Abstract
Population genetic analyses can provide useful data on species’ regional connectivity and diversity which can inform conservation and restoration efforts. In this study, we quantified the genetic connectivity and diversity of Stephanocoenia intersepta corals from shallow (<30 m) to mesophotic (30–45 m) depths across Florida Keys National Marine Sanctuary. We generated single nucleotide polymorphism (SNP) markers to identify genetic structuring of shallow and mesophotic S. intersepta corals. We uncovered four distinct, cryptic genetic lineages with varying levels of depth-specificity. Shallow-specific lineages exhibited lower heterozygosity and higher inbreeding relative to depth-generalist lineages found across both shallow and mesophotic reefs. Estimation of recent genetic migration rates demonstrated that mesophotic sites are more prolific sources than shallow sites, particularly in the Lower Keys and Upper Keys. Additionally, we compared endosymbiotic Symbiodiniaceae among sampled S. intersepta using the ITS2 region and SymPortal analysis framework, identifying symbionts from the genera Symbiodinium, Breviolum, and Cladocopium. Symbiodiniaceae varied significantly across depth and location and exhibited significant, but weak correlation with host lineage and genotype. Together, these data demonstrate that despite population genetic structuring across depth, some mesophotic populations may provide refuge for shallow populations moving forward and remain important contributors to the overall genetic diversity of this species throughout the region. This study highlights the importance of including mesophotic as well as shallow corals in population genetic assessments and informs future science-based management, conservation, and restoration efforts within Florida Keys National Marine Sanctuary.
Similar content being viewed by others
Introduction
Mesophotic coral ecosystems (MCEs) are relatively light-limited coral reefs that typically occur between 30–150 m, characterized by the presence of light-dependent biota including many scleractinian coral species (Lesser et al. 2009). Mesophotic coral ecosystems, like shallow coral reefs, are abundant throughout the tropics and subtropics (Pyle and Copus 2019). Community composition among MCEs is distinct from shallow coral reefs; yet many species, including some corals, are depth-generalists with broad distribution spanning across both shallow and upper mesophotic depth zones (30–60 m; Bongaerts et al. 2010). In the Caribbean Sea, there is as much as 25–40% scleractinian coral species overlap between shallow reefs and upper mesophotic reefs (Bongaerts et al. 2010).
Coral reefs around the world are facing devastating declines in coral cover from an abundance of stressors including, but not limited to, overfishing, terrestrial runoff and pollution, coral disease outbreaks, and global climate change (Hughes 1994; Gardner et al. 2003; Mumby et al. 2006; De’ath et al. 2012; França et al. 2020; Pinheiro et al. 2023). Mesophotic coral ecosystems have the potential to be buffered from anthropogenic stressors due to their depth and/or relative isolation from shore in many regions (Glynn 1996; Bongaerts et al. 2010). The deep reef refuges hypothesis (DRRH) posits that MCEs may function as refuges for depth-generalist coral species, providing viable larvae to degraded shallow reefs after isolated episodic disturbance events, thereby aiding in the recovery of shallow reefs (Glynn 1996; Bongaerts et al. 2010).
Understanding the degree of connectivity and similarity of coral populations and their endosymbiotic dinoflagellate microalgae (Family Symbiodiniaceae) across shallow and mesophotic depth zones is useful for developing effective ecosystem management or restoration practices (Palumbi 2003; Bongaerts et al. 2010). Population genetic approaches and internal transcribed spacer 2 (ITS2) metagenomics have often been used to estimate these levels of similarity and connectedness between coral populations and their in hospite Symbiodiniaceae on shallow reefs and MCEs (Bongaerts et al. 2017; Sturm et al. 2022). The potential reseeding ability of MCEs varies across both region and coral species (Bongaerts et al. 2017; Studivan and Voss 2018). In this aspect, the DRRH, which is already limited to depth-generalist corals species, is further limited in its universal applicability. Although MCEs are not completely isolated from disturbance events common to shallow reefs and the degree of connectivity between mesophotic and shallow populations varies widely, the DRRH remains an important framework for understanding where MCEs have the potential to function as refuges within local and regional coral metapopulations (Smith et al. 2016; Rocha et al. 2018).
Many studies of coral population genetics have uncovered morphologically similar yet genetic distinct lineages (i.e., cryptic lineages), which often segregate along environmental gradients but can occur sympatrically (Burgess et al. 2021; Grupstra et al. 2024). Cryptic diversity may drive functional differences including potential physiological and ecological significance (Burgess et al. 2021). Previous work on shallow and mesophotic conspecific corals has found population structure varies widely, not only across species but across region as well (Bongaerts et al. 2017; Sturm et al. 2023). Two coral species in Bermuda exhibit differing levels of population structure across depth, between 12 m and 40 m, likely influenced by their reproductive modes. Across the same sites, Agaricia fragilis, a brooding species believed to lack long-distance dispersal abilities, demonstrated structure by depth while Stephanocoenia intersepta, a broadcast spawning species with greater potential for dispersal, was panmictic among depths in Bermuda (Bongaerts et al. 2017). Whereas M. cavernosa are structured by depth in parts of Florida and Belize, this species is completely panmictic across depths in the northwestern Gulf of Mexico (Sturm et al. 2023). Depth has also been a segregating factor in divergent cryptic coral lineages and their endosymbionts (Bongaerts et al. 2015; Rippe et al. 2021; Grupstra et al. 2024). Symbiodiniaceae often exhibit depth zonation, especially in lower MCE habitats (≥ 60 m) and may associate with specific lineages in some cases (Lesser et al. 2010; Bongaerts et al. 2015; Rose et al. 2021; Johnston et al. 2022).
Since the 1970s, coral reefs in Florida Keys National Marine Sanctuary (FKNMS) have experienced catastrophic declines in coral cover, with as much as a 50% reduction between 1998 and 2011 alone (Ruzicka et al. 2013; Toth et al. 2014). Coral reefs within FKNMS have been heavily studied, but research in the mesophotic zone is historically limited in this region, consistent with the relative proportion of MCE to shallow coral reef research worldwide (Jaap et al. 2008; Turner et al. 2017). Previous studies on MCEs within Florida have been largely focused on Pulley Ridge, a deep ridge to the southwest of FKNMS that supports the deepest known scleractinian coral reef in the United States (59–105 m; Reed et al. 2019). Studies not specific to Pulley Ridge have often focused on the genetic connectivity of MCE benthic invertebrate populations (Bernard et al. 2018; Studivan and Voss 2018). Notably, the majority of vertical connectivity studies within MCEs at FKNMS have focused on a single coral species, Montastraea cavernosa, and have demonstrated that levels of vertical connectivity are highly variable across the Florida Keys region (Serrano et al. 2014; Sturm et al. 2021).
Stephanocoenia intersepta, like M. cavernosa, is a depth-generalist scleractinian coral species found throughout the Caribbean Sea and the Gulf of Mexico (Lang 2003). Stephanocoenia intersepta is typically not as numerically dominant on shallow coral reefs as M. cavernosa, but as depth increases S. intersepta increases in abundance and cover on many tropical western Atlantic MCEs (Buckel et al. 2014; Goodbody-Gringley et al. 2019). Despite the increased prevalence of S. intersepta on MCEs, there has only been one population genetic analysis published to date for S. intersepta (Bongaerts et al. 2017). In Bermuda, S. intersepta was found to have a lack of population structure and to host similar Symbiodiniaceae across deep and shallow reefs, likely attributed to its reproductive mode (Bongaerts et al. 2017). Stephanocoenia intersepta implements a gonochoric broadcast spawning reproductive strategy, in which male and female colonies release gametes, and eggs are fertilized externally allowing the potential for dispersal through ocean currents. As in many broadcast spawning species, S. intersepta has horizontal transmission of Symbiodiniaceae, whereby larvae or newly settled recruits uptake algal symbionts from the surrounding environment (Bongaerts et al. 2015). However, unlike many other broadcasting species, S. intersepta females have been observed to hold multiple eggs in their tentacles rather than their oral disc, often only releasing eggs after nearby (<2 m) males have released sperm (Hagman, Gittings, and Vize 1998; Vermeij et al. 2010). Given this reproductive mode, and S. intersepta’s relative abundance, particularly at mesophotic depths, S. intersepta is a useful, yet understudied candidate species to investigate connectivity between shallow reefs and MCEs.
To better understand the genetic differences among shallow and mesophotic coral as well as the genetic structuring of corals across depth we sampled S. intersepta colonies from paired shallow/mesophotic reefs throughout FKNMS. We investigated patterns of host genetic diversity, differentiation, and connectivity as well as in hospite Symbiodiniaceae through single nucleotide polymorphism (SNP) genotyping and ITS2 sequencing analyses. Using similar approaches and sampling designs across the same sites as previous population genetics work in FKNMS (Sturm et al. 2021) we investigated MCE refuge potential in this region and uncovered cryptic coral diversity which should be considered in future management decisions for FKNMS.
