydrate-binding modules [CBM], carbohydrate esterases [CE], glycoside hydrolases [GH], glycosyltransferases [GT], and polysaccharide lyases [PL]) in the fecal metagenome of the P9 group compared to the placebo group at days 0, 28, and 42. The color scale represents the relative distribution, ranging from −2 (low abundance) to 2 (high abundance). *P < 0.05, **P < 0.01, ***P < 0.001, evaluated by the two-sided Wilcoxon rank-sum test. The boxplots represent the interquartile range of each group’s distribution of values; the lines inside the boxes represent the median values; the whiskers denote the lowest and highest values within 1.5 times the interquartile range of each group; the dots above/below the upper/lower line are the outliers. Source data can be found in the Source Data file.
We then predicted changes in potential gut bioactive metabolites after the P9 intervention using the MelonnPan pipeline. We found no significant difference in the alpha diversity of the predicted gut bioactive metabolite profile between the P9 and placebo groups at all time points. (P > 0.05; Fig. 5b). The results of the alpha diversity analysis were consistent with those obtained from the beta diversity analysis using principal coordinate analysis and the adonis test. Significant differences were observed in the predicted gut bioactive metabolite profile between the P9 and placebo groups at day 28 (R2 = 0.028, P = 0.013), but not at day 0 (R2 = 0.004, P = 0.538) and day 42 (R2 = 0.008, P = 0.226; Fig. 5c). Moreover, a total of 18 differential gut-active metabolites between the P9 and placebo groups were probiotic-responsive; these predicted metabolites became significantly different between groups only after 28 days of P9 intervention, but not at baseline (Fig. 5d, Supplementary Table 5). Several of these probiotic-responsive metabolites, such as cholate, chenodeoxycholate, C16 carnitine, creatine, and bilirubin, were significantly enriched in the P9 group compared to the placebo group, while deoxycholate and glutamate were significantly depleted (P < 0.05; Fig. 5d).
To further explore the enzyme repertoire for complex polysaccharide metabolism encoded by the patients’ fecal microbiota, CAZyme genes were annotated using dbCAN2. A total of 26,170 CAZyme-encoding genes were found among the 629 SGBs (Supplementary Data 1), and most of these genes encoded the family glycoside hydrolases (GHs, 14,598 genes), followed by glycosyltransferases (GTs, 6,108 genes), carbohydrate esterases (CEs, 2,726 genes), carbohydrate-binding modules (CBMs, 1,690 genes), polysaccharide lyases (PL, 810 genes), and auxiliary activities (AAs, 238 genes). In addition, a comparative analysis of CAZyme-encoding subfamilies between the P9 and placebo groups revealed that 15 subfamilies were enriched in the P9 group at day 28 or day 42, including GHs (GH108, GH13_13, GH13_2, GH13_27, GH158, GH64, GH5_18), CBMs (CBM4, CBM56), glycosyltransferases (GT21, GT60, GT74), PLs (PL1_3, PL10_2), and CE16 (Fig. 5e). Taken together, these results indicated that P9 administration can lead to an enrichment of CAZyme-encoding genes within the gut microbiota, potentially contributing to an expanded carbohydrate utilization capacity.
P9 supplementation modulated patients’ fecal metabolome
Next, we analyzed specific changes in the fecal metabolome of the patients after the P9 intervention. On the principal component analysis score plot of the fecal metabolome, the symbols representing the quality control samples formed a tight cluster (Fig. 6a), indicating good stability of instrumental conditions and reliability of analysis. Partial least squares-discriminant analysis of the fecal metabolomes of samples from the two groups showed that there was a moderate degree of group-based separation at all three-time points, although no significant difference was detected by the adonis test (P > 0.05; Fig. 6b). We further analyzed metabolite-level differences in the fecal metabolomes between the two groups based on the variable importance in projection (VIP) scores generated by the partial least squares-discriminant analysis models (VIP > 2.0) and the P-values calculated by the two-sided Wilcoxon rank-sum test (P < 0.05). A total of 21 significant differential metabolites were identified at days 28 and 42 (Fig. 6c; Supplementary Table 6). Notably, these metabolites did not exhibit significant differences between the two groups at baseline but became differentially abundant only during or after the P9 intervention. Of particular interest were caffeic acid, taurine, 3-hydroxypentanoic acid, hexacosanoic acid, and cerotic acid, which were significantly enriched in the P9 group compared to the placebo group after the probiotic intervention (P < 0.05; Fig. 6d). We further explored the associations between the differentially abundant metabolites and clinical indicators of diarrhea using two-tailed Pearson correlation analysis. Interestingly, the results showed that 3-hydroxypentanoic acid was significantly and negatively correlated with the stool consistency score (r = −0.22, P = 0.048; Fig. 6e) and the diarrhea symptom severity score (r = −0.23, P = 0.039; Fig. 6e).
a Principal component analysis (PCA) and (b) partial least squares-discriminant analysis (PLS-DA) score scatter plots of the fecal metabolome in the two groups at days 0 (0 d), 28 (28 d), and 42 (42 d) of the intervention. Adonis test results are shown in the lower right corner of the PLS-DA score plots. QC represents quality control samples. c Significant differential fecal metabolites identified between the two groups at days 28 and 42. Variable importance in projection (VIP) scores were generated by PLS-DA to identify metabolites contributing to the discrimination, while P-values were calculated using the two-sided Wilcoxon rank-sum test (cut-off level: VIP > 2; P < 0.05). d Boxplots showing relevant responsive fecal metabolites to the P9 intervention. The boxplots represent the interquartile range of each group’s distribution of values; the lines inside the boxes represent the median values; the whiskers denote the lowest and highest values within 1.5 times the interquartile range of each group; the dots above/below the upper/lower line are the outliers. Statistically significant differences are indicated by the P-values, evaluated using the two-sided Wilcoxon rank-sum test, with P < 0.05 considered statistically significant. e Pearson correlation scatter plots showing the correlation between differential fecal metabolites and clinical indicators of diarrhea in the P9 group after 28 days of intervention. The gray error band represents the fitted values, and the shaded area indicates the 95% confidence interval. The blue dots represent the values on the X and Y axes, respectively. Source data can be found in the Source Data file.
P9 administration affected patients’ fecal SCFA and BA levels
Targeted metabolomics was applied to confirm the post-intervention changes in fecal SCFA and BA levels using gas chromatography-mass spectrometry (GC-MS) and liquid chromatography-mass spectrometry (LC-MS). After 28 days of intervention, more acetic acid and butyric acid were detected in the P9 group compared to the placebo group (P < 0.05), but the fecal levels of isovaleric acid, valeric acid, propionic acid, and isobutyric acid exhibited no significant differences between groups (P > 0.05; Fig. 7a). In addition, compared to the placebo group, the P9 intervention significantly reduced the fecal level of deoxycholic acid (P < 0.05), while there was no significant difference between the two groups for cholic acid and chenodeoxycholic acid (P > 0.05; Fig. 7b). These results confirmed that the P9 intervention could increase the patients’ functional intestinal metabolites, such as acetic acid and butyric acid while lowering the deoxycholic acid level.
a, b Concentrations of fecal SCFAs and BAs in the two groups at day 28. The P-values were generated using the two-sided Wilcoxon rank-sum test; P < 0.05 was considered statistically significant. c Schematic diagram showing P9 intervention-associated changes in the host gut microbiota and metabolites, which may have contributed to the relief of diarrhea. Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. CA = cholic acid; CDCA = chenodeoxycholic acid; 7α-HSDH = 7α-hydroxysteroid dehydrogenase; LCA = lithocholic acid; DCA = Deoxycholic acid. The boxplots represent the interquartile range of each group’s distribution of values; the lines inside the boxes represent the median values; the whiskers denote the lowest and highest values within 1.5 times the interquartile range of each group; the dots above/below the upper/lower line are the outliers. Source data can be found in the Source Data file.
