Abstract
In the search for new antibiotic compounds, fractionation of Pseudomonas protegens UP46 culture extracts afforded several known Pseudomonas compounds, including 2,4-diacetylphloroglucinol (DAPG), as well as two new antibacterial alkaloids, 6-(pyrrolidin-2-yl)DAPG (1) and 6-(piperidin-2-yl)DAPG (2). The structures of 1 and 2 were determined by nuclear magnetic resonance spectroscopy and mass spectrometry. Compounds 1 and 2 were found to have antibacterial activity against the Gram-positive bacteria Staphylococcus aureus and Bacillus cereus, with minimal inhibitory concentration (MIC) 2 and 4 μg ml−1, respectively, for 1, and 2 μg ml−1 for both pathogens for 2. The MICs for 1 and 2, against all tested Gram-negative bacteria, were >32 μg ml−1. The half maximal inhibitory concentrations against HepG2 cells for compounds 1 and 2 were 11 and 18 μg ml−1, respectively, which suggested 1 and 2 be too toxic for further evaluation as possible new antibacterial drugs. Stable isotope labelling experiments showed the pyrrolidinyl group of 1 to originate from ornithine and the piperidinyl group of 2 to originate from lysine. The P. protegens acetyl transferase (PpATase) is involved in the biosynthesis of monoacetylphloroglucinol and DAPG. No optical rotation was detected for 1 or 2, and a possible reason for this was investigated by studying if the PpATase may catalyse a stereo-non-specific introduction of the pyrrolidinyl/piperidinyl group in 1 and 2, but unless the PpATase can be subjected to major conformational changes, the enzyme cannot be involved in this reaction. The PpATase is, however, likely to catalyse the formation of 2,4,6-triacetylphloroglucinol from DAPG.
Similar content being viewed by others
Introduction
The spread of multiresistant bacterial pathogens is a major threat to global public health. Consequently, there is a huge need for new antibiotic drugs, especially drugs with novel modes of action [1]. A great majority of the antibiotics used today and in the past were developed from secondary metabolites produced by bacteria or fungi. Many microorganisms produce antimicrobial compounds to gain advantages over competing organisms, and the great diversity of bacteria and fungi have during evolution developed an impressive array of bioactive compounds to increase their chances to thrive in environments with intense microbial competition.
Among bacteria, the genus Pseudomonas is well known to produce a broad range of antimicrobial compounds [e.g 2, 3], for example, antifungal metabolites such as pyoluteorin, pyrrolnitrins, phenazines, rhizoxins, and 2,4-diacetylphloroglucinol (DAPG), which all have been implicated in the biocontrol of various fungal phytopathogens [4,5,6,7,8,9]. These compounds have also been described to have antibacterial activities as well, for example the polyketide DAPG, which is active against methicillin-resistant and vancomycin-resistant Staphylococcus aureus (MRSA and VRSA) as well as against certain vancomycin-resistant Enterococcus spp. (VRE [10]). DAPG is mainly produced by isolates of the Pseudomonas fluorescens complex [11,12,13,14], and the most effective DAPG and pyoluteorin producing isolates of P. fluorescens are now classified as the species P. protegens [15].
In a project aimed at finding new antibiotic drug candidates, we are currently investigating bacterial and fungal strains for the production of antibiotic compounds. In the present paper, we describe the isolation and characterization of two antibacterial pyrrolidinyl and piperidinyl substituted DAPGs, 6-(pyrrolidin-2-yl)DAPG (1) and 6-(piperidin-2-yl)DAPG (2), from P. protegens UP46. These compounds are the first examples of alkaloids containing a DAPG moiety, and we propose a biosynthetic route for the formation of 1 and 2 based on stable isotope labelling experiments.
Materials and methods
General experimental procedures
UV-spectra were recorded on a Hitachi U-2001 spectrophotometer in MeOH at room temperature. 1H and 13C NMR data were acquired at 303 K in MeOH-d4 or acetone-d6 on a Bruker Avance III 600 MHz NMR spectrometer (Bruker Biospin GmBH, Rheinstetten, Germany) equipped with a 5-mm cryo-probe (1H, 13C, 15N, 31P). Standard pulse sequences supplied by Bruker were used for determination of 1H and 13C chemical shifts and connectivities. For structure elucidation, 1D 1H, 1D 13C, COSY, DEPT-HSQC and HMBC were applied. Chemical shifts were determined relative to signals from MeOH-d4 (δC 49.15) or residual MeOH-d3 (δH 3.31), and acetone-d6 (δC 29.92) or residual acetone-d5 (δH 2.05). HPLC-MS was performed on an Agilent 1100 HPLC (Agilent, Palo Alto, CA, USA) connected to a maXis Impact ESI-QTOF MS (Bruker Daltonic GmbH., Bremen, Germany), and UHPLC-MS was performed on an Agilent 1290 Infinity II UHPLC connected to the same mass spectrometer. Preparative HPLC was performed on a Gilson 306/306 pump system (Gilson Inc., Middleton, WI, USA) with a Gilson 119 UV/VIS detector monitoring at 210 nm. MeCN of HPLC gradient grade (Sigma-Aldrich, St. Louis, MO, USA) and deionized filtered water (Millipore, Billerica, MA, USA) were used for mobile phases.
