Abstract
Aim To determine the accuracy of using saliva and oral cytology swabs in the diagnosis of oral squamous cell carcinoma (OSCC) by detecting aberrantly hypermethylated DNA.
Data sources Electronic databases including PubMed/Medline, Embase, Cochrane Library, Scopus and Web of Science with no language or article restrictions. Additionally, LILACS database, OpenGrey and Google Scholar were searched.
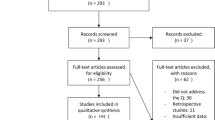
Study selection Studies published since the first report proposing DNA hypermethylation for head and neck carcinomas in 2001 until 2020 were included. The diagnoses of oral cancer were limited to OSCC. Authors screened titles and abstracts for relevance, before further screening of full texts and a consensus for inclusion was reached. Qualitative analysis was conducted on 22 studies, and 11 were selected for meta-analysis.
Data analysis Diagnostic test accuracy meta-analysis was performed using a series of investigations including the Haldane-Anscombe correction, forest plots, receiver operator characteristic (ROC) curves and Deeks' funnel plots. Differences in the pooled estimates of the application of both single and combined hypermethylation markers were assessed using Cochran's Q test and Higgins' I2 test. Random-effects meta-regression analysis was used to evaluate the effects of variation in sensitivity and false-positive rates, and to identify sources of heterogeneity. Sensitivity analyses removed outliers.
Results All studies suggest that DNA hypermethylation can discriminate between OSCC and premalignant conditions or normal mucosa. Using summary receiver operator characteristic (SROC) curves, the sensitivity of combined markers was higher than single markers, and specificity of both combined and single markers was comparable. The biomarkers evaluated had fair-to-excellent sensitivity and good-to-excellent specificity for discriminating OSCC from premalignant and normal mucosa. Four studies included in the review suggest that this method of detection is more applicable to patients who smoke due to increased hypermethylation rates.
Conclusion Hypermethylation markers using saliva and oral swabs are more specific than sensitive for OSCC diagnosis. Combining different genes within the biomarker panel can improve diagnostic test accuracy. However, more blinded evaluation study designs with less bias which replicate real-world application are required to endorse the use of saliva sampling and oral swabs in oral oncology.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.A Commentary on
Adeoye J, Alade A A, Zhu W Y, Wang W, Choi S W, Thomson P.
Efficacy of hypermethylated DNA biomarkers in saliva and oral swabs for oral cancer diagnosis: Systematic review and meta-analysis. Oral Dis 2021; DOI: 10.1111/odi.13773.

GRADE rating
Commentary
Oral cancer is the ninth most common cancer in the UK, and accounts for 2% of all cancers diagnosed.1 Risk factors are linked to social determinants and include smoking or chewing tobacco, drinking alcohol and human papillomavirus (HPV) infection.2 Guidance produced by the National Institute for Health and Care Excellence (NICE) for the recognition and pathways for patients with suspected oral cancer have provided clinicians with a sound criteria for onward referral, including signs and symptoms such as unexplained oral ulceration lasting for more than three weeks, or a persistent and unexplained lump in the neck.3 In patients with suspected oral cancer on the basis of symptoms and clinical presentation, diagnosis is confirmed through biopsy.4 Recent advances in diagnostic tests for oral cancers have included tissue-fluorescence imaging and optical coherence tomography, which have shown promising results.5 However, unfortunately, most oral cancer cases diagnosed in the UK are of late-stage disease, which reduces prognosis.6 Thus, early detection is vital to improve disease prognosis and further research is welcomed in this field.
Adeoye et al. recognised the value of biomarkers present in fluids such as serum, plasma, saliva and urine, in the diagnosis of disease. They considered that oral swabs and saliva are shown to give more promising specimens of DNA, RNA, protein and metabolites that are specific to oral cancer detection. However, it has also been recognised that evidence to suggest efficacy in the early detection of oral cancer using these biomarkers is currently lacking. Therefore, research to determine the diagnostic test accuracy of these biomarkers detected in oral swabs and saliva classifying oral squamous cell carcinoma (OSCC) from normal mucosa and potentially malignant disease was undertaken.
This systematic review and meta-analysis had a focused PICOS question with clearly defined inclusion and exclusion criteria, using only case-controlled or nested-cohort studies. An appropriate range of databases were searched and non-English language studies were included. Three authors separated studies for quantitative and qualitative analysis, and assessed risk of bias using the QUADAS-2 tool. Due to the case-control design of most studies, and the lack of blinding, a high bias concern was concluded by the authors. The results clearly demonstrate that DNA hypermethylation measurement from oral swabs and saliva samples can discriminate between OSCC and premalignant conditions or normal oral mucosa. The biomarkers reported had acceptable sensitivity and specificity rates. Studies included in this systematic review and meta-analysis show high risk of bias, and there is variation between the gene panels evaluated. The authors conclude that results be interpreted with caution due to high bias concerns and differences in study design and diagnostic outcomes.
The findings of this study support further research into saliva samples and oral swabs for early detection of OSCC. This technique has the potential for use as a non-invasive early screening tool which may be of particular benefit in the assessment of special care and medically compromised patients. However, this technique is unlikely to replace oral examination and histopathological assessment for OSCC diagnosis.
References
Oral Health Foundation. State of Mouth Cancer UK Report 2018/2019. 2019. Available at https://www.dentalhealth.org/stateofmouthcancer (accessed February 2022).
NHS. Overview: Mouth Cancer. 2019. Available at https://www.nhs.uk/conditions/mouth-cancer/ (accessed February 2022).
NICE. Suspected cancer: recognition and referral. 2015. Available at https://www.nice.org.uk/guidance/ng12/evidence/full-guideline-pdf-2676000277 (accessed February 2022).
NHS. Diagnosis: Mouth cancer. 2019. Available at https://www.nhs.uk/conditions/mouth-cancer/diagnosis/ (accessed February 2022).
Chen P-H, Lee H-Y, Chen Y-F et al. Detection of Oral Dysplastic and Early Cancerous Lesions by Polarization-Sensitive Optical Coherence Tomography. Cancers (Basel) 2020; 12: 2376.
Public Health England. Oral cancer in England: A report on incidence, survival and mortality rates of oral cancer in England, 2012 to 2016. London: Public Health England, 2020.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gaw, G., Gribben, M. Can we detect biomarkers of oral squamous cell carcinoma from saliva or mouth swabs?. Evid Based Dent 23, 32–33 (2022). https://doi.org/10.1038/s41432-022-0248-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41432-022-0248-9
- Springer Nature Limited