Discussion
Chronic diarrhea is a multifactorial disorder that causes significant psychological distress to patients and economic burden to the healthcare system28. Probiotics are increasingly being prescribed for the management of chronic diarrhea. However, previous intervention studies of probiotic-based diarrhea management have mostly focused on reporting clinical outcomes29,30,31, without elucidating the role of the host gut microbiota in diarrhea relief using systematic multi-omics approaches. Therefore, we conducted a large, randomized, double-blind, placebo-controlled clinical trial with 189 subjects to investigate the potential beneficial effect of P9 administration in alleviating chronic diarrhea (Chinese Clinical Trial Registry, ChiCTR2000038410), and our results indicated that P9 intervention had a modest diarrhea-alleviating effect. Through multi-omics analysis, our preliminary investigation suggests that the mechanism of the diarrhea-relieving effect of P9 is via modulating of certain gut bacteria (Lactiplantibacillus plantarum, Ruminococcus_A faecicola, and Paraprevotella xylaniphila), bacteriophages (Herelleviridae and Microviridae), and microbial metabolites (acetic acid, butyric acid and deoxycholic acid) through function-driven changes and co-interactions in the treatment process (Fig. 7c).
Our first finding is that P9 administration improved diarrhea symptoms in patients in both the ITT and PP analyses. Significant reductions in diarrhea severity scores were observed at day 28 in both the placebo and P9 groups. However, the magnitude of score reduction in diarrhea severity score was significantly greater in the P9 group compared to the placebo group. The stool consistency score was significantly reduced in the PP analysis but was not significant in the ITT analysis (P = 0.052). These results suggest that P9 administration had a modest diarrhea-alleviating effect. On the other hand, the reduction in diarrhea severity and improvement in stool consistency in the non-probiotic group could be a placebo effect, which is often observed in clinical trials, especially with subjective assessment parameters such as the stool consistency score32. Our results showed that P9 administration only modestly reduced the frequency and urgency of bowel movements in patients. The small magnitude of reduction in bowel movement frequency and urgency may be attributed to the relatively low baseline levels of bowel movement frequency and urgency in this cohort of patients, leaving little room for improvement in these aspects. Nevertheless, our results are consistent with some previous studies finding that probiotic use can alleviate chronic diarrhea. For example, Bai et al. reported that probiotic administration could alleviate IBS-D, but the beneficial effect was only short-term33. A separate study found that Lactiplantibacillus plantarum CCFM1143 supplementation for four weeks reduced defecation frequency and Bristol score in patients with chronic diarrhea18. Similarly, a placebo-controlled study reported a reduction in loose stool frequency after consuming Lactiplantibacillus plantarum CJLP243 for 2 months20. Importantly, no serious adverse events occurred in any of the patients during the intervention period. Thus, the use of P9 as a dietary intervention appears to be a safe alternative strategy for alleviating chronic diarrhea.
Probiotics have been shown to reduce diarrhea through two known mechanisms: improving the balance of the gut microbiota and preventing the colonization of harmful bacteria34. Therefore, we analyzed changes in the host gut microbiota during the P9 intervention, which revealed no significant post-intervention alterations in the alpha diversity (Shannon and Simpson diversity indices) of the patients’ gut bacterial and bacteriophage microbiota. Significant post-intervention changes in the fecal microbiota of subjects in the P9 group were, however, identified by beta diversity analysis (evaluated by principal coordinate analysis and the adonis test) at day 28. We further explored our dataset to identify P9-responsive gut bacteria and bacteriophages, and found that P9 administration could substantially enrich taxa like Acutalibacteraceae sp000431775, Paraprevotella xylaniphila, Ruminococcus_A faecicola, Butyricicoccus_A sp002395695, and Streptococcus thermophilus in the subjects’ gut microbiota, while significantly decreasing the levels of Mediterraneibacter torques, Coprococcus sp., Enterocloster sp000431375, Podoviridae, and Myoviridae.
Some of these differentially abundant taxa have been shown to play a part in diarrhea. Acutalibacteraceae is a novel and relatively undescribed family, and most of its members are involved in BA metabolism, ameliorating stressful intestinal conditions (such as stress-induced diarrhea) by improving bile tolerance and facilitating host mucin glycan degradation35,36. Previous studies have reported that SCFA (especially butyric acid and acetic acid) producers, such as Paraprevotella xylaniphila, Ruminococcus_A faecicola, and Butyricicoccus_A sp002395695, could substantially raise colonic SCFA levels in patients with inflammatory bowel disease and antibiotic-associated diarrhea37,38,39,40. Colonic SCFAs have been found to correlate positively with diarrhea alleviation41,42. Streptococcus thermophilus is a common yogurt starter species, and yogurt consumption has been shown to effectively alleviate diarrhea in a randomized clinical trial in children43. On the other hand, Mediterraneibacter torques, also known as Ruminococcus torques, is linked to intestinal permeability and gastrointestinal diseases and has been found to be enriched in the gut microbiota of patients with IBS-D44,45. We also found that bowel frequency showed a significant positive correlation with Mediterraneibacter torques and Herelleviridae, while crAss-phage and Microviridae exhibited a significant positive correlation with fecal urgency score. These results suggest that high levels of gut crAss-phage, Microviridae, and Herelleviridae sequences may be linked to diarrhea symptoms.
Several studies have reported an association between increased abundance of Coprococcus and Enterocloster and conditions such as antibiotic-associated diarrhea in mice46,47, Crohn’s disease, and IBS-D in patients48,49. Notably, these genera, along with Mediterraneibacter, Coprococcus, and Enterocloster, belong to the phyla Firmicutes and Bacteroidetes, which are commonly targeted by various phage families, including Herelleviridae, Microviridae, Podoviridae, Myoviridae, and crAss-phage. Multiple studies also reported that patients with gastrointestinal diseases (such as ulcerative colitis50, IBS-D51, and colorectal cancer52) had significantly more crAss-phage, Microviridae, Podoviridae, Myoviridae, and Herelleviridae than healthy subjects. These results suggest that probiotic administration may affect the intestinal phageome composition by modulating the gut microbiota profile. However, it is worth noting that we found only weak associations between family-level phageome and diarrhea relief, which could be attributed to the lack of available high-resolution viral genome assembly, annotation, and database tools and resources. Interestingly, the strength of the correlations between diarrhea relief and individual vOTUs varied, with some vOTUs exhibiting stronger correlations with diarrhea symptom scores than the family-level phageome. This suggests that certain vOTUs may play a more critical role in symptom relief than others. Thus, how the gut phageome and individual vOTUs affect diarrhea requires further clarification. In summary, our results suggest that the post-intervention changes in the interactions between the host’s gut bacteria and phage microbiota may represent a potential mechanism for diarrhea relief.
Gut microbiota-derived metabolites, such as BAs, can influence disease severity53. Approximately 5% of unabsorbed primary BAs (cholate and chenodeoxycholate) in the ileum are deconjugated by gut bacteria on reaching the colon to form secondary BAs, such as deoxycholic acid and lithocholic acid54. Malabsorption or overproduction of BAs can lead to chronic watery diarrhea55. One study reported that patients with IBS-D had more fecal primary BAs, which correlated with stool consistency and frequency56. Our study found that administering P9 lowered the predicted levels of deoxycholate while increasing that of cholate and chenodeoxycholate. These results are concordant with those found by targeted metabolomics to monitor fecal BA levels. Previous studies have reported that deoxycholate can lead to diarrhea through stimulating fluid secretion, increasing mucosal permeability, and inducing high-amplitude contractions in the colon54,57. Therefore, our results suggest that the P9 intervention may alleviate diarrhea by reducing patients’ intestinal deoxycholate content.