Strain origin, identity and maintenance
The isolate P. protegens strain UP46 was isolated from a soil sample collected near the farm Olsbo, Runhällen, located around 65 km west of Uppsala, Sweden. The strain was isolated on Nutrient Broth-Casamino Acids agar [NBCA; 1 g Nutrient Broth (BD, Difco Ltd), 1 g Casamino Acids Broth (BD, Difco Ltd), 14 g Bacto Agar (Saveen & Werner) in 1 l deionized water] supplemented with 1% soil extract (SE), two fungicides (50 mg cycloheximide; 10 mg nystatin) and three antibiotics (20 mg chloramphenicol; 20 mg erythromycin and 0.5 mg rugulosin in 1 l NBCA agar). The SE was made by shaking 100 g of the soil sample with 200 ml of sterile water in a 500-ml Erlenmeyer flask (130 rpm and 20 °C, 2 h) followed by sterile filtration of the liquid phase 0.2 µm filter (SartoriousTM). Fungicides, antibiotics and SE were added to NBCA agar precooled to 50 °C. The strain UP46 was further purified on Vegetable Peptone Broth Agar [VPA, 10 g Vegetable Peptone Broth (VPB, Oxoid Ltd), 15 g Bacto Agar (BD Difco Ltd) in 1 l deionized water] and identified as belonging to the genus Pseudomonas and the group fluorescent pseudomonads by sequencing of the 16 S rRNA (GenBank accession number MN994375). According to sequencing of the full gene of 16 S rRNA the isolate was more than 99.9% identical to the strains of P. protegens Pf-5 and CHA0. The isolate was maintained as deep-frozen (−70 °C) stock culture. When appropriate, cultures were transferred to VPA plates.
Isolates of human pathogens Escherichia coli LMG15862, Acinetobacter baumannii LMG1041T, Enterobacter cloacae LMG2783T, Klebsiella pneumoniae LMG20218, Pseudomonas aeruginosa LMG6395, S. aureus LMG15975 were purchased from Belgian Co-ordinated Collections of Micro-organisms (Gent, Belgium) and Bacillus cereus CCUG7414 originates from Culture Collection of University of Gothenburg (Sweden). The origin and cell/spore production of the strains of the yeast Candida albicans (Robin) Brekhout and the fungus Aspergillus fumigatus Fres., were as previously described [16]. All strains were maintained as advised by the Culture Collections and cells/spores were stored as deep-frozen (−70 °C) stock cultures.
Culture conditions and metabolite sampling
UP46 cultures (150 ml in 500-ml Erlenmeyer flasks) were grown in two substrates: (A) Half-strength VPB [15 g of VPB (Oxoid Ltd.) in 1000 ml of deionized H2O] and B MM-TCA, prepared by mixing 800 ml modified mineral medium (MM) for Pseudomonas [17, 18], 100 ml of tryptone broth [TB; 10 g of tryptone (BD Difco Ltd.), 1 g of l-tryptophan, 5 g of NaCl, 1 l of deionized water] and 100 ml Casamino Acids-dextrose broth [CADB; 10 g of Casamino Acids (BD Difco Ltd.), 2 g of dextrose, 1.25 g of K2HPO4, 1 g of yeast extract (BD Difco Ltd.), 1 l of deionized water]. The MM, TB, and CADB media were sterilized separately prior to mixing, and the resulting MM-TCA medium was supplemented with 2% glycerol. The 150 ml VPB cultures were started either by transferring 100 μl of deep-frozen strain stock per 150 ml of VPB or by transferring a loop (10 μl) of 24−48 h old cell material grown on VPA plates. The MM-TCA cultures were started by transferring 5 ml of 24 h old VPB cultures per 150 ml of MM-TCA. Cultures were incubated on a rotary shaker (120-130 rpm) for 4−7 days at 20 °C in the dark. Extracellular metabolites were collected using sterile nylon bags with a polymeric resin, Amberlite XAD16 (Sigma-Aldrich; one bag with ~8 g per bag for 150 ml cultures), as previously described [19]. The resulting extracts were subjected to HPLC-MS (below), and then dried for preparative HPLC as described below.
For upscaling the production of compounds 1 and 2, the strain UP46 was cultured in Yeast Extract- Malt Extract Medium (ISP2 [20]) supplemented with L-proline (1 g l−1). Ten 150-ml cultures in 500-ml Erlenmeyer flasks were incubated for 4-7 days on a rotary shaker at 20 °C in the dark. Extracellular metabolites were collected using adsorbents, as above.
For labelling with 13C labelled substrates, 10-ml UP46 ISP2 cultures in 100-ml Erlenmeyer flasks were started by inoculating a loop of UP46 cells growing on VPA plates. After incubation overnight at 20 °C in the dark, 10 ml of ISP2 medium containing 20 mg of 13C5 ornithine (CIL, Cambridge Isotope Laboratories Inc., MA, USA) or 13C6 lysine (CIL) was added to the cultures. Each culture was immediately provided one sterile nylon bag with ca 2 g Sepabeads® SP850 adsorbent (Mitsubishi Chemical Co., Tokyo, Japan). The cultures were incubated on a rotary shaker (130 rpm) for 3 days at 20 °C in the dark. At the harvest, the adsorbent bags were washed with deionized H2O to remove attached bacterial cells and then extracted with 2 ml of MeOH prior to UHPLC-MS analysis (as below).