Materials and methods
Sample collection and DNA extraction
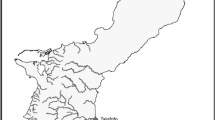
Stephanocoenia intersepta colonies were sampled from shallow (<30 m) and upper mesophotic (30–45 m) depths by SCUBA and technical divers, respectively, from four sites across FKNMS (Supplementary Table S1; Fig. 1). Samples were collected opportunistically within ~10 m of target GPS waypoints where divers were deployed (Fig. 1). Small fragments (~5 cm2) of coral skeleton and tissue were removed from colonies with a hammer and clean masonry chisel. Sampling depths were recorded and scaled photographs were taken for each sampled colony. At the surface, samples were placed into absolute molecular-grade ethanol and frozen at −20 °C while at sea. Samples were transported on dry ice to FAU Harbor Branch where the ethanol was replaced with fresh molecular-grade ethanol and samples were stored at −80 °C prior to genomic DNA extraction. Small tissue subsamples (~1 cm2) were transferred into 500 µL of TRI reagent (Invitrogen) and allowed to soak overnight at 4 °C. Genomic DNA was extracted using a chaotropic buffer extraction optimized for increased DNA yield and quality from marine benthic invertebrates (Sturm et al. 2021). DNA extracts were purified with the Zymo DCC-5 kit and DNA quality/quantity were measured with NanoDrop (Thermo Fisher) and Qubit 4.0 fluorometer (Thermo Fisher). DNA extracts were normalized to 25 ng µL−1 and used for both coral host genotyping and algal symbiont molecular characterization.
Color denotes sampling site and shape denotes depth zone (shallow, mesophotic). Donut plots in insets show site lineage assignment proportion on outer ring and average sampling depth inside ring (sampling depth ranges in Supplementary Table S1), with the top and bottom plot of each pair representing the shallow and mesophotic sites, respectively. FKNMS and Sanctuary Preservation Areas boundaries are shown as black and purple polygons, respectively. Bathymetric contours are shown as gray lines in insets.
Stephanocoenia intersepta SNP genotyping
SNP genotyping libraries were prepared with the endonuclease BcgI (New England BioLabs, Inc.) and 100 ng of genomic DNA per sample, using an updated protocol for the 2bRAD-seq method (Wang et al. 2012). Wet lab protocols were adapted from the current 2bRAD GitHub repository (Matz 2020; Sturm et al. 2021) and detailed lab protocols are found in this manuscript’s accompanying GitHub repository (Eckert 2024). Pooled 2bRAD libraries were size selected and sequenced at The University of Texas Genome Sequencing and Analysis Facility on Illumina NovaSeq (SR100 S1) with 20% PhiX spike-in.
Returned sequencing reads were demultiplexed, deduplicated, filtered and trimmed with custom perl scripts and cutadapt v3.4 (Martin 2011). As there was no S. intersepta genome available we employed a de novo reference analysis pipeline (Matz 2020; Rippe et al. 2021). Reads from each sample were first aligned to a concatenated Symbiodiniaceae reference consisting of representative genomes for four genera (Symbiodinium, Breviolum, Cladocopium, and Durusdinium) using Bowtie2 v1.2.2 (Langmead and Salzberg 2012; Shoguchi et al. 2013, 2021; Aranda et al. 2016; Liu et al. 2018). After excluding the Symbiodiniaceae aligned reads, resulting high-quality reads were used to create a de novo RAD tag reference assembly for S. intersepta consisting of clustered (91% similarity) unique sequences found in at least 10% of all samples using custom perl scripts and CD-HIT v4.8.1 (Fu et al. 2012). Potential contamination sequences were removed from the clustered sequences with Kraken2 (standard Kraken database with added Symbiodiniaceae genomes) and remaining RAD tags were concatenated into a de novo reference and formatted as 30 equally-sized pseudo-chromosomes to be used for mapping (Wood et al. 2019; Rippe et al. 2021). Sample reads were aligned to the de novo reference with Bowtie2 (Langmead and Salzberg 2012). The program angsd v0.933 was used to determine SNP loci from sequencing reads, generate genotype likelihoods, and create an identity-by-state (IBS) matrix (Korneliussen et al. 2014). SNP loci underwent stringent filtering, requiring a minimum mapping quality of 20, minimum base quality score of 30, minimum allele frequency of 0.05, minimum p-value of 10−5 for deviation from Hardy-Weinberg equilibrium, minimum p-value of 10−5 for strand bias, p-value of 10−5 for heterozygosity bias, remove triallelic SNPs, p-value that a locus is variable of 10−6, and had to be present in ≥ 75% of samples. No clonal multi-locus genotypes were identified through hierarchical clustering using an IBS threshold determined by the technical triplicate samples (Supplementary Fig. S1; Manzello et al. 2019). Two of each of the three technical replicates were removed and angsd was run on the clone-free sample set with the previously described filters.
Stephanocoenia intersepta population genetic structure
Principal component analysis based on IBS distances was conducted using pcangsd (Meisner and Albrechtsen 2018). ngsadmix was used to calculate population structure models for cluster values from K = 1–11 (sample populations +3; Skotte et al. 2013). In addition to calculating clusters with pcangsd, the most likely value of K was evaluated using the Evanno method through Clumpak (Evanno et al. 2005; Kopelman et al. 2015). We tested for isolation by distance using a Mantel Test (9999 permutations; Dray and Dufour 2007). Because we discovered distinct lineages in sympatry, we used individual genetic distances from the IBS matrix from angsd, which has been shown to be an effective method for calculating genetic distance in instances of variable and low coverage sequencing samples (Korneliussen et al. 2014; Manzello et al. 2019).
Stephanocoenia intersepta lineage demographics
To avoid ascertainment bias from the presence of four putative genetic lineages within the sample set, angsd was run within each lineage using the above filters.
Only samples with ≥75% assignment to a single lineage were retained for these analyses, excluding any individuals with recent hybridization between lineages to avoid confounding lineage specific analyses (Fifer et al. 2022). angsd was used to calculate SNPs within each lineage using the above filters with the following modifications: minimum base quality score of 25, p-value that a locus is variable of 10−5, and present in ≥75% of samples. SNPs were refined to only sites found across all lineages with the -sites flag in angsd (4027 SNPs retained). Inbreeding coefficients were calculated for all samples with the program ngsrelate (Hanghøj et al. 2019).
For heterozygosity and site frequency spectra (SFS) calculations angsd was rerun using only filters which did not affect allelic frequencies, retaining all loci, variant and invariant (-uniqueOnly 1 -remove_bads 1 -skipTriallelic 1 -minMapQ 30 -minQ 35 -doHWE 1 -sb_pval 1e-5 -hetbias_pval 1e-5 -maxHetFreq 0.5 -setMinDepthInd 3; Rippe et al. 2021). Heterozygosities were calculated for each sample using custom R scripts (Matz 2020). Due to heterogeneity of variance within the data and uneven sample sizes, Welch’s ANOVA was used to test for differences between heterozygosity, mean inbreeding coefficients, and depth distribution within each lineage. Pairwise comparisons for significant ANOVA results were implemented with non-parametric Games-Howell tests, which are effective as a post-hoc test when variance is heterogeneous and sample sizes are uneven, which is often the case when coral population genetics work unveils cryptic lineages (Games and Howell 1976; Midway et al. 2020; Grupstra et al. 2024). Site frequency spectra (SFS) were calculated for each lineage with realSFS and were used to calculate nucleotide diversity (π) and pairwise weighted FST among lineages. Nucleotide diversity was averaged across the 30 pseudochromosomes used as a de novo reference. We tested the relationship between mean nucleotide diversity and mean depth of each lineage with linear regression. Effective population size was calculated using the formula \(N_{e}=\frac{\pi}{4\mu}\), where the mutation rate, µ, was estimated to be 2 × 10−8 per base per generation (Matz et al. 2018; Rippe et al. 2021).
To understand the relative differences in lineage diversity throughout time, population sizes of each lineage were reconstructed using stairwayplot v2 on folded SFS (Liu and Fu 2020). Prior to running stairwayplot, FST outliers were detected with bayescan v2.1 (Foll and Gaggiotti 2008). To ensure only neutral loci were used for analysis we used conservative filtering, removing all loci with q-value < 0.5 prior to SFS calculations (101 loci removed; Rippe et al. 2021). stairwayplot runs were run using suggested default parameter estimates and an estimated mutation rate (2 × 10−8 per base per year; Matz et al. 2018; Rippe et al. 2021) with an assumed generation time of 15 years, based on previous estimates and growth rates of massive hermatypic corals (Szmant 1991; Soong 1993). Because these parameters are largely unknown for many coral species including S. intersepta the absolute values derived from these calculations must considered with caution, however the relative values may be taken as accurate representations of putative patterns in the data (Rippe et al. 2021).
Population genetic connectivity
Genetic connectivity between sampled sites was measured through recent migration rate estimates (immigrant individuals from the previous 2–3 generations) with the BA3SNP function of the program bayesass v3.0.4.2 (Wilson and Rannala 2003). bayesass was run only on the most abundant, Blue lineage. Ten independent runs were conducted with random start seeds for 30 million Markov-Chain Monte Carlo (MCMC) model simulations with a 10 million burn-in and 1000 run sampling frequency. Mixing parameters (migration rate [m] = 0.25, allele frequencies [a] = 0.85, inbreeding coefficient [f] = 0.05) were adjusted to allow adequate mixing within model runs (acceptance rates = 0.2–0.6; Wilson and Rannala 2003). Trace files from independent runs were visualized with tracer v1.7 to ensure model convergence and consistency between independent model runs (Rambaut et al. 2018). Bayesian deviance was calculated for each independent run and the run with the lowest deviance was used for further analysis (Faubet et al. 2007). Migration rate estimates (m) were calculated as the mean of the posterior distribution and their uncertainty as 95% high posterior density (HPD) intervals.