Interestingly, this study found that the P9 intervention decreased the glutamate level in the gut metabolome, while the taurine level increased. Glutamate has been implicated in reshaping the gut microbiota and the overall colonic environment in ways that can be detrimental to host health. Dietary glutamate has been associated with exacerbating symptoms in patients with IBS-D by increasing intestinal dysmotility and pain sensitivity58,59. In contrast, taurine increased in the fecal metabolome of patients receiving P9. Previous animal studies have shown that taurine can increase intestinal epithelial barrier integrity via enhancing the expression of tight junction proteins60. Furthermore, taurine supplementation has shown promise in attenuating diarrhea severity61. These results suggest that the regulation of amino acid metabolism may represent a potential pathway for P9 to alleviate diarrhea. Additionally, there were substantial increases in other bioactive metabolites (such as carnitine and bilirubin) and fecal metabolites (such as caffeic acid and long-chain fatty acids like hexacosanoic acid and cerotic acid) following the P9 intervention. Previous in vitro and animal studies have shown that both carnitine and bilirubin can lessen colonic oxidative stress and inflammation, thereby alleviating colitis62,63. Thus, high intestinal carnitine and bilirubin levels can be part of the mechanism underlying diarrhea relief after P9 administration. Moreover, caffeic acid supplementation has been shown to reverse gut dysbiosis, intestinal barrier damage, and inflammatory cell infiltration in colonic tissues in colitis mice64. Additionally, a traditional Mexican phytomedicine containing caffeic acid has been used to treat diarrhea and dysentery65. An earlier animal study found that bacterial long-chain fatty acids can alleviate colitis and inflammation in mice by activating peroxisome proliferator-activated receptor gamma66.
The human gut microbiota genome encodes numerous CAZYmes that enable the hydrolysis of complex carbohydrates into functional metabolites67. Thus, the profile of gut microbiota-encoded CAZYmes reflects its carbohydrate metabolic capacity. Our study identified a panel of differentially abundant metabolic modules between the P9 and placebo groups after the intervention, including 15 significantly increased subfamilies of CAZymes, primarily GHs, GTs, PLs, and CBMs. These are key enzymes for decomposing complex carbohydrates to generate SCFAs and other metabolites that promote host energy acquisition68,69. Consistently, most fecal SGBs in the P9 group contained more metabolic modules of SCFAs compared to the placebo group. The post-intervention increases in fecal acetic acid and butyric acid in the P9 group were confirmed by GC-MS analysis. Our observations are in line with a previous study, which demonstrated that Lactiplantibacillus plantarum CCFM1143 administration significantly raised the levels of acetic acid and propionic acid in diarrhea patients compared to placebo18. Short-chain fatty acids, including acetate, butyrate, and propionate, are major gut microbiota-derived metabolites responsible for microbial-host interactions70,71. Oral administration of sodium acetate or sodium butyrate has been shown to alleviate diarrhea in mice by enriching beneficial bacteria and suppressing pathogens72. Butyric acid not only serves as a vital source of energy for enterocytes but also has anti-inflammatory and protective effects on the intestinal epithelial barrier73,74. We also confirmed that the fecal content of 3-hydroxypentanoic acid was significantly higher in the P9 group compared to the placebo group and was negatively correlated with stool consistency score and diarrhea severity score. 3-Hydroxypentanoic acid is an SCFA that is biochemically related to valeric acid, but its function requires further investigation. Overall, the significant increase in intestinal SCFAs, particularly acetic acid and butyric acid, subsequent to P9 intervention may play an important role in promoting intestinal barrier repair and enhancing intestinal immune function, thereby alleviating diarrhea.
This study has several limitations. Firstly, the Rome IV criteria were implemented as the inclusion criteria to identify patients with chronic diarrhea in this study. Although multiple methods were employed, including a diarrhea population questionnaire, interviews with clinicians and healthcare professionals, and routine stool tests, the recent recommendations from the United European Gastroenterology and European Society for Neurogastroenterology and Motility (June 2022) suggest the inclusion of additional diagnostic tests such as complete blood count, biochemical parameters like C-reactive protein and fecal calprotectin, and celiac disease serological analysis for the diagnosis of IBS-D75. However, conducting this study during the COVID-19 pandemic constrained access to medical resources, leading to the exclusion of patients with IBS-D based solely on clinical history rather than colonoscopy results and other laboratory-based tests. Consequently, the definitive exclusion of IBS-D could not be ensured in this study. To address this limitation, we adopted a broader term, ‘chronic diarrhea’, instead of the more specific term, “functional diarrhea”, specified in the Rome IV criteria to describe the study cohort. We propose that future research endeavors utilize more stringent inclusion criteria, incorporating peripheral blood routine, blood biochemistry, medical imaging, and endoscopy, for a more comprehensive assessment and accurate differentiation between different subtypes of chronic diarrhea. In addition, this study initially aimed to recruit 200 subjects for the ITT population. However, due to the constraints posed by the COVID-19 pandemic, our enrollment was reduced to 189 subjects. However, after evaluation and approval from the Ethics Committee, we could meet the statistical requirements of the minimum sample size.
Secondly, both the probiotic and placebo groups showed a reduction in diarrhea severity and improvement in stool consistency. However, the changes were more pronounced in the probiotic group, suggesting a potential placebo effect. Additionally, only the PP analysis, but not the ITT analysis, demonstrated a significantly lower mean stool consistency score in the P9 group compared to the placebo group at day 28. The marginal functional effect on improving stool consistency is likely attributed to the low baseline symptom level in the current young adult cohort. High intragroup variability in diarrhea symptom severity and stool consistency scores was noted, possibly due to the diverse individual responses to the intervention. This variability is not unexpected, as our study focused on evaluating the beneficial effects of a probiotic strain rather than conventional medications. Probiotics primarily function by modulating the overall balance of the gut microbiota to restore and maintain colonic homeostasis. Thus, dramatic post-probiotic intervention responses are not always anticipated, and a longer duration of intervention may be needed for more substantial effects. In fact, non-responders are common in probiotic intervention trials, particularly in large-scale studies76. Future probiotic intervention trials can be strengthened by clearly defining the endpoint, maximizing positive responses through implementing precise and objective clinical assessment scales, and carefully selecting appropriate study cohorts. These measures can improve the reliability and validity of the findings and provide a more comprehensive understanding of the effects of probiotics in similar interventions.
Thirdly, this study faced challenges in obtaining blood samples to monitor physiological parameters, gut barrier indicators, and inflammatory responses. Additionally, this study also lacked complete data on the impact of dietary habits on clinical outcomes. The diary recorded incidents of dietary changes, but detailed dietary surveys were not implemented to quantify the effect of food intake and eating habits on clinical presentation, symptom improvement, and changes in the gut metagenome and metabolome. Future studies should incorporate detailed dietary surveys to better understand and quantify the influence of these factors.
In conclusion, this study provides evidence for the modest alleviating effect of the probiotic P9 on chronic diarrhea. The observed improvement in symptoms was accompanied by modulation of specific gut bacteria, bacteriophages, and metabolites, playing a role in reducing inflammation and enhancing intestinal barrier function. These findings support the existing body of literature on the use of probiotics in managing chronic diarrhea.
Methods
Sample size calculation, subject enrollment, and trial design
Details of the sample size determination have been published in the study protocol77. Briefly, we estimated the sample size using the diarrhea symptom severity score as the primary outcome variable. During the trial protocol design, we conducted a thorough literature review and comparison. This probiotic intervention study was the first large-scale randomized controlled trial conducted in young adults with chronic diarrhea. Given the limited availability of reference data for calculating sample size based on the primary outcome, we had to employ an alternative approach. To estimate the required sample size, we pooled data from several previous studies78,79,80 on functional gastrointestinal diseases and sought the guidance of clinical gastroenterologists. Based on the combined information, it was estimated that the difference in diarrhea symptom severity score between the P9 and placebo groups on day 28 could be as high as three points, with a standard deviation of 6.8. With a two-sided 5% significance level and 80% power, a sample size of 81 subjects per group was required, with an expected dropout rate of 20%.