Analysis by HPLC-MS and UHPLC-MS
VPB and MM-TCA culture extracts (above), as well as HPLC fractions (below) were analyzed by HPLC-MS on a reversed phase HPLC column (3.0 × 50 mm, 2.6 μm, Accucore RP-MS, Thermo Scientific, Waltham, MA, USA), using a gradient of MeCN in water (10–95% MeCN in 3 min, 95% MeCN for 4 min, at 0.8 ml min−1, with 0.2% formic acid). Culture extracts from 13C-labelling experiments were analyzed by UHPLC-MS on a reversed phase UHPLC column (2.1 × 50 mm, 1.5 μm, Thermo Accucore Vanquish RP-MS, Thermo Scientific), using a gradient of MeCN in water (10–95% MeCN in 3 min, 95% MeCN for 1.2 min, at 0.9 ml min−1, with 0.2% formic acid). For both HPLC-MS and UHPLC-MS, the mass spectrometer was operated in positive mode with scanning of m/z 50–1500, and calibration of the mass spectra was obtained by using sodium formate clusters.
Isolation of compounds 1 and 2
VPB and MM-TCA culture extracts were dried under nitrogen and re-dissolved in 1.5 ml 50% MeCN, and fractionated by gradient reversed phase preparative HPLC (21.2 × 100 mm, 5 μm, Hypersil Gold, Thermo Scientific, Waltham, MA, USA) by applying a linear gradient aqueous 10-95% MeCN in 10 min, followed by a 10 min hold at aqueous 95% MeCN (flow 10 ml min−1). Collected fractions were assayed for antibacterial activity and were analysed by HPLC-MS as described above.
To obtain more of compounds 1 and 2, ten liquid cultures of P. protegens UP46 (each 150 ml) were extracted using adsorbents (Amberlite XAD 16) as above. The combined and dried extracts were re-dissolved in 9 ml 50% MeCN, centrifuged and subsequently fractionated by gradient preparative HPLC (8 × 1 ml injected, same column and conditions as above). Fractions containing a mixture of compounds 1 and 2 (fractions 23–26) as indicated by HPLC-MS (as above), were pooled and dried under reduced pressure. The dry sample was dissolved in 1 ml aqueous 25% MeCN before further fractionation using reversed phase preparative HPLC (column and flow as above) using aqueous 25% MeCN. Following analysis by HPLC-MS (as above), fractions containing compounds 1 and 2 (fractions 51–56 and 58–63, respectively) were pooled and dried, and the purification was repeated once on the same column and conditions, to finally yield 1.42 mg of compound 1 and 0.75 mg of compound 2.
6-(pyrrolidin-2-yl)DAPG (1). Compound 1 was obtained as white amorphous solid. UV λmax (MeOH) nm (log ε): 287 (4.55); NMR-data, see Table 1; HRMS: m/z 280.1178 [M + H]+ (calcd. for C14H18NO5, 280.1179).
6-(piperidin-2-yl)DAPG (2). Compound 2 was obtained as white amorphous solid. UV λmax (MeOH) nm (log ε): 287 (4.53); NMR-data, see Table 1; HRMS: m/z 294.1331 [M + H]+ (calcd. for C15H20NO5, 294.1336).
In vitro bioassay
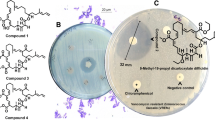
Chromatographic fractions were assayed for antimicrobial activity using an earlier developed protocol (“Microtiter plate assay 2”) [21], with the following organisms: E. coli LMG15862, A. baumannii LMG1041T, E. cloacae LMG2783T, K. pneumoniae LMG20218, P. aeruginosa LMG6395, S. aureus LMG15975, C. albicans and A. fumigatus. Further steps of purification of compounds 1 and 2, were done with S. aureus LMG15975 as activity indicator.
MIC determination
Minimal inhibitory concentration (MIC) was measured by means of a broth micro-dilution method in 96-wells microtiter plates as previously described [19], using E. coli LMG15862, A. baumannii LMG1041T, E. cloacae LMG2783T, K. pneumoniae LMG20218, P. aeruginosa LMG6395, S. aureus LMG15975, C. albicans and A. fumigatus, as test organisms. The MIC was defined as the lowest concentration of each compound with no visible growth of pathogen. All MIC tests were performed in triplicate and repeated at least twice.
In vitro cytotoxicity determination
In vitro cytotoxicity, half maximal inhibitory concentration (IC50), was determined by a fluorometric microculture cytotoxicity assay, based on hydrolysis of fluorescein diacetate to fluorescein by HepG2 cells with intact plasma membranes, as previously described [22].
Studies of PpATase crystal structures
The programs Coot LIDIA 0.8.9 Ligand Sketcher and AceDRG (version: 212) [23,24,25] of the CCP4 Program Suite (version 7.0.077; [26]) were used to generate atom coordinate and cif dictionary files for the ligands piperidine and 1-piperideine, and the R- and S-enantiomer, respectively, of compounds 1 and 2. Atom coordinate and structure factor files were retrieved from the Protein Data Bank for protein crystal structures of P. protegens PpATase, without ligand (PDB code 5M3K) and soaked with MAPG (MAPG; PDB code 5MG5) [27]. Coot 0.8.9.2 [23, 24] was used for examination of the structures. The PhlC subunits from both structures (2 in 5M3K; 8 in 5MG5) were superposed with each other.