Symbiodiniaceae ITS2 sequencing
Extracted and purified genomic DNA was used to generate ITS2 amplicon libraries to identify in hospite putative Symbiodiniaceae taxa and to determine if algal symbionts differed with respect to host genetic lineage, location, and depth. Symbiodiniaceae ITS2 sequences were amplified using Symbiodiniaceae-specific primers with Illumina MiSeq adapter sequences at the 5′ ends (SYM_VAR_5.8SII/SYM_VAR_REV; Hume et al. 2018). PCR products were cleaned using the Zymo DCC-5 kit and normalized to 10 ng µL−1. Unique double identifying indices were incorporated into each sample with an additional PCR using 50 ng PCR product and 7 cycles of PCR. Detailed lab protocols are found in this manuscript’s accompanying GitHub repository (Eckert 2024). Libraries were pooled equimolarly and sequenced on Illumina MiSeq V2 (500 cycle) with 20% PhiX spike-in.
Symbiodiniaceae SymPortal ITS2 analyses
Demultiplexed Symbiodiniaceae ITS2 .fastq files generated from the sequencing facility were run remotely through the SymPortal analytical framework (Hume et al. 2019). Sequences occurring in a sufficient number of samples within both the S. intersepta dataset and the entire database of samples run through SymPortal were identified as “defining intragenomic variants” (DIVs) which were used to characterize ITS2 type profiles, representative of putative Symbiodiniaceae taxa (Hume et al. 2019).
The betadisper function in the package vegan was used to calculate multivariate homogeneity of dispersion (PERMDISP) using Bray–Curtis distances (Oksanen et al. 2020). Pairwise comparisons for significant factors were calculated with permutation tests using the permutest function in vegan (9999 permutations). Permutational multivariate analysis of variance (PERMANOVA) was implemented with the adonis2 function in vegan to test for differences in Symbiodiniaceae ITS2 type profiles with depth, sampling site, and host lineage assignment used as fixed factors from Bray–Curtis distances (9999 permutations). Pairwise PERMANOVA tests were conducted on significant factors with the package pairwiseAdonis using false discovery rate corrected p-values (Martinez Arbizu 2017). Procrustes analysis was used to test for correlation between algal symbionts (ITS2 type profile) and host genotype (IBS) as well as host genetic lineage assignment (admixture analysis).
Additionally, we validated the accuracy of using 2bRAD Symbiodiniaceae sequence alignments as a proxy for in hospite Symbiodiniaceae genera, as proposed in Manzello et al. (2019). 2bRAD sequences which mapped to each of the four Symbiodiniaceae genomes were counted for the genus they mapped to for each sample and SymPortal ITS2 type profile counts were separated by the genera they represented. Both 2bRAD and ITS2 counts were normalized and Bray–Curtis distances were calculated in vegan. The protest function in vegan was used to conduct a symmetric Procrustes analysis (9999 permutations) to test for similarities between principal coordinates analyses of each dataset.
Results
Stephanocoenia intersepta SNP data processing
2bRAD sequencing returned a total of ~796.1 million reads, an average of ~3.52 million reads per sample. After deduplicating, filtering, and trimming a total of ~443.8 million reads remained, with an average of ~2.01 million reads per sample (excluding technical replicates). On average, 2.16% of 2bRAD reads mapped to the Symbiodiniaceae metagenome, with a total of 9.79 million reads removed, or an average of 44,510 reads per sample. After removing technical replicates, a total of 220 samples remained for genotyping (i.e., no natural clones were detected). Samples had an average of 90.54% mapping efficiency to the de novo S. intersepta reference, resulting in a suite of 22,837 high-quality SNPs after filtering and processing with angsd. Filtering SNPs to those common among independent angsd runs within lineage resulted in a total of 4027 sites.
Stephanocoenia intersepta population genetic structure
Clumpak and pcangsd both determined K = 4 to be the most likely number of genetic clusters (i.e., lineages). Most samples had little admixture (<10%), with a large majority of assignment to a single genetic lineage (Fig. 2D). Three colonies were considered to be admixed (<75% assignment to any single lineage) and removed from lineage-specific analyses (Tortugas Bank Mesophotic, Upper Keys Shallow, and Upper Keys Mesophotic; Fig. 2D). There were no overt morphological differences among lineages that could be identified in situ. At all four regions there was evidence of structure across depth zones (Fig. 2). All four lineages were present in shallow sites, although the Yellow lineage was not found in the shallow Upper Keys population, except for within one admixed individual (Fig. 2D). The Blue and Teal lineages appear to be depth-generalists and were the dominant lineages within mesophotic sites comprising 81 and 15% of all mesophotic samples, respectively. The Green and Yellow lineages were not observed in mesophotic sites, except for two individuals at Riley’s Hump and within some admixed individuals in Tortugas Bank and Lower Keys populations and accounted for 24.17 and 12.5% of all shallow individuals, respectively (Fig. 2D). A mantel test indicated a lack of isolation by distance (r = −0.01, p = 0.721).
A Principal component analysis. Color and shape denote sampling site and depth zone, respectively. Larger, opaque shapes represent sample population centroids and smaller, transparent shapes are individuals. B Same plot as A but with samples colored by putative lineage assignment. Samples with <75% assignment to a single lineage were considered admixed. C Same plot as B but showing axis 2 and 3. D Structure plot from ngsadmix analysis. Each bar represents an individual sample and the proportion of each color is the probability of membership to that genetic lineage. White asterisks denote admixed individuals. E Proportion of normalized ITS2 type profiles calculated from SymPortal analysis. Each vertical bar represents one S. intersepta colony and color corresponds to ITS2 type profiles. DIVs in a type profile name are listed in order of abundance with ‘/’ separating DIVs with co-dominance in samples. Bars below each facet indicate the lineage assignment of the coral host.
Stephanocoenia intersepta lineage diversity and demography
There was a significant depth distribution of S. intersepta lineages, with the Blue lineage being most abundant and found predominantly in mesophotic sites (Welch’s ANOVA; F(3, 67.5) = 49.01, p < 0.001; Fig. 3A). Two shallow specific S. intersepta lineages (i.e., Green and Yellow) exhibited lower heterozygosity than the deeper, depth-generalist blue lineage across all loci (variant and invariant; Welch’s ANOVA; F(3, 60.9) = 85.15, p < 0.001; Fig. 3B). Shallow lineages also had higher inbreeding coefficients (Welch’s ANOVA; F(3, 49.9) = 232.04, p < 0.001) in comparison to mesophotic lineages (Fig. 3C). Nucleotide diversity (π) ranged from 4.84 × 10−3 to 5.83 × 10−3 (Table 1), and there was a positive correlation between nucleotide diversity and depth (R2 = 0.69). Reconstruction of effective population size throughout time revealed stark differences between lineages. The Blue lineage underwent a drastic expansion ~100 kyr before present and was by far the largest population in the region throughout the recent past. Most lineages also experienced slight expansion followed by contraction in the last ~50–25 kyr (Fig. 3D). FST among lineage ranged from 0.048 to 0.211. Most lineages were highly differentiated from one another, with the exception of the depth-generalist lineages (Blue, Teal; FST = 0.048) which included 96% of all mesophotic samples (Fig. 3E).
A Distribution of lineages across depth. B Lineage heterozygosity across all RAD loci (variant and invariant sites). C Inbreeding estimates from SNPs by genetic lineage. Significant Welch’s ANOVA results between population depth are listed and letters denote significant differences among lineages on A–C. D Lineage effective population size through time. X-axis is thousands of years ago (KYA). Bounding ribbons are 75% confidence intervals. E Pairwise fixation index (FST) heatmap among lineages. Darker coloration indicates higher FST. Colors in A–E depict lineage assignments.
Genetic connectivity across FKNMS
Analysis of recent migration with bayesass was low overall (mean ± SEM = 4.10 ± 0.76%) but indicated that mesophotic populations were greater sources than shallow populations (mean ± SEM = 6.70 ± 1.37%; 1.51 ± 0.08%, respectively; Supplementary Table S2). Additionally, mesophotic to shallow subsidy was greater than shallow to mesophotic (mean ± SEM = 6.46 ± 1.22% ; 1.38 ± 0.12%, respectively). Many shallow sites provided little substantial migration (i.e., HPD range ≯ 0) to other sites (e.g., Shallow Riley’s Hump and Upper Keys; Fig. 4, Supplementary Fig. S2). The largest sources were mesophotic sites in Upper Keys (2.6–12.2% of recent migrants into sink populations) and mesophotic sites in Lower Keys (8.8–26.4% of recent migrants into sink populations; Supplementary Table S2, Fig. 4, Supplementary Fig. S2). It is important to note that there are many unsampled sites throughout this range in our design. Parentage analyses and assignment tests have been found to suffer from lack of inclusion of all source populations, as migrants from an unsampled site get incorrectly assigned to a source population (Christie et al. 2017). BayesAss assigns migration rates using a Bayesian approach, accounts for multiple generations of geneflow, and our overall sampling range is relatively small (Wilson and Rannala 2003; Meirmans 2014; Christie et al. 2017). Therefore, these estimates should provide a baseline for understanding the relative connectivity between these habitats.
