Abstract
Cytokine storms are crucial in the development of various inflammatory diseases, including sepsis and autoimmune disorders. The immunosuppressive cytokine INTERLEUKIN (IL)-37 consists of five isoforms (IL-37a-e). We identified IL-37a as a nuclear cytokine for the first time. Compared to IL-37b, IL-37a demonstrated greater efficacy in protecting against Toll-like receptor-induced cytokine hypersecretion and lethal endotoxic shock. The full-length (FL) form of IL-37a and the N-terminal fragment, which is processed by elastase, could translocate into cell nuclei through a distinctive nuclear localization sequence (NLS)/importin nuclear transport pathway. These forms exerted their regulatory effects independent of the IL-1R8 receptor by transcriptionally upregulating the nuclear receptor peroxisome proliferator-activated receptor (PPARγ). This process involved the recruitment of the H3K4 methyltransferase complex WDR5/MLL4/C/EBPβ and H3K4me1/2 to the enhancer/promoter of Pparg. The receptor-independent regulatory pathway of the nuclear IL-37a–PPARγ axis and receptor-dependent signaling by secreted IL-37a maintain homeostasis and are potential therapeutic targets for various inflammatory diseases, including sepsis.
Similar content being viewed by others
Introduction
A cytokine storm, or cytokine release syndrome, is a prevalent pathogenic syndrome observed in various inflammatory, autoimmune, and infectious diseases, including coronavirus disease 2019 (COVID-19) and septic shock [1,2,3,4]. This syndrome is characterized by excessive production of proinflammatory cytokines, such as interleukin (IL)-1α, IL-1β, IL-6, and tumor necrosis factor (TNF)-α, leading to systemic inflammation, organ failure, and potential death [1,2,3,4,5]. Effective clinical management of cytokine storms poses a significant challenge [2, 5]. Insights derived from the human immunoregulatory system could aid in identifying safe and efficacious immunoregulators and signaling molecules for treating cytokine storms and inflammatory diseases.
IL-37 is a recently identified regulatory cytokine that is exclusively expressed in humans. It consists of five isoforms (IL-37a–e) [6,7,8,9,10,11]. IL-37 isoforms have been found in various tissues, including the spleen, lung, bladder and immune cells, such as macrophages, dendritic cells, and T cells [6, 8,9,10,11,12,13]. Despite these findings, the specific expression patterns and regulatory mechanisms of IL-37 isoforms, especially in disease contexts, remain poorly understood.
IL-37 isoforms are initially synthesized as full-length (FL) (pro)-proteins that can undergo cleavage, resulting in the generation of mature forms and short N-terminal peptides. All isoforms except for IL-37a possess a caspase-1 cleavage site encoded by common exon 1. However, IL-37a features a potential elastase cleavage site encoded by exon 3 [7]. Exon 3 also encodes a putative nuclear localization sequence (NLS) spanning residues 5 to 21 in the N-terminus of IL-37a [7]. This NLS facilitates the nuclear import of various proteins, including IL-1α [14,15,16]. The transport of proteins across the nuclear membrane via the NLS is mediated by an importin-receptor complex composed of importin α and importin β [14, 15]. Exons 4-6 encode the IL-1-like domain, which is composed of 12 β-strands that are essential for receptor binding and cytokine activity [7]. IL-37a, b, and d possess all 12 β-strands and are thought to exhibit cytokine bioactivity, while IL-37c and IL-37e lack the first three β-strands [7, 17].
Only the function of IL-37b has been studied in detail. IL-37b exerts immunosuppressive effects on innate and adaptive immune cells and controls the development of inflammatory disorders [6, 7, 18,19,20,21,22,23,24,25]. In contrast to IL-1 and IL-18, both the precursor and mature forms of IL-37b are bioactive [6, 7, 26]. Evidence suggests that IL-37b and IL-37d attenuate inflammatory diseases, including endotoxemia, in vivo through a receptor complex composed of IL-18Ra and IL-1R8 [17, 26,27,28]. Mature IL-37b but not FL IL-37b can translocate into the nucleus via Smad3 [6, 29], but it is still unclear whether nuclear IL-37b can control the inflammatory response via a receptor-independent mechanism. IL-37 genes are evolutionarily absent in mice, but their receptors IL-1R8 and IL-18Ra are present and can respond to human IL-37, which provides a unique model to study the functions of human IL-37 in vivo [27].
The biology and functions of other IL-37 isoforms, particularly IL-37a, in both normal physiological conditions and disease states remain poorly understood. Moreover, whether IL-37b is the dominant isoform in immune responses and disease processes is unknown. This study presents novel findings demonstrating that human IL-37a possesses distinctive characteristics separate from IL-37b regarding its biological properties, nuclear localization, and immunoregulatory activities. Notably, IL-37a is a unique transcriptional regulator within the nucleus, suggesting that it primarily serves as a nuclear factor and cytokine. Through intracrine, autocrine, and paracrine mechanisms, IL-37a effectively restricts the occurrence of lethal cytokine storms and endotoxin shock. This research sheds light on the critical role of IL-37a in immune regulation and suggests its potential therapeutic applications.
Materials and methods
Study patients
The clinical study was approved by the Research Ethics Board of the Children’s Hospital of Fudan University in Shanghai, China. Written informed consent was obtained from the parents of each study participant.
Twenty healthy children and patients suffering from sepsis (aged from 1 month to 4 years) were recruited from the pediatric intensive care unit of the Children’s Hospital of Fudan University (Table S1). Sepsis was diagnosed based on the International Pediatric Sepsis Consensus Conference in 2005 [30]. The exclusion criteria for patients were as follows: (i) suffering from immunocompromise (e.g., infection with the human immunodeficiency virus or acquired immune deficiency syndrome); (ii) hematological malignancies; (iii) solid tumors necessitating immunosuppressive medications within the last 3 months; and (iv) other diseases involving the immune system.
Two milliliters of whole blood was collected from the children within 24 h of the diagnosis of sepsis. Age- and sex-matched healthy children were recruited from the Children’s Hospital of Fudan University as controls.
Peripheral blood mononuclear cells (PBMCs) were purified by Ficoll-gradient centrifugation. The cells were then cultured in RPMI 1640 (#11875119; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (#10099-141; Gibco), L-glutamine (2 mM), penicillin/streptomycin (100 units/mL; #15070063; Gibco), and 2-mercaptoethanol (50 μM). PBMCs were stimulated with different doses of the Toll-like receptor (TLR) ligands Pam3CSK4 (#tlrl-kit1hw; InvivoGen, San Diego, CA, USA) or lipopolysaccharide (LPS) (#L6011; Sigma, Burlington, MA, USA) for different times. Cells were collected for qPCR.
Animals
All animal experiments were approved (ILAS-GC-2015-001) by the Animal Care and Use Committee of the Institute of Laboratory Animal Science of Peking Union Medical College (Beijing, China).
The mice used in this study had a C57BL/6 J background and were purchased from Beijing Vital River Laboratory Animal Technology (Beijing, China). The mice were housed in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care with ad libitum access to food and water.
FLIL-37a, NIL-37a, and FLIL-37b transgenic mice were generated. Briefly, complementary (c) DNA for FLIL-37a, NIL-37a (63 bp), and FLIL-37b was cloned from human PBMCs by reverse transcription polymerase chain reaction (RT‒PCR). The sequences were 100% homologous with the IL-37a (NM_173205.1) and IL-37b (NM_014439.3) sequences published in GenBank (www.ncbi.nlm.nih.gov/). The primers used to clone the cDNA of human FLIL-37a, NIL-37a, and FLIL-37b are shown in the Supplementary Information (Table S2).
To generate transgenic mice, FLIL-37a, NIL-37a, and FLIL-37b cDNA was inserted downstream of the ubiquitin C (UBC) promoter in the pUBC2(+)vector between the EcoRI and XhoI cloning sites for FLIL-37a and NIL-37a and the HindIII and XhoI cloning sites for FLIL-37b. Plasmids were linearized, and gel-purified DNA fragments were injected into the pronucleus of fertilized zygotes harvested from C57BL/6 J mice using conventional methods to generate transgenic mice [31].
Il1r8−/−, FLIL-37a tg/Il1r8−/−, and NIL-37a tg/Il1r8−/− mice were also generated. Il1r8−/− mice were generated by CRISPR/Cas9 as described by Hall and colleagues [32]. Briefly, Cas9 mRNA and two single guide (sg) RNAs (Table S3) targeting exon 3 of murine IL-1R8 (Gene ID: 24058) were prepared with a MEGA short script T7 Transcription Kit (#AM1354; Ambion, Austin, TX, USA) using a pUC57-sgRNA vector (#51132; Addgene, Watertown, MA, USA). The Cas9 mRNA and sgRNA mixture was microinjected into fertilized eggs at the one-cell stage. The injected zygotes were transferred to pseudopregnant C57BL/6 J mice. The pups were genotyped by PCR using genomic DNA from the tails.
Homozygous Il1r8−/− mice were crossed with FLIL-37a tg mice or NIL-37a tg mice on the same C57BL/6 J background for five generations to generate homozygous FLIL-37a tg/Il1r8−/− mice and NIL-37a tg/Il1r8−/− mice.
Genotyping of transgenic mice was carried out. Mouse genomic DNA was extracted from tail biopsies. Genomic PCR was performed in a reaction system containing genomic DNA and specific primer pairs. The primer pairs for identifying FLIL-37a tg, NIL-37a tg and Il1r8−/− mice are shown in Table S4. PCR products were examined by electrophoresis on 1% agarose gels.
Antibodies
Antibodies against green fluorescent protein (GFP) (#598) were purchased from MBL. Antibodies against H3K4me1 (#5326), H3K4me2 (#9725), PPARγ (#2443), and WDR5 (#13105) and normal rabbit immunoglobulin-G (#2729) were purchased from Cell Signaling Technology. Antibodies against β-tubulin (#ab6046) and IL-37 (#ab101376) were purchased from Abcam. Antibodies against α-tubulin (#sc-69969) were purchased from Santa Cruz. Antibodies against histone 3 (#H0164) were purchased from Sigma. Antibodies against IL-1R8 (#MAB1092) were purchased from R&D Systems.
Production of rIL-37a and rIL-37b in Escherichia coli
FL and mature human IL-37a and IL-37b cDNA was cloned from an LPS-stimulated human THP-1 cell line by RT‒PCR using specific primer pairs. The confirmed IL-37 cDNA sequences were inserted into the pQE expression vector and fused with a His-tag (Qiagen, Stanford, VA, USA). The construct was then transferred into the E. coli strain BL21. IL-37 protein expression was induced by IPTG (#I6758; Sigma) and purified using Ni-NTA affinity chromatography, followed by size-exclusion chromatography using a polymyxin-B column to remove endotoxin. The purity of the IL-37 protein was >95%, and the endotoxin level was <0.01 EU/μg of protein, as determined by the Limulus amebocyte lysate QCL-1000 pyrogen test (Bio-Whittaker, Walkersville, MD, USA).