The piperidine and 1-piperideine molecules were manually placed in the active site, between Cys 88 and the PG ligand in the crystal structures. The ligand ring nitrogen was positioned at the oxyanion hole, i.e., at the position of the oxygen of the acetyl moiety bound to Cys 88. The molecules were then moved around manually to try and find position(s) where they might fit. The product molecules were placed in the active site, and their aromatic rings were aligned flat with that of the PG ligand, with the piperidine moiety near the oxyanion hole. The piperidine moiety was rotated and flipped from chair to boat/skew conformation to see if it might fit. Thereafter the molecules were rotated within the plane of the ring to try and find orientation(s) where they might fit. Similarly, compounds 1 and 2 were placed in the active site of superimposed PhlC subunits, aligned with the PG ring in the ligand complexes, to test if they might fit. The active site of the PhlC subunits was also examined in MacPyMol (PyMOL v1.8.6.0 Enhanced for Mac OS X, Schrödinger LLC), in surface representation to identify cavities adjacent to the PG ring that could potentially accommodate acetyl (or other) substituents on the ring.
Results and discussion
Isolation of compounds
Analysis of P. protegens UP46 culture extracts, using HPLC-MS, followed by comparison of MS-data with databases, indicated the presence of several well-known Pseudomonas produced compounds, including DAPG (Fig. 1), pyrrolnitrin and pyoluteorin. The data analysis further suggested the presence of two possibly unknown substances with m/z 280.1178 (1) and 294.1331 (2). Gradient HPLC fractionation of the culture extracts, followed by bioassays against a panel of bacteria and fungi, gave a number of fractions with antibacterial and/or antifungal activities. Subsequently, HPLC-MS analysis showed the presence of DAPG, pyrrolnitrin and pyoluterin in active fractions, but also that compounds 1 and 2 have activity against S. aureus and A. baumannii. The presence of DAPG in active fractions were subsequently verified by NMR analysis followed by comparison with literature data [28]. Following culture upscaling, the tentative new compounds 1 and 2 were isolated by a combination of gradient and isocratic preparative HPLC, which yielded 1.42 mg of 1 and 0.75 mg of 2, and the structures were characterized by NMR and MS, as described below.
Structure analysis
Based on MS-data, the molecular formula C14H17NO5 was proposed for compound 1 and C15H19NO5 for compound 2 (unsaturation index seven for both), i.e., the compounds differed by a -CH2- group. When analysed by one-dimensional 1H NMR spectroscopy, compound 1 and 2 both had two methyl signals at ca δH 2.6 (Table 1) and in HMBC experiments (Fig. 2), these methyl signals displayed correlations to sp2 carbons at δC ca 203-206 and δC ca 101–109, suggesting two aromatic acetyl groups in each compound just as in DAPG. This proposed both compounds to be DAPG derivatives, and since the compounds lacked the aromatic H-6 signal of DAPG, this suggested 1 and 2 to be C-6 substituted DAPGs. Analysis of COSY and HSQC-data, showed the presence of a CH-CH2-CH2-CH2 spin-system in 1 and a CH-CH2-CH2-CH2-CH2 spin-system in 2 (Table 1). For both 1 and 2, the CH proton had an HMBC correlation to the methylene carbon at the other end of the spin-system (Fig. 2), suggesting these spin-systems to be part of ring-closed structure elements of the compounds. According to the 13C chemical shifts for the terminal methylene groups (δC ca 46), the rings should be closed via a nitrogen atom, in agreement with a pyrrolidin-2-yl group in 1 and a piperidin-2-yl group in 2. In both 1 and 2, the respective CH proton further had HMBC cross-peaks to one carbon atom at δC ca 101-104, and to two carbon atoms at ca δC 168 and ca δC 181. This suggested linkage of the putative pyrrolidinyl/piperidinyl group to a carbon atom in an aromatic ring, situated between two oxygen substituted carbons, supporting the hypothesis that 1 and 2 indeed are 6-substituted DAPG derivatives. After adding the two acetyl groups to the structures of the proposed DAPG derivatives, the unsaturation index seven was fulfilled for both 1 and 2, and after adding hydroxyl functions to the open positions (C-1, C-3, and C-5), the proposed structures for 1 and 2 (Fig. 1) were in accord with their respective molecular formula. When compound 2 was analysed by NMR in acetone-d6, a sharp OH-signal appeared at δH 15.34, along with two broad OH-signals at δH 11.49 and δH 7.14, suggesting one of the three hydroxyl functions (δH 15.34) to be strongly hydrogen bonded to a neighbouring acetyl group. The sharp OH-signal shared correlations in HMBC with the piperidinyl CH and with one of the acetyl methyl groups (Fig. 2), which positioned the hydrogen bonded OH between these two groups. The presence of this hydrogen bond in the structure, also explained why C-1/C-5, C-2/C-4, the C-2/C-4 acetyl groups, and the C-1/C-5 hydroxyl groups were not equivalent, but gave individual signals in NMR. Compound 1 was thus concluded to be 6-(pyrrolidin-2-yl)DAPG [or 1,1’-(2,4,6-trihydroxy-5-(pyrrolidin-2-yl)-1,3-phenylene)bis(ethan-1-one)] and compound 2 to be 6-(piperidin-2-yl)DAPG [or 1,1’-(2,4,6-trihydroxy-5-(piperidin-2-yl)-1,3-phenylene)bis(ethan-1-one)].