In hospite Symbiodiniaceae
ITS2 sequencing was successful for 219 samples, returning a total of ~7.38 million reads, or an average of 33,723 reads per sample. Following SymPortal quality filtering, a total of ~3.28 million sequences were retained and used in ITS2 type profile calculation. SymPortal analysis calculated 19 unique ITS2 types. 78.45% of type profile sequences were assigned to the genus Cladocopium, 21.33% to Symbiodinium, 0.22% to Breviolum, and none to Durusdinium (Fig. 2E). In hospite Symbiodiniaceae beta diversity was greater in shallow populations (PERMDISP; F1,217 = 10.815, p = 0.0013) but did not vary across sampling site or lineage (PERMDISP; F3,215 = 0.772, p = 0.516; F3,212 = 2.021, p = 0.112, respectively). Symbiodiniaceae were significantly different across lineage, depth, and site with significant interactions except for the interaction of lineage and depth (PERMANOVA; depth: F1,192 = 7.8398, p < 0.0001; site: F3,192 = 9.1605, p < 0.0001; lineage: F3,192 = 10.8777, p < 0.0001; Supplementary Table S3). All pairwise comparisons of lineage and of site were significant (pairwise PERMANOVA; all p < 0.05) and only three site by depth zone comparisons were not significantly different (Riley’s Hump Shallow:Riley’s Hump Mesophotic, Riley’s Hump Mesophotic:Upper Keys Mesophotic, Tortugas Bank Mesophotic:Lower Keys Mesophotic; Supplementary Table S3; Fig. 2E). The majority of Symbiodinium type profiles were found within shallow S. intersepta populations, namely Tortugas Bank and Lower Keys (Fig. 2E). There was a strong correlation between Symbiodiniaceae genera calculated using 2bRAD reads and SymPortal results grouped to genera (t0 = 0.9175, p = 0.0001; Fig. 5). Procrustes analyses indicated a correlation between S. intersepta host genotype and in hospite Symbiodiniaceae (t0 = 0.2283, p = 0.0001) as well as between S. intersepta genetic lineage and in hospite Symbiodiniaceae (t0 = 0.2531, p = 0.0001), though neither relationship was very strong.
A Proportion of Symbiodiniaceae derived from ITS2 DIVs from SymPortal analysis. Each vertical bar represents one S. intersepta colony and color corresponds to Symbiodiniaceae genera, with gray spaces indicating lack of sample. B Same as A but with genera derived from 2bRAD loci mapped to concatenated Symbiodiniaceae metagenomic reference. Colored bars below each facet denote the lineage assignment of the coral host.
Discussion
Four cryptic S. intersepta lineages throughout FKNMS
This study uncovered four putative cryptic lineages of S. intersepta throughout the Florida Keys. Shallow-specific lineages (<30 m) exhibited lower genetic diversity than depth-generalist lineages, which were more abundant on mesophotic reefs (Fig. 3). Our findings, along with the depth specificity of certain lineages, is very similar to patterns previously observed for M. cavernosa and Siderastrea siderea in the Florida Keys (Rippe et al. 2021). The abundance and diversity of the S. intersepta depth-generalist Blue lineage, which dominated the sampled mesophotic sites has persisted throughout the recent past, with a large population expansion circa 100 kyr before present and contraction shortly thereafter. This pattern is also similar to M. cavernosa and S. siderea in the region, where multiple lineages undergo similar population flux. While in S. intersepta, M. cavernosa, and S. siderea population changes appear to occur during glacial cycles, the estimation of mutation rate and generation time make the absolute values of Ne and time uncertain (Rippe et al. 2021).
The presence of these distinct cryptic lineages and the lack of much admixture between lineages despite the sympatric distribution across FKNMS is likely indicative of reproductive isolation between lineages. As these lineages have diverged through time it is possible that some pre- or postzygotic barriers have arisen and are responsible for the lack of admixture we observe. For corals, spawning timing is crucial to ensure fertilization success. Differences in spawning timing may act as a pre-zygotic barrier, thereby avoiding hybridization through the prevention of gametic mixing between two sister species or cryptic lineages (Levitan et al. 2004). Such differences in timing may have evolutionary benefits if hybrid individuals are unviable between different lineages (Grupstra et al. 2024). The lowest pairwise FST value between lineages was the Blue to Teal comparison. Therefore, it is likely that there is more recent divergence or relatively recent introgression between these two depth-generalist lineages and therefore admixture in proceeding generations may be possible (i.e., a semi-permeable reproductive barrier). It is possible that hybrids could still be viable and propagated through assisted gene flow for restoration. In Acropora cerviconrnis, hybridization was possible and viable between cryopreserved gametes from Florida and Curaçao (Hagedorn et al. 2021). It may also be the case that interbreeding between lineages is occurring and offspring are viable but they do not successfully recruit due to changing ocean currents or a lack of suitable recruitment substrate or settlement cues (Kuffner et al. 2006; Ritson-Williams et al. 2009; Wakwella et al. 2020). If admixed individuals are mainly emigrating from FKNMS we may expect to see more admixed individuals in other unsampled downstream populations, but as we see few recently admixed individuals across the range of this study, it seems unlikely to be the case. It is also possible that there are genomic underpinnings of depth partitioning among S. intersepta lineages, leaving Yellow and Green lineages unable to successfully settle/survive at greater depths due to some phenotype-environment mismatch (Marshall et al. 2010). Future studies and ex situ experimentation on reproduction/recruitment would provide valuable insight on what pre- or postzygotic barriers are driving this apparent reproductive isolation in FKNMS S. intersepta and if inter-lineage crosses produce viable offspring.
The observed lack of diversity within shallow lineages is expectedly correlated with a greater prevalence of more highly inbred individuals. While clonality of coral populations is typically assessed in population genetic analyses, many studies have not explicitly examined inbreeding within populations. Inbreeding may have fitness and evolutionary consequences and has often been considered rare in marine environments, due to the perceived lack of barriers and a large potential for larval dispersal (Knowlton and Jackson 1993; Olsen et al. 2020). However, inbreeding may be more common in benthic marine invertebrates than previously thought, with many species showing inbreeding frequencies as high as that of terrestrial plants (Olsen et al. 2020). The high levels of inbreeding detected in shallow lineages may be driven by individuals from discrete founder events (i.e., “sweepstakes reproductive success”; Hedgecock 1994). If cohorts of larvae from a cross between closely related parents successfully co-recruit to the same reef, we would expect patterns of closely related and inbred groups of individuals within a reef, as we see here with shallow S. intersepta populations. Although collective dispersal of larvae has been more commonly applied to fish species, there is recent evidence in benthic marine species, including corals (Barfield et al. 2020). Collective dispersal, and thereby co-recruitment patterns are influenced by larval swimming behavior, which may be similar among inbred and related larvae (Burgess et al. 2022). Together these behavioral similarities and settlement preferences may contribute to the patterns observed in shallow S. intersepta lineages. Stephanocoenia intersepta exhibits high rates of fertilization (>95%) among collected eggs, thought to be a consequence of colonies holding multiple eggs within polyp tentacles that are not released until after fertilization (Hagman et al. 1998; Vermeij et al. 2010). This reproductive strategy may contribute to high rates of inbreeding if nearest neighbors are also close relatives or clones, as has been found in some coral species (Lord et al. 2023; Shilling et al. 2023). Phenotypic settlement behavior, co-recruitment patterns, and elevated rates of local larval retention and recruitment may all have an effect on the lower diversity observed in shallow S. intersepta lineages in FKNMS.
The detection of these cryptic lineages and the potential of reproductive isolation among S. intersepta in FKNMS highlights an important need to evaluate multiple populations of critical coral species for genetic diversity, especially in the context of conservation biology (Beger et al. 2014). Mesophotic corals in FKNMS have so far been less impacted by SCTLD and thereby may have the potential to provide genetic diversity to shallow reefs throughout the region (Reed et al. 2021). Even if mesophotic S. intersepta populations are unable to successfully provide viable larvae to degraded shallow reefs in the future, they may be useful moving forward for “seed bank” collection or colony “rescue” and rearing/breeding within land-based nurseries to aid in ongoing and future restoration efforts (Boström-Einarsson et al. 2020). Though collection of colonies and/or gametes from mesophotic depths presents unique challenges in and of itself. However, if S. intersepta lineage differentiation is due to gametic incompatibility or hybrid unviability greater issues and loss of diversity may occur given the evidence of inbreeding we find in shallow lineages and the present and persistent threats to reefs across FKNMS.