Construction of the pEGFP-N1-IL-37a plasmid
The gene sequences for FLIL-37a, NIL-37a and mutants, and mIL-37a with XhoI and AgeI or BamHI restriction sites at the 5’ and 3’ ends were synthesized. Then, they were inserted into pEGFP-N1 vectors.
Generation of bone marrow-derived macrophages
Bone marrow-derived macrophages (BMDMs) were generated using the method described by Kurowska-Stolarska and colleagues [33]. Cell culture was conducted using RPMI 1640 medium supplemented with 10% fetal bovine serum, penicillin/streptomycin, and glutamine (complete medium). In brief, femurs from wild-type (WT), FLIL-37 tg, NIL-37 tg or Il1r8−/− mice were flushed with complete medium. The cells were then cultured in 10-cm plastic plates with complete medium containing recombinant human CSF-1 (100 U/mL) for 7 days in a 37 °C incubator with 5% CO2. For in vitro macrophage activation, BMDMs were plated in 24-well plates at a density of 1 × 106 cells/well in complete medium supplemented with CSF-1 and incubated overnight. Subsequently, the cells were treated with LPS or different TLR ligands for various times. Cells and supernatants were collected for further analysis.
To overexpress FLIL-37a, NIL-37a, mIL-37a, and the mutants, lentiviruses containing the respective IL-37a DNA sequences were constructed (GeneChem, Shanghai, China). In brief, RAW264.7 cells (1 × 106) or BMDMs (1 × 106) were cultured in six-well plates for 18 h before being infected with the lentiviruses. After 12 h, the cells were cultured in fresh medium for 3–4 days. To establish a stable cell line, puromycin (4 μg/mL) was used to select positive cells. The cells were transfected with lentiviruses and expressed GFP for 2–3 days as previously described [34].
siRNA knockdown
A549, THP-1, and RAW264.7 cells were transfected with siRNAs targeting total IL-37 (targeting exon 5), IL-37a (targeting exon 3), Pparg, Wdr5, Mll4, or Cebpb or control siRNA. The specific siRNA oligonucleotides were synthesized (GenePharm, Pallini, Greece), and their sequences are listed in Table S5. Cells were transfected with the siRNAs using Lipofectamine® RNAiMAX (#13778075; Thermo Fisher Scientific, Waltham, MA, USA) for 48 h according to the manufacturer’s instructions.
LPS shock
Age- and sex-matched FLIL-37a tg, NIL-37a tg, Il1r8−/−, FLIL-37a tg/Il1r8−/−, NIL-37a tg/Il1r8−/−, FLIL-37b tg, and control WT C57BL/6 J mice were injected (i.p.) with a minimal lethal dose of LPS (40 mg/kg bodyweight). In the rFLIL-37-treatment experiments, the mice were first treated with rFLIL-37a or rFLIL-37b and then challenged 2 h later with LPS (40 mg/kg bodyweight). The animals were observed regularly for general health and mortality according to guidelines. In some cases, age- and sex-matched WT, FLIL-37a tg, and NIL-37a tg mice (n = 7–10/group) were injected (i.p.) with the PPARγ antagonist GW9662 (1.5 mg/kg bodyweight; #70785; Cayman Chemicals, Ann Arbor, MI, USA) or vehicle control for two consecutive days, and LPS-induced mortality was examined as described above.
Cytokine analysis
Cytokine expression was quantified using enzyme-linked immunosorbent assay (ELISA) or cytometric bead array (CBA) according to the manufacturer’s protocols. ELISA kits for mouse IL-1α (#MLA00), human IL-1α (#SLA50), mouse IL-1β (#DY401), human IL-1β (#SLB50), mouse IL-6 (#DY406), human IL-6 (#S6050), and human IL-37 (#EA100823) were purchased from R&D Systems. The ELISA kit for IL-37 contains an antibody against the common C-terminus of the IL-37 isoforms to detect all IL-37 isoforms.
CBA kits for mouse IL-1α (#560157), mouse IL-1β (#560232), mouse IL-6 (#558301), mouse IL-10 (#558300), mouse IL-17A (#560283), mouse TNF (#558299), and mouse IFN-γ (#558296) were purchased from BD Biosciences.
qPCR
RNA was extracted from cultured cells or tissue samples using the RNeasy Mini Kit (#74104; Qiagen) according to the manufacturer’s instructions. Reverse transcription was performed using the High-Capacity cDNA Reverse Transcription Kit (#K1691; Thermo Fisher Scientific). Real-time PCR was conducted using Fast SYBR™ Green Master Mix on a Prism 7900HT Sequence Detection system (Thermo Fisher Scientific) and analyzed using Fast System Software (Applied Biosystems, Carlsbad, CA, USA). All primers used for the target genes, including the IL-37 isoforms, were designed to span exons and did not cross-react with genomic DNA. The primer sets for human and mouse PCR are listed in Table S6.
Immunofluorescence analysis and confocal microscopy
A549 cells were cultured in DMEM supplemented with 10% FBS, penicillin/streptomycin, and glutamine. The cells were transfected with plasmid constructs encoding GFP, GFP-fusion FLIL-37a, NIL-37a, or mIL-37a using Lipofectamine 2000 according to the manufacturer’s instructions (Thermo Fisher Scientific). After being transfected, the cells were cultured for 24 h in Lab-Tek II Chamber Slides (Nunc) before being stimulated with LPS (500 ng/mL) or left untreated for 18 h. The cells were then washed with phosphate-buffered saline (PBS) and fixed for 10 min with 4% polyformaldehyde, and the nuclei were stained with DAPI (4,6-diamidino-2-phenylindole; Invitrogen). Visualization was performed using a Leica TCS SPE laser scanning confocal microscope (Leica Microsystems GmbH).
Western blot analysis and immunoprecipitation assays
Transfected RAW264.7 cells expressing FLIL-37a, FLIL-37b, mutant IL-37a (del aa18-19, del aa18-22, K19/R22S), or control were lysed in RIPA buffer (Sigma). Cytoplasmic and nuclear proteins were then separated using the Nuclear and Cytoplasmic Protein Extraction Kit (Thermo Fisher Scientific). The proteins were subjected to SDS‒PAGE, transferred onto nitrocellulose membranes, and probed with the respective antibodies.
For the immunoprecipitation assay, RAW264.7 cells (1 × 106) were transfected with 8 μg of plasmid DNA expressing GFP-fused FLIL-37a or NIL-37a for 24 h. The cells were lysed using 1× cell lysis buffer (Cell Signaling Technology, Danvers, USA), followed by sonication. The lysates were then subjected to immunoprecipitation using protein A/G beads coupled with antibodies against GFP (0.5 μg) or normal rabbit IgG. The immunoprecipitated proteins, including WDR5 and C/EBPβ (Cell Signaling Technology, Danvers, USA), bound to the beads were subsequently fractionated using SDS‒PAGE.
Transcriptomics and bioinformatics analysis
Splenocytes from WT, FLIL-37a, and FLIL-37a tg/Il1r8−/− mice (n = 3/group) were stimulated with LPS (500 ng/mL) for 4 h. Total RNA was isolated, reverse-transcribed into cDNA, and labeled. Microarray analysis was performed (Aksomics) using the whole mouse genome 4 × 44 K gene expression microarray v2 (Agilent Technologies, Santa Clara, CA, USA). The chips were scanned using the Axon GenePix 4000B Microarray Scanner (Molecular Devices, Silicon Valley, CA, USA). Raw data was normalized for among-sample/group comparisons. Normalized data were log2 transformed (Aksomics). Differentially expressed genes (P < 0.05, fold-change ≥ 2.0) were identified using GeneSpring GX v12.1 (Agilent Technologies).
Area-proportional Euler diagrams were generated using BioVenn (www.biovenn.nl/) to visualize the overlapping gene sets. Innate immune genes were identified using InnateDB (www.innatedb.com/). Further analysis of signaling pathway enrichment was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (www.genome.jp/) and the Database for Annotation, Visualization, and Integrated Discovery (www.david.abcc.ncifcrf.gov/) [35, 36]. We would like to provide additional information related to the transcriptomic and bioinformatics analysis upon request.
Mass spectrometry
RAW264.7 cells transfected with lentiviruses expressing Flag-tagged FLIL-37a or control were lysed, and immunoprecipitation was performed using an anti-Flag antibody. Equivalent amounts of immunoprecipitates were separated on gels and stained with Coomassie Brilliant Blue to visualize the protein bands. The gels were then subjected to overnight digestion with 100 ng of trypsin. LTQ Orbitrap Velos (Thermo Fisher Scientific) was operated in a data-dependent mode. Subsequently, the spectral data were searched against the mouse protein RefSeq database using Mascot software in Proteome Discoverer 1.3 Suites.
ChIP assays
RAW264.7 cells were transfected with equal amounts of vectors expressing FLIL-37a, NIL-37a, or Wdr5 siRNA, along with the respective controls. Chromatin immunoprecipitation (ChIP) assays were performed using the Simple ChIP Enzymatic Chromatin IP Kit (#9003; Cell Signaling Technology, Danvers, MA, USA) according to the manufacturer’s instructions. In summary, cells were crosslinked with 37% formaldehyde, and the crosslinking was stopped with glycine. Micrococcal nuclease digestion was carried out for 20 min at 37 °C to fragment the chromatin. Complete lysis of cell nuclei was achieved using the Bioruptor Plus Sonicator (Diagenode, Liege, Belgium) with three sonication cycles (20 s on, 30 s off) in high power mode. For each immunoprecipitation (IP), 2% of the input sample was saved, and 10 µL of antibody or negative control IgG was added to the digested and crosslinked chromatin (5–10 µg), followed by overnight incubation at 4 °C with rotation. The IP sample was then incubated with 30 µL of protein G magnetic beads at 4 °C for 2 h and washed with low salt and high salt wash buffers. Chromatin was eluted from the complexes, and the bound DNA was separated and subjected to qPCR analysis.