Neither compound 1 nor compound 2 gave measurable optical rotation. Either the specific rotations were too small to measure, or the compounds were subjected to racemization during sample workup/isolation or by bacterial enzymes in the cultures, or when biosynthesised. When analysed by NMR in MeOH-d4, the H-1”’ signals of 1 and 2 were not decreasing in intensity with time, as would be expected if these hydrogens were easily exchanged. Furthermore, there are many alkaloids with a piperidinyl or pyrrolidinyl group linked to an aromatic ring, without reported problems with racemization during workup/isolation, leaving this possibility less probable. Possible racemization during biosynthesis is further discussed below.
There are some DAPG structural analogues isolated and described from natural sources, including phloroglucinol (PG, benzene-1,3,5-triol), 2-acetylphloroglucinol (monoacetylphloroglucinol, MAPG), and 2,4,6-triacetylphloroglucinol (TAPG), produced by e.g., P. fluorescens [11], and a methylene bridged C-6 dimer of DAPG in P. aurantiaca [29]. Other examples include prenylated MAPGs and DAPG from different plants of the Rutaceae or Hypericaceae [30,31,32,33], and a PG carrying one 3-(3,4-dihydroxyphenyl)propanoyl substituent and one carboxycarbonyl substituent, found in the plant Balanophora involucrata [34], but compounds 1 and 2 are the first examples of alkaloids containing a DAPG moiety. In addition to these natural DAPG analogues, recent synthetic efforts have resulted in a few bis-pyrrolidine functionalized PG analogues [35].
Biological activity of compounds 1 and 2
During bioassay-guided fractionation, compounds 1 and 2 showed activity against S. aureus and A. baumannii, but when the MIC values towards these pathogenic bacteria were determined on isolated compounds, the activity was much better towards S. aureus. The MIC against S. aureus was 2 μg ml−1 for both 1 and 2, whereas the MIC against A. baumannii was >32 μg ml−1. The MIC against B. cereus was 4 and 2 μg ml−1, respectively, for 1 and 2. DAPG was also tested against S. aureus and B. cereus, and it was slightly more potent than 1 and 2 against both S. aureus and B. cereus (MIC 0.5 and 1 μg ml−1, respectively). These MICs are similar to previously reported values for DAPG, e.g., MIC 0.25 to 1 μg ml−1 against ten clinical MRSA isolates [36] and MIC 5-6 μg ml−1 against B. subtilis [8, 37]. Compounds 1, 2 and DAPG, were found to have MIC > 32 μg ml−1 against all tested Gram-negative bacteria, including E. coli, K. pneumoniae and P. aeruginosa. Previously, the MICs for DAPG against these three Gram-negative bacteria were reported to be in the range 12.5 to 25 μg ml−l [37], and these differences could be due to the MIC determination protocols used or due to differences between the tested bacterial strains. The IC50 of the compounds against the human liver cell line HepG2 was determined to be 11, 18 and 7 μg ml−1, for compounds 1, 2 and DAPG, respectively. Thus, regarding the clinically important pathogen S. aureus, the MIC/IC50 ratio for compound 1 and 2 was ~6 and 9, respectively, which suggested the compounds to be too toxic for further evaluation for development into new antibiotic drugs.
DAPG has earlier been indicated to be involved in the biocontrol of various fungal phytopathogens, as shown by decreased biocontrol potency of DAPG knock-out strains [8] and higher frequency of DAPG producing fluorescent pseudomonads in disease suppressive soils compared with conducive soils [9]. The MICs for compounds 1 and 2, and DAPG against two fungi, A. fumigatus and C. albicans, were higher than the highest tested concentration, >32 μg ml−1. This is in line with the reported in vitro MICs for DAPG against a selection of fungal pathogens (64–128 μg ml−1 [8]).
Biosynthetic considerations
It is well-known that most alkaloids containing pyrrolidine or piperidine rings are biosynthesised starting from ornithine or lysine, respectively. To test this, cultures of P. protegens were fed with 13C5-ornithine and 13C6-lysine, respectively, and culture extracts were analysed by UHPLC-MS. Compound 1 from cultures with 13C5-ornithine feeding showed a +4 increase in m/z, whereas compound 2 from cultures with 13C6-lysine feeding showed a +5 increase in m/z. This clearly demonstrated that four carbons from ornithine were incorporated into 1, and that five carbon atoms from lysine were incorporated into 2. This is in line with the expected biosynthesis of pyrrolidine/piperidine alkaloids, including decarboxylation of ornithine/lysine and transamination to the respective aminoaldehyde and formation of cyclic imines (1-pyrroline/1-piperideine), which, after protonation, are good substrates in Mannich-like reactions with an activated aromatic ring such as DAPG, leading to 1 and 2 (Fig. 3).
