Abstract
Adeno-associated virus (AAV) has been investigated to transfer the cystic fibrosis transmembrane conductance regulator (CFTR) to airways. Inhaled AAV2-CFTR in people with cystic fibrosis (CF) is safe, but inefficient. In vitro, AAV2 transduction of human airway epithelia on the apical (luminal) side is inefficient, but efficient basolaterally. We previously selected AAV2.5T, a novel capsid that apically transduces CF human airway epithelia and efficiently restores CFTR function. We hypothesize the AAV receptor (AAVR) is basolaterally localized, and that AAV2.5T utilizes an alternative apical receptor. We found AAVR in human airway epithelia by western blot and RNA-Seq analyses. Using immunocytochemistry we did not find endogenous AAVR at membranes but overexpression localized AAVR to the basolateral membrane, where it preferentially increased transduction. Anti-AAVR antibodies blocked transduction by AAV2 from the basolateral side but not AAV2.5T from the apical side, suggesting a unique apical receptor. Finally, we found infection by AAV2 but not AAV2.5T was blocked by CRISPR knockout of AAVR in cell lines. Our data suggest the absence of apical AAVR is rate limiting for AAV2, and efficient transduction by AAV2.5T is accomplished using an AAVR independent pathway. Our findings inform the development of gene therapy for CF, and AAV vectors in general.
Similar content being viewed by others
Introduction
Cystic fibrosis (CF) lung disease is caused by various mutations in the gene encoding the cystic fibrosis transmembrane conductance regulator (CFTR). CF is a target for gene therapy because it is caused by defects in a single gene, and because the airways are accessible to treatment [1,2,3]. Viral vector-mediated gene transfer is a mutation-agnostic treatment, and adeno-associated viruses (AAVs) are among the preferred vectors due to low immunogenicity, scalability of production, and cell-specific tropism. AAVs have restored CFTR function in an organotypic model comprised of human airway epithelial cells taken from CF lungs, and in relevant animal models [4,5,6,7]. Clinical translation of gene therapy for CF, however, remains just out of reach. In clinical trials, aerosolizing AAV serotype 2 (AAV2) carrying CFTR into the lungs of people with CF was safe [8,9,10], but molecular evidence of CFTR expression was nearly nonexistant [1, 11]. Possible obstacles to transduction include a lack of cellular receptors and co-receptors, impeded endosomal processing and viral trafficking to the nucleus, impaired transcription and translation of the transgene, and the presence of neutralizing antibodies [11,12,13].
AAV serotypes target-specific cell types. This tropism is thought to be conferred by interactions between different amino acids in capsid loop domains and receptor proteins on permissive cells [14]. In 1996, Mizukami et al. [15] showed AAV2 attachment to an unidentified 150 kDa membrane protein in human cells. Subsequent studies in various cell types proposed the protein receptor was either fibroblast growth factor receptor 1, hepatocyte growth factor receptor, or integrin αVβ5 after correlating surface proteins with permissiveness to infection [16,17,18]. Work was done in parallel demonstrating the important role heparan sulfate proteoglycan (HSPG) plays in mediating transduction by AAV2, AAV3, and AAV6 [19]. Later, other glycan moieties were shown to aid transduction by other serotypes. AAV1, AAV4, AAV5, and AAV6 interact with O− or N−linked sialic acid moieties [20,21,22,23], while AAV9 interacts with N-terminal galactose [24]. This led to a model of infection where AAV serotypes are thought to initially attach to distinct proteoglycans in the extracellular matrix, and subsequently bind to a protein receptor that mediates endocytosis.
More recently, the KIAA0319L gene was shown to be required for infection by AAV serotypes 1, 2, 3b, 5, 6, 8, and 9, and was consequently named the AAV receptor (AAVR) [25]. AAVR is a predicted type-1 transmembrane protein detected at 150 kDa [26]. AAVR binds directly to AAV2 in HeLa cells, and translocates between the vicinity of the cellular membrane and the trans-Golgi network [25]. The cellular function of AAVR is unknown, although potential roles in neuronal development and dyslexia have been described [27, 28]. Like other proteins exploited by viruses, AAVR contains extracellular IgG-like polycystic kidney domains (PKD 1–5) that could be used for cell–cell adhesion [29]. AAV2 utilizes PKD2 for transduction, whereas AAV5 uses PKD1, and AAV1 and AAV8 require both [26].
In order to circumvent the obstacles to efficient gene transfer in human airways, our previous collaborative efforts produced a novel vector using directed evolution [5]. We shuffled the capsid sequences of AAV2 and AAV5 and added point mutations using error-prone PCR. The resulting library of chimeric viruses was apically passaged on human airway epithelia cultures and AAV2.5T was selected. AAV2.5T is a chimera of the VP1 region of AAV2 and the VP2 and VP3 regions of AAV5, with a single A581T mutation in VP3. AAV2.5T can efficiently transfer CFTR to human airway epithelia form the apical side, and restore CFTR function [5].
Here, we investigate the localization of AAVR in human airway epithelia, and the influence AAVR has on transduction by AAV2, AAV5, and AAV2.5T. As AAV2 infection is only efficient basolaterally, while AAV2.5T is apically efficient, we hypothesized that AAVR is localized in the basolateral membrane, and that AAV2.5T has evolved to utilize an alternative apical receptor.
Materials and methods
Viruses
Purified, titrated stocks of recombinant self-complimentary AAV2/2, AAV2/5, and AAV2/2.5T carrying an enhanced green fluorescent protein (GFP) transgene driven by the cytomegalovirus (CMV) promoter were purchased from University of Iowa Viral Vector Core, divided into 10 μl aliquots and stored at –80 °C.
