Abstract
The adaptor molecule MAVS forms prion-like aggregates to govern the RIG-I-like receptor (RLR) signaling cascade. Lys63 (K63)-linked polyubiquitination is critical for MAVS aggregation, yet the underlying mechanism and the corresponding E3 ligases and deubiquitinating enzymes (DUBs) remain elusive. Here, we found that the K63-linked polyubiquitin chains loaded on MAVS can be directly recognized by RIG-I to initiate RIG-I-mediated MAVS aggregation with the prerequisite of the CARDRIG-I-CARDMAVS interaction. Interestingly, many K63-linked polyubiquitin chains attach to MAVS via an unanchored linkage. We identified Ube2N as a major ubiquitin-conjugating enzyme for MAVS and revealed that Ube2N cooperates with the E3 ligase Riplet and TRIM31 to promote the unanchored K63-linked polyubiquitination of MAVS. In addition, we identified USP10 as a direct DUB that removes unanchored K63-linked polyubiquitin chains from MAVS. Consistently, USP10 attenuates RIG-I-mediated MAVS aggregation and the production of type I interferon. Mice with a deficiency in USP10 show more potent resistance to RNA virus infection. Our work proposes a previously unknown mechanism for the activation of the RLR signaling cascade triggered by MAVS-attached unanchored K63-linked polyubiquitin chains and establishes the DUB USP10 and the E2:E3 pair Ube2N-Riplet/TRIM31 as a specific regulatory system for the unanchored K63-linked ubiquitination and aggregation of MAVS upon viral infection.
Similar content being viewed by others
Introduction
Innate immunity is the first line of defense against invading pathogens, including viruses, bacteria, and parasites [1, 2]. Pathogen-associated molecular patterns (PAMPs), such as viral nucleic acids, can be sensed by pattern recognition receptors (PRRs) and trigger signal transduction cascades that culminate in the production of type I interferons (IFNs) [3]. IFN-α/β further elicits downstream signal transduction and leads to the transcriptional induction of a wide range of interferon-stimulated genes (ISGs) encoding antiviral molecules, which collectively suppress viral replication and assembly [3].
Viral RNA is sensed by cytosolic RNA sensors, known as retinoic-acid-inducible gene-I (RIG-I)-like receptors (RLRs), including RIG-I, MDA5, and LGP2, all of which contain a DExD-box RNA helicase domain [4, 5]. Previous studies have detailed the molecular mechanisms by which RIG-I recognizes dsRNA with a 5′-triphosphate moiety (5′-pppRNA) and becomes activated [6,7,8]. In the absence of viral RNA, RIG-I adopts an autorepressed state, and its N-terminal tandem caspase activation and recruitment domains (CARDs) form an intramolecular interaction with the HEL2i domain [7]. dsRNA binding and subsequent ATP hydrolysis release the autorepression of CARDRIG-I [7]. CARDRIG-I then recruits MAVS through a homotypic CARDRIG-I-CARDMAVS interaction and nucleates CARDMAVS filament formation [9, 10], which serves as a signaling platform to recruit and activate downstream signaling molecules, such as TBK1 and IRF3 [11].
Lysine63 (K63)-linked polyubiquitin chains are essential for RIG-I to activate MAVS. Short, unanchored K63-linked polyubiquitin chains can directly activate RIG-I after RIG-I binds with ligand RNA and exposes its CARD domain [12]. Biochemical and structural studies have revealed that RIG-I binds to K63-linked polyubiquitin chains to form a CARD tetramer, which serves as a helical template to nucleate MAVS aggregates on the mitochondrial membrane [9, 12, 13]. K63-linked polyubiquitin chains bind the periphery of the core CARD tetramer, bridging the adjacent subunits and promoting its assembly. Several critical ubiquitin-conjugating enzymes (E2s) and ubiquitin ligases (E3s) have been identified for RIG-I activation [14, 15]. Ube2D3-Riplet mediates the covalent conjugation of polyubiquitin chains to RIG-I, while Ube2N-Riplet preferentially catalyzes the formation of unanchored polyubiquitin chains for RIG-I [15]. Both types of ubiquitination of RIG-I are essential for RIG-I-mediated MAVS aggregation [15]. In addition, Riplet is reported to cross-bridge RIG-I filaments on longer dsRNAs to induce RIG-I assembly in a ubiquitin-independent manner [16]. Unlike RIG-I, which requires short K63-linked polyubiquitin chains, long, unanchored K63-linked polyubiquitin chains are reported to mediate tetramerization of the CARDs of MDA5 and bridge the CARDMDA5-CARDMAVS interaction [17]. The works above collectively prove the importance of K63-linked polyubiquitin chains in the RLR pathway. However, the origin of the K63-linked polyubiquitin chains recognized by RIG-I is still unclear, and whether certain molecules can provide polyubiquitin chains for RIG-I is still unknown.
In contrast to the detailed description of the ubiquitination of RIG-I, the function and molecular mechanism of MAVS-loaded ubiquitin chains in regulating RLR pathway activation remain elusive. Previously, we identified TRIM31 as a key E3 ubiquitin ligase catalyzing MAVS K63-linked ubiquitination, which facilitates the formation of prion-like MAVS aggregates [18, 19]. However, the underlying mechanism by which K63-linked polyubiquitination of MAVS regulates the aggregation and activation of MAVS is still totally unknown. In addition, despite being an important regulator for maintaining the ubiquitin balance of target proteins, the direct deubiquitinating enzyme (DUB) toward the K63-linked ubiquitination of MAVS is rarely reported and needs further exploration.
Here, utilizing an in vitro assay to detect RIG-I-mediated MAVS aggregation, we found that K63-linked polyubiquitin chains can be preloaded onto MAVS without inducing its aggregation and then recognized by RIG-I after the CARDRIG-I-CARDMAVS interaction to trigger RIG-I-mediated MAVS aggregation. A large portion of K63-linked polyubiquitin chains attach to MAVS via an unanchored linkage, which can serve as a direct activator for RIG-I after separation in vitro. We propose a previously unknown mechanism by which ubiquitin chains modify and regulate MAVS. In addition, we have identified Ube2N as the major E2 for MAVS K63-linked polyubiquitination and Riplet as a new E3 ligase for MAVS independent of RIG-I. Ube2N cooperates with Riplet or TRIM31 to catalyze the unanchored K63-linked polyubiquitin chains for MAVS and initiate RIG-I-mediated MAVS aggregation. Furthermore, we identified USP10 as a new DUB for MAVS, which mainly removes the unanchored K63-linked polyubiquitin chains loaded on MAVS and inhibits RIG-I-mediated MAVS aggregation. Deficiency of USP10 results in more potent resistance to RNA virus infection in cells and in mice. We propose that the DUB USP10 and E2:E3 pair Ube2N-Riplet/TRIM31 constitute a specific regulatory system for MAVS unanchored K63-linked ubiquitination and aggregation upon viral infection. Our work proposes a previously unknown mechanism of the activation of the RLR signaling cascade triggered by MAVS-attached unanchored K63-linked polyubiquitin chains and unveils corresponding regulators.
Results
K63-linked polyubiquitin chains on mitochondria induce RIG-I-mediated MAVS aggregation
To explore the possible role of K63-linked polyubiquitin chains attached to MAVS in the RIG-I-MAVS signaling cascade, we first developed an in vitro assay using MAVS aggregation as a readout for measuring the activation of RLR signaling (Supplementary Fig. 1A). Full-length Flag-tagged RIG-I was affinity purified from HEK293T cells transfected with the Flag-RIG-I expression plasmid (Supplementary Fig. 1B). Recombinant RIG-I was first incubated with its ligand 5′-pppRNA and free K63/K48-linked polyubiquitin chains (K63/K48-polyUb) or monomeric ubiquitin (Mono-Ub) in reaction buffer containing ATP and Mg2+. Then, the reaction mixture was added to crude mitochondria to trigger MAVS aggregation, which was detected by semidenaturing detergent agarose gel electrophoresis (SDD-AGE). Flag-RIG-I treated with 5′-pppRNA together with free K63-polyUb, but not K48-polyUb or mono-Ub, could induce MAVS aggregation in mitochondria (Supplementary Fig. 1C, Lanes 4 and 5), which is consistent with a previous report that unanchored K63-linked polyubiquitin chains can directly activate the RIG-I-mediated pathway [12] and validates the feasibility of our in vitro assay system.
To investigate whether K63-linked polyubiquitin chains loaded on mitochondria can activate RIG-mediated MAVS activation, we simply transfected HEK293T cells with plasmids expressing hemagglutinin (HA)-tagged ubiquitin mutants K63 (K63-Ub) and K48 (K48-Ub), in which all lysine residues except the lysine at position 63 or position 48, respectively, are replaced with arginine residues (Supplementary Fig. 1D). Then, HEK293T cells overexpressing K63-Ub or K48-Ub were homogenized to obtain crude mitochondria (named mitochondria-K63 or mitochondria-K48, respectively). Immunoprecipitation analysis showed that the mitochondria were efficiently modified with the corresponding polyubiquitination after transfection (Supplementary Fig. 1E). Then, the in vitro assay of RIG-I-mediated MAVS aggregation was performed with these crude mitochondria. As shown in Supplementary Fig. 1F, RIG-I pretreated with 5′-pppRNA efficiently triggered MAVS aggregation in mitochondria-K63 but not in mitochondria obtained without K63-Ub transfection (Lanes 5 and 6). In contrast, RIG-I without 5′-pppRNA treatment failed to trigger MAVS aggregation in mitochondria-K63 (Lane 3). In addition, transfection of K63-Ub alone could not induce MAVS aggregation in mitochondria-K63 (Lane 1). The above results indicate that K63-linked polyubiquitin chains attached by mitochondria can directly induce RIG-I-mediated MAVS aggregation when RIG-I has been activated by RNA ligands. The addition of free K63-polyUb could induce MAVS aggregation in mitochondria without K63-Ub transfection (Lane 8). However, the addition of free K63-polyUb could not induce more MAVS aggregation in mitochondria-K63 (Lane 7), probably due to the saturation of K63-linked ubiquitin chains offered to RIG-I. In addition, RIG-I pretreated with 5′-pppRNA could not promote MAVS aggregation in mitochondria-K48 (Supplementary Fig. 1G, Lane 3) compared to that in mitochondria-K63 (Supplementary Fig. 1G, Lane 1). The K270A mutation in the ATPase domain of RIG-I can abrogate the ability of RIG-I to induce interferon and IRF3 dimerization [12, 20]. We also prepared Flag-RIG-IK270A (Supplementary Fig. 1B) and performed the in vitro assay of MAVS aggregation. We found that RIG-IK270A with 5′-pppRNA could not trigger MAVS aggregation in mitochondria-K63, further proving that RIG-I activation is required for MAVS aggregation (Supplementary Fig. 1G, Lane 4). RIG-I pretreated with other ligands, 5′-ppp-hairpin RNA and poly(I:C), could also induce MAVS aggregation in mitochondria-K63 but not in mitochondria, indicating that activation of RIG-I-mediated MAVS aggregation by the K63-linked polyubiquitin chains on mitochondria has no specificity for RIG-I RNA ligand (Supplementary Fig. 1H).
To further confirm the above findings, we performed the in vitro assay of RIG-I-mediated MAVS aggregation using mitochondria prepared from HEK293T cells transfected with HA-tagged ubiquitin mutants K48R and K63R, which contain a single lysine-to-arginine substitution at position 48 or 63, respectively (Supplementary Fig. 1D and I). RIG-I pretreated with 5′-pppRNA was capable of inducing MAVS aggregation in mitochondria-K48R but not in mitochondria-K63R and mitochondria, further proving that K63-linkage is the major type of polyubiquitin chain on mitochondria activating RIG-I-mediated MAVS aggregation (Supplementary Fig. 1J, Lane 4–6). In summary, utilizing the established in vitro assay, we prove that K63-linked polyubiquitin chains loaded on mitochondria can replace free K63-polyUb to activate RIG-I.