Dual-luciferase assays
DNA fragments containing mouse Pparg1 promoters were PCR-amplified from mouse genomic DNA using the primers listed in the Supplementary Information (Table S7). The promoter fragments (−1500 bp-TSS) were then cloned and inserted into the luciferase reporter pGL3-basic vector (Promega, Madison, WI, USA). Transfection was performed when RAW264.7 cells reached 40–50% confluence using the Pparg1-luciferase reporter gene (pGL3). The Renilla luciferase reporter vector (pRL-TK; Promega) was used as an internal control to determine transfection efficiency. After 48 h of transfection using Lipofectamine 2000 (#11668500; Thermo Fisher Scientific), the transfected cells were stimulated with LPS (500 ng/mL) for 24 h. Luciferase activity was measured using the Dual-Glo™ Luciferase Assay System (Promega) according to the manufacturer’s protocol.
Quantification and statistical analysis
Statistical analysis was performed using Prism 8 software (GraphPad, La Jolla, CA, USA). For in vitro studies, analysis of variance (ANOVA) followed by Tukey’s test or Student’s t test was applied. For in vivo comparisons between individual groups, ANOVA followed by the Student–Newman–Keuls test or Student’s t test was performed. All experiments were repeated at least three times. The data are presented as the mean ± SEM. A significance level of P < 0.05 was considered statistically significant.
Results
Induction and bioactivity of IL-37a in human cells
Specific antibodies for detecting IL-37 isoforms were unavailable. Therefore, we initially evaluated the gene expression of IL-37 in PBMCs from healthy individuals and macrophages using lipopolysaccharide (LPS), which is an agonist of TLR4. IL-37a was highly induced by LPS in PBMCs and macrophages (Fig. 1A, B). Pam3CSK4 stimulation resulted in an earlier increase in the expression of IL-37a than the IL-37b, c, and d isoforms in PBMCs (Fig. S1). To assess the impact of intrinsic IL-37a on the production of proinflammatory cytokines during sepsis, we selectively silenced the expression of IL-37a or total IL-37 in human epithelial cells using specific small interfering RNAs. Silencing the expression of total IL-37 mRNA by targeting the common exon 5 reduced the expression of all IL-37 isoforms induced by LPS and IL-1β (Fig. 1C). Importantly, selective silencing of IL-37a expression by targeting exon 3 or total IL-37 resulted in a 20–40% increase in the secretion of IL-1α, IL-1β, and IL-6 induced by LPS/IL-1β compared to the effect of the control siRNA (Fig. 1D). Consistently, the overexpression of human FLIL-37a in murine macrophages significantly suppressed the production of IL-1β and IL-6 induced by TLR4, TLR1/2, TLR3, TLR5, TLR8, and IL-1β agonists (Fig. 1E, F).
Induction and function of IL-37a in human PBMCs. A, B Human PBMCs (A n = 5 donors) or PMA-differentiated THP1 cells (B) were stimulated with a fixed dose of LPS (1 µg/mL) for the indicated times. Total RNA was extracted, and the expression of five IL-37 isoforms was measured by qPCR. C Human A549 cells were transfected with siRNA targeting total IL-37 and IL-37a or control siRNA for 48 h before being simulated with LPS (1 µg/mL) plus IL-1β (10 ng/mL) for 24 h. Total RNA was extracted, and the expression of IL-37 isoforms was determined by qPCR. D Human A549 cells were transfected with siRNA targeting total IL-37 and IL-37a or control siRNA. The levels of IL-1α, IL-1β, and IL-6 in culture supernatants were measured by ELISA. E, F Murine RAW264.7 cells were stably transduced with recombinant lentiviral particles expressing FLIL-37a or control and then stimulated with LPS (500 ng/mL), Pam3CSK4 (200 ng/mL), poly (I:C) (500 ng/mL), FLA-ST (200 ng/mL), ssRNA40 (500 ng/mL), or IL-1β (10 ng/mL) for 24 h. The levels of IL-1β and IL-6 were determined by ELISA. G rFLIL-37a was purified, and RAW264.7 cells were stimulated with LPS (1 µg/mL) in the presence or absence of different doses of rFLIL-37a or commercial mature IL-37b (novoprotein) for 24 h. The levels of of IL-6 in culture supernatants were measured by ELISA. H, I PBMCs (n = 3 donors) were cultured with optimal doses of Pam3CSK4 (10 ng/mL), poly (I:C) (10 ng/mL), FLA-ST (10 ng/mL), ssRNA40 (500 ng/mL), ODN2006 (5 μM), or IL-1β (10 ng/mL) and rFLIL-37a (50 ng/mL) for 24 h. The production of IL-6 and IL-1β was determined by ELISA. J RAW264.7 cells were cultured with LPS (500 ng/mL) in the presence or absence of different concentrations of rFLIL-37a, rmIL-37a, or rmIL-37b for 15 h. IL-6 levels in culture supernatants were measured by ELISA. K Splenocytes from WT and Il1r8−/− mice were treated with rFLIL-37 or rFLIL-37b (50 ng/mL) in the presence of LPS (500 ng/mL) for 24 h. IL-6 levels in culture supernatants were determined by ELISA. The data are the mean ± SEM and are representative of three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001, compared with the control
Next, we produced FL recombinant human IL-37a (rFLIL-37a) and assessed its bioactivity. rFLIL-37a dose-dependently inhibited LPS-induced IL-6 production in murine macrophages and exhibited greater suppressive effects than mature rIL-37b (IL-37b) (Fig. 1G). Furthermore, rFLIL-37a effectively suppressed the production of IL-1β and IL-6 in PBMCs induced by TLR4, TLR1/2, TLR3, TLR5, TLR8, and IL-1β agonists (Fig. 1H, I).
In murine macrophages, rFLIL-37a more robustly inhibited LPS-induced IL-6 production than mIL-37a or mIL-37b (Fig. 1J). There was no significant difference in IL-6 inhibition between the mIL-37a and mIL-37b groups. These findings suggest that, unlike IL-1 and IL-18, the maturation of IL-37a is not required for its bioactivity as a cytokine.
We examined whether extracellular IL-37a used IL-1R8 for signal transduction. To investigate this, Il1r8−/− mice were generated through CRISPR/Cas9-mediated genome editing [37] (Fig. S2). In WT splenocytes, rFLIL-37a and rFLIL-37b effectively inhibited LPS-induced IL-6 production. However, this inhibitory effect was absent in Il1r8−/− splenocytes (Fig. 1K). These results indicate that IL-1R8 is essential for the protective effect mediated by extracellular IL-37a.
These findings strongly indicate that IL-37a, especially in its FL form, is a novel immunoregulator that can inhibit proinflammatory cytokines. Furthermore, IL-37a significantly contributes to the suppressive effects mediated by IL-37 during the inflammatory response.
IL-37a is more effective than IL-37b in protecting against lethal LPS shock in mice
The expression and function of IL-37 isoforms in children with septic shock are currently unknown. In children with sepsis, the expression of all IL-37 isoforms was significantly downregulated compared to that in healthy children, except for IL-37e, which could not be detected (Fig. 2A).
FLIL-37a is more immunosuppressive than FLIL-37b in vivo. A PBMCs (n = 20) were isolated from children suffering from sepsis and from healthy controls. Total RNA was extracted, and the expression of five IL-37 isoforms was measured by qPCR. B The expression of human IL-37a and IL-37b in the organs of transgenic and WT mice was determined by qPCR with or without IL-37a or IL-37b plasmids as positive/negative (+/−) controls. C WT, FLIL-37a and FLIL-37b tg mice (n = 5 mice/group) were injected (i.p.) with LPS (10 mg/kg), and serum levels of IL-37a or IL-37b were measured by ELISA 18 h post-LPS injection. D, E WT, FLIL-37a and FLIL-37b tg mice (n = 10 mice/group) were challenged with a lethal dose of LPS (40 mg/kg, i.p.), and morality was documented regularly (D). Eight hours after LPS challenge, blood samples were collected to measure cytokine levels in serum by cytometric bead array (CBA) (E). F C57BL/6 J mice (n = 10 mice/group) were pretreated with rFLIL-37a or rFLIL-37b (10 mg/kg) and challenged 2 h later with LPS (40 mg/kg). The survival rate in each group was analyzed. G Cytokine levels in serum at 18 h (F) was measured by CBA. The data are the mean ± SEM and are representative of three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001, compared with the control
To evaluate and directly compare the biology and impact of human IL-37a and IL-37b in various diseases, including LPS shock, transgenic mice expressing FL human IL-37a and IL-37b were generated using an identical procedure (Fig. S3). The FLIL-37a and FLIL-37b transgenic strains exhibited similar expression and distribution of IL-37a and IL-37b mRNA in the immune-related organs that were tested (Fig. 2B). In addition, both strains showed comparably low levels of IL-37a and IL-37b in serum under resting conditions, and these levels were significantly increased following LPS challenge (Fig. 2C).
The effect and distinction between IL-37a and IL-37b in protecting against endotoxic shock have not been determined. First, we compared the impact of FLIL-37a and FLIL-37b on LPS-induced lethal endotoxic shock in vivo. In WT mice, mortality was observed as early as 12 h after LPS challenge, and only 44% of mice survived to 20 h (Fig. 2D). Mortality in FLIL-37b tg mice occurred at 16 h, and 67% of mice survived to 20 h. In contrast, all FLIL-37a tg mice survived to 20 h (Fig. 2D), although they exhibited moderate shock symptoms such as reduced body weight and mobility. The development of shock symptoms and mortality in response to LPS was associated with an increase in proinflammatory cytokines. Additionally, both strains exhibited significantly reduced serum levels of the proinflammatory cytokines IL-1α, IL-6, TNF-α, and interferon (IFN)-γ compared to the WT controls, but the reduction was more pronounced in FLIL-37a tg mice than in FLIL-37b tg mice. FLIL-37a tg mice also exhibited reduced IL-10 expression compared to WT mice (Fig. 2E), indicating that IL-10 was not responsible for the immunosuppressive effect of IL-37a.
Next, C57BL/6 J mice were administered rFLIL-37a, rFLIL-37b protein, or PBS (as a control) and subsequently exposed to a lethal dose of LPS. Consistently, at 18 h post-LPS challenge, the survival rates of mice treated with rFLIL-37a (90%) were significantly higher than those of rFLIL-37b-treated mice (50%) and PBS control mice (0%) (Fig. 2F). The survival rate exhibited an inverse correlation with the serum levels of key proinflammatory cytokines, including IL-1α, IL-1β, IFN-γ, and TNF-α (Fig. 2G).
These in vivo data further suggest that IL-37a, especially when expressed transgenically, is more effective than IL-37b in reducing mortality caused by endotoxin. This effect is attributed, at least partially, to the efficient regulation of the proinflammatory cytokine storm.