Cell culture
Primary human airway epithelial cells (HAE) from bronchus or trachea were harvested, seeded onto collagen-coated, semipermeable membranes (Costar no. 3470, Corning, Corning, NY, USA), grown at an air–liquid interface as previously described [30]. Experiments were performed at least 14 days after seeding, when cells had differentiated into ciliated and non-ciliated pseudo-stratified cells forming an organotypic tissue model. HAE were maintained in Ultroser G media (Pall, Port Washington, NY, USA). Immortalized cell lines were obtained from the University of Iowa Cell Culture Core, seeded at 20,000 cells/well into 96-well black fluorescence plates (Corning) and grown to confluence. Hela cells were maintained in minimal essential medium (MEM), HEK cells in Dulbecco’s modified Eagle’s medium (DMEM). Media were supplemented with 10% fetal bovine serum (FBS) and 1% non-essential amino acids (NEAA) (all from Thermo Fisher Scientific, Waltham, MA, USA). Growth media were supplemented with 100 IU/ml penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO, USA), and cells maintained in a 37 °C humidified incubator at 5% CO2.
RNA-seq
Total RNA was isolated from HAE using the mirVanaTM miRNA isolation kit (Thermo Fisher Scientific). Total RNA was tested on an Agilent Model 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), and samples with an RNA integrity number (RIN) over 7.0 were selected for downstream processing. Libraries were prepared using the TruSeq RNA Sample Prep (Illumina, San Diego, CA, USA). Libraries were submitted to the University of Iowa DNA Facility for deep sequencing, where paired-end DNA libraries were sequenced to an average depth of 29 million (M) read pairs (range: 22–44 M) with 100 base reads. Gene expression differences between nine primary culture donors were then analyzed using Cuffdiff software v2.0.2 (Dr. Cole Trapnell’s lab, University of Washington, Seattle, WA, USA). TPM values (transcripts per million) were extracted from the differential analysis logs to use for further analysis.
Western blots
Cell lines cultured in six-well plates (Corning) or Primary HAE cells were scraped into 25 μl ice-cold RIPA lysis buffer (25 mM Tris•HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) containing protease and phosphatase inhibitors (Thermo Fisher Scientific) and 1% TX-100, rotated 15 m at 4 °C and centrifuged at 10,000 × g for 20 m. Protein in supernatant was quantitated with a microplate BCA assay (Thermo Fisher Scientific). An equal volume of 2 × sample buffer was added and incubated at 55 °C for 30 m while agitating. Samples were run on 4–15% polyacrylamide gels (Bio-Rad, Hercules, CA, USA) with high molecular mass standards (Thermo Fisher Scientific). Electrophoresed gels were transferred to PDF-FL (EMD Millipore, Burlington, MA) overnight. Membranes were blocked in 0.01% casein buffer in Dulbecco’s phosphate-buffered saline (DPBS, Thermo Fisher Scientific), immunostained with AAVR antibodies diluted in DPBS 1:1000 (105385 Abcam, Cambridge, UK) and secondary antibodies (Molecular Probes, Eugene, OR), and visualized and quantified on an Odyssey IR imager (LiCor, Lincoln, NE, USA).
Immunocytochemistry
In preparation for staining, HAE were cooled to 4 °C and fixed in ice-cold 4% paraformaldehyde in DPBS with calcium and magnesium (Thermo Fisher Scientific) for 20 m. Lung tissue was fixed 1 h at room temperature in 2% paraformaldehyde, and kept overnight in ice-cold 30% sucrose before quick-freezing in OCT in an ethanol/dry ice bath and cryosectioning and mounting. After fixation, all samples were permeablized in 0.2% Triton X-100 (Thermo Fisher Scientific) in PBS, and blocked in Super-Block (Thermo Fisher Scientific) with 5% normal goat serum (Jackson Laboratories, Bar Harbor, ME, USA). Tissue sections were incubated for 2 h at 37 °C in AAVR antibodies (Abcam 105385) and either β-catenin (Thermo Fisher Scientific PA5-16762) or ZO-1 (Thermo Fisher Scientific 61-7300) antibodies, all at 1:100 dilution, followed by secondary goat anti-mouse Alexa Fluor 488 and goat anti-rabbit Alexa Fluor 568 antibodies (Molecular Probes A11034 and A11036), at 1:1000 dilution. Samples were mounted with Vectashield containing DAPI (Vector Laboratories, Burlingame, CA, USA) to visualize nuclei. Images were acquired on a Fluoview FV1000 confocal microscope with a UPLSAPO × 60 oil lens (Olympus, Shinjuku, TYO, JP) and scanned sequentially at 2 ms/pixel. Signal obtained from identical HAE absent primary antibody treatment was subtracted as background. Epithelia and tissues were visualized with Imaris imaging software (Bitplane, Zurich, CH).
Quantitation of infection
The post excitement emission signal from GFP was utilized as a reporter for infection. Vectors carrying a GFP expression cassette were diluted in DPBS with calcium and magnesium (Thermo Fisher Scientific) and added to the apical or basolateral surface of human airway epithelia, or added to cell lines at a multiplicity of infection of 104. Vectors on HAE were incubated at 37 °C for 4 h, followed by two washes with PBS. GFP expression was visualized with an IX71 fluorescence microscope (Olympus), and data were calculated as GFP expressing cells per high-power magnification field (GFP + /HPF). Vectors on immortalized cell lines were allowed to infect overnight. The infection of immortalized cells was quantified with a SpecrtraMax i3x microplate reader (Molecular Devices, San Jose, CA, USA). The fluorescence signal from uninfected cells was subtracted as background. When noted in figure legends transduction efficiency was measured as relative fluorescence units (RFU). RFU is GFP signal plotted as a percentage of nuclear Hoechst fluorescence to normalize cell density. Where AAVR antibodies (Abcam 105385) were used to block infection they, or mouse IgG isotype control (Millipore), were diluted in media and added to cells 30 m prior to vector administration.