The planned sample size was 200 subjects (100 per group), while the enrollment was slowed by COVID-19 pandemic, and the final sample size had to be reduced to 189 subjects. After a rigorous evaluation and unanimous approval from the Ethics Committee, the decision was made to proceed with the smaller sample size, rather than delaying the work to wait for the last 11 qualified subjects to appear. And the number of subjects reached the minimum sample size of statistical requirement. After each subject completed four weeks of intervention and two weeks of follow-up, the study was terminated, and 170 subjects successfully completed the trial.
A total of 189 patients with chronic diarrhea were enrolled in this study based on the Rome IV criteria and previous reports18,81. Inclusion criteria for patients with chronic diarrhea were: 1) males and females aged 18–65 years; 2) diagnosis based on the Rome IV criteria82, which required the presence of loose or watery stools accounting for more than 25% of bowel movements in the last three months, Bristol stool scale types 5, 6, and 7, and symptom onset at least six months prior to enrollment; 3) normal routine fecal tests (including occult blood) in subjects aged 18–50 years. Exclusion criteria were: 1) history of colon cancer, celiac disease, organic bowel disease, or inflammatory bowel disease; 2) abnormal results in routine fecal examination; 3) pregnancy or lactation; 4) allergy to any of the ingredients in the intervention material; 5) treatment with antibiotics or probiotics within the last two weeks, or antidepressants, antianxiety agents, and other psychotropic drugs one month prior to enrollment; 6) long-term medication to improve diarrhea; 7) concurrent serious illness (such as myocardial infarction, cerebral infarction, malignant tumor, and so on); 8) major mental illness, inability to control behavior, or inability to cooperate; 9) illiteracy, unable to understand or independently sign the informed consent. Each participant received a modest reimbursement to offset transportation expenses and acknowledge their time commitment to the study.
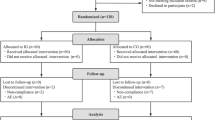
Following a two-week run-in period, participating patients were randomized into two groups: P9 (probiotic) and placebo in 1:1 ratio using a simple randomization method. The randomization process was performed by two independent project administrators using a computer-generated random number of codes to assign participants to their respective groups. The participants, physicians, and investigators involved in this study were blinded, and the group information, concealed in an opaque envelope, was opened just before the primary data analysis. The ITT population analysis was performed on all 189 subjects who were enrolled in the study, and the PP population analysis was also performed for 170 subjects who demonstrated good compliance with the protocol (n = 85 subjects per group; Figs. 1, 2a). Patient gender was self-reported and was not specifically accounted for in the study design. However, upon analysis, no statistically significant differences in gender distribution were observed between the two groups. The male-to-female ratio in the probiotic group was n = 44/49, and in the placebo group, it was n = 48/48.
The probiotic and placebo materials used in the study were provided by Jinhua Yinhe Biological Technology Co., Ltd. (Zhejiang, China). Once the probiotic powder was manufactured in the factory, it was transported to the study center and refrigerated. Each subject received the probiotics once a week and was instructed to store them in a cool, dry place to ensure the probiotic stability and efficacy. The P9 powder was prepared as a dry powder of 1.0 × 1011 colony-forming units in 2 g. It was taken once daily, 30 min after meals, and mixed with warm water. The placebo powder was maltodextrin. The probiotic and placebo materials were identical in shape, texture, and appearance. The intervention lasted 28 days from day 0 to day 28, followed by a 14-day post-intervention follow-up period until day 42. Each subject received a 30-day supply of either P9 or placebo, including two spare doses provided as a precautionary measure to account for potential loss or damage. Compliance with the probiotic intervention was assessed by counting returned doses. The compliance rate was calculated as [(Number of actual dispensed dose − Number of returned dose) / Number of prescribed doses] x 100%.
Fecal samples were collected from the participants using sterile stool samplers on days 0, 28, and 42, with the addition of a DNA protection solution (Guangdong Longsee Biomedical Co., Ltd., Guangzhou, China). The collected samples were stored at −80 °C until further analysis.
Endpoint definitions
The primary endpoint of this study was the assessment of diarrhea symptom severity using three items selected from the Gastrointestinal Symptom Rating Scale: increased passage of stools, loose stools, and urgent need for defecation83,84, specifically focusing on diarrhea symptoms. The first item was “increased passage of stools”, which was scored on a scale of 0 to 3. Scores of 0, 1, 2, and 3 represented stool passage frequencies of one, three, five, and seven or more times per day, respectively. The second item, “loose stools”, was assessed using the Bristol Stool Scale as a reference. The score ranged from 0 to 3, with 0 indicating normal stools (Bristol stool types 1–4), 1 indicating loose stools (Bristol stool type 5), 2 indicating runny stools (Bristol stool type 6), and 3 indicating watery stools (Bristol stool type 7). The third item, “urgent need for defecation”, was scored on a scale of 0–3. A higher score indicated more severe symptoms, with 0 representing no urgency, 1 indicating mild urgency, 2 denoting moderate urgency, and 3 indicating severe urgency. The diarrhea symptom severity score was calculated as the sum of the average weekly scores for these three items85.
Secondary endpoints included stool consistency (evaluated using the Bristol Stool Form scale, score 1 to 7), number of bowel movements, fecal urgency score, Depression Anxiety Stress Scales-21 (DASS-21)86, and fecal metagenome and metabolome87. The fecal urgency score was calculated from a four-point scale based on symptom severity (0 = none, 1 = mild, 2 = moderate, and 3 = severe)88. Data were collected before and during the intervention using a daily electronic stool diary. The mean of each dimension at each time point (days 14, 28, and 42) was then calculated from the relevant data to obtain the Gastrointestinal Symptom Rating Scale scores.
Fecal and serum miRNAs expression profiling and differential analysis were prespecified secondary outcomes in this study. However, these miRNA-related analyses were planned to be conducted separately by our colleagues at Jiangxi University of Chinese Medicine. As a result of this institutional collaboration, the miRNA components of the study have not been included in the current manuscript but will be reported in a future publication.
Adverse events, including systemic infection, deleterious metabolic activities, excessive immune stimulation, and gastrointestinal side effects89, were recorded throughout the intervention period.
Fecal DNA extraction, shotgun metagenomic sequencing, and data quality control
Fecal metagenomics analysis was performed using samples from the PP population, with a total of 169 subjects contributing stool samples (84 in the P9 group and 85 in the placebo group). The stool samples were subjected to sequencing on an Illumina Novaseq 6000 (Illumina Inc., San Diego, CA, USA). Metagenomic DNA was extracted from these samples using the Magnetic Soil and Stool DNA Kit (DP712; TIANGEN Biotech Co., Ltd., Beijing, China) following the corresponding procedures27,90.
Notably, metagenomic sequencing failed for 12 samples from 12 different subjects at one of the time points (either day 28 or day 42), resulting in a total of 495 successfully sequenced samples (Supplementary Table 7). A total of 11.5 Tbp of high-quality clean data were generated, with an average of 23.2 Gbp/sample. The clean data obtained from the samples were then assembled into contigs using MEGAHIT91, and contigs larger than 2,000 bp were selected for binning using MetaBAT292, VAMB93, and DAS Tool94 with default parameters. Finally, all the bins were combined to generate metagenome-assembled genomes using custom scripts.
The quality of the metagenome-assembled genomes was examined using CheckM95. Subsequently, they were categorized into high-, medium-, and partial-quality genomes based on their levels of completeness and contamination. The high-quality genomes were clustered, and the most representative genomes from each replicate set were selected to obtain SGBs using dRep (parameters: -pa 0.95 and -sa 0.95)96. Taxonomic annotation of the SGBs was performed by aligning the genomes to a curated set of 6,530 representative bacterial genomes from the National Center for Biotechnology Information Non-Redundant Protein Sequence Database using Kraken2. The relative abundance of each SGB was calculated using CoverM (https://github.com/wwood/CoverM, parameters: -min-read-percent-identity 0.95 -min-covered-fraction 0.4).