The biosynthesis of DAPG has been extensively studied during the last twenty years [27, 38,39,40,41], and the phl biosynthetic gene cluster has been shown to be responsible for synthesis of DAPG, via the synthesis of PG and subsequent acetylation to obtain DAPG [39]. This gene cluster was found to be present in all investigated DAPG producing Pseudomonas strains, in a study using bacterial strains collected world-wide [12]. Acetylation of PG to MAPG and DAPG requires the presence of phlA-C [38] and the functional acetyltransferase (ATase) is a multicomponent enzyme composed of PhlA, PhlB, and PhlC [40], which catalyzes the disproportionation of two MAPG molecules to one PG and one DAPG molecule [40]. Recently, Pavkov-Keller et al. [27] described the crystal structure of the functional multicomponent P. protegens ATase (PpATase), and they showed that the PhlC unit is the protein catalysing the actual acetylation step. The acetyl group from one MAPG was proposed to be relocated to the sulfhydryl group of a cysteine residue of PhlC, and then transferred to another MAPG in a Friedel-Craft type of acylation [27] (Fig. 4, top). This multicomponent PpATase has been shown to accept several different acetyl donors, but also different acetyl acceptors, and in addition also to catalyse N-acetylation or O-acetylation instead of C-acetylation of some substrates [42, 43]. P. protegens UP46, used in the current study, is 99% identical to P. protegens Pf-5 (16 S rRNA), which has phlA-H in the genome [44], and thus, UP46 is highly likely to have phlA-H in the genome.
Top: Acetylation of MAPG to DAPG [27], or acetylation of DAPG to TAPG, by a Pseudomonas protegens acetyltransferase (PpATase). Bottom: Hypothetic biosynthesis of 6-(pyrrolidin-2-yl)DAPG (1, n = 1) and 6-(piperidin-2-yl)DAPG (2, n = 2), from 1-pyrroline/1-piperideine and DAPG, by PpATase. Depicted is initial attack from the deprotonated sulfhydryl group onto the imine from below, but the attack can also be from above, from an intact sulfhydryl group, and the imine may also be protonated. In addition, to enable the bottom reaction, major conformational changes of the PpATase is required
As mentioned above, no optical rotation was measured for compounds 1 and 2, and one possible explanation is that 1-piperideine/1-pyrroline is enzymatically added to PG/MAPG/DAPG in a non-stereo specific reaction. Such a reaction could hypothetically be catalyzed by the promiscuous PpATase, if the central acetyl delivering cysteine of PpATase may accept a 1-piperideine/1-pyrroline instead of an acetyl group, for subsequent transfer to PG/MAPG/DAPG in a nucleophilic substitution reaction (SN2 type; Fig. 4, bottom). However, studies of the crystal structures of the PpATase [27] indicated that the cavity for acetyl transfer to/from Cys 88 appears to be too small to accommodate a 1-piperideine molecule without clashing into surrounding residues, Gly385, Phe148, Tyr298 and/or His144. In addition, the R- and the S-enantiomers of compounds 1 and 2 were placed at the active site of the superposed PhlC subunits, aligned with the PG ring in the ligand complexes, and were rotated and moved around at the active site to try and find any orientation where they might fit. However, no such position could be found. In particular, the cavity that would hold the pyrrolidinyl/piperidinyl group if added by the enzyme (between Cys88 and Gly385), is well defined by the folded structure, without obvious signs of flexibility. The outer carbon atoms of the pyrrolidinyl/piperidinyl group clash with the protein at Gly385 and will be in the way for interaction between Gly385 and the ring nitrogen. Thus, without substantial conformational changes of the PpATase, the enzyme is not likely to catalyse this reaction, and this is thus not the reason for the absence of measurable optical rotation for 1 and 2.
In the current study, DAPG was found in supernatants of P. protegens, but also small amounts of MAPG as well as traces of TAPG were detected, as based on HRMS data from UHPLC analysis. TAPG has previously been isolated from P. fluorescens [11], but its biosynthesis has not been described. As suggested by the promiscuity of PpATase, we investigated, using the crystal structures of PpATase [27], if PpATase also may accept DAPG as an acetyl acceptor to produce TAPG (Fig. 4, top). Using this static model, there is not enough space to accommodate both acetyl groups of DAPG to allow transfer of a cysteine bound acetyl group. However, one acetyl group should fit in a cavity lined by the side chain of Trp 211, which seems to act as a lid covering the active site and has been observed in closed and open conformation [27]. The other acetyl group may fit in another cavity after conformational changes of the protein, involving a short alfa-helix between two surface loops. These conformational adaptations of PpATase are likely to allow the binding of DAPG to the active site, for subsequent acetylation, to produce TAPG. In addition, we tested if the PpATase may acetylate piperidyl-PG/MAPG to piperidyl-MAPG/2. As mentioned above, assuming conformational changes of a short alfa-helix between two surface loops and flexibility of the Trp211 side chain, the enzyme should be able to accommodate a DAPG for acetylation to TAPG. The cavity, which is lined by the flexible side chain of Trp211, may also be able to hold a piperidyl group, which would enable the acetylation of piperidyl-PG/MAPG to piperidyl-MAPG/2.