CRISPR genome editing
AAVR knock-out cells were generated by disrupting AAVR alleles with CRISPR/Cas9 technology [31]. Guide RNAs (gRNA 48: 5′-CCGTCGCTGTCCAGGGAGGC-3′, and gRNA 4690: 5′-CCATGGTTATCTGGGTATTG-3′) targeting exons at either end of NCBI reference sequence NM_024874.4 were created using an online design tool (Benchling, San Francisco, CA, USA). Cas9 protein, ATTO550 conjugated linking RNA (tracrRNA), synthesized gRNAs and buffer were purchased from Integrated DNA Technologies (IDT, Coralville, IA, USA). To increase the likelihood of gene disruption, cells were simultaneously transfected with complexes containing gRNA 48 and gRNA 4690. Using a modified IDT Alt-R protocol, ribonucleoprotein (RNP) complexes were assembled by first mixing tracrRNA and gRNA in equimolar concentrations in nuclease-free duplex buffer and heating to 95 °C for 5 m. RNP complexes were formed by mixing room temperature RNA complexes diluted to working concentration with spCas9 nuclease diluted in OptiMEM (Thermo Fisher Scientific) and incubating on benchtop for 20 m. Each RNP was then incubated with 10% Lipofectamine RNAiMAX (Thermo Fisher Scientific) in room temperature OptiMEM for 20 m to create transfection complexes. Fifty microliters of this solution was then placed one well of a 24-well tissue culture plate (Corning). In all, 300,000 cells suspended in 100 μl of antibiotic free growth medium were then added to the well and incubated at 37 °C in 5% CO2. After 2 days ATTO550-positive transfected cells were isolated by fluorescence-activated cell sorting with an Aria II (Becton Dickinson, Franklin Lakes, NJ, USA) housed at the University of Iowa Flow Cytometry Facility, supported by the National Center for Research Resources of the National Institutes of Health under Award Number 1 S10 OD016199-01A1. Positive cells were serially diluted across a 96-well plate and grown under standard conditions. Colonies derived from single cells were subsequently tested for AAVR protein expression to verify successful genomic editing. We confirmed the results with six individual clones. The clone used in this paper had the best infection by AAV2.5T.
Binding assay
Under standard cell culture conditions, AAVRKO HeLa cells were grown to confluence in a 96-well plate and then incubated at 4 °C for 30 m. Then, gradients of either AAV2-CMV > GFP, AAV5-CMV > GFP or AAV2.5T-CMV > GFP were allowed to bind for 1 h, followed by two washes with cold calcium and magnesium supplemented DPBS. Total DNA was extracted using a Quick-DNA universal 96 kit (Zymo Research, Irvine, CA, USA). Quantitative PCR was performed using Syber Premix Ex Taq II (Takara, Kusatsu, Shiga Prefecture, JP) according to the manufacturer’s instructions. qPCR primers were designed for the eGFP transgene (5′-GGTGAACTTCAAGATCCGCC-3′ and 5′-GGGTGTTCTGCTGGTAGTGG-3′) and compared with known concentrations of plasmid DNA standards to calculate the number of bound viral genomes. Electron microscopy of viral preparations verified that < 10% of the capsids lacked the eGFP transgene.
Statistical analysis
Sample size was selected based on previous work, and determined to be appropriate for a power of 80% or greater using G*Power software (University of Düsseldorf). All data were evaluated using Prism software (GraphPad, La Jolla, CA, USA). Variance was similar between compared groups. Error bars represent ± SEM. Data were normally distributed. Unpaired Student’s t-test, linear regression and ANOVA were performed when appropriate. P ≤ 0.05 was set as the threshold for statistical significance.
Results
AAVR is expressed in human airway epithelia
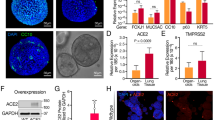
AAV2 has been investigated as a vector for gene therapy in human airway epithelia, but gene transfer was below the level needed for therapeutic efficacy [32, 33]. We have previously shown that AAV2 does not bind to the apical membrane of primary human airway epithelia cultures [34, 35], a finding consistent with preferential infection from the basolateral membrane. We hypothesized that localization of AAVR in human airway epithelia contributes to AAV2 apical inefficiency. Human Protein Atlas RNA sequencing data suggests AAVR is expressed in whole lung tissue at a level similar to other proteins that function as viral receptors. Respiratory syncytial virus receptor CX3CR1 and coxsackievirus and adenovirus receptor CAR transcripts are both present at a frequency similar to AAVR, while the reovirus receptor F11R is more abundant (reads per kilobase of transcript per million mapped reads (RPKM): AAVR 5.392 ± 0.509, CX3CR1 6.402 ± 2.335, CAR 5.243 ± 1.35, F11R 46.265 ± 7.614) [36]. Our RNA sequencing of well differentiated human airway epithelia cultures confirmed relative expression levels for AAVR, CAR, and F11R consistent with whole lung data (accession # GSE106812, referenced by Pezzulo et al. [37]). We detected AAVR message at 31.66 transcripts per million (TPM), CX3CR1 at 0.04164, CAR at 35.93, and F11R at 156.3 TPM (Fig. 1a). The predicted molecular mass of AAVR is 108 kDa, but western blot detected protein at 150 kDa in lysate from HeLa cells and human airway epithelia (Fig. 1b), likely due to glycosidic linkages [26].