FLIL-37a but not FLIL-37b is a nuclear cytokine and translocates to the nucleus via its NLS and the importin pathway
Next, we aimed to elucidate the functional differences between IL-37a and IL-37b. Unlike FLIL-37b, FLIL-37a possesses an NLS at its N-terminal end, and we investigated the cellular localization of IL-37a and IL-37b and the role of the putative NLS in the nuclear translocation of IL-37a. Given that FLIL-37a can be cleaved by elastase into N-terminal (N; aa1–21, containing the NLS) IL-37a and mature (m) IL-37a (aa22–192, lacking the NLS) [7], we also examined the cellular localization of the processed N and m forms. Expression constructs containing GFP fused with the FL, N, and m forms of IL-37a, as well as the GFP control, were transfected into human A549 epithelial cells. FL and NIL-37a, which harbor the NLS, were predominantly observed in the nucleus in transfected cells, as indicated by their colocalization with 4’,6-diamidino-2-phenylindole (DAPI)-stained nuclear DNA (blue), particularly after LPS stimulation. In contrast, mIL-37a exhibited weak fluorescence in the nucleus (Fig. 3A), whereas GFP alone did not localize to the nucleus (Fig. S4). Furthermore, since mice lack the IL-37 gene, the distinct distribution of ectopically expressed human FL and mIL-37a in the cytoplasmic and nuclear compartments of transfected murine macrophages was confirmed and quantified using ELISA to detect the C-terminus of IL-37 (Fig. 3B). However, N-terminal IL-37a (NIL-37a) could not be measured by this ELISA kit due to the absence of an antibody specific to the N-terminal region. It has been previously reported that mature IL-37b but not FL IL-37b can translocate into the nucleus via Smad3 [6, 29]. We further validated that FLIL-37b was exclusively localized in the cytoplasm and not the nucleus under these conditions, as confirmed by western blotting using an antibody against the Flag tag of FLIL-37a and FLIL-37b in transfected macrophages (Fig. 3C).
Human IL-37a is a nuclear cytokine that translocates into the nucleus via its NLS. A A549 cells in chamber slides were transfected with plasmids expressing GFP-fused FLIL-37a, NIL-37a, or mIL-37a for 24 h. Then, the cells were stimulated with LPS (500 ng/mL) or PBS (control) for an additional 24 h before being counterstained with DAPI. Subcellular localization of GFP-fusion proteins was visualized by confocal immunofluorescence microscopy. Scale bar: 100 μm. B, C Cytoplasmic and nuclear fractions were separated from transduced RAW264.7 cells expressing FL or mIL-37a, and the protein expression levels of FL and mIL-37a were measured by an IL-37 ELISA kit (B). Cytoplasmic and nuclear FLIL-37a and FLIL-37b were isolated from transduced cells, separated by SDS‒PAGE and detected by western blotting; α-tubulin and histone 3 (H3) were used as cytoplasmic and nuclear protein controls, respectively (C). D RAW264.7 cells were stably transduced with viral constructs expressing Flag-tagged WT or mutant FLIL-37a (with deletions of aa18–19, 18–22, or K19/R22S sequences) and stimulated with LPS for 24 h. The nuclear and cytoplasmic fractions were separated, and the protein expression of IL-37a was detected by western blotting. E A549 cells were transduced with viral constructs expressing GFP-FLIL-37a for 24 h before the addition of importazole (50 µM) for 6 h. The cellular localization of IL-37a was visualized by confocal microscopy. F Murine RAW264.7 cells stably expressing human FL, N, or mIL-37a were stimulated with LPS (500 ng/mL) for different times. IL-6 levels in the culture were measured by ELISA. G, H RAW264.7 cells expressing FL or NIL-37a were cultured with or without LPS (500 ng/mL), Pam3CSK4 (5 ng/mL), TNF-α (20 ng/mL), IL-17A (20 ng/mL), or IFN-γ (20 ng/mL) for 24 h. The levels of (G) IL-1β and (H) IL-6 in culture supernatants were determined by ELISA. The data are representative of at least three independent experiments. The data are the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, compared with the control
Subsequently, we investigated whether the nuclear translocation of IL-37a occurred through the NLS/importin pathway. Initially, crucial amino acids (aa18–22) within the IL-37a NLS were predicted using the NLStradamus tool (www.moseslab.csb.utoronto.ca/NLStradamus/) and then confirmed by deleting and introducing point mutations to the amino acids within the NLS of FLIL-37a. Deletion of aa18–22 (KKRLR) in FLIL-37a resulted in complete loss of nuclear translocation, whereas mutations at K18/K19 or K19S/R22S had no effect (Fig. 3D). Additionally, treatment with importazole, an inhibitor of importin β [38], disrupted the nuclear translocation of FLIL-37a from the cytoplasm (Fig. 3E). These findings suggest that the NLS of FLIL-37a plays a crucial role in importin-mediated nuclear translocation, and amino acids R20 and L21 may have prominent functions.
To evaluate the differential impact of FLIL-37a and its elastase-processed N and mIL-37a fragments on the TLR response and cytokine production in vitro, murine macrophages were transfected with lentivirus particles expressing the FL, N, and mature forms of IL-37a. Surprisingly, all three forms of IL-37a time-dependently inhibited LPS-induced IL-6 production, and FLIL-37a exhibited the highest efficacy, followed by NIL-37a and mIL-37a at 20 h (Fig. 3F). FLIL-37a and, to a lesser extent, NIL-37a also effectively suppressed the production of IL-1β and IL-6 induced by LPS, several TLR agonists, and proinflammatory cytokines (Fig. 3G, H). These findings suggest that NIL-37a may be involved in the regulatory effect induced by FLIL-37a in the nucleus.
FLIL-37a but not FLIL-37b induces receptor-independent protection against LPS shock in mice
NIL-37a lacks the receptor-binding domain of IL-37. Therefore, we hypothesized that nuclear FL and NIL-37a could regulate inflammation through a mechanism independent of receptor signaling during LPS shock.
To determine the receptor required for the regulatory effect mediated by FL, N, and mIL-37a, BMDMs from WT and Il1r8−/− mice were transfected with FL, N, mIL-37a, or control FLIL-37b constructs and stimulated with LPS, and the production of IL-6 was measured using ELISA. The different forms of endogenously produced IL-37a and IL-37b in WT cells suppressed IL-6 production to varying degrees. Specifically, FL and NIL-37a but not mIL-37a or FLIL-37b produced by Il1r8−/− cells effectively suppressed IL-6 secretion, and NIL-37a and FLIL-37a showed similar effects (Fig. 4A). Furthermore, the FLIL-37a mutant lacking the NLS (deletion of aa18–22) failed to inhibit the effects of LPS on macrophages, suggesting that FL and N IL-37a could regulate the inflammatory response within the nucleus through a receptor-independent mechanism (Fig. 4B).
Transgenic mice expressing FL and N-terminal portions of human IL-37a are resistant to lethal LPS shock independent of IL-1R8. A BMDMs derived from WT and Il1r8−/− mice were transduced with virus constructs expressing FL, N, mature IL-37a, or FLIL-37b. Then, the cells were stimulated with LPS (500 ng/mL) for 24 h, and IL-6 levels were measured by ELISA. B BMDMs derived from Il1r8−/− mice expressing FLIL-37 or mutant FLIL-37a were cultured in the presence of LPS for 24 h. The levels of IL-1β and IL-6 were measured by ELISA. C WT, FLIL-37a tg, Il1r8−/−, and FLIL-37a tg/Il1r8−/− mice (n = 8/group) were injected with LPS (40 mg/kg, i.p.), and survival was recorded. D WT, Il1r8−/−, FLIL-37a tg, and FLIL-37a tg/Il1r8−/− mice were injected with LPS (10 mg/kg, i.p.), and blood samples were collected at 16 h and 20 h (n = 3 mice/group/time point) to measure IL-1β and IL-6 levels by ELISA. E WT, FLIL-37a tg, FLIL-37b tg, FLIL-37a tg/Il1r8−/−, and FLIL-37b tg/Il1r8−/− mice (n = 10/group) were injected with LPS (40 mg/kg, i.p.), and survival was recorded. F WT, NIL-37a tg, and NIL-37a tg/Il1r8−/− mice (n = 8/group) were injected with LPS (40 mg/kg, i.p.), and survival was monitored. G WT, NIL-37a tg, and NIL-37a tg/Il1r8−/− mice (n = 3/group) were injected with LPS (10 mg/kg, i.p.). Sixteen hours later, blood samples were collected to measure cytokine levels by ELISA. The data are representative of at least three independent experiments. The data are the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, compared with the control
To investigate the impact of a receptor-independent regulatory pathway on LPS shock in vivo, WT, Il1r8−/−, FLIL-37a tg, and FLIL-37a tg/Il1r8−/− mice were generated and challenged with a lethal dose of LPS, and the survival rate was monitored. All FLIL-37a tg mice but not WT or Il1r8−/− mice survived 20 h post LPS challenge (Fig. 4C). Notably, 70% of FLIL-37a tg/Il1r8−/− mice also survived to that time point, which suggested that an IL-1R8-independent pathway significantly contributed to FLIL-37a-mediated protection (Fig. 4C). Twenty hours after LPS challenge, serum levels of IL-1β and IL-6 in FLIL-37a tg/Il1r8−/− mice remained significantly lower than those in Il1r8−/− mice but not in WT controls, whereas serum cytokine levels in FLIL-37a tg mice remained significantly lower than those in WT and Il1r8−/− mice (Fig. 4D). Consistent with a previous report [27], we confirmed that FLIL-37b tg mice lacking the receptor IL-1R8 failed to resist LPS shock (Fig. 4E), suggesting that the receptor-independent regulatory effect was specific to IL-37a.
To investigate the receptor-independent effect of NIL-37a, which is the 21-aa fragment, NIL-37a tg mice (Fig. S5), and NIL-37a tg/Il1r8−/− mice were generated by crossbreeding. We compared the protective effect against lethal LPS shock in NIL-37a tg mice with or without IL-1R8 (Fig. 4F). Twenty hours after LPS exposure, no WT control mice survived, but ~50% of NIL-37a tg mice and NIL-37a tg/Il1r8−/− mice were alive. A significant difference was not found between the NIL-37a tg and NIL-37a tg/Il1r8−/− groups in terms of survival or general health (Fig. 4F). The protective effect on NIL-37a tg and NIL-37a tg/Il1r8−/− mice was associated with reduced levels of key proinflammatory cytokines in the serum 16 h after LPS challenge compared with those in WT controls (Fig. 4G). These in vivo results demonstrated that FLIL-37a protected against lethal LPS shock predominantly via IL-1R8-independent activity. However, the IL-1R8-dependent pathway was also required for more robust protection by FLIL-37a.