Samples for this study were collected in August–September 2019, when stony coral tissue loss disease (SCTLD) was present in the Upper Keys and Lower Keys, but before it emerged in the Dry Tortugas (Dobbelaere et al. 2022). The patterns of lower diversity within shallow lineages were not driven by the SCTLD outbreak as they are consistent across all sampling sites, including those within the Dry Tortugas that had not yet been affected by SCTLD at the time of sampling. If a genetic bottleneck event is responsible for these observed patterns, it would be more likely that the decades of coral loss seen throughout shallow reefs in FKNMS contributed to the lack of genetic diversity. Particularly, events like dark spots syndrome outbreaks where S. intersepta experienced mortality of 3.8% yr−1 from 2002–2004 and episodic ocean warming which significantly decreased shallow forereef scleractinian cover between 1999 and 2009 (Porter et al. 2011; Ruzicka et al. 2013).
Population structuring across depth
Population genetic structure analyses indicate S. intersepta are structured by depth throughout FKNMS despite small horizontal distances between depth zones within sampling site (Fig. 1). Similar patterns have been observed in other coral population genetic studies throughout the tropical western Atlantic. Montastraea cavernosa populations are depth stratified on the Belize Barrier Reef, despite extremely small horizontal distances between depth zones (i.e., <10 m; Eckert et al. 2019). In Alacranes Reef, Mexico, M. cavernosa populations are also structured by depth, with some lineages almost exclusively found in mesophotic populations (Sturm et al. 2022). However, in this study we identified two depth-generalist lineages (Blue and Teal; Figs. 2D and 3A) and two lineages mostly endemic to shallow populations (Green and Yellow; Figs. 2D and 3A). In contrast, S. intersepta populations in Bermuda exhibit a total lack of genetic structuring across depth zones (Bongaerts et al. 2017).
The variable patterns of differentiation observed across depth in FKNMS S. intersepta are not unique within FKNMS. In the species M. cavernosa, populations across the same sampling sites also exhibit variable patterns of vertical differentiation (Sturm et al. 2021). Montastraea cavernosa populations in Lower Keys and Tortugas Bank are less differentiated across depth than in Riley’s Hump and Upper Keys (Sturm et al. 2021). Both M. cavernosa and Siderastrea siderea also exhibited ecological divergence between inshore and offshore reefs in FKNMS (Rippe et al. 2021). This variation holds across regional scales where deep and shallow M. cavernosa populations in the northwestern Gulf of Mexico function as a single, panmictic population, and populations in Florida are structured across depth (Studivan and Voss 2018).
The timing of spawning, the dominant current regimes post-fertilization, and abiotic factors influencing settlement selection and recruitment success all may contribute to variable patterns of horizontal or vertical population structuring. Stephanocoenia intersepta typically spawn annually after the full moon in August/September, in a tight temporal window (Hagman, Gittings, and Deslarzes 1998). In some locations, shallow and mesophotic conspecifics, including S. intersepta, spawn within the same hour (Vize 2006). Slight asynchrony in spawning across depth may even be beneficial to mixing of gametes, with earlier spawning at depth allowing positively buoyant gametes or gamete bundles more time to reach the surface and mix with gametes from shallow colonies (Levitan et al. 2004).
Mesophotic S. intersepta may also act as larger source populations throughout the sanctuary. Generation time is relatively long in many scleractinian coral species, thus the two to three generations estimated through bayesass migration rates likely encompass decades. Patterns of migration somewhat agree with previous biophysical models for the region, where dispersal of larvae is strongly eastward across the Florida Keys, but mesoscale coastal counter-currents may emerge from mesoscale eddies and result in episodic westward movements (Lee et al. 1994; Yeung et al. 2001; Frys et al. 2020). Stephanocoenia intersepta at mesophotic sites are predominantly members of the depth-generalist Blue and Teal lineages (Fig. 2D), which also persist on shallow reefs. With current losses of coral cover in FKNMS, mesophotic Blue/Teal lineages have the potential to re-seed shallow reefs, if oceanographic and environmental regimes remain conducive to successful migration and post settlement success (Harley et al. 2006; Matz et al. 2020). Unfortunately, with the strong potential for reproductive isolation and shallow specific cryptic lineages, ongoing coral loss throughout FKNMS suggest the potential risk for extirpation of these shallow lineages.
Variable Symbiodiniaceae associations
We found significantly different Symbiodiniaceae across sampling sites and depths in FKNMS (Figs. 2E, 5), consistent with M. cavernosa Symbiodiniaceae in Mexico, where symbionts differed between Alacranes and Bajos del Norte reefs as well as between depth zones (Sturm et al. 2022). Some ITS2 type profiles are only found in shallow S. intersepta colonies, possibly due to adaptation to shallow reef conditions (e.g., light, nutrients). Since S. intersepta is a broadcast spawning species with a large depth range and potential for long-distance dispersal, acquiring symbionts from the local environment where larvae settle is advantageous, especially if there is prevalent endosymbiont zonation across depth and/or site, such as we identified here (Bongaerts et al. 2015). It is likely that different site and depth zone combinations in FKNMS harbor specific Symbiodiniaceae within the environment, as seen in American Samoa (Cunning et al. 2015). In the case of S. intersepta in FKNMS, there is some correlation between host genotype and Symbiodiniaceae association, these differences across depth and site may potentially avoid any issues with maladapted Symbiodiniaceae associations (Bongaerts et al. 2010). Environmental factors other than depth may be influencing the most abundant taxa within a site, influencing host-symbiont associations, or inhibiting the dispersal of taxa across site (e.g., current), thereby shaping the differences observed here (Cooper et al. 2011; Tonk et al. 2013).
Cladocopium was the most abundant Symbiodiniaceae genus found among S. intersepta across FKNMS, particularly in mesophotic S. intersepta. Similar to M. cavernosa Symbiodiniaceae assemblages in Belize, we found that deeper sites generally harbored fewer Symbiodiniaceae type profiles and shallow sites were more likely to harbor unique profiles (Eckert et al. 2020). This pattern of depth-generalist and shallow-specialist ITS2 type profiles differs from studies of M. cavernosa in Mexico and the Bahamas, where depth-specialist Symbiodiniaceae were found (Lesser et al. 2010; Sturm et al. 2022). The dominance of Cladocopium spp. within S. intersepta colonies is consistent with results from Curaçao; however, S. intersepta in Curaçao only harbored Cladocopium spp. and depth-specialist symbionts within the genus Cladocopium were identified at 50 and 60 m (Bongaerts et al. 2015). The patterns and dominance we observe among in hospite Cladocopium across FKNMS S. intersepta are rather expected, as Cladocopium is a genetically and ecologically diverse genus (LaJeunesse et al. 2018).
RADseq alignments to Symbiodiniaceae spp. genomes were highly concordant with ITS2 sequences at the genus level (Fig. 5). While using markers such as ITS2 or psbAncr allows a much finer level of differentiation (i.e., Symbiodiniaceae species or genotype), these methods require additional lab work and sequencing costs. Despite the reference genomes coming from congener algal taxa, and despite the high level of diversity and homology across Symbiodiniaceae, using RADseq SNPs to identify algal symbionts, as first proposed by Manzello et al. (2019) was highly effective and significantly correlated to the ITS2 assignment at the genus level. Identifying algal endosymbionts via RADseq-based approaches can provide critical time and cost savings if genus-level resolution is sufficient to answer the research questions at hand.
Conclusions
Stephanocoenia intersepta in FKNMS are comprised of distinct, cryptic lineages that show variable depth fidelity, with shallow specific (Green and Yellow) and depth-generalist (Blue and Teal) lineages. The lower diversity and higher levels of inbred individuals across shallow specific lineages are quite worrisome and may portend potential risks moving forward, especially for conservation and restoration efforts. Successful migration or hybridization from sources such as mesophotic S. intersepta, with greater genetic diversity, may allow shallow populations to persist if there are losses of the shallow-specific lineages due to genetic bottleneck events (Ingvarsson 2001). Yet the evidence for potential reproductive isolation between lineages and inbreeding within shallow lineages may contribute to a loss of overall diversity for this coral species with increasing stressors and repeated losses of corals across shallow FKNMS reefs (Toth et al. 2014). Additionally, the presence of cryptic lineages in sympatry and the potential of reproductive isolation between them highlights the importance of genetic management for coral outplanting and assisted sexual reproduction restoration strategies. Mesophotic reefs have been relatively understudied to date, yet they are critical to assessments of ecosystem biodiversity and genetic diversity (Turner et al. 2017). Throughout FKNMS, mesophotic reefs harbor greater genetic diversity of S. intersepta with depth-generalist cryptic lineages able to persist across both shallow and mesophotic depths. In FKNMS mesophotic genets may offer an opportunity to maintain lost shallow S. intersepta diversity through restoration-based fragmentation and/or reproductive propagation, thereby increasing the likelihood of long-term population persistence in the region (Reynolds et al. 2012).
Data availability
Sequence data from this manuscript can be accessed at the NCBI BioProject Accession PRJNA884416. All wet lab, bioinformatic, and data analysis procedures used in this study are available in the GitHub repository associated with this manuscript (Eckert 2024).