The apical membrane of human airway epithelia is devoid of AAVR
We investigated the subcellular localization of AAVR in human airway epithelia and native tissue using immunofluorescent probes. Previous work in HeLa cells suggested an association between AAVR and the Golgi network, and that AAVR localization at the cellular membrane is only apparent when endocytotic recycling is halted [25]. Therefore, we lowered the temperature of human airway epithelia to 4 °C to prevent recycling, and fixed cells prior to staining with anti-AAVR antibodies (AAVRAb) labeled green by secondary antibodies. Nuclei were co-stained with DAPI (blue), the lateral membrane with β-catenin (red), and actin filaments especially apparent in the apical membrane were stained with phalloidin (gray). Human airway epithelia are shown in the X-Z plane (Fig. 2a), and an en face slice tilted at an angle (Fig. 2b). Endogenous AAVR never co-localized with apical markers, but was exclusive to perinuclear and basolateral compartments. We also stained human trachea with AAVRAb (green), and co-stained tight junctions with ZO-1 (white) and nuclei with DAPI (blue). The representative image (Fig. 2c) shows a slice where AAVR is localized below the luminal surface. In summary, while AAVR is expressed in human airway epithelia, we cannot detect AAVR in the apical membrane. Endogenous AAVR was found in a perinuclear compartment and was difficult to detect in the lateral membrane.
AAVR is subapically localized. a Confocal image reconstruction of in vitro human airway epithelia in the X-Z plane: phalloidin (white), AAVR (green), β-catenin (red), and DAPI (blue). b Tilted en face image at three different depths numbered corresponding to: apical membrane (1), lateral membrane (2), and basolateral membrane (3). c Immunocytochemistry of native human tracheal epithelia. AAVR (green), ZO-1 (white), and DAPI (blue). Representative images from >25 different donors. Scale bar 20 μm
AAVR mediates basolateral AAV2 infection of human airway epithelia
AAV2 infection is inefficient on apical human airway epithelia, but basolaterally efficient [38]. Further, AAVRAb has been used to block AAV2 access to its receptor on HeLa cells [25]. We hypothesized that AAVR is basolaterally localized in human airway epithelia, and that basolateral AAVRAb blocks AAV2 infection. We applied either AAVRAb or nonspecific IgG control to the apical or basolateral sides of human airway epithelia, followed by AAV2-CMV > GFP (Fig. 3). Using GFP expression as a proxy for infection, we quantified GFP-positive cells per high-power microscopic field. We observed little or no apical infection of human airway epithelia by AAV2. However, robust basolateral infection was completely blocked by AAVRAb. These data suggest that AAV2 requires AAVR for infection of human airway epithelia, and that AAVR is localized to the basolateral membrane.
AAV2 utilizes AAVR as its basolateral receptor on human airway epithelia. Vector was applied apically or basolaterally along with 50 μg/ml either anti-AAVR antibodies (orange bar) or IgG isotype control (gray bar). Apical infection is inefficient, and does not differ with blocking antibodies or control. Basolateral infection is robust in the presence of IgG control, but completely blocked by antibodies. n = 12 from three different experiments. P < 0.05
AAV2.5T apically infects human airway epithelia independent of AAVR
AAV2 is inefficient on the apical surface of human airway epithelia [34], and requires AAVR [25]. We previously reported the directed evolution of AAV2.5T, a chimeric AAV2 and AAV5 capsid with one point mutation, which apically infects human airway epithelia with orders of magnitude increased efficiency over parental serotypes [5]. We hypothesized that AAV2.5T infects apical human airway epithelia independently of AAVR, and therefore infection would not be affected by excess AAVRAb. We infected basolateral or apical human airway epithelia with AAV2.5T-CMV > GFP after treatment with either AAVRAb or nonspecific IgG control (Fig. 4). There was no significant difference in apical infection when AAVRAb was applied, while basolateral AAVRAb application partially blocked AAV2.5T infection. Thus, AAV2.5T infects apical human airway epithelia independently of AAVR. AAV2.5T can, however, utilize AAVR for basolateral infection. These data suggest a possible unknown receptor, which AAV2.5T utilizes for gene transfer to apical human airway epithelia.
AAV2.5T utilizes an unknown non-AAVR receptor apically on human airway epithelia. Vector was applied apically or basolaterally along with 50 μg/ml either anti-AAVR antibodies (blue bar) or IgG isotype control (gray bar). Apical infection is robust, and does not significantly differ with blocking antibodies or control. Basolateral infection is robust in the presence of IgG control, and only partially blocked with antibodies. n = 12 from three different experiments. P < 0.05
AAV2.5T can infect HeLa independent of AAVR
We compared the relative infectivity of AAV2, AAV5, and AAV2.5T on HeLa and HEK293T cells. The rank order of infection on HEK293T cells was AAV2 > AAV2.5T > AAV5 (Fig. 5a). On HeLa cells, however, AAV2.5T infected best, resulting in AAV2.5T > AAV2 > AAV5 (Fig. 5b). These data show that HeLa cells are more permissive than HEK293T cells to AAV2.5T infection. As anticipated, AAVR affinities calculated from dose response curves were similar for all three serotypes, and Hill coefficients were > 1 consistent with cooperative binding (data not shown). AAVRAb blocked HeLa and HEK293T cell's infection by all three serotypes in a dose-dependent manner. We calculated the concentration of AAVRAb that would reduce the inhibitory response by half (IC50 apparent). The IC50 apparent on Hela cells was: 9.3 for AAV2, 2.6 for AAV5, and 6.3 for AAV2.5T. On HEK293T cells the IC50 apparent was: 11.2 for AAV2, 6.4 for AAV5, and 2.4 for AAV2.5T. We hypothesized that the increased efficiency of AAV2.5T on HeLa might be attributed to a novel receptor. AAV2 infection of HeLa cells is known to be blocked by AAVRAb in a dose-dependent manner [25]. Competition with AAVRAb (50 µg/ml) decreased infection of HEK293T cells by AAV2, AAV5, and AAV2.5T alike, with GFP levels converging under 25 relative fluorescence units (Fig. 5c). On HeLa cells, AAV2 and AAV5 responses to AAVRAb block was analogous to the responses seen on HEK293T cells, but AAV2.5T was less susceptible to AAVRAb block on HeLa cells (Fig. 5d). These data are reminiscent of the incomplete block of AAV2.5T infection seen on basolateral human airway epithelia. They suggest (1) AAV2 and AAV5 utilize AAVR to infect HeLa and HEK293T cells, (2) directed evolution did not disable AAVR utilization in AAV2.5T, and (3) directed evolution conferred transduction via an AAVR independent pathway in human airway epithelia and HeLa cells, but not in HEK293T cells.