Published literature and the MetaCyc metabolic database were used to predict SGBs encoding related GMMs97,98. Open reading frames were compared against the KEGG database to annotate the key metabolic modules of each SGB. The distribution of synthesis or degradation modules in the SGBs was identified by Omixer-RPM (parameter: -c 0.66). In addition, the distribution of gut bioactive metabolites was predicted using the MelonnPan-predict workflow99. Genes encoding CAZymes were further detected using dbCAN2100.
Phageome identification and abundance analysis
All assembled contigs were first assessed by CheckV to assess viral genome quality. Then, potential viral sequences were identified by VIBRANT with default options101,102. Proposed viral contigs with a length greater than 5000 bp were further clustered into vOTUs with 95% nucleotide identity over 80% of the sequence using CD-HIT (https://github.com/weizhongli/cdhit)103.
The resulting 145,589 vOTUs were compared against a curated set of 189,680 viral genomes from the Metagenomic Gut Virus catalog (accessed in July 2021) to assess the novelty of vOTUs in this dataset104. The average abundance of vOTUs across the viral population was finally calculated using the CoverM contig pipeline (https://github.com/wwood/CoverM, parameters: -min-read-percent-identity 0.95, -min-read-aligned-percent 0.5, -proper-pairs-only, and –exclude-supplementary).
Untargeted fecal metabolomics analysis by LC-MS
Untargeted metabolomic analysis was performed on the stool samples collected from patients included in the PP population. However, some subjects presented with unformed stools, which posed challenges for fecal metabolomics analysis. The non-targeted metabolomics analysis included samples from 161 patients (80 and 81 patients in the probiotic and placebo groups, respectively), and successfully analyzed a total of 479 samples across all time points.
Fecal samples were dried and then mixed with 600 µL of a methanol solution containing 2-chlorophenylalanine. The mixture was vortexed for 30 s and then ground for 60 s at 55 Hz in the presence of 100 mg glass beads. After ultrasonication and centrifugation at 12,000 x g for 10 min, the supernatant was filtered through a 0.22 μm ultrafiltration membrane. Chromatographic separations were performed with an Agilent 1290 Infinity LC ultra-high-performance liquid chromatography system (Agilent Technologies, Inc., Santa Clara, CA, USA), and the separations were performed on an ACQUITY UPLC BEH amide column (100 × 2.1 mm, 1.7 μm; Waters Corporation, Milford, MA, USA) at 25 °C with a flow rate of 0.5 mL/min. The mobile phase consisted of a mixture of 25 mM ammonium acetate and 25 mM ammonia in water (A) and acetonitrile (B). The optimized gradient elution (0–0.5 min, 95% B; 0.5–7 min, 95–65% B; 7–8 min, 65–40% B; 8–9 min, 40% B; 9–9.1 min, 40–95% B; 9.1–12 min, 95% B) was used to separate the different metabolites in the samples. Each sample was analyzed in both positive and negative ionization modes under electrospray ionization. The optimal MS parameters were as follows: auxiliary gas, 60; curtain gas, 30 psi; ion source temperature: 600 °C; spray electricity, ± 5500 V (positive and negative modes). The first-level ion scanning range was m/z 60–1000, and the second-level fragmentation was performed using information-dependent acquisition mode. The cluster-removing voltage was ± 60 V (both positive and negative modes), the collision energy was 35 ± 15 eV, and the information-dependent acquisition was set as follows: dynamic exclusion of isotope ions range of 4 Da, collecting 10 fragment maps per scan.
ProteoWizard software (version 3.0.5047) and the R package XCMS were used to process the raw data generated by ultra-high performance liquid chromatography-quadrupole time-of-flight mass spectrometry. Principal component analysis and partial least squares-discriminant analysis were applied to identify differential biomarker metabolites between groups (cut-off level: VIP value > 2 and P-value < 0.05). Selected biomarkers and potential metabolic pathways were further identified by searching the HMDB, METLIN, Massbank (http://www.massbank.jp), and KEGG databases.
Targeted fecal metabolomics analysis for quantification of fecal SCFAs and BAs
Targeted metabolomic analysis was performed on the stool samples collected from patients included in the PP population. However, some patients had unformed stools, posing challenges for targeted fecal metabolomics analysis. Of these, 42 patients (18 and 24 patients in the probiotic and placebo groups, respectively) had insufficient stool volume. Therefore, the fecal SCFA and BA levels of only 127 patients (66 and 61 in the probiotic and placebo groups, respectively) were determined by targeted metabolomics using GC-MS and LC-MS (BioNovoGene, Suzhou, China).
To detect SCFAs, stool samples (50 mg) were weighed and transferred to clean centrifuge tubes. 15% phosphoric acid (50 μL), 75 μg/mL internal standard (isocaproic acid, 10 μL), and diethyl ether (140 μL) were added to the samples. The solutions were then mixed for 1 min and centrifuged at 12,000 x g for 10 min at 4 °C. An aliquot of 200 μL of the supernatant was transferred to a sample vial for analysis. The SCFA levels were determined by GC-MS using a TRACE™ 1300-ISQ LT GC system (Thermo Fisher Scientific, Waltham, MA, USA)105. The GC was equipped with a capillary column HP-INNOWAX (30 m × 0.25 mm, 0.25 μm, Agilent Technologies, Inc., Santa Clara, CA, USA), and helium was used as the carrier gas at a constant flow rate of 1 mL/min. The initial temperature was maintained at 90 °C and then increased to 120 °C at 10 °C/min, 150 °C at 5 °C/min, and 250 °C at 25 °C/min, with the final temperature held for 2 min. The temperatures of the injector, transfer line, and ion source were set to 250 °C, 250 °C, and 230 °C, respectively. Mass spectrometric detection of the metabolites was performed on an ISQ 7000 (Thermo Fisher Scientific, Waltham, MA, USA) with electron impact ionization of 70 eV in a full-scan mode with an m/z range of 35–780. Acquisition was performed in negative selective ion monitoring mode.
To detect BAs106, 400 μL of methanol was added to the fecal suspensions, and the mixture was vortexed for 60 s. Then, the samples were ground for 60 s at 55 Hz in the presence of 100 mg glass beads. After ultrasonication and centrifugation at 12,000 x g for 10 min, 300 μL supernatant was added with 600 μL water and vortexed for 30 s. The supernatant was diluted with 30% methanol and then filtered through a 0.22 μm ultrafiltration membrane. The separation of BAs in the samples was performed using an ACQUITY UPLC® BEH C18 column (100 × 2.1 mm, 1.7 μm; Waters Corporation, Milford, MA, USA) at 40 °C with a flow rate of 0.25 mL/min. The mobile phase consisted of 0.01% formic acid in water (A) and acetonitrile (B). The optimized gradient elution program was as follows: 0–4 min, 25% B; 4–9 min, 25–30% B; 9–14 min, 30–36% B; 14–18 min, 36–38% B; 18–24 min, 38–50% B; 24–32 min, 50–75% B; 32–33 min, 75–90% B; 33–35.5 min, 90–25% B. Acquisition was performed in electrospray ionization mode. The ion source temperature was set to 500 °C, and scanning was performed using multiple response monitoring.
Statistics & reproducibility
Statistical tests and data visualization were performed using R software (version 4.1.0) and Adobe Illustrator 2021. And the body structure illustration was built with the BioRender graphical tool (http://BioRender.com). Differences in gender, body mass index, allergy history, drug use, and adverse events between the P9 and placebo groups were evaluated using the chi-square test. Inter- and intragroup differences in the primary outcome, number of bowel movements, and frequency and urgency of bowel movements were evaluated using the unpaired or two-sided paired Wilcoxon rank-sum test. To address missing data in the ITT population analysis, the baseline-observation-carried-forward method was utilized for imputation, ensuring a comprehensive assessment of the entire population. Moreover, to accommodate initial variations, we performed a non-parametric covariance analysis on the diarrhea symptom scores using the sm package in R, incorporating the baseline data as a covariate.