References
Aranda M, Li Y, Liew YJ, Baumgarten S, Simakov O, Wilson MC et al. (2016) Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci Rep 6:1–15. https://doi.org/10.1038/srep39734
Barfield S, Davies SW, Matz MV (2020) Co-recruitment of relatives in a broadcast-spawning coral (Acropora hyacinthus) facilitates emergence of an inbred, genetically distinct group within a panmictic population. Evol Biol. https://doi.org/10.1101/2020.02.26.956680
Beger M, Selkoe KA, Treml E, Barber PH, von der Heyden S, Crandall ED et al. (2014) Evolving coral reef conservation with genetic information. Bull Mar Sci 90:159–185. https://doi.org/10.5343/bms.2012.1106
Bernard AM, Finnegan KA, Shivji MS (2018) Genetic connectivity dynamics of the giant barrel sponge, Xestospongia muta, across the Florida reef tract and Gulf of Mexico. Bull Mar Sci 95:161–175. https://doi.org/10.5343/bms.2018.0021
Bongaerts P, Carmichael M, Hay KB, Tonk L, Frade PR, Hoegh-Guldberg O (2015) Prevalent endosymbiont zonation shapes the depth distributions of scleractinian coral species. R Soc Open Sci 2:140297. https://doi.org/10.1098/rsos.140297
Bongaerts P, Ridgway T, Sampayo EM, Hoegh-Guldberg O (2010) Assessing the ‘deep reef refugia’ hypothesis: focus on Caribbean reefs. Coral Reefs 29:309–327. https://doi.org/10.1007/s00338-009-0581-x
Bongaerts P, Riginos C, Brunner R, Englebert N, Smith SR, Hoegh-Guldberg O (2017) Deep reefs are not universal refuges: reseeding potential varies among coral species. Sci Adv 3:e1602373. https://doi.org/10.1126/sciadv.1602373
Boström-Einarsson L, Babcock RC, Bayraktarov E, Ceccarelli D, Cook N, Ferse SCA et al. (2020) Coral restoration – A systematic review of current methods, successes, failures and future directions (CN Bianchi, Ed.). PLOS ONE 15:e0226631. https://doi.org/10.1371/journal.pone.0226631
Buckel CA, Viehman TS, Edwards K, Tester PA, Piniak GA, Muñoz RC, et al. (2014) Benthic Communities of the Coral Reef. In: Clark R, Taylor JC, Buckel CA, Kracker LM (eds) Fish and benthic communities of the Flower Garden Banks National Marine Sanctuary: science to support sanctuary management, NOAA Technical Memorandum NOS NCCOS 179, Silver Spring, MD, pp 21–70
Burgess SC, Bode M, Leis JM, Mason LB (2022) Individual variation in marine larval-fish swimming speed and the emergence of dispersal kernels. Oikos 2022. https://doi.org/10.1111/oik.08896
Burgess SC, Johnston EC, Wyatt ASJ, Leichter JJ, Edmunds PJ (2021) Response diversity in corals: hidden differences in bleaching mortality among cryptic Pocillopora species. Ecology 102:e03324. https://doi.org/10.1002/ecy.3324
Christie MR, Meirmans PG, Gaggiotti OE, Toonen RJ, White C (2017) Disentangling the relative merits and disadvantages of parentage analysis and assignment tests for inferring population connectivity (Hauser L, ed). ICES J Mar Sci 74:1749–1762. https://doi.org/10.1093/icesjms/fsx044
Cooper TF, Ulstrup KE, Dandan SS, Heyward AJ, Kühl M, Muirhead A et al. (2011) Niche specialization of reef-building corals in the mesophotic zone: metabolic trade-offs between divergent Symbiodinium types. Proc R Soc B Biol Sci 278:1840–1850. https://doi.org/10.1098/rspb.2010.2321
Cunning R, Yost DM, Guarinello ML, Putnam HM, Gates RD (2015) Variability of Symbiodinium communities in waters, sediments, and corals of thermally distinct reef pools in American Samoa (CA Chen, Ed.). PLoS One 10:e0145099. https://doi.org/10.1371/journal.pone.0145099
De’ath G, Fabricius KE, Sweatman H, Puotinen M (2012) The 27-year decline of coral cover on the Great Barrier Reef and its causes. Proc Natl Acad Sci USA 109:17995–17999. https://doi.org/10.1073/pnas.1208909109
Dobbelaere T, Holstein DM, Muller EM, Gramer LJ, McEachron L, Williams SD et al. (2022) Connecting the Dots: Transmission of Stony Coral Tissue Loss Disease From the Marquesas to the Dry Tortugas. Front Mar Sci 9:778938. https://doi.org/10.3389/fmars.2022.778938
Dray S, Dufour A-B (2007) The ade4 Package: Implementing the Duality Diagram for Ecologists. J Stat Softw 22. https://doi.org/10.18637/jss.v022.i04
Eckert RJ (2024) Stephanocoenia_FKNMS_PopGen. Zenodo, https://github.com/RyanEckert/Stephanocoenia_FKNMS_PopGenhttps://doi.org/10.5281/zenodo.7130749
Eckert RJ, Reaume AM, Sturm AB, Studivan MS, Voss JD (2020) Depth Influences Symbiodiniaceae Associations Among Montastraea cavernosa Corals on the Belize Barrier Reef. Front Microbiol 11:518. https://doi.org/10.3389/fmicb.2020.00518
Eckert RJ, Studivan MS, Voss JD (2019) Populations of the coral species Montastraea cavernosa on the Belize Barrier Reef lack vertical connectivity. Sci Rep 9:7200. https://doi.org/10.1038/s41598-019-43479-x
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Faubet P, Waples RS, Gaggiotti OE (2007) Evaluating the performance of a multilocus Bayesian method for the estimation of migration rates. Mol Ecol 16:1149–1166. https://doi.org/10.1111/j.1365-294X.2007.03218.x
Fifer JE, Yasuda N, Yamakita T, Bove CB, Davies SW (2022) Genetic divergence and range expansion in a western North Pacific coral. Sci Total Environ 813:152423. https://doi.org/10.1016/j.scitotenv.2021.152423
Foll M, Gaggiotti O (2008) A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180:977–993. https://doi.org/10.1534/genetics.108.092221
França FM, Benkwitt CE, Peralta G, Robinson JPW, Graham NAJ, Tylianakis JM et al. (2020) Climatic and local stressor interactions threaten tropical forests and coral reefs. Philos Trans R Soc B Biol Sci 375:20190116. https://doi.org/10.1098/rstb.2019.0116
Frys C, Saint-Amand A, Le Hénaff M, Figueiredo J, Kuba A, Walker B et al. (2020) Fine-scale coral connectivity pathways in the Florida reef tract: implications for conservation and restoration. Front Mar Sci 7:1–42. https://doi.org/10.3389/fmars.2020.00312
Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. https://doi.org/10.1093/bioinformatics/bts565
Games PA, Howell JF (1976) Pairwise Multiple Comparison Procedures with Unequal N’s and/or Variances: A Monte Carlo Study. J Educ Stat 1:113–125
Gardner TA, Côté IM, Gill JA, Grant A, Watkinson AR (2003) Long-term region-wide declines in Caribbean corals. Science 301:958–960. https://doi.org/10.1126/science.1086050
Glynn PW (1996) Coral reef bleaching: facts, hypotheses and implications. Glob Change Biol 2:495–509. https://doi.org/10.1111/j.1365-2486.1996.tb00063.x
Goodbody-Gringley G, Noyes T, Smith SR (2019) Bermuda. In: Loya Y, Puglise KA, Bridge TCL (eds) Mesophotic Coral Ecosystems, Coral Reefs of the World. Springer International Publishing, Cham, vol 12, pp 31–45. https://doi.org/10.1007/978-3-319-92735-0_2
Grupstra CGB, Gómez-Corrales M, Fifer JE, Aichelman HE, Meyer-Kaiser KS, Prada C et al. (2024) Integrating cryptic diversity into coral evolution, symbiosis and conservation. Nat Ecol Evol 8:622–636. https://doi.org/10.1038/s41559-023-02319-y
Hagedorn M, Page CA, O’Neil KL, Flores DM, Tichy L, Conn T et al. (2021) Assisted gene flow using cryopreserved sperm in critically endangered coral. Proc Natl Acad Sci 118:e2110559118. https://doi.org/10.1073/pnas.2110559118
Hagman DK, Gittings SR, Deslarzes KJP (1998) Timing, Species Participation, and Environmental Factors Influencing Annual Mass Spawning at the Flower Garden Banks (Northwest Gulf of Mexico). Gulf Mex Sci 16. https://doi.org/10.18785/goms.1602.06
Hagman DK, Gittings SR, Vize PD (1998) Fertilization in Broadcast-Spawning Corals of the Flower Garden Banks National Marine Sanctuary. Gulf Mex Sci 16. https://doi.org/10.18785/goms.1602.07
Hanghøj K, Moltke I, Andersen PA, Manica A, Korneliussen TS (2019) Fast and accurate relatedness estimation from high-throughput sequencing data in the presence of inbreeding. GigaScience 8. https://doi.org/10.1093/gigascience/giz034
Harley CDG, Randall Hughes A, Hultgren KM, Miner BG, Sorte CJB, Thornber CS et al. (2006) The impacts of climate change in coastal marine systems: Climate change in coastal marine systems. Ecol Lett 9:228–241. https://doi.org/10.1111/j.1461-0248.2005.00871.x
Hedgecock D (1994). Does variance in reproductive success limit effective population size of marine organisms? In: Beaumont A (ed) Genetics and Evolution of Aquatic Organisms, Chapman & Hall: London, pp 122–134.