K63-linked polyubiquitin chains provided by MAVS activate RIG-I
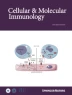
MAVS is located on mitochondria and interacts with RIG-I through the CARDRIG-I-CARDMAVS interaction [21, 22]. Immunoprecipitation analysis showed that MAVS in mitochondria was efficiently modified with polyubiquitination after transfection with K63-Ub, K48-Ub, K48R-Ub and K63R-Ub (Supplementary Fig. 1E and I). Thus, we speculate that K63-linked polyubiquitin chains provided by MAVS may be recognized by and activate RIG-I. To verify that the K63-linked polyubiquitin chains activating RIG-I are indeed provided by MAVS, we purified MAVS from HEK293T cells with or without transfection of K63-Ub as previously reported (named MAVS-K63-Ub and MAVS, respectively) [11] and performed the in vitro assay of RIG-I-mediated MAVS aggregation (Fig. 1A). In addition, we purified MAVS after Sendai virus (SeV) infection from HEK293T cells (MAVS-SeV) (Fig. 1B). The three purified MAVS samples showed a similar predominant band that corresponded to Flag-MAVS detected by silver staining (Fig. 1B). MAVS ubiquitination was easily detected in purified MAVS-K63-Ub and MAVS-SeV, whereas no ubiquitination signal was detected on MAVS (Fig. 1B). In addition, directly purified MAVS-SeV formed aggregates, as detected by SDD-AGE, whereas MAVS and MAVS-K63-Ub showed marginal aggregation signals, suggesting that K63-linked polyubiquitination of MAVS cannot directly trigger MAVS aggregation (Fig. 1C).
K63-linked polyubiquitin chains provided by MAVS activate RIG-I. A Procedures for the in vitro assay of RIG-I-mediated MAVS aggregation with purified MAVS (top) and crude mitochondria expressing MAVS or lysine-mutant MAVS (below). B Immunoblot (top) and silver staining (below) analysis of the ubiquitination of purified MAVS. C SDD-AGE analysis of purified MAVS aggregates. D SDD-AGE analysis of the purified MAVS aggregation in the in vitro assay. RIG-I was pretreated with or without 5′-pppRNA and free K63-polyUb and then incubated with purified MAVS or MAVS-K63-Ub in the in vitro assay, followed by the detection of MAVS aggregation with SDD-AGE (top) and SDS‒PAGE (below). E Sucrose gradient ultracentrifugation of purified MAVS and MAVS-K63-Ub pretreated in the in vitro assay system. Aliquots of the fractions were immunoblotted with MAVS antibody, and the corresponding quantification is shown below (present as % of the first lane). F Immunoprecipitation analysis of the ubiquitination of MAVS-wild type (WT) and its mutants in HEK293T cells transfected with plasmids encoding Myc-tagged MAVS (WT or mutant) and K63-Ub. G SDD-AGE analysis of the aggregation of exogenous MAVS-WT and its mutants K0, K7, K136, K297 and K500 in mitochondria extracted from HEK293T cells cotransfected with K63-Ub in the in vitro assay. H SDD-AGE analysis of the aggregation of exogenous MAVS-WT, mutant K0 and W56R in mitochondria extracted from HEK293T cells cotransfected with K63-Ub and with the addition of free K63-polyUb in the in vitro assay. I Sucrose gradient ultracentrifugation of mitochondrial Myc-tagged MAVS and its mutants in the in vitro assay system. Aliquots of the fractions were immunoblotted with Myc antibody, and corresponding quantification was performed as in (E). J Immunoprecipitation analysis of the ubiquitination of MAVS-WT and its mutants K0 and 4KR in HEK293T cells transfected with plasmids encoding Myc-tagged MAVS (WT or mutant) and K63-Ub. K SDD-AGE analysis of the aggregation of exogenous MAVS in mitochondria extracted from (J) in the in vitro assay. L Immunoprecipitation analysis of the ubiquitination of MAVS-WT and its mutants K0 and 4KR in HEK293T-MAVS−/− cells transfected with plasmids encoding Myc-tagged MAVS (WT or mutant) and K63-Ub. M SDD-AGE analysis of the aggregation of exogenous MAVS-WT and mutant K0 and 4KR in mitochondria extracted as (L) in the in vitro assay with CARDRIG-I-GST or GST. N SDD-AGE and immunoprecipitation analysis of MAVS-WT and its mutants K0 and 4KR in HEK293T-MAVS−/− cells transfected with plasmids encoding Myc-tagged MAVS (WT or mutant) and stimulated with SeV. The phosphorylation of IRF3 was detected in the same input sample. O, P SDD-AGE analysis of exogenous Myc-tagged MAVS aggregation in the mitochondria of HEK293T-MAVS−/− cells triggered by MAVS aggregates formed in vitro and immunoblot analysis of the phosphorylation of IRF3 in the cytosolic extracts prepared from HEK293T cells triggered by MAVS aggregates formed in vitro. MAVS aggregates were formed in the in vitro assay with purified MAVS (O, left panel) or endogenous MAVS in the mitochondria of HEK293T cells (P, left panel). Then, the aggregates were incubated with mitochondria from HEK293T-MAVS−/− cells transfected with Myc-tagged MAVS, followed by the detection of MAVS aggregation with SDD-AGE using a Myc antibody (right panel). The formed aggregates were then incubated with cytosolic extracts prepared from HEK293T cells, followed by the detection of IRF3 phosphorylation by immunoblot (right panel). β-Actin was used as a loading control. EV empty vector
Purified MAVS and MAVS-K63-Ub were used in the following in vitro RIG-I-mediated MAVS aggregation assay. As shown in Fig. 1D, purified RIG-I could not directly trigger MAVS-K63-Ub and MAVS aggregation (Lanes 1–4). Pretreatment of RIG-I with 5’-pppRNA induced aggregation of MAVS-K63-Ub but not MAVS (Lanes 5 and 6). The addition of free K63-polyUb to the in vitro reaction system caused the purified MAVS to aggregate as MAVS-K63-Ub (Lanes 7 and 8). These data indicate that K63-linked polyubiquitin chains provided by MAVS can replace free K63-polyUb to induce RIG-I-mediated MAVS aggregation. We reconfirmed the above results by sucrose gradient ultracentrifugation. Purified MAVS-K63-Ub, but not MAVS, formed a large complex and sedimented to the bottom of a centrifuge tube containing a high concentration of sucrose in the presence of 5′-pppRNA-treated RIG-I (Fig. 1E). The addition of free K63-polyUb induced the purified MAVS to sediment to a similar extent to MAVS-K63-Ub in the high-molecular-weight fractions (Fig. 1E). The results of sucrose gradient ultracentrifugation are consistent with the results of the in vitro MAVS aggregation assay.
To further demonstrate that the K63-linked polyubiquitin chains, which trigger RIG-I-mediated MAVS aggregation in the in vitro assay system, are indeed provided by MAVS, we abolished MAVS ubiquitination by mutating the lysine residues on MAVS as previously reported and detected MAVS aggregation in the in vitro assay [18] (Fig. 1A). First, we constructed the mutant MAVS-K0, in which all of the lysine residues in MAVS were replaced with arginine (Supplementary Fig. 2). Then, we reintroduced individual lysine residues into mutant K0 to generate single-lysine mutants (Supplementary Fig. 2). We transfected wild-type (WT) Myc-tagged MAVS and its mutants together with K63-Ub into HEK293T cells and measured K63-linked polyubiquitination of MAVS through immunoprecipitation. As shown in Fig. 1F, transfection of K63-Ub mainly induced polyubiquitination of MAVS-WT and mutants K7, K136, K297 and K500 in HEK293T cells, whereas K0 and other mutants showed relatively weak ubiquitination signals. Then, we extracted the corresponding crude mitochondria and performed the in vitro MAVS aggregation assay as shown in Fig. 1A. We found that MAVS-WT and MAVS mutants K7, K136, K297 and K500, which showed high levels of MAVS polyubiquitination after transfection with K63-Ub, could potently aggregate with the induction of 5′-pppRNA-treated RIG-I, whereas mutant K0 showed a weak aggregation signal (Fig. 1G). The addition of free K63-polyUb promoted mutant MAVS-K0 aggregation but had no effect on the polymerization-deficient mutant MAVS-W56R [23, 24] (Fig. 1H). The results indicate that although the mutant MAVS-K0 is not sufficiently modified by K63-linked polyubiquitin chains, it retains the capacity to form aggregates induced by the extra addition of free K63-polyUb. We also detected the aggregation of MAVS, mutant MAVS-K0 and MAVS-W56R utilizing sucrose gradient ultracentrifugation (Fig. 1I). The experiment showed that WT MAVS transfected with K63-Ub could sediment at the bottom of the sucrose gradient as a large complex upon treatment with RIG-I and 5′-pppRNA, whereas MAVS-K0 and MAVS-W56R remained at the top of the sucrose gradient, further confirming that MAVS-attached K63-linked polyubiquitin chains can trigger RIG-I-mediated MAVS aggregation (Fig. 1I).
To confirm that K63-linked polyubiquitination of Lysine 7, 136, 297 and 500 was involved in RIG-I-mediated MAVS aggregation, we constructed a mutant MAVS-4KR, in which the lysine residues of MAVS at positions 7, 136, 297 and 500 were replaced with arginine (Supplementary Fig. 2). We transfected MAVS-K0 and MAVS-4KR together with K63-Ub into HEK293T cells, extracted the corresponding mitochondria and detected MAVS aggregation in the in vitro assay. Immunoprecipitation analysis showed that mutants MAVS-K0 and 4KR were modified with fewer K63-linked polyubiquitin chains than MAVS-WT (Fig. 1J). Consistently, RIG-I-induced MAVS aggregation was greatly decreased in mutant MAVS-K0 and 4KR compared to MAVS-WT (Fig. 1K). CARDRIG-I can bind to free K63-Ub and bypass 5’-pppRNA stimulation to directly activate RLR signaling [12]. We expressed CARDRIG-I in E. coli and purified it to near homogeneity (Supplementary Fig. 3). We cotransfected MAVS-WT and mutants K0 and 4KR with K63-Ub into HEK293T-MAVS−/− cells and extracted the corresponding mitochondria. Immunoprecipitation analysis showed that mutants MAVS-K0 and 4KR were modified with impaired K63-linked polyubiquitin chains compared to MAVS-WT, which is consistent with the observations in WT HEK293T cells (Fig. 1L). We incubated CARDRIG-I with the above mitochondria in the in vitro assay. SDD-AGE analysis showed that CARDRIG-I could trigger obvious aggregation of MAVS-WT but not MAVS-K0 and 4KR (Fig. 1M). Together, the results confirmed that K63-linked polyubiquitin chains provided by MAVS can replace free K63-Ub to trigger RIG-I-mediated MAVS aggregation.