Several R packages, including vegan, optparse, mixOmics, ggplot2, and ggpubr, were used to calculate the Shannon and Simpson’s diversity indices and to execute principal component analysis, principal coordinate analysis, partial least squares-discriminant analysis, adonis test, and Procrustes analysis. The two-sided Wilcoxon rank-sum test and t-test were used to evaluate statistical differences in various variables between groups. A P-value of less than 0.05 was considered statistically significant. Two-tailed Pearson correlation analysis was used to analyze the associations between clinical indicators, fecal bacterial microbiota, and metabolites.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The demographic characteristics are provided as Supplementary Information. The private clinical information of each patient is available under restricted access to ensure patients’ rights to privacy and anonymity; access may be obtained by contacting the first authors Teng Ma (18447054019@163.com) and Ni Yang (yangni@crjz.com). Access to the data is granted for academic purposes only, and there are no restrictions on how long data will be made available. The sequencing data generated in this study have been deposited in the NCBI genome database (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1130110). The metabolomics data generated in this study have been deposited in the MassIVE database under accession code MSV000095296 (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=5b6dd91f6d974fa1a39f4e4836fc28f9). The clinical protocol of this study has been published77. The remaining data are available within the manuscript and the Source Data files. Source data are provided with this paper.
Code availability
The analysis pipeline and custom scripts used in this study are available in the GitHub repository (https://doi.org/10.5281/zenodo.11194887)107.
References
Chu, C., Rotondo-Trivette, S. & Michail, S. Chronic diarrhea. Curr. Probl. Pediatr. Adolesc. Health Care 50, 100841 (2020).
Burgers, K., Lindberg, B. & Bevis, Z. J. Chronic diarrhea in adults: evaluation and differential diagnosis. Am. Fam. physician 101, 472–480 (2020).
Longstreth, G. F. et al. Functional bowel disorders. Gastroenterology 130, 1480–1491 (2006).
Schiller, L. R., Pardi, D. S. & Sellin, J. H. Chronic diarrhea: diagnosis and management. Clin. Gastroenterol. Hepatology. 15, 182–193.e183 (2017).
Ma, H. et al. The effect of clonidine on gastrointestinal side effects associated with ultra-rapid opioid detoxification. Anesth. Analg. 96, 1409–1412 (2003).
Wang, Y. et al. Probiotics therapy for adults with diarrhea-predominant irritable bowel syndrome: a systematic review and meta-analysis of 10 RCTs. Int. J. Colorectal Dis. 37, 2263–2276 (2022).
Wang, Y. et al. Antidepressants can induce mutation and enhance persistence toward multiple antibiotics. Proc. Natl Acad. Sci. USA 120, e2208344120 (2023).
Li, Y. et al. Gut microbiota and diarrhea: an updated review. Front. Cell. Infect. Microbiol. 11, 625210 (2021).
Rouhani, S. et al. Diarrhea as a potential cause and consequence of reduced gut microbial diversity among undernourished children in Peru. Clin. Infect. Dis. 71, 989–999 (2020).
Zhan, K. et al. Gut microbiota-bile acid crosstalk in diarrhea-irritable bowel syndrome. BioMed. Res. Int. 2020, 3828249 (2020).
Ma, T. et al. Targeting gut microbiota and metabolism as the major probiotic mechanism—an evidence-based review. Trends Food Sci. Technol. 138, 178–198 (2023).
Cao, Z. et al. The gut virome: a new microbiome component in health and disease. EBioMedicine 81, 104113 (2022).
Duan, Y., Young, R. & Schnabl, B. Bacteriophages and their potential for treatment of gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 19, 135–144 (2022).
Hill, C. et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514 (2014).
Bron, P. A. et al. Can probiotics modulate human disease by impacting intestinal barrier function? Br. J. Nutr. 117, 93–107 (2017).
Gu, Y. et al. Saccharomyces boulardii, a yeast probiotic, inhibits gut motility through upregulating intestinal serotonin transporter and modulating gut microbiota. Pharmacol. Res. 181, 106291 (2022).
Wu, Y. et al. Effect of a multispecies probiotic mixture on the growth and incidence of diarrhea, immune function, and fecal microbiota of pre-weaning dairy calves. Front. Microbiol. 12, 681014 (2021).
Yang, B. et al. Lactobacillus plantarum CCFM1143 alleviates chronic diarrhea via inflammation regulation and gut microbiota modulation: a double-blind, randomized, placebo-controlled study. Front. Immunol. 12, 746585 (2021).
Ki Cha, B. et al. The effect of a multispecies probiotic mixture on the symptoms and fecal microbiota in diarrhea-dominant irritable bowel syndrome: a randomized, double-blind, placebo-controlled trial. J. Clin. Gastroenterol. 46, 220–227 (2012).
Jung, M. et al. A randomized, double-blind, placebo-controlled trial to assess the efficacy and safety of Lactiplantibacillus plantarum CJLP243 in patients with functional diarrhea and high fecal calprotectin levels. Nutrients 14, 389 (2022).
Simrén, M. et al. Clinical trial: the effects of a fermented milk containing three probiotic bacteria in patients with irritable bowel syndrome - a randomized, double-blind, controlled study. Aliment. Pharmacol. Ther. 31, 218–227 (2010).
Hemalatha, R. et al. A community-based randomized double blind controlled trial of Lactobacillus paracasei and Bifidobacterium lactis on reducing risk for diarrhea and fever in preschool children in an urban slum in India. Eur. J. Nutr. Food Saf. 4, 325–341 (2014).
Waller, P. A. et al. Dose-response effect of Bifidobacterium lactis HN019 on whole gut transit time and functional gastrointestinal symptoms in adults. Scand. J. Gastroenterol. 46, 1057–1064 (2011).
Li, C. et al. Screening for lactobacillus plantarum strains that possess organophosphorus pesticide-degrading activity and metabolomic analysis of phorate degradation. Front. Microbiol. 9, 2048 (2018).
Hai, H. J. et al. Screening for Lactobacillus plantarum with potential inhibitory activity against enteric pathogens. Ann. Microbiol. 65, 1257–1265 (2015).
Liu, W. et al. Lactiplantibacillus plantarum P9 improved gut microbial metabolites and alleviated inflammatory response in pesticide exposure cohorts. iScience 25, 104472 (2022).
Ma, T. et al. Effect of the probiotic strain, Lactiplantibacillus plantarum P9, on chronic constipation: a randomized, double-blind, placebo-controlled study. Pharmacol. Res. 191, 106755 (2023).
Schiller, L. R. Evaluation of chronic diarrhea and irritable bowel syndrome with diarrhea in adults in the era of precision medicine. Am. J. Gastroenterol. 113, 660–669 (2018).
Maity, C. & Gupta, A. K. Therapeutic efficacy of probiotic Alkalihalobacillus clausii 088AE in antibiotic-associated diarrhea: A randomized controlled trial. Heliyon 7, e07993 (2021).
McFarland, L. V. & Goh, S. Are probiotics and prebiotics effective in the prevention of travellers’ diarrhea: a systematic review and meta-analysis. Travel Med. Infect. Dis. 27, 11–19 (2019).
Szajewska, H., Kołodziej, M. & Zalewski, B. M. Systematic review with meta-analysis: Saccharomyces boulardii for treating acute gastroenteritis in children-a 2020 update. Aliment. Pharmacol. Ther. 51, 678–688 (2020).
Morozov, A. et al. A systematic review and meta-analysis of placebo effect in clinical trials on chronic prostatitis/chronic pelvic pain syndrome. Prostate 82, 633–656 (2022).
Bai, T. et al. The short-term efficacy of bifidobacterium quadruple viable tablet in patients with diarrhea-predominant irritable bowel syndrome: potentially mediated by metabolism rather than diversity regulation. Am. J. Gastroenterol. 118, 1256–1267 (2023).