Conclusion and outlook
The new antibacterial alkaloids 6-(pyrrolidin-2-yl)DAPG (1) and 6-(piperidin-2-yl)DAPG (2), were isolated and characterized from P. protegens UP46, and these compounds are the first examples of alkaloids containing a DAPG structure element. Stable isotope labelling experiments showed the pyrrolidinyl/piperidinyl group of these compounds to originate from ornithine and lysine, respectively, as expected for these types of alkaloids. No optical rotation was observed for 1 and 2. The published crystal structure of the enzyme PpATase was studied to investigate if this enzyme could introduce the pyrrolidinyl/piperidinyl group of 1 and 2 in a non-stereospecific fashion. The results indicated, however, that this is not the case, unless the PpATase can undergo substantial conformational changes. Thus, the pyrrolidinyl/piperidinyl groups of 1 and 2 are likely to be transferred to the aromatic core of the compounds by another enzyme, and future studies may show if this process may be non-stereospecific or not. On the other hand, the studies showed that the PpATase, which has been shown to biosynthesise DAPG, also may be involved in the biosynthesis of TAPG.
Compounds 1 and 2 were both found to have good MICs against S. aureus and B. cereus, but their IC50 against human cells were slightly too high to merit further studies as putative future antibiotic drugs. It is however possible, that future chemical optimization of the compounds may result in compounds with retained or improved antibacterial properties along with decreased toxicity.
References
Theuretzbacher U, Gottwalt S, Beyer P, Butler M, Czaplewski L, Lienhardt C, et al. Analysis of the clinical antibacterial and antituberculosis pipeline. Lancet Infect Dis. 2019;19:E40–E50.
Walsh UF, Morrissey JP, O’Gara F. Pseudomonas for biocontrol of phytopathogens: from functional genomics to commercial exploitation. Curr Opin Biotechnol. 2001;12:289–95.
Gross H, Loper JE. Genomics of secondary metabolite production by Pseudomonas spp. Nat Prod Rep. 2009;26:1408–46.
Maurhofer M, Keel C, Haas D, Défago G. Pyoluteorin production by Pseudomonas fluorescens strain CHA0 is involved in the suppression of Pythium damping-off of cress but not of cucumber. Eur J Plant Pathol. 1994;100:221–32.
Homma Y, Sato Z, Hirayama F, Konno K, Shirahama H, Suzui T. Production of antibiotics by Pseudomonas cepacia as an agent for biological control of soilborne pathogens. Soil Biol Biochem. 1989;21:723–28.
Thomashow LS, Weller DM. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J Bacteriol. 1988;170:3499–508.
Takeuchi K, Noda N, Katayose Y, Mukai Y, Numa H, Yamada K, et al. Rhizoxin analogs contribute to the biocontrol activity of a newly isolated Pseudomonas strain. Mol Plant-Microbe Interact. 2015;28:333–42.
Keel C, Schnider U, Maurhofer M, Voisard C, Laville J, Burger U, et al. Suppression of root diseases by Pseudomonas fluorescens CHA0: importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol Plant–Microbe Interact. 1992;5:4–13.
Raaijmakers JM, Weller DM, Thomashow LS. Frequency of antibiotic-producing Pseudomonas spp. in natural ecosystems. Appl Environ Microbiol. 1997;63:881–7.
Isnansetyo A, Cui L, Hiramatsu K, Kamei Y. Antibacterial activity of 2,4-diacetylphloroglucinol produced by Pseudomonas sp. AMSN isolated from a marine alga, against vancomycin-resistant Staphylococcus aureus. Int J Antimicrob Agents. 2003;22:545–7.
Reddi TK, Borovkov AV. Mono-, di- and triacetylphloroglucinols from Pseudomonas fluorescens. Khimiya Prirodnykh Soedinenii. 1969;5:133.
Keel C, Weller DM, Natsch A, Défago G, Cook RJ, Thomashow LS. Conservation of the 2,4-Diacetylphloroglucinol Biosynthesis Locus among Fluorescent Pseudomonas Strains from Diverse Geographic Locations. Appl Environ Microbiol. 1996;62:552–63.
Raaijmakers JM, Weller DM. Natural plant protection by 2,4-diacetylphloroglucinol–producing Pseudomonas spp. in take-all decline soils. Mol Plant–Microbe Interact. 1998;11:144–52.
Loper JE, Hassan KA, Mavrodi DV, Davis EW II, Lim CK, Shaffer BT, et al. Comparative genomics of plant-associated Pseudomonas spp.: Insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012;8:1–27. e1002784.
Ramette A, Frapolli M, Saux MF-L, Gruffaz C, Meyer J-M, Défago G, et al. Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst Appl Microbiol. 2011;34:180–8.
Levenfors JJ, Hedman R, Thaning C, Gerhardson B, Welch CJ. Broad-spectrum antifungal metabolites produced by the soil bacterium Serratia plymuthica A 153. Soil Biol Biochem. 2004;36:677–85.
Pohanka A, Broberg A, Johansson M, Kenne L, Levenfors JJ. Pseudotrienic acid A and B, two novel bioactive metabolites from Pseudomonas sp. MF381-IODS. J Nat Prod. 2005;68:1380–5.
Stanier RY, Palleroni NJ, Doudoroff MJ. The aerobic pseudomonads: a taxonomic study. Gen Microbiol. 1966;43:159–271.
Bjerketorp J, Levenfors JJ, Sahlberg C, Nord C, Andersson PF, Guss B, et al. Antibacterial 3,6-disubstituted 4-hydroxy-5,6-dihydro-2H-pyran-2-ones from Serratia plymuthica MF371-2. J Nat Prod. 2017;80:2997–3002.
Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Bacteriol. 1966;16:313–40.