Hughes TP (1994) Catastrophes, phase shifts, and large-scale degradation of a Caribbean coral reef. Science 265:1547–1551. https://doi.org/10.1126/science.265.5178.1547
Hume BCC, Smith EG, Ziegler M, Warrington HJM, Burt JA, LaJeunesse TC et al. (2019) SymPortal: a novel analytical framework and platform for coral algal symbiont next-generation sequencing ITS2 profiling. Mol Ecol Resour 19:1063–1080. https://doi.org/10.1111/1755-0998.13004
Hume BCC, Ziegler M, Poulain J, Pochon X, Romac S, Boissin E et al. (2018) An improved primer set and amplification protocol with increased specificity and sensitivity targeting the Symbiodinium ITS2 region. PeerJ 2018:1–22. https://doi.org/10.7717/peerj.4816
Ingvarsson PK (2001) Restoration of genetic variation lost – the genetic rescue hypothesis. Trends Ecol Evol 16:62–63. https://doi.org/10.1016/S0169-5347(00)02065-6
Jaap WC, Szmant A, Jaap K, Dupont J, Clarke R, Somerfield P et al. (2008) A perspective on the biology of Florida Keys coral reefs. Coral Reefs USA 1:75–125
Johnston EC, Cunning R, Burgess SC (2022) Cophylogeny and specificity between cryptic coral species (Pocillopora spp.) at Mo′orea and their symbionts (Symbiodiniaceae). Mol Ecol 31:5368–5385. https://doi.org/10.1111/mec.16654
Knowlton N, Jackson JBC (1993) Inbreeding and outbreeding in marine invertebrates. In: Thornhill NW (ed.) The Natural history of inbreeding and outbreeding: theoretical and empirical perspectives. University of Chicago Press, Chicago, pp 200–249
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191. https://doi.org/10.1111/1755-0998.12387
Korneliussen TS, Albrechtsen A, Nielsen R (2014) ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinforma 15:356. https://doi.org/10.1186/s12859-014-0356-4
Kuffner I, Walters L, Becerro M, Paul V, Ritson-Williams R, Beach K (2006) Inhibition of coral recruitment by macroalgae and cyanobacteria. Mar Ecol Prog Ser 323:107–117. https://doi.org/10.3354/meps323107
LaJeunesse TC, Parkinson JE, Gabrielson PW, Jeong HJ, Reimer JD, Voolstra CR et al. (2018) Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr Biol 28:2570–2580.e6. https://doi.org/10.1016/j.cub.2018.07.008
Lang JC (2003) Status of Coral Reefs in the Western Atlantic: Results of Initial Surveys, Atlantic and Gulf Rapid Reef Assessment (Agrra) Program. Washington, D.C.: Smithsonian Institution. https://doi.org/10.5479/si.00775630.496-00.630
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Lee TN, Clarke M, Williams E, Szmant AF, Berger T (1994) Evolution of the Tortugas Gyre and its influence on recruitment in the Florida Keys. Bull Mar Sci 54:621–646
Lesser MP, Slattery M, Leichter JJ (2009) Ecology of mesophotic coral reefs. J Exp Mar Biol Ecol 375:1–8. https://doi.org/10.1016/j.jembe.2009.05.009
Lesser MP, Slattery M, Stat M, Ojimi M, Gates RD, Grottoli A (2010) Photoacclimatization by the coral Montastraea cavernosa in the mesophotic zone: light, food, and genetics. Ecology 91:990–1003. https://doi.org/10.1890/09-0313.1
Levitan DR, Fukami H, Jara J, Kline D, McGovern TM, McGhee KE et al. (2004) Mechanisms of reproductive isolation among sympatric broadcast-spawning corals of the Montastraea annularis species complex. Evolution 58:308–323. https://doi.org/10.1111/j.0014-3820.2004.tb01647.x
Liu X, Fu Y-X (2020) Stairway Plot 2: demographic history inference with folded SNP frequency spectra. Genome Biol 21:280. https://doi.org/10.1186/s13059-020-02196-9
Liu H, Stephens TG, González-Pech RA, Beltran VH, Lapeyre B, Bongaerts P et al. (2018) Symbiodinium genomes reveal adaptive evolution of functions related to coral-dinoflagellate symbiosis. Commun Biol 1:95. https://doi.org/10.1038/s42003-018-0098-3
Lord KS, Lesneski KC, Buston PM, Davies SW, D’Aloia CC, Finnerty JR (2023) Rampant asexual reproduction and limited dispersal in a mangrove population of the coral Porites divaricata. Proc R Soc B Biol Sci 290:20231070. https://doi.org/10.1098/rspb.2023.1070
Manzello DP, Matz MV, Enochs IC, Valentino L, Carlton RD, Kolodziej G, et al. (2019) Role of host genetics and heat-tolerant algal symbionts in sustaining populations of the endangered coral Orbicella faveolata in the Florida Keys with ocean warming. Glob Change Biol 1016–1031. https://doi.org/10.1111/gcb.14545
Marshall DJ, Monro K, Bode M, Keough MJ, Swearer S (2010) Phenotype-environment mismatches reduce connectivity in the sea. Ecol Lett 13:128–140. https://doi.org/10.1111/j.1461-0248.2009.01408.x
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. https://doi.org/10.14806/ej.17.1.200
Martinez Arbizu P (2017) pairwiseAdonis: Pairwise multilevel comparison using adonis. See https://github.com/pmartinezarbizu/pairwiseAdonis. Accessed 10 February 2019
Matz MV (2020) Whole genome de novo genotyping with 2bRAD. See https://github.com/z0on/2bRAD_denovo
Matz MV, Treml EA, Aglyamova GV, Bay LK (2018) Potential and limits for rapid genetic adaptation to warming in a Great Barrier Reef coral (HE Hoekstra, Ed.). PLOS Genet 14:e1007220. https://doi.org/10.1371/journal.pgen.1007220
Matz MV, Treml EA, Haller BC (2020) Estimating the potential for coral adaptation to global warming across the Indo-West Pacific. Glob Change Biol 26:3473–3481. https://doi.org/10.1111/gcb.15060
Meirmans PG (2014) Nonconvergence in Bayesian estimation of migration rates. Mol Ecol Resour 14:726–733. https://doi.org/10.1111/1755-0998.12216
Meisner J, Albrechtsen A (2018). Inferring Population Structure and Admixture Proportions in Low-Depth NGS Data. Genetics 210: 719–731. https://doi.org/10.1534/genetics.118.301336
Midway S, Robertson M, Flinn S, Kaller M (2020) Comparing multiple comparisons: practical guidance for choosing the best multiple comparisons test. PeerJ 8:e10387. https://doi.org/10.7717/peerj.10387
Mumby PJ, Dahlgren CP, Harborne AR, Kappel CV, Micheli F, Brumbaugh DR et al. (2006) Fishing, trophic cascades, and the process of grazing on coral reefs. Science 311:98–101. https://doi.org/10.1126/science.1121129
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. (2020) vegan: Community Ecology Package. See https://CRAN.R-project.org/package=vegan
Olsen KC, Ryan WH, Winn AA, Kosman ET, Moscoso JA, Krueger-Hadfield SA et al. (2020) Inbreeding shapes the evolution of marine invertebrates. Evolution 74:871–882. https://doi.org/10.1111/evo.13951
Palumbi SR (2003) Population genetics, demographic connectivity, and the design of marine reserves. Ecol Appl 13:146–158
Pinheiro HT, MacDonald C, Santos RG, Ali R, Bobat A, Cresswell BJ et al. (2023) Plastic pollution on the world’s coral reefs. Nature 619:311–316. https://doi.org/10.1038/s41586-023-06113-5
Porter JW, Torres C, Sutherland KP, Meyers MK, Callahan MK, Ruzicka R et al. (2011) Prevalence, severity, lethality, and recovery of dark spots syndrome among three Floridian reef-building corals. J Exp Mar Biol Ecol 408:79–87. https://doi.org/10.1016/j.jembe.2011.07.027
Pyle RL, Copus JM (2019) Mesophotic Coral Ecosystems: Introduction and Overview. In: Loya Y, Puglise KA, Bridge TCL (eds.) Mesophotic Coral Ecosystems of the World, Springer New York, pp 3–27. https://doi.org/10.1007/978-3-319-92735-0_1
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst Biol 67:901–904. https://doi.org/10.1093/sysbio/syy032
Reed JK, Farrington S, David A, Harter S, Pomponi SA, Cristina Diaz M, et al. (2019) Pulley Ridge, Gulf of Mexico, USA. 57–69. https://doi.org/10.1007/978-3-319-92735-0_4
Reed JK, Farrington S, Diaz MC, Pomponi SA, Hanisak MD, Voss JD (2021) Characterization of the Mesophotic Coral Reefs in the Florida Keys National Marine Sanctuary. Harbor Branch Oceanographic Institution Technical Report Number 198. http://www.cioert.org/expeditions/mesophotic-reef-ecosystems
Reynolds LK, McGlathery KJ, Waycott M (2012) Genetic Diversity Enhances Restoration Success by Augmenting Ecosystem Services (RKF Unsworth, Ed.). PLoS ONE 7:e38397. https://doi.org/10.1371/journal.pone.0038397