Xu, H. et al. Metagenomic analysis revealed beneficial effects of probiotics in improving the composition and function of the gut microbiota in dogs with diarrhoea. Food Funct. 10, 2618–2629 (2019).
Lin, L. et al. Genome-centric investigation of bile acid metabolizing microbiota of dairy cows and associated diet-induced functional implications. ISME J. 17, 172–184 (2023).
Xia, B. et al. Multi-omics unravel the compromised mucosal barrier function linked to aberrant mucin O-glycans in a pig model. Int. J. Biol. Macromol. 207, 952–964 (2022).
Morotomi, M., Nagai, F., Sakon, H. & Tanaka, R. Paraprevotella clara gen. nov., sp. nov. and Paraprevotella xylaniphila sp. nov., members of the family ‘Prevotellaceae’ isolated from human faeces. Int. J. Syst. Evolut. Microbiol. 59, 1895–1900 (2009).
Gu, X. et al. Gut Ruminococcaceae levels at baseline correlate with risk of antibiotic-associated diarrhea. iScience 25, 103644 (2022).
Liu, Z. et al. Bacillus coagulans in combination with chitooligosaccharides regulates gut microbiota and ameliorates the DSS-induced colitis in mice. Microbiol. Spectr. 10, e0064122 (2022).
Geirnaert, A. et al. Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci. Rep. 7, 11450 (2017).
Wullt, M., Johansson Hagslätt, M. L., Odenholt, I. & Berggren, A. Lactobacillus plantarum 299v enhances the concentrations of fecal short-chain fatty acids in patients with recurrent clostridium difficile-associated diarrhea. Dig. Dis. Sci. 52, 2082–2086 (2007).
Ren, S., Wang, C., Chen, A., Lv, W. & Gao, R. The probiotic Lactobacillus paracasei ameliorates diarrhea cause by Escherichia coli O(8) via gut microbiota modulation(1). Front. Nutr. 9, 878808 (2022).
Szajewska, H. et al. Probiotics for the prevention of antibiotic-associated diarrhea in children. J. Pediatr. Gastroenterol. Nutr. 62, 495–506 (2016).
Lyra, A. et al. Diarrhoea-predominant irritable bowel syndrome distinguishable by 16S rRNA gene phylotype quantification. World J. Gastroenterol. 15, 5936–5945 (2009).
Crost, E. H., Coletto, E., Bell, A. & Juge, N. Ruminococcus gnavus: friend or foe for human health. FEMS Microbiol. Rev. 47, fuad014 (2023).
Li, S., Qi, Y., Ren, D., Qu, D. & Sun, Y. The structure features and improving effects of polysaccharide from Astragalus membranaceus on antibiotic-associated diarrhea. Antibiotics 9, 8 (2019).
Pan, H. Y. et al. A galacturonan from Dioscorea opposita Thunb. regulates fecal and impairs IL-1 and IL-6 expression in diarrhea mice. Glycoconj. J. 39, 131–141 (2022).
Auer, I. O., Röder, A., Wensinck, F., van de Merwe, J. P. & Schmidt, H. Selected bacterial antibodies in Crohn’s disease and ulcerative colitis. Scand. J. Gastroenterol. 18, 217–223 (1983).
Chen, H. et al. Alternation of the gut microbiota in irritable bowel syndrome: an integrated analysis based on multicenter amplicon sequencing data. J. Transl. Med. 21, 117 (2023).
Yang, H. et al. Quyushengxin formula causes differences in bacterial and phage composition in ulcerative colitis patients. Evid. Based Complement. Altern. Med. 2020, 5859023 (2020).
Mihindukulasuriya, K. A. et al. Multi-omics analyses show disease, diet, and transcriptome interactions with the virome. Gastroenterology 161, 1194–1207.e1198 (2021).
Zuo, W., Michail, S. & Sun, F. Metagenomic analyses of multiple gut datasets revealed the association of phage signatures in colorectal cancer. Front. Cell. Infect. Microbiol. 12, 918010 (2022).
Min, Y. W., Rezaie, A. & Pimentel, M. Bile acid and gut microbiota in irritable bowel syndrome. J. Neurogastroenterol. Motil. 28, 549–561 (2022).
Camilleri, M. & Vijayvargiya, P. The role of bile acids in chronic diarrhea. Am. J. Gastroenterol. 115, 1596–1603 (2020).
Farrugia, A. & Arasaradnam, R. Bile acid diarrhoea: pathophysiology, diagnosis and management. Frontline Gastroenterol. 12, 500–507 (2021).
Duboc, H. et al. Increase in fecal primary bile acids and dysbiosis in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol. Motil. 24, e246–e517 (2012).
Vijayvargiya, P. & Camilleri, M. Current practice in the diagnosis of bile acid diarrhea. Gastroenterology 156, 1233–1238 (2019).
Mishima, Y. & Ishihara, S. Molecular mechanisms of microbiota-mediated pathology in irritable bowel syndrome. Int. J. Mol. Sci. 21, 8664 (2020).
Li, P. et al. Characterization of kynurenine pathway in patients with diarrhea-predominant irritable bowel syndrome. Eur. J. Histochem. 64, 3132 (2020).
Ahmadi, S. et al. A human-origin probiotic cocktail ameliorates aging-related leaky gut and inflammation via modulating the microbiota/taurine/tight junction axis. JCI Insight 5, e132055 (2020).
Shimizu, M., Zhao, Z., Ishimoto, Y. & Satsu, H. Dietary taurine attenuates dextran sulfate sodium (DSS)-induced experimental colitis in mice. Adv. Exp. Med. Biol. 643, 265–271 (2009).
Fortin, G. L-Carnitine and intestinal inflammation. Vitam. Horm. 86, 353–366 (2011).
Kim, S. Y., Seo, Y. S., Lee, E. S. & Kim, K. N. Total bilirubin is associated with small intestinal bacterial overgrowth in diarrhea predominant irritable bowel syndrome. Ann. Clin. Lab. Sci. 49, 344–352 (2019).
Wan, F. et al. Caffeic acid supplement alleviates colonic inflammation and oxidative stress potentially through improved gut microbiota community in mice. Front. Microbiol. 12, 784211 (2021).
Calzada, F., Correa-Basurto, J., Barbosa, E., Mendez-Luna, D. & Yepez-Mulia, L. Antiprotozoal constituents from Annona cherimola miller, a plant used in Mexican traditional medicine for the treatment of diarrhea and dysentery. Pharmacogn. Mag. 13, 148–152 (2017).
Pujo, J. et al. Bacteria-derived long chain fatty acid exhibits anti-inflammatory properties in colitis. Gut 70, 1088–1097 (2021).
Wardman, J. F., Bains, R. K., Rahfeld, P. & Withers, S. G. Carbohydrate-active enzymes (CAZymes) in the gut microbiome. Nat. Rev. Microbiol. 20, 542–556 (2022).
El Kaoutari, A. et al. Development and validation of a microarray for the investigation of the CAZymes encoded by the human gut microbiome. PloS One 8, e84033 (2013).
Lan, Q. et al. Association of gut microbiota and SCFAs with finishing weight of Diannan small ear pigs. Front. Microbiol. 14, 1117965 (2023).
Tan, J. et al. The role of short-chain fatty acids in health and disease. Adv. Immunol. 121, 91–119 (2014).
Martin-Gallausiaux, C., Marinelli, L., Blottière, H. M., Larraufie, P. & Lapaque, N. SCFA: mechanisms and functional importance in the gut. Proc. Nutr. Soc. 80, 37–49 (2021).
Chen, X. et al. Sodium acetate/sodium butyrate alleviates lipopolysaccharide-induced diarrhea in mice via regulating the gut microbiota, inflammatory cytokines, antioxidant levels, and NLRP3/Caspase-1 signaling. Front. Microbiol. 13, 1036042 (2022).