To test the effect of MAVS ubiquitination on the activation and transduction of RLR signaling, we reconstituted WT MAVS and mutant MAVS-K0 and 4KR in MAVS−/− cells infected with SeV and detected MAVS ubiquitination, aggregation, and downstream effectors. As shown in Fig. 1N, mutation of the lysine residues on MAVS inhibited SeV-triggered MAVS ubiquitination and aggregation and reduced IRF3 phosphorylation, indicating that the K63-linked ubiquitination of MAVS is crucial for RLR signal transduction. MAVS forms a prion-like structure to mediate downstream signaling [11]. A hallmark of prions is their ability to convert proteins from their native conformation to prion-like aggregation [11, 25]. To determine whether the MAVS aggregates induced by the K63-linked polyubiquitin chains of MAVS have prion-like activity, we checked the ability of MAVS aggregates prepared in vitro to induce native MAVS aggregation and downstream IRF3 activation. We first prepared MAVS aggregates with purified MAVS and MAVS-K63-Ub as shown in Fig. 1D. Then, we incubated these MAVS aggregates with mitochondria extracted from HEK293T-MAVS−/− cells transfected with Myc-tagged MAVS (Fig. 1O). Similar to the free unanchored K63-polyUb, the MAVS aggregates induced by the MAVS attached K63-linked polyubiquitin chains could convert the Myc-tagged MAVS on mitochondria from native conditions to prion-like aggregates (Fig. 1O, right panel). In addition, the above aggregates directly triggered IRF3 phosphorylation via incubation with cytosolic extracts prepared from HEK293T cells as previously reported [12] (Fig. 1O, right panel). We also prepared MAVS aggregates with mitochondria and mitochondria-K63. As shown in Fig. 1P, MAVS aggregates induced by mitochondria-attached K63-linked polyubiquitin chains could also induce the aggregation of native Myc-MAVS and IRF3 phosphorylation. These results prove that the K63-linked polyubiquitin chains on MAVS can mediate the formation of functional MAVS aggregates with prion-like activity and transduce the signal to downstream effectors.
CARDRIG-I-CARDMAVS interaction is the prerequisite for RIG-I recognition of K63-linked polyubiquitin chains attached by MAVS
To further investigate how the K63-linked polyubiquitin chains are recognized by RIG-I to induce MAVS activation, we measured the interaction between MAVS and RIG-I through co-immunoprecipitation analysis. As shown in Fig. 2A, purified RIG-I showed a relatively weak association with MAVS and MAVS-K63-Ub (Lanes 2 and 3), whereas RIG-I pretreated with 5′-pppRNA pulled down significant amounts of MAVS and MAVS-K63-Ub (Lanes 4 and 5). Notably, RIG-I bound more MAVS-K63-Ub than MAVS after treatment with 5′-pppRNA (Lanes 4 and 5), indicating that MAVS-attached ubiquitin chains are captured by RIG-I in a manner dependent on RIG-I activation by RNA ligands and strengthen the association between RIG-I and MAVS. However, RIG-I pretreated with free K63-polyUb bound approximately equal amounts of MAVS and MAVS-K63-Ub (Lanes 6 and 7). These results indicate that K63-linked polyubiquitin chains in MAVS, but not in RIG-I, strengthen the interaction between MAVS and RIG-I. Interestingly, RIG-I preincubated with free K63-polyUb (Lane 7) pulled down less MAVS-K63-Ub than RIG-I without free K63-polyUb (Lane 5). These results suggest that the formed RIG-I-Ub complex (Lane 7) cannot further capture the K63-linked polyubiquitin chains provided by MAVS and exhibits less capacity for binding with MAVS-K63-Ub than RIG-I without K63-polyUb pretreatment (Lane 5). We infer that pretreatment with free K63-polyUb exhausts the ubiquitin-binding capacity of RIG-I and induces a conformational change in RIG-I, which decreases the association between RIG-I and MAVS-K63-Ub. To further validate the data, we carried out co-immunoprecipitation analysis with inactive mutant RIG-IK270A and MAVS. RIG-IK270A exhibited no binding affinity with MAVS and MAVS-K63-Ub, and the K63-linked polyubiquitin chains on MAVS could not enhance this interaction in the presence of 5′-pppRNA, confirming that the function of K63-linked polyubiquitin chains of MAVS in strengthening the association between MAVS and RIG-I relies on RIG-I activation (Fig. 2B). We further found that RIG-I pretreated with 5′-pppRNA bound with more MAVS-K63-Ub than MAVS in a dose-dependent manner (Fig. 2C). In addition, we purified MAVS-WT and MAVS mutant 4KR from HEK293T cells transfected with MAVS or its mutant MAVS-4KR in the presence of K63-Ub. MAVS-4KR was modified with fewer K63-linked polyubiquitin chains than MAVS-WT (Fig. 2D). As expected, RIG-I with 5′-pppRNA pulled down more MAVS-K63-Ub than MAVS-4KR-K63-Ub, further proving that the K63-linked polyubiquitin chains on MAVS mediate the enhancement of the interaction between RIG-I and MAVS (Fig. 2D). Based on our results in Fig. 1F, the major lysine residues mediating MAVS ubiquitination are K7, K136, K297 and K500. K7 is located in the MAVS CARD domain, while K136, K297 and K500 are located in the other regions of MAVS (Supplementary Fig. 2). Then, we wondered whether the ubiquitin chains on the CARD or other regions of MAVS can specifically affect the association between MAVS and RIG-I. We constructed the MAVS mutant K136/K297/K500 (noted as 3K for short), as shown in Supplementary Fig. 2, which only conserves the identified lysine residues on MAVS except the CARD domain. Then, we purified MAVS-K7 and MAVS-3K in the presence or absence of K63-Ub and detected their association with RIG-I by co-immunoprecipitation. As shown in Fig. 2E, K63-linked polyubiquitin chains on either MAVS-K7 or MAVS-3K could enhance the association between MAVS and RIG-I, indicating that RIG-I can recognize the K63-linked polyubiquitin chains on CARD and other regions of MAVS.
The CARDRIG-I-CARDMAVS interaction is the prerequisite for RIG-I recognition of K63-linked polyubiquitin chains attached by MAVS. A Co-immunoprecipitation analysis of the interaction between RIG-I and MAVS or MAVS-K63-Ub. MAVS or MAVS-K63-Ub was incubated with RIG-I in the in vitro assay with or without 5′-pppRNA and free K63-polyUb, followed by immunoprecipitation with RIG-I antibody and immunoblot analysis of RIG-I and MAVS. B Co-immunoprecipitation analysis of the interaction between RIG-IK270A and MAVS or MAVS-K63-Ub. C Co-immunoprecipitation analysis of the interaction between RIG-I and increasing amounts of MAVS or MAVS-K63-Ub. D Co-immunoprecipitation analysis of the interaction between RIG-I and MAVS, MAVS-K63-Ub or MAVS-4KR-K63-Ub. E Co-immunoprecipitation analysis of the interaction between RIG-I and MAVS-K7, MAVS-K7-K63-Ub, MAVS-3K or MAVS-3K-K63-Ub. The reactions and analysis in (B–E) were the same as in (A). F Depiction of MAVS functional domains. G Immunoblot analysis of purified CARDMAVS and ∆CARDMAVS with or without K63-Ub. H Co-immunoprecipitation analysis of the interaction between RIG-I and CARDMAVS or CARDMAVS-K63-Ub. I Co-immunoprecipitation analysis of the interaction between RIG-I and ∆CARDMAVS or ∆CARDMAVS-K63-Ub. The reaction and analysis of (H) and (I) were the same as in (A). J SDD-AGE analysis of the aggregation of CARDMAVS, CARDMAVS-K63-Ub, ∆CARDMAVS and ∆CARDMAVS-K63-Ub in the in vitro assay with RIG-I in the presence or absence of 5′-pppRNA. K SDD-AGE analysis of the aggregation of MAVS and MAVS-K63-Ub with or without ∆CARDMAVS-K63-Ub and free K63-polyUb in the in vitro assay
To further determine which domain of MAVS is essential for the K63-linked polyubiquitin chains mediating RIG-I and MAVS interaction, we constructed the Flag-tagged MAVS truncation mutants CARDMAVS (amino acids 1-91) and ∆CARDMAVS (amino acids 92-540) and purified them with or without K63-Ub transfection (Fig. 2F and G). Both domains could be efficiently modified with K63-linked polyubiquitin chains (Fig. 2G). We performed co-immunoprecipitation analysis of MAVS truncation mutants and RIG-I in the in vitro assay and found that RIG-I pretreated with RNA ligand bound more CARDMAVS-K63-Ub than CARDMAVS (Fig. 2H), indicating that K63-linked polyubiquitin chains on CARDMAVS can be recognized by RIG-I after RNA ligand treatment and strengthen the interaction between RIG-I and MAVS. In contrast, RIG-I could not bind ∆CARDMAVS with or without K63-linked polyubiquitin chains (Fig. 2I). Although RIG-I has been reported to bind free K63-polyUb [12], the above data indicate that RIG-I cannot recognize the K63-linked polyubiquitin chains loaded on MAVS without the CARDMAVS domain, suggesting that the interaction between CARDRIG-I and CARDMAVS is the prerequisite for RIG-I recognition of the K63-linked polyubiquitin chains on MAVS. In addition, both MAVS-K63-Ub and CARDMAVS-K63-Ub formed aggregates with RIG-I in the in vitro assay, whereas ∆CARDMAVS could not form aggregates with or without K63-linked polyubiquitin chains (Fig. 2J). We added ∆CARDMAVS-K63-Ub into the in vitro assay and found that ∆CARDMAVS-K63-Ub could not replace free K63-polyUb to induce RIG-I-mediated MAVS aggregation (Fig. 2K). These results further prove that RIG-I cannot recognize the K63-linked polyubiquitin chains loaded on ∆CARDMAVS. In summary, these data demonstrate that the CARDRIG-I-CARDMAVS interaction is required for RIG-I to recognize the K63-linked polyubiquitin chains loaded on MAVS and that K63-linked polyubiquitination of MAVS can further strengthen the interaction between MAVS and RIG-I.
Unanchored K63-linked polyubiquitin chains from MAVS activate RIG-I
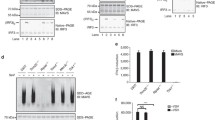
The above data show that the K63-linked polyubiquitin chains attached to MAVS can be recognized by RIG-I to induce MAVS aggregation. RIG-I has been reported to bind unanchored K63-linked polyubiquitin chains to activate RLR signaling [12]. Thus, we investigated whether the K63-linked polyubiquitin chains that activate RIG-I are covalently or noncovalently attached to MAVS. We first prepared two deubiquitination enzymes, viral OTU (vOTU) and isopeptidase T (IsoT) (Supplementary Fig. 3). The vOTU destroys polyubiquitin chains covalently linked to a target protein as well as unanchored polyubiquitin chains [12, 26, 27]. IsoT only cleaves unanchored polyubiquitin chains [12, 28]. Then, we treated purified MAVS-K63-Ub with vOTU and IsoT. We found that either IsoT or vOTU could potently reduce the ubiquitination signal on the purified MAVS-K63-Ub (Fig. 3A), suggesting that a large portion of polyubiquitin chains existing on MAVS are unanchored linkages. Following treatment with vOTU or IsoT, purified MAVS-K63-Ub was tested in the in vitro assay for its ability to trigger RIG-I-mediated MAVS aggregation. SDD-AGE analysis showed that both vOTU and IsoT treatment abolished MAVS aggregation (Fig. 3B). These data indicate that unanchored K63-linked polyubiquitin chains on MAVS are involved in RIG-I-mediated MAVS aggregation. To further confirm that unanchored polyubiquitin chains are attached to MAVS, we applied reimmunoprecipitation (Re-IP) experiments to investigate unanchored ubiquitin chains on MAVS. MAVS was first immunoprecipitated from HEK293T cells transfected with HA-K63-Ub or K48-Ub and then subjected to heat denaturation to disrupt noncovalent interactions. Immunoblot analysis showed that most of the K63-linked ubiquitination signal on MAVS was lost after heat denaturation, while the K48-ubiquitination signal remained relatively stable (Fig. 3C, see lanes labeled Re-IP). To further clarify the types of MAVS-loaded K63-linked polyubiquitin chains under physiological conditions, we detected MAVS ubiquitination after viral infection. As shown in Fig. 3D, SeV infection induced K63-linked polyubiquitination of MAVS, and the ubiquitination signal was greatly decreased after heat denaturation (see lanes labeled Re-IP). The results prove that viral infection triggers the attachment of a large number of noncovalently K63-linked polyubiquitin chains to MAVS.