Rippe JP, Dixon G, Fuller ZL, Liao Y, Matz M (2021) Environmental specialization and cryptic genetic divergence in two massive coral species from the Florida Keys Reef Tract. Mol Ecol 30:3468–3484. https://doi.org/10.1111/mec.15931
Ritson-Williams R, Arnold S, Fogarty N, Steneck RS, Vermeij M, Paul VJ (2009) New perspectives on ecological mechanisms affecting coral recruitment on reefs. Smithson Contrib Mar Sci 437–457. https://doi.org/10.5479/si.01960768.38.437
Rocha LA, Pinheiro HT, Shepherd B, Papastamatiou YP, Luiz OJ, Pyle RL et al. (2018) Mesophotic coral ecosystems are threatened and ecologically distinct from shallow water reefs. Science 361:281–284. https://doi.org/10.1126/science.aaq1614
Rose NH, Bay RA, Morikawa MK, Thomas L, Sheets EA, Palumbi SR (2021) Genomic analysis of distinct bleaching tolerances among cryptic coral species. Proc R Soc B Biol Sci 288:20210678. https://doi.org/10.1098/rspb.2021.0678
Ruzicka RR, Colella MA, Porter JW, Morrison JM, Kidney JA, Brinkhuis V et al. (2013) Temporal changes in benthic assemblages on Florida Keys reefs 11 years after the 1997/1998 El Niño. Mar Ecol Prog Ser 489:125–141. https://doi.org/10.3354/meps10427
Serrano XM, Baums IB, O’Reilly K, Smith TB, Jones RJ, Shearer TL et al. (2014) Geographic differences in vertical connectivity in the Caribbean coral Montastraea cavernosa despite high levels of horizontal connectivity at shallow depths. Mol Ecol 23:4226–4240. https://doi.org/10.1111/mec.12861
Shilling EN, Eckert RJ, Sturm AB, Voss JD (2023) Porites astreoides coral populations demonstrate high clonality and connectivity in southeast Florida. Coral Reefs https://doi.org/10.1007/s00338-023-02417-0
Shoguchi E, Beedessee G, Hisata K, Tada I, Narisoko H, Satoh N et al. (2021) A New Dinoflagellate Genome Illuminates a Conserved Gene Cluster Involved in Sunscreen Biosynthesis (S Phadke, Ed.). Genome Biol Evol 13:evaa235. https://doi.org/10.1093/gbe/evaa235
Shoguchi E, Shinzato C, Kawashima T, Gyoja F, Mungpakdee S, Koyanagi R et al. (2013) Draft Assembly of the Symbiodinium minutum Nuclear Genome Reveals Dinoflagellate Gene Structure. Curr Biol 23:1399–1408. https://doi.org/10.1016/j.cub.2013.05.062
Skotte L, Korneliussen TS, Albrechtsen A (2013) Estimating Individual Admixture Proportions from Next Generation Sequencing Data. Genetics 195:693–702. https://doi.org/10.1534/genetics.113.154138
Smith TB, Gyory J, Brandt ME, Miller WJ, Jossart J, Nemeth RS (2016) Caribbean mesophotic coral ecosystems are unlikely climate change refugia. Glob Change Biol 22:2756–2765. https://doi.org/10.1111/gcb.13175
Soong K (1993) Colony size as a species character in massive reef corals. Coral Reefs 12:77–83. https://doi.org/10.1007/BF00302106
Studivan MS, Voss JD (2018) Population connectivity among shallow and mesophotic Montastraea cavernosa corals in the Gulf of Mexico identifies potential for refugia. Coral Reefs 37:1183–1196. https://doi.org/10.1007/s00338-018-1733-7
Sturm AB, Eckert RJ, Carreiro AM, Klein AM, Studivan MS, Dodge Farelli D et al. (2023) Does depth divide? Variable genetic connectivity patterns among shallow and mesophotic Montastraea cavernosa coral populations across the Gulf of Mexico and western Caribbean. Ecol Evol 13:e10622. https://doi.org/10.1002/ece3.10622
Sturm AB, Eckert RJ, Carreiro AM, Simões N, Voss JD (2022) Depth-Dependent Genetic Structuring of a Depth-Generalist Coral and Its Symbiodiniaceae Algal Communities at Campeche Bank, Mexico. Front Mar Sci 9:835789. https://doi.org/10.3389/fmars.2022.835789
Sturm AB, Eckert RJ, Carreiro AM, Voss JD (2021) Population genetic structure of the broadcast spawning coral, Montastraea cavernosa, demonstrates refugia potential of upper mesophotic populations in the Florida Keys. Coral Reefs https://doi.org/10.1007/s00338-021-02112-y
Szmant AM (1991) Sexual reproduction by the Caribbean reef corals Montastrea annularis and M. cavernosa. Mar Ecol Prog Ser 74:13–25. https://doi.org/10.3354/meps074013
Tonk L, Sampayo EM, Weeks S, Magno-Canto M, Hoegh-Guldberg O (2013) Host-Specific Interactions with Environmental Factors Shape the Distribution of Symbiodinium across the Great Barrier Reef (CR Voolstra, Ed.). PLoS ONE 8:e68533. https://doi.org/10.1371/journal.pone.0068533
Toth LT, van Woesik R, Murdoch TJT, Smith SR, Ogden JC, Precht WF et al. (2014) Do no-take reserves benefit Florida’s corals? 14 years of change and stasis in the Florida Keys National Marine Sanctuary. Coral Reefs 33:565–577. https://doi.org/10.1007/s00338-014-1158-x
Turner JA, Babcock RC, Hovey R, Kendrick GA (2017) Deep thinking: A systematic review of mesophotic coral ecosystems. ICES J Mar Sci 74:2309–2320. https://doi.org/10.1093/icesjms/fsx085
Vermeij MJA, Barott KL, Johnson AE, Marhaver KL (2010) Release of eggs from tentacles in a Caribbean coral. Coral Reefs 29:411–411. https://doi.org/10.1007/s00338-010-0595-4
Vize PD (2006) Deepwater broadcast spawning by Montastraea cavernosa, Montastraea franksi, and Diploria strigosa at the Flower Garden Banks, Gulf of Mexico. Coral Reefs 25:169–171. https://doi.org/10.1007/s00338-005-0082-5
Wakwella A, Mumby PJ, Roff G (2020) Sedimentation and overfishing drive changes in early succession and coral recruitment. Proc R Soc B Biol Sci 287:20202575. https://doi.org/10.1098/rspb.2020.2575
Wang S, Meyer E, McKay JK, Matz MV (2012) 2b-RAD: a simple and flexible method for genome-wide genotyping. Nat Methods 9:808–810. https://doi.org/10.1038/nmeth.2023
Wilson GA, Rannala B (2003) Bayesian Inference of Recent Migration Rates Using Multilocus Genotypes. Genetics 163:1177–1191
Wood DE, Lu J, Langmead B (2019) Improved metagenomic analysis with Kraken 2. Genome Biol 20:257. https://doi.org/10.1186/s13059-019-1891-0
Yeung C, Jones DL, Criales MM, Jackson TL, Richards William J (2001) Influence of coastal eddies and counter-currents on the influx of spiny lobster, Panulirus argus, postlarvae into Florida Bay. Mar Freshw Res 52:1217. https://doi.org/10.1071/MF01110
Acknowledgements
Funding for this research was awarded to J. Voss by NOAA Ocean Exploration and Research under award NA14OAR4320260 through the Cooperative Institute for Ocean Exploration, Research, and Technology, and by the NOAA National Center for Coastal Ocean Science under award NA18NOS4780166 To J. Voss and S. Herrera through the Connectivity of Coral Ecosystems in the Northwest Gulf of Mexico project. We thank the participants of the 2019 FAU Harbor Branch CIOERT Florida Keys Expedition including M. Studivan, J. Emmert, M. McCallister, E. Shilling, I. Combs, J. Beal, C. Haymaker, S. Farrington, and the crew of R/V F.G. WALTON SMITH. The University of Texas at Austin’s Genome Sequencing and Analysis Facility provided sequencing support and computation capacity was provided by Research Computing Services at Florida Atlantic University. We thank Dr. Hannah Aichelman and two anonymous reviewers for their thoughtful feedback which greatly improved our manuscript. This is contribution 2322 from Harbor Branch Oceanographic Institute.
Author information
Authors and Affiliations
Contributions
JDV and RJE conceived and designed the study. JDV served as chief scientist for the expedition and secured permits. JDV, RJE, and ABS collected the samples. RJE, ABS, AMC, and AMK performed lab work. RJE performed bioinformatic analyses, analyzed the data and prepared figures. All authors contributed to the final version of the manuscript prepared by RJE.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval
All corals were collected under permit FKNMS-2019-088 from Florida Keys National Marine Sanctuary.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Sam Banks
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Eckert, R.J., Sturm, A.B., Carreiro, A.M. et al. Cryptic diversity of shallow and mesophotic Stephanocoenia intersepta corals across Florida Keys National Marine Sanctuary. Heredity 133, 137–148 (2024). https://doi.org/10.1038/s41437-024-00698-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-024-00698-x
- Springer Nature Switzerland AG