AAV2.5T infects HeLa cells better than HEK293T cells. HEK293T cells (a) and HeLa cells (b) were transduced by AAV2, AAV5, or AAV2.5T at MOI 104. Data were quantified as a percentage of GFP over Hoechst fluorescence. Effects of competition with neutralizing antibody (AAVRAb) on AAV2 (orange), AAV5 (white), or AAV2.5T (blue) transduction of HEK293T cells (c) and HeLa cells. Asterisk shows P < 0.05 comparing AAV2.5T to AAV2 at 50 µg/ml AAVRAb. (d). n = 12 from three different experiments. P < 0.05
AAV2.5T infects AAVRKO HeLa cells
In order to verify AAV2.5T utilization of an AAVR independent pathway in HeLa cells, we employed clustered regularly interspaced short palindromic repeat (CRISPR) technology to remove AAVR from the HeLa genome [31]. Western blot of lysate from a mixed population of edited cells confirmed a high-percentage deletion (Fig. 6a). AAVR knockout HeLa cell (AAVRKO) colonies grown from single-cell dilutions were infected with AAV2 and AAV5. AAVRKO were refractory to AAV2 (Fig. 6b) and AAV5 (data not shown). Adenovirus expressing an AAVR transgene driven by the CMV promoter (Ad5-CMV > AAVR) rescued AAV2 and AAV5 infection of AAVRKO (Fig. 6c). In contrast to AAV2 and AAV5, AAV2.5T can infect AAVRKO, albeit at lower levels. This suggests AAV2.5T can utilize both AAVR and an alternative pathway of infection. Moreover, AAVRAb was unable to block AAV2.5T infection of AAVRKO further suggesting an AAVR independent mechanism (Fig. 6d). We then investigated the binding specificity of serotypes on AAVRKO. For AAV2 and AAV5, binding curves showed a linear relationship between the number of viruses applied to AAVRKO and the number of viruses bound, suggesting ubiquitous nonspecific interactions. In contrast, AAV2.5T binding plateaued, saturating available binding partners and suggesting a specific receptor (Fig. 6e). Similar binding experiments on parental HeLa cells showed higher total binding of AAV2.5T than on AAVRKO, however the binding failed to plateau, suggesting two receptors are present but they are not saturated at the highest dose tested (data not shown). The dissociation constant for AAV2.5T on AAVRKO was 5.53 × 104 + /− 4.9 × 103, indicating an interaction >8.5 times stronger than those calculated for AAV2 and AAV5. These data demonstrate the ability of AAV2.5T to bind a non-AAVR receptor with high affinity, and utilize a non-AAVR-mediated pathway for transduction.
AAV2.5T does not require AAVR. a Western blot shows AAVR in control HeLa cells (left lane) and a mixed population of CRISPR AAVR deleted cells with a reduced intensity band (right lane). b AAVRAb significantly decreases AAV2 infection of HeLa cells in a dose-dependent manner (circles), and AAV2 does not infect AAVR knockout HeLa cells (AAVRKO) (orange squares). Data were quantified as a percentage of GFP over Hoechst fluorescence, and normalized to AAV2.5T infection of HeLa cells in the absence of antibodies. c In a dose-dependent manner, adenovirus-mediated AAVR overexpression complements AAVRKO grown from a single cell. d AAV2.5T infection of HeLa cells is partially blocked by AAVRAb (circles), and AAV2.5T infection of AAVRKO HeLa is unaffected by AAVRAb (blue squares). e As more virus is added to AAVRKO HeLa, more bound virus is measured by qPCR. The AAV2.5T binding curve (blue) has a high amplitude and plateaus indicating specific high-affinity binding, unlike curves for AAV5 (white) and AAV2 (orange). n = 12 from three different experiments. P < 0.05
Recombinant AAVR is expressed in the basolateral membrane of human airway epithelia
As reported above, basolaterally applied AAVR antibodies block infection of human airway epithelia by AAV2. This suggests AAVR is localized in the basolateral membrane. Nonetheless, unambiguous basolateral signal fell below the level of detection by immunocytochemistry (Fig. 2). Therefore, we infected human airway epithelia with Ad5-CMV > AAVR, 2 days later cooled the cells to 4 °C, and then stained for overexpressed AAVR. An X-Z confocal reconstruction of human airway epithelia (Fig. 7a) shows basolateral AAVR at adherens junctions (green arrow), abundant AAVR in the cytosol, and no AAVR in the apical membrane (white arrow). We hypothesized these findings might be recapitulated functionally, and infected AAVR overexpressing human airway epithelia either apically or basolaterally with AAV2 or AAV2.5T. We observed increased basolateral, but not apical, infection from both serotypes (Fig. 7b). These results suggest a preferential basolateral localization of overexpressed AAVR.