Unanchored K63-linked polyubiquitin chains from MAVS activate RIG-I. A Immunoprecipitation analysis of the ubiquitination of MAVS-K63-Ub with the treatment of IsoT or vOTU. Purified MAVS-K63-Ub was pretreated with or without IsoT or vOTU before immunoprecipitation. B SDD-AGE analysis of the aggregation of purified MAVS-K63-Ub with the treatment of IsoT or vOTU in the in vitro assay. MAVS-K63-Ub was pretreated with IsoT or vOTU before affinity purification. Then, the purified MAVS-K63-Ub was incubated with RIG-I with or without 5’-pppRNA in the in vitro assay, followed by the detection of MAVS aggregation with SDD-AGE (top) and SDS‒PAGE (below). C Re-immunoprecipitation analysis of the ubiquitination of MAVS in HEK293T cells transfected with WT/K48/K63-Ub. MAVS was pulled down from HEK293T cells after ubiquitin transfection, and then the immunopurified MAVS was boiled in 1% SDS, re-immunoprecipitated, and analyzed for ubiquitination by immunoblotting. (D). Re-immunoprecipitation analysis of the ubiquitination of MAVS in HEK293T cells infected with SeV for 8 h. The experimental procedure was the same as in (C). E Procedures for the isolation of unanchored polyubiquitin chains on MAVS. F Immunoblot analysis with ubiquitin antibody of the supernatant in (E). The supernatant treated with CYLD was used as a control. G SDD-AGE analysis of MAVS aggregation triggered by the supernatant of (F) in the in vitro assay. RIG-I was pretreated with 5’-pppRNA and the supernatant and then incubated with purified MAVS (without ubiquitin), followed by the detection of MAVS aggregation with SDD-AGE (top) and SDS‒PAGE (below). H Immunoblot analysis with K63-ubiquitin antibody of the supernatant isolated from immunoprecipitated MAVS. MAVS was immunoprecipitated from HEK293T-DDX58−/− cells transfected with MAVS and K63-Ub, and then the supernatant was isolated as in (E). Supernatants treated with IsoT, vOTU and CYLD were used as controls. I SDD-AGE analysis of MAVS aggregation triggered by the supernatant of (H) in the in vitro assay. J Immunoblot analysis with K63-ubiquitin antibody of the supernatant isolated from immunoprecipitated MAVS and its mutants K0 and 4KR. The experimental procedure was the same as in (H). K SDD-AGE analysis of MAVS aggregation triggered by the supernatant of (J) in the in vitro assay. The reaction and analysis in (I) and (K) were the same as those in (G). L Co-immunoprecipitation analysis of the binding between purified MAVS and ubiquitin chains with different lengths and linkages. M Co-immunoprecipitation analysis of the binding between purified MAVS or its mutants and K63-linked ubiquitin chains with 5 moieties (Ub5). β-Actin was used as a loading control
To further confirm that unanchored K63-linked polyubiquitin chains are attached to MAVS, we attempted to release the unanchored K63-linked polyubiquitin chains on MAVS and check whether these ubiquitin chains can directly activate RIG-I. It has been reported that ubiquitin chains remain relatively stable at high temperature [12]. We followed the protocol described in Fig. 3E to prepare unanchored K63-linked polyubiquitin chains on MAVS from HEK293T cells after SeV infection. Immunoblot analysis of the supernatant with the ubiquitin antibody clearly showed the presence of unanchored polyubiquitin chains and that the major bands were in the range of 40–120 kDa, indicating that these unanchored ubiquitin chains contain ~5–12 ubiquitin moieties (Fig. 3F). We treated the supernatant with the K63-targeted deubiquitination enzyme CYLD [12] and found that CYLD diminished most of the bands, suggesting that most of the ubiquitin chains in the supernatant were K63-linked (Fig. 3F and Supplementary Fig. 3). The addition of the above supernatant to the in vitro assay with RIG-I and MAVS directly triggered RIG-I-mediated MAVS aggregation, and the CYLD-treated supernatant abolished this effect (Fig. 3G). Because RIG-I has been reported to bind unanchored K63-linked polyubiquitin chains [12], we also separated the polyubiquitin chains on MAVS purified from RIG-I-deficient (DDX58−/−) HEK293T cells transfected with MAVS and K63-Ub to rule out the possible effect of unanchored ubiquitin chains on RIG-I. As shown in Fig. 3H, the K63-linked polyubiquitin chains were separated from the immunoprecipitated MAVS, and these polyubiquitin chains could be cleaved by IsoT, vOTU and CYLD. Importantly, the separated supernatant potently induced RIG-I-mediated MAVS aggregation in the in vitro assay (Fig. 3I). In addition, there were clearly fewer polyubiquitin chains in the supernatant separated from MAVS-K0 and 4KR than from MAVS-WT (Fig. 3J), and the supernatants from the mutants showed a weak capacity to induce MAVS aggregation in the in vitro assay (Fig. 3K). The above results indicate that the unanchored K63-linked polyubiquitin chains separated from MAVS can serve as direct activators of RIG-I-mediated MAVS aggregation.
To further test whether MAVS and K63-linked polyubiquitin chains directly interact, we incubated purified MAVS with a panel of ubiquitin chains of different lengths and linkages. As shown in Fig. 3L, co-immunoprecipitation analysis showed that MAVS bound to K63-linked polyubiquitin chains containing 3–8 ubiquitin moieties (K63-Ub3-8) but showed weak affinity for K63-linked polyubiquitin chains with 2 ubiquitin moieties. In contrast, MAVS did not interact with monomeric ubiquitin and K48-linked polyubiquitin chains (Fig. 3L). In addition, we incubated MAVS mutants K0 and 4KR with K63-Ub5 and performed co-immunoprecipitation. The experiment showed that MAVS-K0 and 4KR bound to less K63-Ub5 than MAVS-WT, suggesting that the mutation may cause a conformational change in MAVS that blocks its interaction with polyubiquitin chains (Fig. 3M). Together, these results indicate that unanchored polyubiquitin chains are the major form of K63-linked polyubiquitination on MAVS and that these ubiquitin chains isolated from MAVS can directly activate RIG-I-mediated MAVS aggregation.
Ube2N-Riplet and Ube2N-TRIM31 catalyze the formation of unanchored K63-linked polyubiquitination on MAVS
Ube2N and Ube2D3 (also known as Ubc13 and UbcH5c, respectively) have been reported as critical E2s for K63-linked polyubiquitination and play essential roles in the RLR pathway [12, 15]. To test whether these E2s are required for MAVS ubiquitination, small interfering RNAs (siRNAs) against these E2s were transfected into HEK293T cells to knock down the expression of Ube2N and Ube2D3. siRNA knockdown of UBE2N expression significantly abolished the K63-linked polyubiquitination of MAVS in HEK293T cells after SeV infection, whereas knockdown of UBE2D3 showed no effect (Supplementary Fig. 4A and B), suggesting that Ube2N is the major E2 for K63-linked polyubiquitination of MAVS. Several E3s, including Riplet and TRIM25, can catalyze the synthesis of polyubiquitin chains with Ube2N and Ube2D3 and play critical roles in the RLR pathway [15, 16, 29]. To determine if these E3s engage in MAVS ubiquitination, siRNAs targeting RIPLET and TRIM25 were transfected into HEK293T cells to knock down the expression of RIPLET and TRIM25. We found that knockdown of RIPLET but not TRIM25 abrogated the K63-linked polyubiquitination of MAVS upon SeV infection (Supplementary Fig. 4A and C). Furthermore, we showed that siRNA knockdown of UBE2N and RIPLET expression, but not UBE2D3 and TRIM25, reduced the K63-linked polyubiquitination of exogenous MAVS in HEK293T cells transfected with Myc-tagged MAVS and K63-Ub (Supplementary Fig. 4D). In addition, siRNA knockdown of UBE2N and RIPLET in HeLa and RAW264.7 cells led to a reduction in the K63-linked polyubiquitination of MAVS (Supplementary Fig. 4E–G). Ube2N and Riplet have been reported to catalyze the K63-linked ubiquitination of RIG-I[15]. To eliminate the possibility that the above changes in MAVS K63-linked polyubiquitin signal are from RIG-I, we transfected UBE2N and RIPLET siRNA into HEK293T-DDX58-/- cells. We found that siRNA knockdown of UBE2N and RIPLET in RIG-I-deficient cells could also lead to a significant reduction in K63-linked polyubiquitination of MAVS, indicating that this function of Ube2N and Riplet on MAVS polyubiquitination is independent of RIG-I and should be effective in various cell lines (Supplementary Fig. 4H).
To confirm the function of Ube2N and Riplet in MAVS ubiquitination, HEK293T-UBE2N−/− and RIPLET−/− cells were employed in the following experiments. Knockout of UBE2N and RIPLET clearly reduced K63-linked polyubiquitination of MAVS, especially at lysine residues K7, K297 and K500 (Fig. 4A). Importantly, reconstitution of UBE2N and RIPLET expression in HEK293T-UBE2N−/− and RIPLET−/− cells restored K63-linked polyubiquitination of MAVS (Fig. 4B). Having established the critical roles of Riplet and Ube2N in MAVS ubiquitination, we next investigated whether these E2 and E3 directly interact with MAVS. We performed co-immunoprecipitation experiments in HEK293T-WT and DDX58−/− cells and found that MAVS coimmunoprecipitated with a significant amount of Riplet in both cell lines, indicating that these proteins interact with each other independent of RIG-I (Fig. 4C). In addition, the interaction between MAVS and Riplet was enhanced with SeV infection (Fig. 4C). We did not detect interaction between MAVS and Ube2N, which may be attributable to the instantaneous action of E2 on the substrate. In contrast, Riplet could directly pull down Ube2N (Fig. 4D). Furthermore, we examined the colocalization between MAVS and Riplet, and immunofluorescence experiments showed that endogenous Riplet colocalized with MAVS after SeV infection (Supplementary Fig. 5). The above data establish Ube2N-Riplet as a new E2–E3 pair for the K63-linked polyubiquitination of MAVS.