Gąsiorowska, A. et al. Effects of microencapsulated sodium butyrate, probiotics and short chain fructooligosaccharides in patients with irritable bowel syndrome: a study protocol of a randomized double-blind placebo-controlled trial. J. Clin. Med. 11, 6587 (2022).
Plöger, S. et al. Microbial butyrate and its role for barrier function in the gastrointestinal tract. Ann. N. Y. Acad. Sci. 1258, 52–59 (2012).
Savarino, E. et al. Functional bowel disorders with diarrhoea: clinical guidelines of the United European Gastroenterology and European Society for Neurogastroenterology and Motility. U. Eur. Gastroenterol. J. 10, 556–584 (2022).
Reid, G. et al. Responders and non-responders to probiotic interventions: how can we improve the odds? Gut Microbes 1, 200–204 (2010).
Liu, W. et al. Effect of Lactobacillus plantarum P9 on defecation, quality of life and gut microbiome in individuals with chronic diarrhoea: Protocol for a randomized, double-blind, placebo-controlled clinical trial. Contemp. Clin. Trials Commun. 32, 101085 (2023).
Kalman, D. S. et al. A prospective, randomized, double-blind, placebo-controlled parallel-group dual site trial to evaluate the effects of a Bacillus coagulans-based product on functional intestinal gas symptoms. BMC Gastroenterol. 9, 85 (2009).
Francavilla, R. et al. Clinical and Microbiological Effect of a Multispecies Probiotic Supplementation in Celiac Patients With Persistent IBS-type Symptoms: A Randomized, Double-Blind, Placebo-controlled, Multicenter Trial. J. Clin. Gastroenterol. 53, e117–e125 (2019).
La Monica, M. B., Raub, B., Lopez, H. L. & Ziegenfuss, T. N. A probiotic amylase blend reduces gastrointestinal symptoms in a randomised clinical study. Benef. Microbes 14, 459–476 (2023).
O’Brien, L., Wall, C., Wilkinson, T. J. & Gearry, R. B. Chronic diarrhoea in older adults and the role of dietary interventions. Nutr. Healthy Aging 7, 39–50 (2022).
Mearin, F. et al. Bowel disorders. Gastroenterology 150, 1393–1407 (2016).
Svedlund, J., Sjödin, I. & Dotevall, G. GSRS-a clinical rating scale for gastrointestinal symptoms in patients with irritable bowel syndrome and peptic ulcer disease. Dig. Dis. Sci. 33, 129–134 (1988).
Pilotto, A. et al. The prevalence of diarrhea and its association with drug use in elderly outpatients: a multicenter study. Am. J. Gastroenterol. 103, 2816–2823 (2008).
Wilson, B. et al. Prebiotic galactooligosaccharide supplementation in adults with ulcerative colitis: exploring the impact on peripheral blood gene expression, gut microbiota, and clinical symptoms. Nutrients 13, 3598 (2021).
Lovibond, P. F. & Lovibond, S. H. The structure of negative emotional states: comparison of the Depression Anxiety Stress Scales (DASS) with the Beck Depression and Anxiety Inventories. Behav. Res. Ther. 33, 335–343 (1995).
Skrzydło-Radomańska, B. et al. The effectiveness and safety of multi-strain probiotic preparation in patients with diarrhea-predominant irritable bowel syndrome: a randomized controlled study. Nutrients 13, 756 (2021).
Plasse, T. F. et al. Bimodal release ondansetron improves stool consistency and symptomatology in diarrhea-predominant irritable bowel syndrome: a randomized, double-blind, trial. Am. J. Gastroenterol. 115, 1466–1473 (2020).
Doron, S. & Snydman, D. R. Risk and safety of probiotics. Clin. Infect. Dis. 60, S129–S134 (2015).
Sun, B. et al. Bifidobacterium lactis Probio-M8 adjuvant treatment confers added benefits to patients with coronary artery disease via target modulation of the gut-heart/-brain axes. mSystems 7, e0010022 (2022).
Li, D., Liu, C. M., Luo, R., Sadakane, K. & Lam, T. W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Kang, D. D. et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7, e7359 (2019).
Nissen, J. N. et al. Improved metagenome binning and assembly using deep variational autoencoders. Nat. Biotechnol. 39, 555–560 (2021).
Sieber, C. M. K. et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 3, 836–843 (2018).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).
Olm, M. R., Brown, C. T., Brooks, B. & Banfield, J. F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 11, 2864–2868 (2017).
Skonieczna-Żydecka, K. et al. Gut biofactory-neurocompetent metabolites within the gastrointestinal tract. a scoping review. Nutrients 12, 3369 (2020).
James, S. C., Fraser, K., Young, W., McNabb, W. C. & Roy, N. C. Gut microbial metabolites and biochemical pathways involved in irritable bowel syndrome: effects of diet and nutrition on the microbiome. J. Nutr. 150, 1012–1021 (2020).
Liu, A. et al. Adjunctive probiotics alleviates asthmatic symptoms via modulating the gut microbiome and serum metabolome. Microbiol. Spectr. 9, e0085921 (2021).
Zhang, H. et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic acids Res. 46, W95–w101 (2018).
Kieft, K., Zhou, Z. & Anantharaman, K. VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome 8, 90 (2020).
Nayfach, S. et al. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol. 39, 578–585 (2021).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012).
Nayfach, S. et al. Metagenomic compendium of 189,680 DNA viruses from the human gut microbiome. Nat. Microbiol. 6, 960–970 (2021).
Su, S. H. et al. Fecal microbiota transplantation and short-chain fatty acids protected against cognitive dysfunction in a rat model of chronic cerebral hypoperfusion. CNS Neurosci. Ther. 29, 98–114 (2023).
Xu, J. et al. Protective effects of taurocholic acid on excessive hepatic lipid accumulation via regulation of bile acid metabolism in grouper. Food Funct. 13, 3050–3062 (2022).
Yang N., et al. Lactiplantibacillus plantarum P9 for chronic diarrhea in young adults: a large double-blind, randomized, placebo-controlled trial. https://doi.org/10.5281/zenodo.11194887 (2024).
Acknowledgements
This study was supported by the Research Fund for the National Key R&D Program of China (2022YFD2100700 [H.Z.]), the National Natural Science Foundation of China (U22A20540 [H.Z.]), and the earmarked fund for CARS36. The funding source had no role in study design, data collection, and analysis, decision to submit, or preparation of the manuscript. The authors would like to express their gratitude to Inner Mongolia Tongfang Discovery Tech. Co., Ltd. for providing storage space and computing resources, Suzhou PANOMIX Biomedical Tech. Co., Ltd. for assisting with metabolite detection, and Novogene Co., Ltd. for their help with metagenomic sequencing.
Author information
Authors and Affiliations
Contributions
N.L., W.L., H.Z. conceived and designed the trial. Y.X., Yingmeng.L. collected the clinical data. N.Y., T.M., Q.L., Yalin.L, Z.S. analyzed and interpreted the data. L.Z., Q.X., K.Z. performed the screening of enrolled patients. N.Y., T.M. wrote the manuscript, with contributions from all authors. L.Y.K. critically revised the manuscript and provided advice.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This was a comprehensive randomized, double-blind, placebo-controlled study conducted in collaboration with the First Affiliated Hospital of Nanchang University from October 2020 to May 2021. The study was registered at the Chinese Clinical Trial Registry under the registration number ChiCTR2000038410 (https://www.chictr.org.cn/showproj.html?proj=56542) and approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University (No. IIT [2020] Clinical Ethics Review No. 002).
Informed consent
Informed consent was required from all subjects prior to their participation.
Peer review
Peer review information
Nature Communications thanks Ashwin Ananthakrishnan, Elizabeth Ryan, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, N., Ma, T., Xie, Y. et al. Lactiplantibacillus plantarum P9 for chronic diarrhea in young adults: a large double-blind, randomized, placebo-controlled trial. Nat Commun 15, 6823 (2024). https://doi.org/10.1038/s41467-024-51094-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51094-2
- Springer Nature Limited