Overexpressed AAVR prefers the basolateral membrane. a AAVR overexpression establishes AAVR signal at the basolateral (green arrow), and not the apical (white arrow) membrane in a human airway epithelium viewed in the X-Z plane. b AAVR overexpression preferentially increases AAV basolateral infection of human airway epithelia from multiple donors. Scalebar 20 μm. n = 5 for AAV2.5T and n = 6 for AAV2, from three different experiments. P < 0.05
Discussion
AAVR has been described as the putative AAV receptor, and shown to bind directly to AAV2 [25]. In this manuscript, we used anti-AAVR blocking antibodies (AAVRAb) to show that AAV2 and the novel capsid AAV2.5T can utilize AAVR for basolateral infection of primary human airway epithelia. However, AAV2.5T can infect apically and AAVRAb does not block this infection. The dual-transduction-modality of AAV2.5T, along with the exclusively basolateral AAVRAb block, functionally substantiate AAVR localization in the basolateral membrane of human airway epithelia. When we overexpressed AAVR we detected it on the basolateral membrane. We then functionally verified that AAVR localization determines the polarity of infection.
Despite significant progress, our understanding of AAV transduction remains incomplete. Previous studies have described the influence of membrane polarity on AAV transduction in the lung [38, 39], and proposed a role for differential expression of host cell membrane-associated factors [40]. We earlier demonstrated that the lack of apical coxsackievirus and adenovirus receptor (CAR) for adenovirus, and HSPG for AAV2, contribute to the inefficiency of gene transfer in human airways [34, 41]. We add to these explanations by suggesting the basolateral localization of AAVR in human airway epithelia determines the apical inefficiency of AAV2. We speculate that AAVR translocates between the trans-Golgi network and the basolateral membrane of human airway epithelia, where it can be exploited by AAV. While the absence of AAVR in apical human airway epithelia likely leads to the luminal inefficiency of AAV2, we cannot discount additional factors. Second strand genome conversion, impaired endosomal processing, and viral un-coating may also contribute.
AAVR is clearly the receptor for AAV2 in human airway epithelia, however we know little about its cellular function. Like AAVR, the reovirus receptor (F11R) and CAR are localized to the basolateral adherens junction. Along with location, AAVR shares structural IgG-like elements with F11R and CAR. This raises the possibly that AAVR also shares cellular function with these viral receptors, and plays a role in cell–cell adhesion. Interestingly, we observed a western blot band for AAVR which was more diffuse in human airway epithelia than that seen in HeLa cells, perhaps due to various stages of glycosylation. Pillay et al. [26] showed that AAV serotypes interact with specific PKD domains on AAVR, but glycosylation does not affect binding and infection. We speculate that glycosylation may play a role in the non-receptor function of AAVR.
Our data confirm the findings of Carette and colleagues [25] indicating AAVR is an essential receptor for AAV2 and AAV5 in HeLa cells. AAVR can also serve as a receptor for AAV2.5T. We also independently corroborate the conclusions of Vandenberghe et al., who recently reported an AAVR independent pathway of infection for a subset of AAVs on HeLa, A549, and Huh7 cells [42]. The increased transduction efficiency of AAV2.5T on HeLa cells compared to HEK293T cells could be explained by both an AAVR-dependent and independent pathway. However, AAV2 infects HEK293T cells better than AAV2.5T. This suggests that the increased efficiency in HeLa cells is not due to AAVR. Our manuscript adds clinical significance to previous findings by showing that the basolateral localization of AAVR in human airway epithelia contributes to the inefficiency of apical gene transfer. Furthermore, we advance the field by demonstrating that a novel AAV employs a non-AAVR pathway for efficient gene transfer on the apical surface of human airway epithelia.
Cystic fibrosis research provided early evidence that AAV vectors can be safely administered to human lungs [8,9,– 10]. Recently the FDA approved AAV-mediated transfer of retinoid isomerohydrolase (RPE65) to the eye to treat Leber’s congenital amaurosis [43, 44]. There, the target cell population is small and immunoprivileged, and localized low-dose AAV administration is therapeutic. In cystic fibrosis, however, relatively vast airways lined with immunocompetent target cells may require cost-prohibitive repeat administration at high-doses. These obstacles to gene transfer and gene editing can be overcome by employing more efficient vectors. Attempts to increase vector efficiency have addressed stages throughout the viral-cycle, often focusing on the manipulation of cell-surface receptors and the capsid elements they recognize [45,46,47]. Our previous efforts used directed evolution to produce AAV2.5T, a novel vector with tropism for human airway epithelia [5].
Like AAV2 and AAV5 from which it is derived, AAV2.5T can utilize AAVR. However, our previous studies demonstrated AAV2.5T binding to a specific receptor on apical human airway epithelia, unlike AAV2 and AAV5 [48]. Here, we establish that this receptor is not AAVR. Binding assays on AAVR knockout HeLa cells revealed proportionality between the number of capsids applied and the number of bound capsids recovered for AAV2 and AAV5, similar to our reports on apical human airway epithelia [4, 48]. This linearity indicates nonspecific attachment to ubiquitous apical partners. In contrast, AAV2.5T binding curves plateau at a maximum number of bound capsids recovered, even as more capsids are applied. This indicates AAV2.5T has specific apical binding partners that eventually become saturated. Furthermore, AAV2.5T can infect AAVR knockout HeLa cells, whereas AAV2 and AAV5 cannot. These data indicate AAV2.5T can utilize a specific non-AAVR receptor. We speculate that the efficiency of AAV2.5T on apical human airway epithelia can be attributed to the use of a non-AAVR receptor, and that the same receptor is utilized by AAV2.5T in AAVR knockout HeLa cells. We have reported that AAV2.5T and AAV5 have similar binding affinities for 2,3 N-linked sialic acid on human airway epithelia, and that enzymatic removal of sialic acid significantly decreases viral infection [48]. Surprisingly, the amount of sialic acid binding did not correlate with internalization or infection for AAV5. However, binding correlated with internalization and infection for AAV2.5T. These results, combined with the current study, are consistent with a model of infection where sialic acid is a co-receptor facilitating proximity between AAV2.5T and a non-AAVR receptor protein that mediates internalization from the apical side. The present study suggests binding to non-AAVR partners may be a requisite for efficient luminal airway transduction, and provides an important step toward the realization of gene therapy for lung disease.