Transcriptomic assessment of the IL-1R8-dependent and -independent effects of FLIL-37a
To elucidate the receptor-dependent and receptor-independent mechanisms through which IL-37a effectively protects against LPS shock, we performed transcriptomic analysis of LPS-induced splenocytes from WT (w), FLIL-37a tg (a), and FLIL-37a tg/Il1r8−/− (a/R) mice. The goal was to identify the genes that were specifically regulated by IL-37a in IL-1R8-dependent and IL-1R8-independent pathways using a reported method [27].
A total of 1436 genes were downregulated, and 1373 genes were upregulated by IL-37a in an IL-1R8-dependent manner. Additionally, a total of 1468 genes were downregulated, and 1508 genes were upregulated by IL-37a in an IL-1R8-independent manner (Fig. 5A, B). The full list of genes that were dysregulated by FLIL-37a via IL-1R8-dependent or independent pathways and descriptive and comparative statistics are provided in Table S8. Gene ontology analysis of these differentially regulated genes using the Innate database (InnateDB) [39] and KEGG [35] identified several biological pathways affected by changes in the expression of these genes. Some pathways were associated with receptor-dependent mechanisms (Fig. 5C, D, G, H, and Table S9), while others were linked to receptor-independent mechanisms (Fig. 5E, F, I, J, and Table S10).
Transcriptomic analysis of IL-1R8-dependent and -independent effects of IL-37a. Splenocytes from WT (w), FLIL-37a tg (a), and FLIL-37a tg/Il1r8−/− (a/R) mice (n = 3/group) were stimulated with LPS (500 ng/mL) for 4 h. Total RNA from each mouse was analyzed by microarray. Euler diagram separated genes that were (A) downregulated (a < w and a/R) or (B) upregulated (a > w and a/R cells) by FLIL-37a in IL-1R8-dependent (overlapping genes) and independent pathways (the remaining genes in yellow and green areas). Innate immune genes were selected by using InnateDB, and KEGG analysis identified the signaling pathways associated with the immune genes that were down- and upregulated by IL-37a via IL-1R8-dependent (C, D) and IL-1R8-independent effects (E, F). Bars correspond to the statistical significance [-log(10)FDR] of the enrichment of the regulated pathways, and dots correspond to the number of DEGs involved in each of the pathways. Hierarchical clustering of the genes that were (G) downregulated (a < w ≈ a/R) or (H) upregulated (a > w ≈ a/R) by IL-37a via an IL-1R8-dependent pathway. I, J IL-1R8-independent genes (I) inhibited by IL-37a (w > a ≈ a/R) or (J) enhanced by IL-37a (w < a ≈ a/R). Normalized expression levels were used to create the heatmaps that depict the relative abundance of each of the genes among the genes of interest
Overall, the transcriptomic analysis provides valuable insights into the potential global regulatory impact of IL-37a on proinflammatory genes and pathways. Notably, it implicated various pathways, including TLR, cytokines/chemokines, mitogen-activated protein kinase (MAPK), and Janus kinase-signal transducer and activator of transcription (JAK-STAT), in the protective effects of IL-37a against lethal endotoxic shock. Furthermore, these findings suggest a molecular mechanism by which IL-37a confers protection through IL-1R8-dependent and IL-1R8-independent pathways (Fig. S6).
Transcriptomic analysis revealed that through an IL-1R8-dependent regulatory pathway, IL-37a significantly repressed genes associated with eight inflammatory signaling pathways (Fig. 5C, G; Table S9). These included several important pathogen pattern recognition receptors and the related genes Tlr2, Tlr7, Cd14, and Ddx58 (encoding RIG-I-like receptor) in the RLR pathway and the downstream signaling molecules Ikbke and Irf7. Consistent with our results, IL-37a inhibited a number of inflammatory cytokine/receptor genes, including Il15, Il18, Ifnb1, Kdr (encoding VEGF receptor), Csf1r (encoding CSF1 receptor), Ltbr (encoding lymphotoxin B receptor) and a chemokine (Cxcl10). Furthermore, IL-37a inhibited the expression of key genes in several signaling pathways, including JAK-STAT (Jak3) and MAPK (Pla2g4a, encoding cytosolic phospholipase A2 for the metabolic production of leukotrienes and prostaglandins) [40]. IL-37a also enhanced the expression of several regulatory genes (Fig. 5D, H). Cryab encodes alpha B-crystallin, which is in the protein processing in the endoplasmic reticulum (PPIER) pathway and has a therapeutic effect on inflammatory diseases [41]. Sirt1 and Tollip are well-known inhibitors of inflammation and the TLR signaling pathway [42, 43].
The unique receptor-independent regulatory effect of IL-37a prompted us to conduct a more comprehensive analysis of the differentially expressed genes, particularly those upregulated by IL-37a. Notably, IL-37a upregulated several immunoregulatory genes, such as Fzd1 (encoding Frizzled-1), Tsc22d3 (encoding Gilz), Serpinb2 (encoding SerpinB2), and Pparg (encoding peroxisome proliferator-activated receptor-gamma, PPARγ) [44,45,46,47,48,49] (Fig. 5J). Of particular interest among these immunoregulatory genes is Pparg. PPARγ can interfere with various proinflammatory pathways and may serve as a mechanistic link to the regulatory function of IL-37a. PPARγ functions as both a nuclear hormone receptor and a crucial regulator of lipid metabolism and plays a pivotal role in a wide range of proinflammatory diseases, including LPS shock, by targeting multiple proinflammatory and metabolic pathways [50,51,52].
Therefore, considering the significance and functional similarity between PPARγ and IL-37a in regulating inflammation, we conducted further investigations to explore the induction mechanism and the impact of PPARγ on nuclear IL-37a-mediated inhibition of the inflammatory response, including LPS shock. These studies were conducted in vitro and in vivo.
FL and N-terminal IL-37a inhibit LPS-induced production of proinflammatory cytokines via transcriptional upregulation of Pparg expression in vitro
Consistent with the transcriptomic analysis of splenocytes from WT and FLIL-37a tg mice, the mRNA and protein expression levels of Pparg in FLIL-37a- and NIL-37a-expressing but not mIL-37a-expressing murine macrophage lines and primary macrophages were markedly increased compared with those in the WT control group (Fig. 6A). Using specific Pparg siRNA, we also demonstrated the inhibitory effects of PPARγ on the expression of proinflammatory cytokines (Il6, Il1b, Tnfa) in macrophages, as reported previously [47, 48] (Fig. 6B). More importantly, knockdown of PPARγ expression abolished the anti-inflammatory effect of FL and NIL-37a on murine and human macrophages following LPS stimulation (Fig. 6C–I and Fig. S7). Thus, these findings suggest that FL and NIL-37a in the nucleus inhibit the inflammatory response by modulating PPARγ expression.
FL and N-terminal IL-37a inhibit LPS-induced production of proinflammatory cytokines by enhancing Pparg transcription. A In RAW264.7 cells stably expressing FLIL-37a, NIL-37a, or mIL-37a, Pparg mRNA expression was measured by qPCR and western blotting. Bone marrow-derived macrophages (BMDMs) from C57BL/6 J mice were transduced with lentiviral particles expressing FLIL-37a, NIL-37a, or control for 72 h. PPARγ expression was measured by western blotting using β-tubulin as a control. B RAW264.7 cells were transfected with siRNA targeting Pparg or control siRNA for 48 h, followed by stimulation with LPS for 24 h. The mRNA expression of Il6, Il1b and Tnfa was measured by qPCR. C–F BMDMs derived from WT, FLIL-37a tg, and NIL-37a tg mice were transfected with Pparg or control siRNA, followed by stimulation with LPS for 24 h. The levels of PPARγ and proinflammatory cytokines were measured by qPCR and western blotting. G–I PMA-differentiated THP1 cells were transfected with or without plasmids expressing FLIL-37a or NIL-37a and siRNA targeting Pparg or control siRNA as indicated for 48 h, followed by LPS treatment for 24 h. The expression of Pparg, Il1b, Il6, and Tnfa was measured by qPCR (G, H), and the protein expression of PPARγ was measured by western blotting (I). J ChIP‒qPCR of the Pparg1 enhancer/promoter region in RAW264.7 cells transfected with GFP-FLIL-37a or GFP-NIL-37a plasmids. Cell lysates were subjected to ChIP with bead-coupled antibodies against GFP. K, L Dual-luciferase assays of Pparg1 promoter activity in RAW264.7 cells transfected with (K) FLIL-37a or (L) NIL-37a plasmids, followed by LPS stimulation for 24 h. The data are representative of at least three independent experiments. The data are the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, compared with the control
Moreover, using a bioinformatics algorithm (Cistrome Data Browser, http://cistrome.org/db), FL and NIL-37a were predicted to bind to the promoter/enhancer region of Pparg1. We conducted a chromatin immunoprecipitation assay (ChIP) and demonstrated that FL and NIL-37a could bind to the distal promoter/enhancer region (−1300 to −1500) of Pparg1 (Fig. 6J). Next, to confirm whether FL and NIL-37a directly activated the transcription of Pparg1, we performed luciferase reporter assays on FL- or NIL-37a-overexpressing cells. The results revealed that the activity of the Pparg1 promoter was significantly increased by FL and NIL-37a (Fig. 6K, L) in cells with Pparg1 luciferase reporter constructs, including the enhancer elements and proximal promoter sequences of Pparg1 [47]. Overall, these findings suggested that FL and NIL-37a regulated the response to LPS, at least in part, via transcriptional upregulation of PPARγ expression.
IL-37a induces Pparg transcription by recruiting the H3K4 methyltransferase complex to the Pparg enhancer/promoter
Recent studies have suggested that MLL4 (KMT2D), a major mammalian H3K4 mono- and di-methyltransferase, interacts with the transcription factor CCAAT-enhancer-binding protein beta (C/EBPβ) to initiate PPARγ expression by methylating H3K4 on the Pparg enhancer [53]. To investigate the involvement of MLL4 and C/EBPβ in the effects of IL-37a, macrophages overexpressing FL or NIL-37a were transfected with Mll4 or Cebpb siRNA, and Pparg expression levels were measured. Neither IL-37a isoform affected the expression of Cebpb or Mll4, but knockdown of C/EBPβ and MLL4 expression abolished FL- and NIL-37a-induced Pparg expression (Fig. 7A–D and Fig. S8a–d).