Ube2N-Riplet and Ube2N-TRIM31 catalyze the formation of unanchored K63-linked polyubiquitination on MAVS and promote RIG-I-mediated MAVS aggregation. A Immunoprecipitation analysis of the ubiquitination of exogenous MAVS in HEK293T-wild type (WT), UBE2N −/− or RIPLET −/− cells cotransfected with K63-Ub and Myc-tagged MAVS-WT or mutant K7, K136, K297 and K500. B Immunoprecipitation analysis of the ubiquitination of MAVS in HEK293T-WT, UBE2N−/− and RIPLET−/− cells transfected with K63-Ub and reintroduced Ube2N and Riplet. C Co-immunoprecipitation analysis of the binding between endogenous MAVS and Riplet or Ube2N in HEK293T-WT or DDX58−/− cells with or without SeV infection for 8 h. D Co-immunoprecipitation analysis of the binding between endogenous Riplet and Ube2N in HEK293T-WT cells. E Immunoprecipitation analysis of the ubiquitination of exogenous MAVS in HEK293T cells transfected with Flag-tagged TRIM31/TRIM31C53A C56A, Myc-tagged MAVS/MAVS-3KR/MAVS-4KR and K63-Ub. F Immunoprecipitation analysis of the ubiquitination of MAVS in HEK293T-UBE2N−/− cells transfected with K63-Ub and Riplet or TRIM31. G Immunoprecipitation analysis of the ubiquitination of exogenous MAVS and mutant K0 in the mitochondria of HEK293T-MAVS−/− cells transfected with Myc-MAVS and mutant K0. After transfection, the mitochondria were extracted and reacted with purified E1, Ube2N, Riplet or TRIM31, followed by treatment with IsoT or vOTU. H SDD-AGE analysis of MAVS aggregation in the mitochondria of (B) in the in vitro assay. Mitochondria were extracted as (B) and then incubated with RIG-I with or without 5′-pppRNA, followed by the detection of MAVS aggregation with SDD-AGE (top) and SDS‒PAGE (below). I SDD-AGE analysis of MAVS aggregation in the mitochondria of (E) in the in vitro assay. The reaction and analysis were the same as in (H). J SDD-AGE analysis of MAVS aggregation in the mitochondria of (G) in the in vitro assay. Mitochondria in (G) were terminated by NEM and purified by centrifugation. Then, the mitochondria were incubated with RIG-I with or without 5′-pppRNA in the in vitro assay, followed by the detection of MAVS aggregation with SDD-AGE (top) and SDS‒PAGE (below). β-actin and GAPDH were used as loading controls
Previously, our group reported that TRIM31 catalyzes K63-linked polyubiquitination of MAVS and facilitates MAVS aggregation upon viral infection [18]. We identified K10, K311 and K461 as the major ubiquitination sites catalyzed by TRIM31 on MAVS [18]. The polyubiquitination mediated by TRIM31 was greatly decreased in MAVS-3KR (K10R, K311R, K461R), in which the three lysine residues were replaced with arginine [18] (Supplementary Fig. 2). However, the E2 responsible for TRIM31-mediated MAVS ubiquitination is unknown. As reported, TRIM31 could enhance the K63-linked polyubiquitination of MAVS but not MAVS-3KR, while the catalytically inactive TRIM31C53A C56A had no effect (Fig. 4E). Moreover, overexpression of TRIM31 could still elevate the K63-linked polyubiquitination of MAVS-4KR to some extent, indicating that TRIM31 does not primarily target the lysine residues of MAVS-K7/K136/K297/K500 (Fig. 4E). Similar to Riplet, TRIM31 also uses Ube2N as an E2 to catalyze K63-linked ubiquitination of MAVS because TRIM31-induced MAVS ubiquitination was greatly decreased in UBE2N−/− cells (Fig. 4F).
To further validate the function of Ube2N, Riplet and TRIM31 in MAVS ubiquitination, we carried out an in vitro ubiquitination assay with purified E1, E2, E3s, and crude mitochondria from HEK293T-MAVS−/− cells exogenously expressing Myc-tagged MAVS-WT and mutant MAVS-K0. We found that MAVS was modified with K63-linked polyubiquitin chains catalyzed by Ube2N-Riplet and Ube2N-TRIM31 (Fig. 4G, Lane 3, 7), which could be removed by IsoT and vOTU (Fig. 4G, Lane 4, 5, 8, 9), indicating that these ubiquitin chains noncovalently bind MAVS. MAVS-K0 showed little ubiquitination signal after the reaction (Fig. 4G, Lanes 6 and 10), which was consistent with the results in the HEK293T exogenous expression system. Taken together, these data reveal that Riplet and TRIM31 can work together with Ube2N to catalyze the formation of unanchored K63-linked polyubiquitin chains on MAVS.
Unanchored K63-linked polyubiquitination of MAVS catalyzed by Ube2N-Riplet and Ube2N-TRIM31 promotes RIG-I-mediated MAVS aggregation
The above data show that Ube2N-Riplet and Ube2N-TRIM31 catalyze the formation of unanchored K63-linked polyubiquitin chains on MAVS. Next, we investigated the function of Ube2N-Riplet and Ube2N-TRIM31 in RIG-I-mediated MAVS aggregation. We prepared crude mitochondria from HEK293T-WT, UBE2N−/− and RIPLET−/− cells transfected with K63-Ub and performed the in vitro assay of RIG-I-mediated MAVS aggregation. We found that knockout of UBE2N and RIPLET abrogated RIG-I-induced MAVS aggregation (Fig. 4H). However, restoration of UBE2N and RIPLET rescued MAVS aggregation, suggesting that the K63-linked polyubiquitin chains catalyzed by Ube2N and Riplet on MAVS trigger RIG-I-mediated MAVS aggregation (Fig. 4H). Similar to Riplet, we found that mitochondria overexpressing TRIM31, but not TRIM31C53A C56A, increased RIG-I-mediated MAVS aggregation, while TRIM31 showed no effect on mutant MAVS-3KR (Fig. 4I). These results indicate that the K63-linked polyubiquitin chains added to MAVS by TRIM31 can also trigger RIG-I-mediated MAVS aggregation.
To further validate the function of Ube2N, Riplet and TRIM31 in MAVS aggregation, we first carried out an in vitro ubiquitination assay, as shown in Fig. 4G. We centrifuged and purified the mitochondria after terminating the ubiquitination reaction with N-ethylmaleimide (NEM), an organic compound that inactivates E1 and E2 by modifying their active cysteine residues. We detected MAVS aggregation with the above mitochondria in the in vitro assay. As shown in Fig. 4J, Ube2N-Riplet and Ube2N-TRIM31 promoted the RIG-I-mediated aggregation of MAVS-WT (Lanes 3, 4, 9, 10) but not MAVS-K0 (Lanes 7, 8, 13, 14). Pretreatment of the mitochondria with IsoT completely abrogated this effect (Fig. 4J, Lanes 5, 6, 11, 12). Taken together, the above results indicate that the unanchored polyubiquitin chains on MAVS produced by Ube2N-Riplet and Ube2N-TRIM31 can directly induce RIG-I-mediated MAVS aggregation.
USP10 removes the unanchored K63-linked polyubiquitin chains of MAVS to block RIG-I-mediated MAVS aggregation
Since MAVS attaches K63-linked polyubiquitin chains to activate RIG-I-mediated MAVS aggregation, we hypothesize that some deubiquitinating enzymes (DUBs) may manipulate MAVS-loaded polyubiquitin chains to regulate RLR signal transduction. We transfected a total of 27 Flag-tagged DUBs into HEK293T cells together with Myc-tagged MAVS and HA-tagged K63-Ub, followed by immunoprecipitation and immunoblot analysis. The experiments showed that USP10, USP25, USP39 and OTUB2 potently attenuated the K63-linked polyubiquitination of MAVS, and at the same time, their input HA-K63-Ub levels in total cell lysates were almost unchanged, suggesting that these DUBs may be special enzymes targeting MAVS ubiquitination (Fig. 5A and Supplementary Fig. 6). Given that USP25, USP39 and OTUB2 have been reported as regulators of the RLR signaling pathway [30,31,32], we focused on exploring the functions and mechanisms by which USP10 regulates MAVS ubiquitination and the RLR signaling cascade.
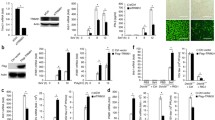
USP10 removes the unanchored K63-linked polyubiquitin chains of MAVS to block RIG-I-mediated MAVS aggregation. A Screening of deubiquitinating enzyme (DUB) targeting MAVS. Ctrl, the group without DUB transfection. B Immunoprecipitation analysis of the ubiquitination of exogenous MAVS in HEK293T cells transfected with Myc-tagged MAVS and HA-tagged K63-Ub or K63R-Ub in the presence of Flag-tagged USP10. C Immunoprecipitation analysis of the ubiquitination of exogenous MAVS in HEK293T cells transfected with Myc-MAVS and HA-K63-Ub in the presence of Flag-USP10 or USP10-C424A. D Immunoprecipitation analysis of the ubiquitination of exogenous MAVS in HEK293T-MAVS-/- cells transfected with Myc-MAVS or mutant MAVS-4KR and HA-K63-Ub under conditions of overexpressing or knocking down USP10. E SDD-AGE analysis of the aggregation of exogenous MAVS in mitochondria extracted from HEK293T cells treated as (C) in the in vitro assay. F Immunoprecipitation analysis of the ubiquitination of purified MAVS-K63-Ub with the treatment of IsoT and USP10. MAVS-K63-Ub was pretreated with or without IsoT and USP10 before immunoprecipitation. G SDD-AGE analysis of the aggregation of MAVS treated as (F) in the in vitro assay. H Immunoprecipitation analysis of the ubiquitination of exogenous Myc-tagged MAVS in the mitochondria of HEK293T-MAVS−/− cells. After transfection, the mitochondria were extracted and reacted with purified E1, Ube2N, Riplet or TRIM31, followed by USP10 treatment. I SDD-AGE analysis of the aggregation of MAVS treated as (H) in the in vitro assay. J Immunoprecipitation analysis of the ubiquitination of endogenous MAVS in HEK293T cells upon SeV infection with or without USP10 knockdown. K SDD-AGE analysis of the aggregation of MAVS and immunoblot of phosphorylated IRF3 in (J). L Immunoprecipitation analysis of the ubiquitination of endogenous MAVS in wild-type or Usp10-knockout RAW264.7 cells upon SeV infection. M SDD-AGE analysis of the aggregation of MAVS and immunoblot of phosphorylated IRF3 in (L). N Immunoprecipitation analysis of the ubiquitination of endogenous MAVS in Usp10+/+ or Usp10+/− primary peritoneal macrophages (PMs) upon SeV infection. O SDD-AGE analysis of the aggregation of MAVS and immunoblot of phosphorylated IRF3 in (N). P Co-immunoprecipitation analysis of the interaction between exogenous Flag-tagged USP10 and MAVS-Myc, RIG-I-Myc, IRF3-Myc, MAVS-HA or TBK1-HA. HEK293T cells were transfected with the indicated plasmids for 36 h. The cell lysates were subjected to immunoprecipitation with anti-Flag antibody and then analyzed by immunoblot. Q, R Co-immunoprecipitation analysis of the interaction between endogenous MAVS and USP10 upon SeV infection in HEK293T cells (Q) or RAW264.7 cells (R). The cells were infected with SeV for the indicated times, and then the cell lysates were subjected to immunoprecipitation with anti-MAVS antibody and immunoblot analysis. The quantification of USP10/MAVS (IP) is shown in the right panel (normalized to the SeV group, 0 h). S Colocalization between endogenous USP10 (red) and MAVS (green) in SeV-infected HEK293T cells. Scale bars, 10 μm. Line profiling of USP10 and MAVS and the intensity of each line were quantified by ImageJ software and are shown below. β-Actin was used as a loading control. Data in (Q, R) are shown as the mean ± S.D. *p < 0.05, **p < 0.01, ***p < 0.001; two-tailed Student’s t test. Ctrl control
We transfected WT Myc-tagged MAVS and HA-Ub mutant K63 or K63R into HEK293T cells with or without USP10 and measured K63-linked polyubiquitination of MAVS through immunoprecipitation. We found that USP10 abrogated the ubiquitination signal of K63-Ub but not K63R-Ub, suggesting that K63-linked polyubiquitin chains are the major form of ubiquitination targeted by USP10 on MAVS (Fig. 5B). Then, we examined whether the impact of USP10 on MAVS ubiquitination relies on its enzymatic activity. We constructed an inactive USP10 mutant, C424A (mutation of cysteine to alanine at position 424), which abolished the deubiquitination activity of USP10 [33]. We cotransfected Myc-MAVS, HA-K63-Ub and Flag-tagged USP10 or mutant C424A into HEK293T cells. Immunoprecipitation analysis showed that WT USP10, but not the mutant C424A, decreased MAVS ubiquitination, proving that the enzymatic activity of USP10 is indispensable for its deubiquitination of MAVS (Fig. 5C). Furthermore, we compared the effect of USP10 on exogenous MAVS and its mutant 4KR in HEK293T-MAVS−/− cells. As shown in Fig. 5D, overexpression of USP10 decreased the ubiquitination of MAVS, and knockdown of USP10 enhanced the MAVS ubiquitination signal. In contrast, USP10 did not affect the ubiquitination of MAVS-4KR (the panel of long-term exposure). These results indicate that USP10 mainly targets the polyubiquitin chains loaded by the K7, K136, K297 and K500 sites of MAVS. We also extracted the mitochondria of HEK293T cells expressing MAVS, K63-Ub, USP10 or USP10-C424A, as shown in Fig. 5C, and performed the in vitro assay of RIG-I-mediated MAVS aggregation. We found that USP10, but not USP10-C424A, could substantially reduce the aggregation triggered by MAVS-attached K63-linked polyubiquitin chains (Fig. 5E). The above data demonstrate that USP10 negatively regulates RIG-I-mediated MAVS aggregation by removing MAVS-attached K63-linked polyubiquitin chains.