References
Griesenbach U, Alton EW. Moving forward: cystic fibrosis gene therapy. Hum Mol Genet. 2013;22(R1):R52–58.
Griesenbach U, Alton EW. Progress in gene and cell therapy for cystic fibrosis lung disease. Curr Pharma Design. 2012;18:642–62.
Welsh MJ. Gene transfer for cystic fibrosis. J Clin Invest. 1999;104:1165–6.
Steines B, Dickey DD, Bergen J, Excoffon KJ, Weinstein JR, Li X et al. CFTR gene transfer with AAV improves early cystic fibrosis pig phenotypes. JCI Insight. 2016;1:e88728.
Excoffon KJ, Koerber JT, Dickey DD, Murtha M, Keshavjee S, Kaspar BK et al. Directed evolution of adeno-associated virus to an infectious respiratory virus. Proc Natl Acad Sci USA. 2009;106:3865–70.
Yan Z, Sun X, Feng Z, Li G, Fisher JT, Stewart ZA et al. Optimization of recombinant adeno-associated virus-mediated expression for large transgenes, using a synthetic promoter and tandem array enhancers. Hum Gene Ther. 2015;26:334–46.
Flotte TR, Afione SA, Zeitlin PL. Adeno-associated virus vector gene expression occurs in nondividing cells in the absence of vector DNA integration. Am J Resp Cell Mol Biol. 1994;11:517–21.
Moss RB, Milla C, Colombo J, Accurso F, Zeitlin PL, Clancy JP et al. Repeated aerosolized AAV-CFTR for treatment of cystic fibrosis: a randomized placebo-controlled phase 2B trial. Hum Gene Ther. 2007;18:726–32.
Moss RB, Rodman D, Spencer LT, Aitken ML, Zeitlin PL, Waltz D et al. Repeated adeno-associated virus serotype 2 aerosol-mediated cystic fibrosis transmembrane regulator gene transfer to the lungs of patients with cystic fibrosis: a multicenter, double-blind, placebo-controlled trial. Chest. 2004;125:509–21.
Aitken ML, Moss RB, Waltz DA, Dovey ME, Tonelli MR, McNamara SC et al. A phase I study of aerosolized administration of tgAAVCF to cystic fibrosis subjects with mild lung disease. Hum Gene Ther. 2001;12:1907–16.
Virella-Lowell I, Zusman B, Foust K, Loiler S, Conlon T, Song S et al. Enhancing rAAV vector expression in the lung. J Gene Med. 2005;7:842–50.
Guggino WB, Cebotaru L. Adeno-Associated Virus (AAV) gene therapy for cystic fibrosis: current barriers and recent developments. Exp Opin Biol Ther. 2017;17:1265–73.
Sanders N, Rudolph C, Braeckmans K, De Smedt SC, Demeester J. Extracellular barriers in respiratory gene therapy. Adv Drug Deliv Rev. 2009;61:115–27.
Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–80.
Mizukami H, Young NS, Brown KE. Adeno-associated virus type 2 binds to a 150-kilodalton cell membrane glycoprotein. Virology. 1996;217:124–30.
Kashiwakura Y, Tamayose K, Iwabuchi K, Hirai Y, Shimada T, Matsumoto K et al. Hepatocyte growth factor receptor is a coreceptor for adeno-associated virus type 2 infection. J Virol. 2005;79:609–14.
Qing K, Mah C, Hansen J, Zhou S, Dwarki V, Srivastava A. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat Med. 1999;5:71–7.
Summerford C, Bartlett JS, Samulski RJ. AlphaVbeta5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat Med. 1999;5:78–82.
Summerford C, Samulski RJ. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–45.
Seiler MP, Miller AD, Zabner J, Halbert CL. Adeno-associated virus types 5 and 6 use distinct receptors for cell entry. Hum Gene Ther. 2006;17:10–19.
Wu Z, Miller E, Agbandje-McKenna M, Samulski RJ. α2,3 and α2,6 N-linked sialic acids facilitate efficient binding and transduction by adeno-associated virus types 1 and 6. J Virol. 2006;80:9093–103.
Walters RW, Yi SM, Keshavjee S, Brown KE, Welsh MJ, Chiorini JA et al. Binding of adeno-associated virus type 5 to 2,3-linked sialic acid is required for gene transfer. J Biol Chem. 2001;276:20610–6.
Kaludov N, Brown KE, Walters RW, Zabner J, Chiorini JA. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J Virol. 2001;75:6884–93.
Bell CL, Vandenberghe LH, Bell P, Limberis MP, Gao GP, Van Vliet K et al. The AAV9 receptor and its modification to improve in vivo lung gene transfer in mice. J Clin Invest. 2011;121:2427–35.
Pillay S, Meyer NL, Puschnik AS, Davulcu O, Diep J, Ishikawa Y et al. An essential receptor for adeno-associated virus infection. Nature. 2016;530:108–12.