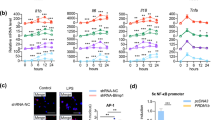
N-terminal IL-37a epigenetically activates Pparg expression in a WDR5/C/EBPβ-dependent manner. A–D The expression of Pparg, Cebpb, or Mll4 in BMDMs from (A, B) FLIL-37a tg or (C, D) NIL-37a tg mice that were transfected with or without siCebpb (A, C) or siMll4 (B, D), followed by LPS treatment for 24 h, was measured by qPCR. E, F RAW264.7 cells were transfected with plasmids expressing GFP-fused (E) FLIL-37a or (F) NIL-37a for 24 h. Cell extracts were subjected to immunoprecipitation with bead-coupled antibodies against GFP, WDR5 or C/EBPβ. Proteins bound to the beads were fractionated by SDS‒PAGE. G–J Protein and mRNA expression levels of PPARγ and WDR5 in BMDMs from (G, H) FLIL-37a tg or (I, J) NIL-37a tg mice with or without siWdr5 transfection, followed by LPS stimulation for 24 h. K, L Dual-luciferase analysis of Pparg1 promoter activity in (K) FLIL-37a- or (L) NIL-37a-stably expressed RAW264.7 cells with or without siWdr5 transfection, followed by LPS treatment for 24 h. M, N ChIP‒qPCR analysis the expression of H3K4me1 and H3K4me2 in the Pparg1 enhancer/promoter region in RAW264.7 cells transfected with or without FLIL-37a- or NIL-37a-expressing plasmids or siWdr5. Cell lysates were subjected to ChIP with bead-coupled antibodies against H3K4me1 and H3K4me2. The data are representative of at least three independent experiments. The data are the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, compared with the control
To further elucidate the mechanism by which nuclear IL-37a enhances Pparg transcription, a series of proteomics, immunoprecipitation, and luciferase reporter analyses were performed to identify nuclear IL-37a-interacting proteins in macrophages expressing FL or NIL-37a tagged with GFP. First, proteomics analysis showed that WD repeat domain 5 (WDR5) was a major protein that coprecipitated with FLIL-37a (Table S11). WDR5 is a core subunit of MLL4 methyltransferase complexes that mediates H3K4 methylation during gene transactivation [54], but its role in Pparg transcription is unknown. Subsequently, we investigated whether FL or NIL-37a could directly bind to the WDR5/MLL4/C/EBPβ complex in macrophages. Coimmunoprecipitation experiments revealed marked enrichment in the protein levels of WDR5 and C/EBPβ in FL- and NIL-37a-overexpressing cells relative to the IgG control, suggesting an interaction between IL-37a and the WDR5/C/EBPβ complex in macrophages (Fig. 7E, F). Knockdown of WDR5 expression diminished the effect of FL- and NIL-37a-enhanced PPARγ expression in macrophages (Fig. 7G–J and Fig. S8e–h). Furthermore, luciferase reporter assays showed that knockdown of WDR5 expression significantly decreased the FL- and NIL-37a-enhanced activity of Pparg1 promoters in murine macrophages (Fig. 7K, L), suggesting the important role of WDR5 in nuclear IL-37a-mediated Pparg transcription.
To determine whether WDR5/MLL4 mediated H3K4me1/2 levels on the Pparg1 enhancer/promoter in cells with FL- and NIL-37a-induced Pparg1 transactivation, we performed ChIP assays using H3K4me1- or H3K4me2-specific antibodies and macrophages. We observed that FL and NIL-37a significantly enhanced the level of H3K4me1 and H3K4me2 on the enhancer/promoter region of Pparg1. However, this enhancement was abolished by the knockdown of WDR5 expression (Fig. 7M, N), suggesting the regulatory role of WDR5 in modulating H3K4me1/2 levels on the Pparg1 enhancer/promoter.
Finally, we identified the interacting residues between IL-37a and WDR5, MLL4, and C/EBPβ using UCSF Chimera software [55] with the “Viewdock” and “Find Hbond” options. Our findings indicated that NIL-37a may exert its regulatory effects on Pparg expression via Asn15, leading to an interaction with WDR5/MLL4/C/EBPβ (Fig. S9).
Collectively, these data suggest that FL and NIL-37a are novel inducers of Pparg that promote the recruitment of the H3K4 methyltransferase complex WDR5/MLL4/C/EBPβ, as well as H3K4me1 and H3K4me2, to the Pparg enhancer/promoter region, thereby enhancing Pparg transcription.
PPARγ plays a critical role in FLIL-37a-mediated protection against LPS shock in vivo
We further investigated the impact of PPARγ on IL-37a-mediated beneficial effects in vivo using the well-defined PPARγ antagonist GW9662 [47, 48]. WT, N, and FLIL-37a tg mice were pretreated with or without GW9662 before a lethal LPS challenge, and mortality was determined as described previously. Consistent with the results shown in Fig. 4, more FLIL-37a tg mice and, to a lesser extent, NIL-37a tg mice survived than WT control mice. Importantly, the protective effects of N and FLIL-37a were significantly decreased by the PPARγ antagonist compared to groups that were not given the inhibitor (Fig. 8A). The increased mortality observed in response to PPARγ inhibition was accompanied by elevated expression levels of the cytokines IL-1α, IL-1β, IL-6, and TNF-α (Fig. 8B). GW9662 administration also normalized the expression of genes induced (Angpl4, Dusp5, and Tmem258) or inhibited (Nrp2, Ltbr, and Nad2) by PPARγ [48] in spleen tissues compared to the levels in untreated controls (Fig. 8C, D). These findings suggest that PPARγ plays a critical role in N- and FLIL-37a-mediated protection against lethal endotoxic shock and cytokine storms.
PPARγ plays a critical role in FLIL-37a-mediated protection against LPS shock in vivo. A WT, FLIL-37a tg and NIL-37a tg mice (n = 10/group) were pretreated (i.p.) with 100 μL of GW9662 (1.5 mg/kg) or vehicle control on Day 0 and challenged with LPS (40 mg/kg, i.p.) on Day 3, and the survival rate and wellbeing were monitored regularly up to 24 h. B–D WT, FLIL-37a tg and NIL-37a tg mice (n = 3 mice/group) were pretreated (i.p.) with 100 μL of GW9662 (1.5 mg/kg) or solvent control on Day 0 and challenged with LPS (10 mg/kg, i.p.) on Day 3. Sixteen hours after LPS challenge, the mice were sacrificed, and samples of blood and spleen tissue were harvested. The serum levels of proinflammatory cytokines were measured by CBA (B), and expression of PPARγ-enhanced (C) and -inhibited (D) genes was measured by qPCR. The data are the mean ± SEM and are representative of three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001. E Schematic representation of the IL-1R8-dependent and -independent pathways mediated by nuclear and cytokine IL-37a
Discussion
The evidence presented in this study reveals the previously unknown function of IL-37a, specifically its nuclear form. The nuclear form of IL-37a could act as both a nuclear factor and a cytokine through IL-1R8-dependent and -independent mechanisms (Fig. 8E). These mechanisms are clearly distinct from those of IL-37b.
One of the remarkable characteristics of IL-37a is its nuclear localization and receptor-independent regulatory property, which is shared by both FLIL-37a and NIL-37a. Complete inhibition of the nuclear translocation of FLIL-37a was achieved by a mutation in the NLS or by using an importin inhibitor. This led us to hypothesize that FLIL-37a enters the nucleus through its NLS, particularly at amino acids 20 and 21, via the classic nuclear importin pathway, in which is common to many transcription factors [15, 56]. Surprisingly, similar to FLIL-37a, the NIL-37a fragment, which consists of 21 amino acids, also exerted protective effects against LPS-induced mortality independent of IL-1R8. This unique phenomenon suggests that the regulatory effect of FLIL-37a in the nucleus may be attributed to NIL-37a. Notably, the NIL-37a fragment can be generated through elastase-mediated cleavage between amino acids L21 and R22 of FLIL-37a. Consequently, the NIL-37a fragment could naturally occur in various cell types that produce both IL-37a and elastase, including macrophages and neutrophils [57]. Thus, under inflammatory conditions, the induction of FLIL-37a and NIL-37a could regulate proinflammatory genes independent of a receptor. It is important to note that the expression of the IL-1R8 receptor can be rapidly downregulated by various inflammatory signals or posttranscriptional modifications in diseases [50, 58, 59]. Therefore, an IL-1R8-independent regulatory mechanism may hold greater relevance in these contexts.
Compared to FL and NIL-37a, only a small portion of mature IL-37a was found in the nucleus. The mature IL-37a protein (R22-D192), which lacks the NLS, shares 98% homology with caspase-1-cleaved IL-37b. Therefore, it is likely that after being processed, mature IL-37a undergoes nuclear translocation via Smad3, as observed for mature IL-37b [6, 7]. However, both endogenous and exogenous mIL-37a failed to inhibit the response to LPS in the absence of IL-1R8, suggesting that, similar to IL-37b [27], mIL-37a primarily regulates the response to LPS through a receptor-dependent mechanism.
Despite their nuclear localization, IL-37a proteins, similar to IL-37b proteins, are also abundant in the cytosol [16, 60]. The mechanisms by which IL-37a is released from cells are poorly understood, although ATP has been shown to promote the release of IL-37b and nuclear IL-33 [61]. Moreover, our findings demonstrate that both the recombinant FL and mature forms of IL-37a effectively inhibit the response to LPS in an IL-1R8-dependent manner. Given that the C-terminus of IL-37a possesses the same IL-1-like domain as IL-37b, these two isoforms of IL-37 are expected to share some similarities in receptor binding, signaling, and function [7]. Consequently, FLIL-37a can act as both a nuclear factor and a cytokine, suppressing ongoing inflammatory responses through intracrine, autocrine, and paracrine mechanisms (Fig. 8E).
In mice, the rFLIL-37a protein and transgenic FLIL-37a more robustly inhibited the TLR response than IL-37b in vitro and in vivo. In FLIL-37a transgenic mice, this effect can be attributed to its unique nuclear-based, receptor-independent regulatory property. However, it is still unknown why rFLIL-37a exhibited higher suppressive activity than rFLIL-37b in response to LPS. It has been shown that mature IL-37b can form head-to-head homodimers, which abolish its suppressive activity [62, 63]. It remains unknown whether FLIL-37a can effectively form homodimers. It would be intriguing to investigate whether the unique N-terminal sequence of the IL-37a protein influences its dimerization to potentially enhance protein stability, receptor-binding affinity, and signaling. Further studies are necessary to test this hypothesis.