To test whether USP10 removes the unanchored polyubiquitin chains of MAVS, we immunoprecipitated purified MAVS and MAVS-K63-Ub after treatment with USP10 and IsoT (Supplementary Fig. 3) and detected MAVS ubiquitination. As shown in Fig. 5F, USP10 potently reduced the ubiquitination signal of MAVS (Lane 3) but showed a marginal effect after pretreatment with IsoT (Lane 4, the panel of long-term exposure). Then, we detected the corresponding MAVS aggregation in the in vitro assay. Treatment of MAVS-K63-Ub with USP10 significantly inhibited RIG-I-mediated MAVS aggregation but showed little effect after pretreatment with IsoT (Fig. 5G). The above results indicate that USP10 mainly targets the unanchored K63-linked polyubiquitin chains of MAVS to regulate RIG-I-mediated MAVS aggregation.
We also performed an in vitro ubiquitination assay with purified E1, Ube2N, Riplet/TRIM31, and crude mitochondria from HEK293T-MAVS−/− cells transfected with Myc-tagged MAVS. The addition of USP10 abrogated the ubiquitination of MAVS catalyzed by Ube2N-Riplet and Ube2N-TRIM31 (Fig. 5H). Moreover, treatment with USP10 reduced the corresponding MAVS aggregation in the in vitro assay (Fig. 5I). The results prove that USP10 can cleave the polyubiquitin chains of MAVS catalyzed by Ube2N-Riplet and Ube2N-TRIM31 and inhibit the triggering of RIG-I-mediated MAVS aggregation by these polyubiquitin chains.
Then, we examined endogenous MAVS ubiquitination and aggregation with viral infection in the presence or absence of USP10. We transfected USP10-siRNA into HEK293T cells, stimulated the cells with SeV and tested the ubiquitination of MAVS by immunoprecipitation. We observed that the K63-linked polyubiquitination of MAVS emerged after treatment with SeV, while knockdown of USP10 potently enhanced the ubiquitination signal under this condition (Fig. 5J). Consistently, knockdown of USP10 also promoted MAVS aggregation (Fig. 5K). Then, we established Usp10-knockout (KO) RAW264.7 cells and examined MAVS ubiquitination with SeV infection. DNA sequencing and immunoblot experiments confirmed the success of Usp10 KO in RAW264.7 cells (Supplementary Fig. 7). WT and Usp10-KO RAW264.7 cells were treated with SeV, and MAVS ubiquitination and aggregation were then detected. As shown in Fig. 5L and M, SeV infection induced MAVS ubiquitination and aggregation, and Usp10 KO further enhanced both behaviors. To further test the function of USP10 on MAVS under physiological conditions, we established a Usp10 knockout mouse (C57BL/6 N) by CRISPR/Cas9-mediated genome engineering (Supplementary Fig. 8A). The Usp10 knockout mice (Usp10-/-) were born at the expected Mendelian frequency (Usp10+/+/ Usp10+/−/ Usp10−/− = 11:18:9) and looked normal at birth. However, the Usp10−/− mice died within 1 week after birth. The results were consistent with a previous report that USP10 is essential for survival [34]. The heterozygous Usp10+/− mice appeared healthy and survived longer than 300 days, and USP10 protein expression was significantly lower in Usp10+/− mice than in WT Usp10+/+ littermates (Supplementary Fig. 8B). Thus, heterozygous Usp10+/− mice were used in the following study. We prepared primary mouse macrophages (PMs) from USP10-deficient (Usp10+/−) mice and their WT littermates (Usp10+/+) and infected them with SeV. Deficiency of USP10 in Usp10+/− PMs led to an increase in MAVS K63-linked ubiquitination and aggregation compared to Usp10+/+ PMs with SeV infection (Fig. 5N and O). In addition, in HEK293T and RAW264.7 cells and PMs, USP10 deficiency led to enhanced IRF3 phosphorylation, suggesting that USP10 negatively regulates the activation of the RLR signaling pathway in these cells (Fig. 5K, M and O). Thus, we propose that USP10 removes the unanchored K63-linked polyubiquitin chains of MAVS to block RIG-I-mediated MAVS aggregation.
To further elucidate the mechanism by which USP10 regulates MAVS, the interaction between USP10 and MAVS was examined. We transfected MAVS-Myc, RIG-I-Myc, IRF3-Myc, MAVS-HA or TBK1-HA together with Flag-tagged USP10 into HEK293T cells and then carried out co-immunoprecipitation. The experiments showed that USP10 could pull down MAVS but not RIG-I, IRF3 or TBK1 (Fig. 5P). Furthermore, we examined the interaction between USP10 and MAVS under physiological conditions. In HEK293T cells, MAVS consistently pulled down USP10, and the interaction was slightly decreased after treatment with SeV for 6 h (Fig. 5Q). A similar phenomenon was also observed in RAW264.7 cells (Fig. 5R). We also investigated the colocalization between USP10 and MAVS. The immunofluorescence experiments showed that endogenous USP10 colocalized with MAVS with or without SeV infection (Fig. 5S). In summary, these data indicate that USP10 directly associates with MAVS.
USP10 negatively regulates the RLR signaling cascade upon RNA virus infection in vitro and in vivo
Next, we investigated the physiological role of USP10 in innate immunity against RNA virus infection. We reintroduced MAVS and mutant MAVS-K0 into HEK293T-MAVS−/− cells with or without USP10 transfection and measured the expression of IFNB1 mRNA after SeV infection. As shown in Fig. 6A, transfection of USP10 significantly reduced IFNB1 mRNA with reconstitution of WT MAVS upon viral infection. In contrast, although reconstitution of mutant MAVS-K0 in HEK293T-MAVS−/− cells could also induce a certain amount of IFNB1 expression after SeV treatment, overexpression of USP10 could not regulate IFNB1 expression in this condition (Fig. 6A). The results demonstrated that USP10 mainly targets the ubiquitination of MAVS to regulate RLR signal transduction. Furthermore, we designed a siRNA that targeted human USP10 and transfected it into HEK293T cells. We infected HEK293T cells with GFP-tagged VSV (VSV-GFP) and performed immunofluorescence and flow cytometric analysis. The results showed that VSV-GPF replication was decreased with siRNA-mediated USP10 knockdown and increased with USP10 overexpression (Fig. 6B and Supplementary Fig. 9A). Consistently, we observed that the siRNA-mediated knockdown of USP10 expression in HEK293T and THP-1 cells increased SeV-, VSV- and 5′-pppRNA-induced expression of IFNB1 (Supplementary Fig. 9B and C).
USP10 negatively regulates the RLR signaling cascade upon RNA virus infection in vitro and in vivo. A Quantitative PCR (qPCR) analysis of IFNB1 in HEK293T-MAVS−/− cells reintroducing MAVS and mutant MAVS-K0 with or without cotransfection of USP10, followed by SeV infection for 6 h. B Fluorescence microscopy of VSV-GFP replication in HEK293T cells with USP10 overexpression or knockdown, followed by VSV-GFP infection for 12 h. The corresponding quantification is shown in the right panel. Scale bars, 100 μm. C qPCR analysis of Ifnb1 and VSV mRNA in Usp10-knockout RAW264.7 cells infected with VSV for the indicated times. D qPCR analysis of Ifnb1, Cxcl10, Ccl5, and Ifna4 and ELISA quantification of IFN-β in Usp10+/+ and Usp10+/− primary mouse macrophages (PMs) with SeV or VSV infection for the indicated times. E qPCR analysis of VSV mRNA and plaque assay of VSV titers in Usp10+/+ and Usp10+/− PMs infected with VSV for the indicated times. F Immunoblot analysis of VSV glycoprotein (VSV-G) in lysates of Usp10+/+ and Usp10+/− PMs infected with VSV for 0–6 h. The quantification of VSV bands is normalized to individual β-actin and shown in the right panel. G Fluorescence microscopy of VSV-GFP replication in Usp10+/+ and Usp10+/− PMs. The PMs were infected with VSV-GFP for 12 h before detection. The corresponding quantification is shown in the right panel. Scale bars, 100 μm. H qPCR analysis of Ifnb1 and Ifna4 in Usp10+/+ and Usp10+/− PMs infected with HSV-1 for the indicated times. I qPCR analysis of Ifnb1 mRNA in the liver, lung, and spleen from Usp10+/+ and Usp10+/− mice (5 per group) infected by intraperitoneal injection of VSV (1 × 108 PFU per mouse) for 24 h. J ELISA analysis of IFN-β in serum from Usp10+/+ and Usp10+/− mice infected by intraperitoneal injection of VSV (1 × 108 PFU per mouse) for 24 h. K Plaque assay of VSV titers of the liver, lung, and spleen from Usp10+/+ and Usp10+/− mice infected by intraperitoneal injection of VSV (1 × 108 PFU per mouse) for 48 h. L Hematoxylin-and-eosin-stained images of lung sections of Usp10+/+ and Usp10+/− mice infected by intraperitoneal injection of VSV (1 × 108 PFU per mouse) for 36 h. Scale bars, 100 μm. M Survival of Usp10+/+ and Usp10+/− mice infected intravenously with VSV (1 × 108 PFU per mouse). Data are shown as the mean ± S.D. *p < 0.05, **p < 0.01, ***p < 0.001; n.s. no significance; the log-rank Mantel‒Cox test for (M) and two-tailed Student’s t test for (A–K). Ctrl control, EV empty vector, OE overexpression, MFI mean fluorescence intensity
To confirm the function of USP10, we tested the mRNA expression level of Ifnb1 in Usp10-KO RAW264.7 cells after VSV infection with or without USP10 reconstitution (Fig. 6C and Supplementary Fig. 7). Similar to the above results in HEK293T and THP-1 cells, the mRNA expression level of Ifnb1 was upregulated in Usp10-KO RAW264.7 cells after VSV infection, and the reintroduction of USP10 recovered Ifnb1 mRNA (Fig. 6C). Consistently, the replication of VSV was attenuated in Usp10-KO RAW264.7 cells, which could also be recovered by overexpression of USP10 (Fig. 6C).