Pillay S, Zou W, Cheng F, Puschnik AS, Meyer NL, Ganaie SS et al. AAV serotypes have distinctive interactions with domains of the cellular receptor AAVR. J Virol. 2017;91:00391–17.
Poon MW, Tsang WH, Waye MM, Chan SO. Distribution of Kiaa0319-like immunoreactivity in the adult mouse brain--a novel protein encoded by the putative dyslexia susceptibility gene KIAA0319-like. Histol Histopathol. 2011;26:953–63.
Platt MP, Adler WT, Mehlhorn AJ, Johnson GC, Wright KA, Choi RT et al. Embryonic disruption of the candidate dyslexia susceptibility gene homolog Kiaa0319-like results in neuronal migration disorders. Neuroscience. 2013;248:585–93.
Bhella D. The role of cellular adhesion molecules in virus attachment and entry. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140035.
Karp PH, Moninger TO, Weber SP, Nesselhauf TS, Launspach JL, Zabner J et al. An in vitro model of differentiated human airway epithelia. Methods for establishing primary cultures. Methods Mol Biol (Clifton, N.J.). 2002;188:115–37.
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protocol. 2013;8:2281–308.
Flotte TR, Solow R, Owens RA, Afione S, Zeitlin PL, Carter BJ. Gene expression from adeno-associated virus vectors in airway epithelial cells. Am J Resp Cell Mol Biol. 1992;7:349–56.
Griesenbach U, Alton EW. Current status and future directions of gene and cell therapy for cystic fibrosis. BioDrugs. 2011;25:77–88.
Zabner J, Seiler M, Walters R, Kotin RM, Fulgeras W, Davidson BL et al. Adeno-associated virus type 5 (AAV5) but not AAV2 binds to the apical surfaces of airway epithelia and facilitates gene transfer. J Virol. 2000;74:3852–8.
Walters RW, Duan D, Engelhardt JF, Welsh MJ. Incorporation of adeno-associated virus in a calcium phosphate coprecipitate improves gene transfer to airway epithelia in vitro and in vivo. J Virol. 2000;74:535–40.
Uhlén MFL, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å et al. The human protein atlas. Mol Cell Proteomics. 2005;4:1920–32.
Pezzulo AA, Tudas RA, Stewart CG, Buonfiglio LGV, Lindsay BD, Taft PJ et al. HSP90 inhibitor geldanamycin reverts IL-13- and IL-17-induced airway goblet cell metaplasia. J Clin Invest. 2019;129:744-758.
Duan D, Yue Y, Yan Z, McCray PB Jr., Engelhardt JF. Polarity influences the efficiency of recombinant adenoassociated virus infection in differentiated airway epithelia. Hum Gene Ther. 1998;9:2761–76.
Duan D, Yue Y, Yan Z, Yang J, Engelhardt JF. Endosomal processing limits gene transfer to polarized airway epithelia by adeno-associated virus. J Clin Invest. 2000;105:1573–87.
Boyle MP, Enke RA, Reynolds JB, Mogayzel PJ Jr., Guggino WB, Zeitlin PL. Membrane-associated heparan sulfate is not required for rAAV-2 infection of human respiratory epithelia. Virol J. 2006;3:29.
Walters RW, Grunst T, Bergelson JM, Finberg RW, Welsh MJ, Zabner J. Basolateral localization of fiber receptors limits adenovirus infection from the apical surface of airway epithelia. J Biol Chem. 1999;274:10219–26.
Dudek AM, Pillay S, Puschnik AS, Nagamine CM, Cheng F, Qiu J et al. An alternate route for adeno-associated virus entry independent of AAVR. J Virol. 2018;92:1–15.
Lee H, Lotery A. Gene therapy for RPE65-mediated inherited retinal dystrophy completes phase 3. Lancet (London, England). 2017;390:823–4.
Carroll J. FDA experts offer a unanimous endorsement for pioneering gene therapy for blindness. Science Magazine: Science; 13 October, 2017.
Qing K, Bachelot T, Mukherjee P, Wang XS, Peng L, Yoder MC et al. Adeno-associated virus type 2-mediated transfer of ecotropic retrovirus receptor cDNA allows ecotropic retroviral transduction of established and primary human cells. J Virol. 1997;71:5663–7.
Wickham TJ, Roelvink PW, Brough DE, Kovesdi I. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat Biotechnol. 1996;14:1570–3.
Bartlett JS, Kleinschmidt J, Boucher RC, Samulski RJ. Targeted adeno-associated virus vector transduction of nonpermissive cells mediated by a bispecific F(ab’gamma)2 antibody. Nat Biotechnol. 1999;17:181–6.
Dickey DD, Excoffon KJ, Koerber JT, Bergen J, Steines B, Klesney-Tait J et al. Enhanced sialic acid-dependent endocytosis explains the increased efficiency of infection of airway epithelia by a novel adeno-associated virus. J Virol. 2011;85:9023–30.
Acknowledgements
This work was supported by grants from the NIH (H678200-G) and the University of Iowa Center for Gene Therapy (DK054759).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Dr. Zabner is a founder and holds equity in TaleeBio. The other authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hamilton, B.A., Li, X., Pezzulo, A.A. et al. Polarized AAVR expression determines infectivity by AAV gene therapy vectors. Gene Ther 26, 240–249 (2019). https://doi.org/10.1038/s41434-019-0078-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41434-019-0078-3
- Springer Nature Limited
This article is cited by
-
Serotype-specific transduction of canine joint tissue explants and cultured monolayers by self-complementary adeno-associated viral vectors
Gene Therapy (2023)
-
Tissue and cell-type-specific transduction using rAAV vectors in lung diseases
Journal of Molecular Medicine (2021)