Thaning C, Welch CJ, Borowicz JJ, Hedman R, Gerhardson B. Suppression of Sclerotinia sclerotiorum apothecial formation by the soil bacterium Serratia plymuthica: identification of a chlorinated macrolide as one of the causal agents. Soil Biol Biochem. 2001;33:1817–26.
Lindhagen E, Nygren P, Larsson R. The fluorometric microculture cytotoxicity assay. Nat Protoc. 2008;3:1364–69.
Emsley P, Lohkamp B, Scott W, Cowtan K. Features and development of coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501.
Debreczeni JE, Emsley P. Handling ligands with coot. Acta Crystallogr D Biol Crystallogr. 2012;68:425–30.
Long F, Nicholls RA, Emsley P, Graaeulis S, Merkys A, Vaitkus A, et al. AceDRG: a stereochemical description generator for ligands. Acta Crystallogr D Struct Biol. 2017;73:112–22.
Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–42.
Pavkov-Keller T, Schmidt NG, Żądło-Dobrowolska A, Kroutil W, Gruber K. Structure and catalytic mechanism of a bacterial friedel–crafts acylase. ChemBioChem. 2019;20:88–95.
Marchand PA, Weller DM, Bonsall RF. Convenient synthesis of 2,4-diacetylphloroglucinol, a natural antibiotic involved in the control of take-all disease of wheat. J Agric Food Chem. 2000;48:1882–87.
Esipov SE, Adanin VM, Baskunov BP, Kiprianova EA, Garagulya AD. New fluoroglucid with antibiotic activity from Pseudomonas aurantiaca. Antibiotiki. 1975;20:1077–81.
Li SG, Tian HY, Ye WC, Jiang RW. Benzopyrans and furoquinoline alkaloids from Melicope pteleifolia. Biochem Syst Ecol. 2011;39:64–67.
Hu LH, Khoo CW, Vittal JJ, Sim KY. Phloroglucinol derivatives from Hypericum japonicum. Phytochemistry. 2000;53:705–9.
Su CR, Kuo PC, Wang ML, Liou MJ, Damu AG, Wu TS. Acetophenone derivatives from acronychia pedunculata. J Nat Prod. 2003;66:990–3.
Chan JA, Shultis EA, Carr SA, DeBrosse CW, Eggleston DS, Francis TA, et al. Novel Phloroglucinols from the Plant Melicope sessiliflora (Rutaceae). J Org Chem. 1989;54:2098–103.
She GM, Zhang YJ, Yang CR. A new phenolic constituent and a cyanogenic glycoside from Balanophora involucrata (Balanophoraceae). Chem Biodiv. 2013;10:1081–7.
Smolobochkin AV, Gazizov AS, Burilov AR, Pudovik MA, Khuzyasheva DG, Amirov RR. Synthesis of 2-Arylpyrrolidines by Reaction of γ-Ureidoacetals with Benzene-1,3,5-triol. Russ J Org Chem. 2016;52:538–40.
Isnansetyo A, Horikawa M, Kamei Y. In vitro anti-methicillin-resistant Staphylococcus aureus activity of 2,4-diacetylphloroglucinol produced by Pseudomonas sp. AMSN isolated from a marine alga. J Antimicrob Chemother. 2001;47:724–25.
Syed B, Nagendra Prasad MN, Mohan Kumar K, Satish S. Bioconjugated nano-bactericidal complex for potent activity against human and phytopathogens with concern of global drug resistant crisis. Sc Tot Environ. 2018;637–638:274–81.
Bangera MG, Thomashow LS. Identification and Characterization of a Gene Cluster for Synthesis of the Polyketide Antibiotic 2,4-Diacetylphloroglucinol from Pseudomonas fluorescens Q2-87. J Bacteriol. 1999;181:3155–63.
Achkar J, Xian M, Zhao H, Frost JW. Biosynthesis of phloroglucinol. J Am Chem Soc. 2005;127:5332–33.
Hayashi A, Saitou H, Mori T, Matano I, Sugisaki H, Maruyama K. Molecular and catalytic properties of monoacetylphloroglucinol acetyltransferase from Pseudomonas sp. YGJ3. Biosci Biotechn Biochem. 2014;76:559–66.
Yang F, Cao Y. Biosynthesis of phloroglucinol compounds in microorganisms—review. Appl Microbiol Biotechnol. 2012;93:487–95.
Schmidt NG, Pavkov-Keller T, Richter N, Wiltschi B, Gruber K, Kroutil W. Biocatalytic friedel–crafts acylation and fries reaction. Angew Chem Int Ed 2017;56:7615–9.
Żądło-Dobrowolska A, Schmidt NG, Kroutil W. Promiscuous activity of C-acyltransferase from Pseudomonas protegens: synthesis of acetanilides in aqueous buffer. Chem Commun 2018;54:3387.
Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FS. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44:D646–D653.
Acknowledgements
Ultupharma AB is gratefully acknowledged for funding of JJL, JB, CN, BG, and AB.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Levenfors, J.J., Nord, C., Bjerketorp, J. et al. Antibacterial pyrrolidinyl and piperidinyl substituted 2,4-diacetylphloroglucinols from Pseudomonas protegens UP46. J Antibiot 73, 739–747 (2020). https://doi.org/10.1038/s41429-020-0318-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41429-020-0318-1
- Springer Japan KK