We prepared PMs from USP10-deficient (Usp10+/−) mice and their WT littermates (Usp10+/+) and infected them with either SeV or VSV. Deficiency of USP10 in Usp10+/− PMs led to increased Ifnb1, Cxcl10, Ccl5 and Ifna4 mRNA expression compared to Usp10+/+ PMs with SeV and VSV infection (Fig. 6D). Moreover, the production of IFN-β from Usp10+/− PMs was elevated compared with that of their Usp10+/+ counterparts (Fig. 6D). Congruently, the replication of VSV, presented as the VSV mRNA titer and VSV glycoprotein (VSV-G) expression, was attenuated in Usp10+/− PMs (Fig. 6E and F). We also infected PMs with VSV-GFP, and immunofluorescence experiments showed that the replication of VSV-GFP was attenuated in Usp10+/− PMs compared to Usp10+/+ PMs (Fig. 6G). In addition, we did not observe a significant difference in HSV-1 infection-induced Ifnb1 and Ifna4 expression between Usp10+/+ and Usp10+/− PMs (Fig. 6H). The results suggest that USP10 mainly regulates the RLR signaling pathway but exerts a marginal effect on the cGAS-STING pathway in PMs.
To test the antiviral effect of USP10 in vivo, we infected Usp10+/− mice and their WT littermates with VSV and extracted the corresponding organs and serum for detection. qPCR analysis showed that Usp10+/− mice had substantially increased Ifnb1 mRNA expression in the lung, liver, and spleen compared with WT mice upon VSV infection (Fig. 6I). Moreover, ELISA data showed that the level of IFN-β protein in serum from Usp10+/− mice was significantly increased compared with that from WT mice (Fig. 6J). Consistently, plaque assays for VSV titers reconfirmed that VSV replication was significantly attenuated in the lung, liver, and spleen in Usp10+/− mice compared to Usp10+/+ mice (Fig. 6K). Hematoxylin and eosin staining showed that USP10 deficiency in Usp10+/− mice alleviated inflammatory cell infiltration, pulmonary fibrosis and lung damage compared to those in Usp10+/+ mice (Fig. 6L). We also observed that Usp10+/- mice exhibited improved survival compared with their WT littermates during viral infection (Fig. 6M). Taken together, these data demonstrate that USP10 negatively regulates the RLR signaling cascade upon RNA virus infection in vitro and in vivo.
Discussion
In this study, we established an in vitro assay system to investigate the function and biochemical mechanism of MAVS-attached K63-linked polyubiquitin chains in the RLR signaling pathway. First, we proved that K63-linked polyubiquitin chains loaded on MAVS can be directly recognized by RIG-I after the CARDRIG-I-CARDMAVS interaction and activate RIG-I-mediated MAVS aggregation. Second, we found that MAVS carries a large portion of unanchored K63-linked polyubiquitin chains, which can serve as direct activators of RIG-I after experimental separation in vitro. Third, we identified Ube2N as the major E2 for MAVS and Riplet as a new E3 ligase for MAVS, independent of RIG-I, and proved that Ube2N-Riplet and Ube2N-TRIM31 trigger RIG-I-mediated MAVS aggregation by catalyzing unanchored K63-linked polyubiquitination of MAVS. Fourth, we identified USP10 as a new DUB for the unanchored K63-linked polyubiquitin chains of MAVS and a regulator of RIG-I-mediated MAVS aggregation. Deficiency of USP10 enhances the innate antiviral response to RNA viral infection both in vitro and in vivo. The DUB USP10 and E2:E3 pair Ube2N-Riplet/TRIM31 constitute a specific regulatory system for MAVS unanchored K63-linked ubiquitination and aggregation upon viral infection. Our work reveals a previously unknown mechanism by which ubiquitin chains modify and regulate MAVS, and we propose a new signal-activation model of the RLR pathway in which downstream MAVS attaches K63-linked polyubiquitin chains to form an entity that functions as a platform for the activation of upstream RIG-I and triggers RIG-I-mediated MAVS aggregation (Supplementary Fig. 10).
The RLR pathway is composed of multiple steps, so it is difficult to delineate the detailed mechanism and rule out the possibility of indirect effects. The in vitro assay with defined components can avoid confusion and pinpoint the precise mechanism. In this study, we established an in vitro assay system with purified protein and specific ligand to depict the reaction from RIG-I activation to MAVS aggregation. With the data obtained from the in vitro assay, we propose a new model for the activation of RIG-I-mediated MAVS aggregation (Supplementary Fig. 10). After viral infection, the binding of viral 5’-pppRNA and ATP hydrolysis convert RIG-I from a “closed” conformation to an “open” form, exposing the RIG-I N-terminal CARD domain [12]. Then, the CARDRIG-I-CARDMAVS homotypic interaction mediates the binding of RIG-I with MAVS and shortens the distance between them. At this time, the K63-linked polyubiquitin chains on MAVS are captured and recognized by CARDRIG-I and bridge MAVS and RIG-I. Then, the K63-linked polyubiquitin chains cause additional conformational changes and/or oligomerization of CARDRIG-I, forming a new signaling platform for MAVS prion-like aggregation. Further structural experiments may reveal the detailed mechanism and conformation of ubiquitin chains linking RIG-I and MAVS. To the best of our knowledge, this is the first study to propose a bottom-up model in which a downstream effector (MAVS) provides an activator signal (ubiquitin chains) for an upstream molecule (RIG-I) to trigger the activation of RLR signal transduction.
Several reports have proven that unanchored K63-linked polyubiquitin chains are essential for RIG-I-mediated MAVS aggregation [12,13,14,15]. However, the origin of unanchored K63-linked polyubiquitin chains remains elusive. Here, we have demonstrated that MAVS can catch unanchored K63-linked polyubiquitin chains under both in vitro experimental conditions and physiological conditions with viral infection. MAVS functions as a K63-linked polyubiquitin chain supplier for RIG-I to ensure its full activation, which clarifies the source of the unanchored K63-linked polyubiquitin chains. Although the unanchored K63-linked polyubiquitin chains on MAVS can directly activate RIG-I (Fig. 3F–K), RIG-I recognizes the unanchored K63-linked polyubiquitin chains loaded on MAVS only after the CARDRIG-I-CARDMAVS interaction (Fig. 2H–K). The polyubiquitin chains on ∆CARDMAVS truncation cannot be recognized by RIG-I (Fig. 2I and K). A previous study reported that ubiquitinated TRIM25 is incapable of activating RIG-I [12]. These data suggest that MAVS is a specific polyubiquitin chain supplier for RIG-I that relies on its CARD domain. The mutant MAVS-K0, which bears few polyubiquitin chains, could still aggregate with the addition of free K63-polyUb in the in vitro assay (Fig. 1H) and induce IFNB1 expression after SeV infection (Fig. 6A), suggesting that there also exist other origins of K63-linked polyubiquitin chains provided for RIG-I. Therefore, we infer that the function of K63-linked polyubiquitin chains on MAVS is to strengthen RIG-I activation and amplify RLR signaling but not to serve as a prerequisite for RIG-I-mediated MAVS aggregation. Nevertheless, our results propose a previously unknown mechanism by which ubiquitin chains modify and regulate MAVS.
Previously, we found that TRIM31 enhances the polyubiquitination of mitochondrial MAVS in the absence of infection with SeV but promotes MAVS aggregation only after viral infection [18], suggesting that viral infection is the prerequisite for the K63-linked polyubiquitination of MAVS to facilitate MAVS aggregation. Here, we elucidate the underlying mechanism with the in vitro assay. Our experiments indicate that the K63-linked polyubiquitin chains on MAVS cannot directly nucleate MAVS prion-like aggregation, which requires recognition by RIG-I. RIG-I recognizes the K63-linked polyubiquitin chains of MAVS only after the liberation of its CARD domain by RNA ligand treatment. In addition, the CARDRIG-I-CARDMAVS interaction is essential for RIG-I to recognize K63-linked polyubiquitin chains on MAVS. The sequential manner of reaction determines that the K63-linked polyubiquitin chains of MAVS exerting their function in promoting MAVS aggregation requires the prerequisite that RIG-I has already been activated by the virus.
Here, we identify Ube2N as a new E2 and Riplet as a new E3 ligase for MAVS K63-linked polyubiquitination independent of RIG-I. Ube2N is closely involved in inflammation and immune response pathways, largely through its modulation of NF-κB signaling [35, 36]. This is the first study to identify E2 for MAVS. We found that Ube2N can cooperate with Riplet or TRIM31 to catalyze the unanchored polyubiquitin chains for MAVS and trigger RIG-I-mediated MAVS aggregation. These results contribute to our understanding of the mechanism of Ube2N, Riplet and TRIM31 in regulating RLR signal transduction. Moreover, we cannot exclude the possibility that Ube2N may cooperate with other E3 ligases to catalyze anchored or unanchored ubiquitin chains on MAVS, which needs further exploration.
USP10 has been reported as a regulator of several cellular targets, such as p53 [37], IKKγ/NEMO [38], and G3BP1 [33]. However, the role of USP10 in modulating the activation of RLR antiviral signal transduction has not been investigated. Here, we identified USP10 as a direct DUB for MAVS. USP10 mainly removes the unanchored K63-linked polyubiquitin chains attached by MAVS and blocks RIG-I-mediated MAVS aggregation. Since we could not observe the association between USP10 and RIG-I, TBK1 and IRF3 (Fig. 5P) and overexpressing USP10 could not regulate IFNB1 expression in HEK293T-MAVS−/− cells reconstituting lysine-mutant MAVS (Fig. 6A), we infer that MAVS is a specific target for USP10 in the RLR signaling pathway. In addition, USP10 deficiency boosts the capacity of cells and mice to defend against RNA viral infection. These findings establish USP10 as a specific regulator of MAVS and the antiviral response in vitro and in vivo. USP10 partially dissociated from MAVS after SeV infection for 6 h (Fig. 5Q and R), whereas the binding between Riplet/TRIM31 and MAVS was enhanced after viral infection (Fig. 4C) [18]. Based on this phenomenon, we proposed that the DUB USP10 and E2:E3 pair Ube2N-Riplet/TRIM31 dynamically regulate MAVS ubiquitination and aggregation upon viral infection (Supplementary Fig. 10).
In summary, we provide a yet-unknown mechanistic link between K63-linked polyubiquitin chains on MAVS and RIG-I-mediated MAVS aggregation and establish new regulators of MAVS ubiquitination and the RLR antiviral response. Our findings propose that fine-tuning MAVS-attached unanchored K63-linked polyubiquitin chains may be a potential strategy for intervening in human antiviral immunity and treating related diseases.
Methods
Reagents and antibodies
The reagents and antibodies are listed in Supplementary Table. 1.
Cells and viruses
Human HEK293T, HeLa and mouse RAW264.7 cells were cultured in DMEM supplemented with 10% FBS (LONSERA, Shanghai Shuangru Biology Science & Technology Co., Ltd), 100 U/mL penicillin, and 100 mg/mL streptomycin. The materials for cell culture were from SORFA (Beijing, China). The DDX58−/−, UBE2N−/− and MAVS−/− HEK293T cells were kindly provided by FaJian Hou (Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, China). All the cells were cultured at 37 °C under 5% CO2. Sendai virus (SeV) was purchased from the China Center for Type Culture Collection (Wuhan University, China). Vesicular stomatitis virus (VSV), VSV-GFP and HSV-1 were provided by H. Meng (Institute of Basic Medicine, Shandong Academy of Medical Sciences, China). For the viral infection experiments, cells were plated 24 h before infection. Cells were infected with VSV (0.1 MOI), HSV-1 (10 MOI) or SeV for the indicated times. VSV RNA replicates were examined by quantitative RT‒PCR. Primers for VSV were as follows: 5′-ACGGCGTACTTCCAGATGG-3′ (sense) and 5′-CTCGGTTCAAGATCCAGGT-3′ (antisense). VSV plaque assays and VSV replication were performed as previously described [18].