Transcriptomic analysis revealed that IL-37a can regulate a wide range of proinflammatory genes, including cytokines and those involved in various cell signaling pathways, in response to LPS. Additionally, our investigations demonstrated that nuclear IL-37a can induce the expression of PPARγ, a multifaceted nuclear receptor that plays a crucial role in diverse metabolic and inflammatory conditions [48, 64,65,66]. Evidence from animal studies suggests the beneficial effects of PPARγ on LPS-mediated inflammatory conditions [47, 67]. PPARγ exerts its inhibitory effects on the response to LPS by interacting with and suppressing the functions of several key proinflammatory transcription factors, such as NF-κB, STAT, NF-AT, and AP1 [47, 64,65,66,67,68]. Furthermore, PPARγ interferes with the activity of various protein kinases, particularly MAPK [49, 52, 66,67,68]. Our in vitro and in vivo results indicate that IL-37a can downregulate the expression of these transcription factors and protein kinase-related inflammatory effects. Therefore, it is highly likely that the regulatory effects of IL-37a, particularly in response to LPS, are primarily mediated through PPARγ.
Several studies have explored the transcriptional and epigenetic regulation of PPARγ expression [69]. Our ChIP and luciferase reporter assays provided insights into the anti-inflammatory effects of nuclear FL and NIL-37a by enhancing Pparg transcription. Genome-wide profiling using ChIP-Seq has revealed a correlation between histone methylation and gene activation, which depends on the methylation sites and states [70]. WDR5, a component of the H3K4 mono- and di-methyltransferase MLL4 complex associated with gene activation [54], has attracted attention. Notably, previous studies have demonstrated that ectopic expression of C/EBPβ and the methyltransferase MLL4 induces PPARγ expression [53, 71]. In our study, through a combination of ChIP, luciferase reporter assays, and proteomics analyses, we observed that nuclear IL-37a facilitated the recruitment of the H3K4 methyltransferase complex WDR5/MLL4/C/EBPβ to the enhancer/promoter region of the Pparg gene in macrophages. This interaction may initiate enhancer-promoter communication and subsequently lead to gene transcription [53, 72].
The function of PPARγ is also influenced by phosphorylation. For instance, phosphorylation of PPARγ at S112 has been shown to impair IL-10 production and hinder inflammation resolution during bacterial pneumonia [73, 74]. Additionally, IL-37 stimulation has been shown to induce IL-1R8 phosphorylation and degradation [75]. However, it is currently unknown whether IL-37a modulates the phosphorylation of PPARγ.
Septic shock is a leading cause of human mortality, particularly in children, and limited treatment options are available [4, 76]. The potential role of human IL-37a in a clinical setting remains to be investigated. Our findings revealed significantly reduced expression of most IL-37 isoforms, including IL-37a, in children with sepsis. Furthermore, LPS-induced sepsis was associated with reduced expression of PPARγ [77]. Therefore, selective induction of an IL-37a-PPARγ functional axis may be a novel approach to improve immune tolerance against dysregulated inflammatory responses and cytokine storms in various inflammatory and infectious diseases, including septic shock.
There are certain limitations in this study. The detection of IL-37 isoforms was limited to mRNA levels in human cells due to the lack of specific antibodies or ELISA kits for the IL-37 isoforms. Additionally, a larger number of patient samples is necessary to thoroughly evaluate the impact of IL-37 on children suffering from septic shock.
References
Suntharalingam G, Perry MR, Ward S, Brett SJ, Castello-Cortes A, Brunner MD, et al. Cytokine storm in a phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N. Engl J Med. 2006;355:1018–28.
Schulert GS, Grom AA. Pathogenesis of macrophage activation syndrome and potential for cytokine- directed therapies. Annu Rev Med. 2015;66:145–59.
Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. Into the eye of the cytokine storm. Microbiol Mol Biol Rev. 2012;76:16–32.
Emgard J, Bergsten H, McCormick JK, Barrantes I, Skrede S, Sandberg JK, et al. MAIT cells are major contributors to the cytokine response in group a streptococcal toxic shock syndrome. Proc Natl Acad Sci USA. 2019;116:25923–31.
Kopf M, Bachmann MF, Marsland BJ. Averting inflammation by targeting the cytokine environment. Nat Rev Drug Discov. 2010;9:703–18.
Nold MF, Nold-Petry CA, Zepp JA, Palmer BE, Bufler P, Dinarello CA. IL-37 is a fundamental inhibitor of innate immunity. Nat Immunol. 2010;11:1014–22.
Boraschi D, Lucchesi D, Hainzl S, Leitner M, Maier E, Mangelberger D, et al. IL-37: a new anti-inflammatory cytokine of the IL-1 family. Eur Cytokine Netw. 2011;22:127–47.
Banchereau J, Pascual V, O’Garra A. From IL-2 to IL-37: the expanding spectrum of anti-inflammatory cytokines. Nat Immunol. 2012;13:925–31.
Smith DE, Renshaw BR, Ketchem RR, Kubin M, Garka KE, Sims JE. Four new members expand the interleukin-1 superfamily. J Biol Chem. 2000;275:1169–75.
Kumar S, McDonnell PC, Lehr R, Tierney L, Tzimas MN, Griswold DE, et al. Identification and initial characterization of four novel members of the interleukin-1 family. J Biol Chem. 2000;275:10308–14.
Busfield SJ, Comrack CA, Yu G, Chickering TW, Smutko JS, Zhou H, et al. Identification and gene organization of three novel members of the IL-1 family on human chromosome 2. Genomics. 2000;66:213–6.
Rudloff I, Cho SX, Lao JC, Ngo D, McKenzie M, Nold-Petry CA, et al. Monocytes and dendritic cells are the primary sources of interleukin 37 in human immune cells. J Leukoc Biol. 2017;101:901–11.
Papasavva M, Amvrosiou S, Pilala KM, Soureas K, Christodoulou P, Ji Y, et al. Deregulated expression of IL-37 in patients with bladder urothelial cancer: the diagnostic potential of the IL-37e isoform. Int J Mol Sci. 2023;24:9258.
Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204.
Kaffman A, O’Shea EK. Regulation of nuclear localization: a key to a door. Annu Rev Cell Dev Biol. 1999;15:291–339.
Cohen I, Rider P, Carmi Y, Braiman A, Dotan S, White MR, et al. Differential release of chromatin-bound IL-1alpha discriminates between necrotic and apoptotic cell death by the ability to induce sterile inflammation. Proc Natl Acad Sci USA. 2010;107:2574–9.
Li C, Zhao M, Zhao M, Chen N, Guo Y, Du Y, et al. IL-37 isoform D acts as an inhibitor of soluble ST2 to boost type 2 immune homeostasis in white adipose tissue. Cell Death Discov. 2022;8:163.
McNamee EN, Masterson JC, Jedlicka P, McManus M, Grenz A, Collins CB, et al. Interleukin 37 expression protects mice from colitis. Proc Natl Acad Sci USA. 2011;108:16711–6.
Ballak DB, van Diepen JA, Moschen AR, Jansen HJ, Hijmans A, Groenhof GJ, et al. IL-37 protects against obesity-induced inflammation and insulin resistance. Nat Commun. 2014;5:4711.
Ye L, Jiang B, Deng J, Du J, Xiong W, Guan Y, et al. IL-37 alleviates rheumatoid arthritis by suppressing IL-17 and IL-17-triggering cytokine production and limiting Th17 cell proliferation. J Immunol. 2015;194:5110–9.
Henry CJ, Casas-Selves M, Kim J, Zaberezhnyy V, Aghili L, Daniel AE, et al. Aging-associated inflammation promotes selection for adaptive oncogenic events in B cell progenitors. J Clin Invest. 2015;125:4666–80.
Liu JX, Liao B, Yu QH, Wang H, Liu YB, Guo CL, et al. The IL-37-Mex3B-Toll-like receptor 3 axis in epithelial cells in patients with eosinophilic chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol. 2020;145:160–72.
Zhang ZZ, Zhang Y, He T, Sweeney CL, Baris S, Karakoc-Aydiner E, et al. Homozygous IL37 mutation associated with infantile inflammatory bowel disease. Proc Natl Acad Sci USA. 2021;118:e2009217118.
Luo Y, Cai X, Liu S, Wang S, Nold-Petry CA, Nold MF, et al. Suppression of antigen-specific adaptive immunity by IL-37 via induction of tolerogenic dendritic cells. Proc Natl Acad Sci USA. 2014;111:15178–83.
Tsilioni I, Patel AB, Pantazopoulos H, Berretta S, Conti P, Leeman SE, et al. IL-37 is increased in brains of children with autism spectrum disorder and inhibits human microglia stimulated by neurotensin. Proc Natl Acad Sci USA. 2019;116:21659–65.
Li S, Neff CP, Barber K, Hong J, Luo Y, Azam T, et al. Extracellular forms of IL-37 inhibit innate inflammation in vitro and in vivo but require the IL-1 family decoy receptor IL-1R8. Proc Natl Acad Sci USA. 2015;112:2497–502.
Nold-Petry CA, Lo CY, Rudloff I, Elgass KD, Li S, Gantier MP, et al. IL-37 requires the receptors IL-18Ralpha and IL-1R8 (SIGIRR) to carry out its multifaceted anti-inflammatory program upon innate signal transduction. Nat Immunol. 2015;16:354–65.
Jia C, Zhuge Y, Zhang S, Ni C, Wang L, Wu R, et al. IL-37b alleviates endothelial cell apoptosis and inflammation in Kawasaki disease through IL-1R8 pathway. Cell Death Dis. 2021;12:575.
Sharma S, Kulk N, Nold MF, Graf R, Kim SH, Reinhardt D, et al. The IL-1 family member 7b translocates to the nucleus and down-regulates proinflammatory cytokines. J Immunol. 2008;180:5477–82.
Goldstein B, Giroir B, Randolph A, International Consensus Conference on Pediatric S. International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med. 2005;6:2–8.
Zhiguang X, Wei C, Steven R, Wei D, Wei Z, Rong M, et al. Over-expression of IL-33 leads to spontaneous pulmonary inflammation in mIL-33 transgenic mice. Immunol Lett. 2010;131:159–65.
Hall B, Cho A, Limaye A, Cho K, Khillan J, Kulkarni AB. Genome editing in mice using CRISPR/Cas9 technology. Curr Protoc Cell Biol. 2018;81:e57.
Kurowska-Stolarska M, Stolarski B, Kewin P, Murphy G, Corrigan CJ, Ying S, et al. IL-33 amplifies the polarization of alternatively activated macrophages that contribute to airway inflammation. J Immunol. 2009;183:6469–77.
Guo J, Qiu X, Zhang L, Wei R. Smurf1 regulates macrophage proliferation, apoptosis and migration via JNK and p38 MAPK signaling pathways. Mol Immunol. 2018;97:20–6.
Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13.
Ma Y, Chen W, Zhang X, Yu L, Dong W, Pan S, et al. Increasing the efficiency of CRISPR/Cas9-mediated precise genome editing in rats by inhibiting NHEJ and using Cas9 protein. RNA Biol. 2016;13:605–12.
Drutovic D, Duan X, Li R, Kalab P, Solc P. RanGTP and importin beta regulate meiosis I spindle assembly and function in mouse oocytes. EMBO J. 2020;39:e101689.
Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, et al. InnateDB: systems biology of innate immunity and beyond-recent updates and continuing curation. Nucleic Acids Res. 2013;41:D1228–33.
Sun GY, Chuang DY, Zong Y, Jiang J, Lee JC, Gu Z, et al. Role of cytosolic phospholipase A2 in oxidative and inflammatory signaling pathways in different cell types in the central nervous system. Mol Neurobiol. 2014;50:6–14.
Ousman SS, Tomooka BH, van Noort JM, Wawrousek EF, O’Connor KC, Hafler DA, et al. Protective and therapeutic role for alphaB-crystallin in autoimmune demyelination. Nature. 2007;448:474–9.
Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–80.
Burns K, Clatworthy J, Martin L, Martinon F, Plumpton C, Maschera B, et al. Tollip, a new component of the IL-1RI pathway, links IRAK to the IL-1 receptor. Nat Cell Biol. 2000;2:346–51.
Pinheiro I, Dejager L, Petta I, Vandevyver S, Puimege L, Mahieu T, et al. LPS resistance of SPRET/Ei mice is mediated by Gilz, encoded by the Tsc22d3 gene on the X chromosome. EMBO Mol Med. 2013;5:456–70.
Neumann J, Schaale K, Farhat K, Endermann T, Ulmer AJ, Ehlers S, et al. Frizzled1 is a marker of inflammatory macrophages, and its ligand Wnt3a is involved in reprogramming Mycobacterium tuberculosis-infected macrophages. FASEB J. 2010;24:4599–612.
Schroder WA, Le TT, Major L, Street S, Gardner J, Lambley E, et al. A physiological function of inflammation-associated SerpinB2 is regulation of adaptive immunity. J Immunol. 2010;184:2663–70.
Jiang C, Ting AT, Seed B. PPAR-gamma agonists inhibit production of monocyte inflammatory cytokines. Nature. 1998;391:82–86.
Daniel B, Nagy G, Czimmerer Z, Horvath A, Hammers DW, Cuaranta-Monroy I, et al. The nuclear receptor PPARgamma controls progressive macrophage polarization as a ligand-insensitive epigenomic ratchet of transcriptional memory. Immunity. 2018;49:615–26.e616.
Lehrke M, Lazar MA. The many faces of PPARgamma. Cell. 2005;123:993–9.
Zhao J, Bulek K, Gulen MF, Zepp JA, Karagkounis G, Martin BN, et al. Human colon tumors express a dominant-negative form of SIGIRR that promotes inflammation and colitis-associated colon cancer in mice. Gastroenterology. 2015;149:1860–71.e1868.
Abdelrahman M, Sivarajah A, Thiemermann C. Beneficial effects of PPAR-gamma ligands in ischemia-reperfusion injury, inflammation and shock. Cardiovasc Res. 2005;65:772–81.
Desreumaux P, Dubuquoy L, Nutten S, Peuchmaur M, Englaro W, Schoonjans K, et al. Attenuation of colon inflammation through activators of the retinoid X receptor (RXR)/peroxisome proliferator-activated receptor gamma (PPARgamma) heterodimer. A basis for new therapeutic strategies. J Exp Med. 2001;193:827–38.
Lee JE, Wang C, Xu S, Cho YW, Wang L, Feng X, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife. 2013;2:e01503.
Guarnaccia AD, Tansey WP. Moonlighting with WDR5: a cellular multitasker. J Clin Med. 2018;7:21.
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12.
Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5.
Curci JA, Liao S, Huffman MD, Shapiro SD, Thompson RW. Expression and localization of macrophage elastase (matrix metalloproteinase-12) in abdominal aortic aneurysms. J Clin Invest. 1998;102:1900–10.
Molgora M, Barajon I, Mantovani A, Garlanda C. Regulatory role of IL-1R8 in immunity and disease. Front Immunol. 2016;7:149.
Ueno-Shuto K, Kato K, Tasaki Y, Sato M, Sato K, Uchida Y, et al. Lipopolysaccharide decreases single immunoglobulin interleukin-1 receptor-related molecule (SIGIRR) expression by suppressing specificity protein 1 (Sp1) via the Toll-like receptor 4 (TLR4)-p38 pathway in monocytes and neutrophils. J Biol Chem. 2014;289:18097–109.
Bulau AM, Nold MF, Li S, Nold-Petry CA, Fink M, Mansell A, et al. Role of caspase-1 in nuclear translocation of IL-37, release of the cytokine, and IL-37 inhibition of innate immune responses. Proc Natl Acad Sci USA. 2014;111:2650–5.
Kouzaki H, Iijima K, Kobayashi T, O’Grady SM, Kita H. The danger signal, extracellular ATP, is a sensor for an airborne allergen and triggers IL-33 release and innate Th2-type responses. J Immunol. 2011;186:4375–87.
Ellisdon AM, Nold-Petry CA, D’Andrea L, Cho SX, Lao JC, Rudloff I, et al. Homodimerization attenuates the anti-inflammatory activity of interleukin-37. Sci Immunol. 2017;2:eaaj1548.
Eisenmesser EZ, Gottschlich A, Redzic JS, Paukovich N, Nix JC, Azam T, et al. Interleukin-37 monomer is the active form for reducing innate immunity. Proc Natl Acad Sci USA. 2019;116:5514–22.
Montaigne D, Butruille L, Staels B. PPAR control of metabolism and cardiovascular functions. Nat Rev Cardiol. 2021;18:809–23.
Gross B, Pawlak M, Lefebvre P, Staels B. PPARs in obesity-induced T2DM, dyslipidaemia and NAFLD. Nat Rev Endocrinol. 2017;13:36–49.
Daynes RA, Jones DC. Emerging roles of PPARs in inflammation and immunity. Nat Rev Immunol. 2002;2:748–59.
Chung SW, Kang BY, Kim SH, Pak YK, Cho D, Trinchieri G, et al. Oxidized low density lipoprotein inhibits interleukin-12 production in lipopolysaccharide-activated mouse macrophages via direct interactions between peroxisome proliferator-activated receptor-gamma and nuclear factor-kappa B. J Biol Chem. 2000;275:32681–7.
Pascual G, Glass CK. Nuclear receptors versus inflammation: mechanisms of transrepression. Trends Endocrinol Metab. 2006;17:321–7.
Lee JE, Ge K. Transcriptional and epigenetic regulation of PPARgamma expression during adipogenesis. Cell Biosci. 2014;4:29.
Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37.
Wu Z, Xie Y, Bucher NL, Farmer SR. Conditional ectopic expression of C/EBP beta in NIH-3T3 cells induces PPAR gamma and stimulates adipogenesis. Genes Dev. 1995;9:2350–63.
Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–39.
Garg M, Johri S, Sagar S, Mundhada A, Agrawal A, Ray P, et al. Cardiolipin-mediated PPARgamma S112 phosphorylation impairs IL-10 production and inflammation resolution during bacterial pneumonia. Cell Rep. 2021;34:108736.
Choi JH, Banks AS, Kamenecka TM, Busby SA, Chalmers MJ, Kumar N, et al. Antidiabetic actions of a non-agonist PPARgamma ligand blocking Cdk5-mediated phosphorylation. Nature. 2011;477:477–81.
Li L, Wei J, Suber TL, Ye Q, Miao J, Li S, et al. IL-37-induced activation of glycogen synthase kinase 3beta promotes IL-1R8/Sigirr phosphorylation, internalization, and degradation in lung epithelial cells. J Cell Physiol. 2021;236:5676–85.
Fleischmann-Struzek C, Goldfarb DM, Schlattmann P, Schlapbach LJ, Reinhart K, Kissoon N. The global burden of paediatric and neonatal sepsis: a systematic review. Lancet Respir Med. 2018;6:223–30.
Zhou M, Wu R, Dong W, Jacob A, Wang P. Endotoxin downregulates peroxisome proliferator-activated receptor-gamma via the increase in TNF-alpha release. Am J Physiol Regul Integr Comp Physiol. 2008;294:R84–92.
Acknowledgements
We thank Drs. Ruaidhri Carmody and Thomas D Otto at Glasgow University for helping with the transcriptomic microarray data analysis. This study received financial support from Versus Arthritis UK (21327 to D.X.), the National Key R&D Program of China (2021YFC2701800 and 2021YFC2701802 to Y.Z.), the National Natural Science Foundation of China (82241038, 81974248 to Y.Z. and 81900751 to X.H.), the International Joint Laboratory Program of National Children’s Medical Center (EK1125180109 to Y.Z.), the Program for Outstanding Medical Academic Leader (2019LJ19 to Y.Z.), the Shanghai Rising-Star Program (22QA1401500 to X.H.), the Shanghai Committee of Science and Technology (21140902400 to Y.Z., 23ZR1407600, 21ZR1410000 to F.J. and 20ZR1408300 to X.H.), the Shenzhen Science and Technology Peacock Team Project (KQTD20170331145453160), and the National Health Research Institutes, Taiwan, China (EM-109-PP-10).
Author information
Authors and Affiliations
Contributions
D.X., R.W., and Y.Z. designed the study. R.W., X.H., M.Y., C.Z., J.F., G.L., M.I.C., X.X., X.L., and J.L. performed the experiments. L.Z. generated the four genetically modified mouse strains. M.I.C., Y.J., and M.C. performed the bioinformatics analysis. D.X., R.W., C.Q., P.Y., and Y.Z. analyzed the data. D.X., S.K.H., and Y.Z. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wei, R., Han, X., Li, M. et al. The nuclear cytokine IL-37a controls lethal cytokine storms primarily via IL-1R8-independent transcriptional upregulation of PPARγ. Cell Mol Immunol 20, 1428–1444 (2023). https://doi.org/10.1038/s41423-023-01091-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-023-01091-0
- Springer Nature Limited
Keywords
This article is cited by
-
PPARγ attenuates cellular senescence of alveolar macrophages in asthma-COPD overlap
Respiratory Research (2024)
-
Thirteen years to get from b to a: one of the neglected isoforms of IL-37 enters the stage
Cellular & Molecular Immunology (2023)