Mice
USP10-deficient mice were established by Cyagen Biosciences Inc. (Guangzhou, China) via CRISPR/Cas-mediated genome engineering. The regions upstream and downstream of exons 3–12 of Usp10 were inserted into the Cas9/sgRNA expression plasmid and targeting vector. Cas9 and gRNA were coinjected into fertilized eggs for knockout mouse production. Genotyping was performed by PCR using the following primers:
F1: 5′- ACAAGGAAGGTGACGTTAAACCAT -3′;
R1: 5′- CCAGTCTCTTAGGGCTCTTCAAAA -3′;
F2: 5′- AGCCAGACCTATGTTGTTTTCTTC-3′;
R2: 5’-TACAAAGGATGGGAGTCAACAGG-3′;
The mouse experiments were performed under the general guidelines published by the Association for Assessment and Accreditation of Laboratory Animal Care. All mice were on the C57BL/6 background and fed under specific-pathogen free conditions with the approval of the Scientific Investigation Board of the Medical School of Shandong University.
Plasmid constructs and small interfering RNA
Plasmids encoding full-length MAVS, TRIM31, Riplet, Ube2N, USP10, ubiquitin and ubiquitin mutants were constructed as previously reported [18]. Truncations and point mutations of MAVS were generated by QuikChange site-directed mutagenesis kits according to the manufacturer’s manual (Toyobo, SMK-101). Lipofectamine 2000 (Invitrogen) reagent was used for the transfection of plasmids or siRNA into cells.
The small interfering sequences were as follows:
human UBE2N: 5′-CCAGACAUCUUCAGUCCUUTT-3′;
human RIPLET: 5′-CCAUCCAUCCAACCUUUAATT-3′;
human TRIM25: 5′-GGAAAAGAAAUCCAAGAAATT-3′;
human UBE2D3: 5’-GCCUGCUUUAACAAUUUCUTT-3′;
human USP10: 5′-GCAGGUUGAAGUCAAAGAATT-3′;
mouse Ube2N: 5′-CCUUCCAGAAGAAUACCCATT-3′;
mouse Riplet: 5′-CCUGAGGACCCUCUCCUAUTT-3′;
mouse Usp10: 5′-UCAACGCCCAUGAUAGAUATT-3′;
The scrambled control sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′.
Total RNA was extracted and purified by an RNA extraction kit (Fastagen Biotech Co., RNAfast200) and reverse-transcribed with a PrimeScript RT‒PCR kit (Takara RR014B) according to the manufacturer’s instructions. Real-time PCR was performed with SYBR Green PCR Master Mix (Roche) on an ABI 7300 Detection System (Applied Biosystems). The qPCR primers are listed in Supplementary Table 2. The 2−ΔΔCt method was used to calculate relative expression changes. Data were normalized to the level of β-actin expression in each sample.
Recombinant protein preparation and purification
Flag-tagged proteins were purified from HEK293T cells as previously described with minor modifications [11]. Flag-tagged RIG-I, MAVS, USP10 and CYLD were cloned into pcDNA3.1, expressed in HEK293T cells and then lysed in IP buffer containing 1.0% (vol/vol) NP-40, 50 mM Tris-HCl (pH 7.4), 50 mM EDTA, 150 mM NaCl, and a protease inhibitor cocktail (Merck, Darmstadt, Germany). After centrifugation for 30 min at 20000 g, the supernatant was collected and incubated with anti-Flag M2 agarose for 6 h and eluted by 3 × Flag peptide (200 mg/mL). CARDRIG-I, IsoT and vOTU were cloned into pGEX (GE healthcare) with an N-terminal GST tag and expressed in BL21 at 18 °C for 6 h. Bacterial cultures were collected and sonicated in buffer (10 mM Tris-HCl [pH 7.4], 0.3 M NaCl, 0.5 mM DTT and 1 mM PMSF). The recombinant proteins were purified by Glutathione-Sepharose affinity gel and further polished by size-exclusion chromatography.
Extraction of mitochondria, separation of cytosolic extracts and semidenaturing detergent agarose gel electrophoresis (SDD-AGE)
Cells were isolated and homogenized in buffer (10 mM Tris-HCl [pH 7.5], 10 mM KCl, 1.5 mM MgCl2, 0.25 M D-mannitol, and protease inhibitor cocktail). The cell homogenates were centrifuged at 800 × g for 10 min at 4 °C. Next, the supernatant was extracted and centrifuged again at 11,000 × g for 10 min at 4 °C to form a pellet. The corresponding supernatant was used as the cytosolic extract for the in vitro assay of IRF3 phosphorylation. The pellet was then resuspended in mitochondria resuspension buffer (20 mM HEPES-KOH [pH 7.4], 10% glycerol, 0.5 mM EGTA, and protease inhibitor cocktail) and centrifuged at 11,000 × g for 10 min, and the pellet was collected again as crude mitochondria for further experiments. For SDD-AGE, the crude mitochondria after reaction were resuspended in 1× SDD sample buffer (0.5× TBE, 10% glycerol, 2% SDS, and 0.0025% bromophenol blue) and loaded onto a vertical 1.5% agarose gel (Bio-Rad). Electrophoresis with a constant voltage of 100 V was performed in running buffer (1 × TBE and 0.1% SDS) for 35 min at 4 °C, and then the proteins were transferred to an Immobilon membrane (Millipore) for immunoblot analysis.
In vitro assay of RIG-I-mediated MAVS aggregation and IRF3 phosphorylation
Purified RIG-I (10 ng) was incubated with or without 5′-pppRNA (0.1 μg/mL) and free K63-polyUb (10 ng) at 30 °C for 20 min in buffer containing 20 mM Tris-HCl (pH 7.4), 2 mM ATP, 5 mM MgCl2, and 0.1 mM DTT. Then, the reaction mixture was reacted with crude mitochondria for 10 min at 30 °C. For each reaction, crude mitochondria were extracted from 2 × 106 HEK293T cells. The final reaction volume was 50 μL. After incubation, the crude mitochondria were isolated by centrifugation (11,000 × g, 10 min) and subjected to SDD-AGE. Purified MAVS (500 ng for each reaction) was reacted in the same way as crude mitochondria.
For the in vitro IRF3 phosphorylation experiments, crude mitochondria, which were incubated with MAVS aggregates at 4 °C for 16 h, were added to cytosolic extracts (separated as described above) in a buffer containing 20 mM HEPES-KOH (pH 7.0), 2 mM ATP, and 5 mM MgCl2, incubated at 30 °C for 2 h, and then IRF3 phosphorylation was detected by immunoblot analysis. For each reaction, crude mitochondria were prepared from HEK293T-MAVS−/− cells (2 × 106) transfected with Myc-tagged MAVS, and cytosolic extracts were separated from 2 × 106 HEK293T cells.
Immunoprecipitation and immunoblot analysis
For immunoprecipitation (IP), cells after transfection or viral infection were collected and lysed in IP buffer containing 1.0% (vol/vol) NP-40, 50 mM Tris-HCl (pH 7.4), 50 mM EDTA, 150 mM NaCl, and a protease inhibitor cocktail (Merck, Darmstadt, Germany). Then, centrifugation (15 min at 15,000 × g) was performed to isolate the supernatant. The supernatant was incubated with protein A/G agarose beads (Cell Signaling Technology) together with corresponding antibodies for 8 h at 4 °C, and then the beads were washed five times with IP buffer. The immunoprecipitates were eluted by boiling with 1% (wt/vol) SDS sample buffer (60 mM Tris-HCl [pH 6.8], 1% [wt/vol] SDS, 5% [vol/vol] glycerol, 0.005% [wt/vol] bromophenol blue and 1% [vol/vol] 2-mercaptoethanol). For immunoprecipitation with purified RIG-I and MAVS, RIG-I (1 μg) was incubated with or without 5′-pppRNA (0.5 μg/mL) and free K63-polyUb (1 μg) at 30 °C for 20 min in buffer containing 20 mM Tris-HCl (pH 7.4), 2 mM ATP, 5 mM MgCl2, and 0.1 mM DTT. Then, the reaction mixture was reacted with the purified MAVS (50 ng) for 10 min at 30 °C. The volume of the reaction was 1 mL. After the reaction, immunoprecipitation was performed with an anti-RIG-I antibody as mentioned above.
For immunoblot analysis, immunoprecipitates or whole-cell lysates were loaded and subjected to SDS‒PAGE, transferred onto Immobilon membranes (Millipore), and then blotted with specific antibodies. The original immunoblot images are attached in the Supplementary materials.
Ubiquitination and deubiquitination assay
For analysis of the ubiquitination of proteins in the exogenous expression system, cells were transfected with the indicated plasmids, and then the whole-cell extracts were immunoprecipitated with the corresponding antibody and analyzed by immunoblot. For analysis of the ubiquitination of endogenous proteins in cells, cells were treated with the indicated stimulation, and then the whole-cell extracts were immunoprecipitated with an antibody and analyzed by immunoblotting. For the in vitro ubiquitination assay, E1, Ube2N and 2V1, MAVS (in crude mitochondria), reaction buffer, ubiquitin, ATP, and MgCl2 were mixed with E3 ligase (TRIM31 or Riplet). Then, the reaction mix was incubated at 30 °C for 1 h followed by the addition of NEM or 6 × SDS loading buffer boiled at 95 °C for 5 min to stop the reaction. For the deubiquitination assay, ubiquitinated MAVS was incubated with recombinant Flag-tagged USP10 in deubiquitylation assay buffer (50 mM Tris-HCl [pH 7.5], 5 mM DTT, and 50 mM NaCl) at 37 °C for 1 h, followed by immunoprecipitation and immunoblot analysis.
Enzyme-linked immunosorbent assay (ELISA)
The concentrations of IFN-β were measured by using ELISA kits (R&D Systems) according to the manufacturer’s instructions.
Sucrose gradient ultracentrifugation
Crude mitochondria or purified MAVS was incubated in the in vitro MAVS aggregation assay and then resuspended in buffer (20 mM HEPES [pH 7.4], 10% glycerol, 0.5 mM EGTA, and 0.25 M D-mannitol) and loaded on top of a centrifuge tube with 0.5 mL of 50% sucrose in PBS on the bottom layer and 0.5 mL of 40% sucrose in PBS on the top layer. Then, centrifugation was performed at 10,000 × g for 1 h, and mitochondria or protein enriched at the interface of two layers was collected and solubilized with PBS containing 1% DDM. After that, the mitochondria or protein were added into a sucrose gradient (10%–60%) and centrifuged at 20,000 × g for 8 h. The fractions with equal volume were extracted from the top to bottom of the tube and loaded to the immunoblot analysis.
Generation of Usp10-KO RAW264.7 cells through CRISPR‒Cas9
RAW264.7 cells with Usp10 knockout were established by a lenti-CRISPR/Cas9-V2 system, and the sequence of the target Usp10 guide RNA was as follows: 5′- CTACACCAAACTCGATCCTC-3′. The isolated single clonal knockout cells, which survived after puromycin treatment, were confirmed by immunoblot analysis (Supplementary Fig. 7).
Confocal microscopy
HEK293T cells were cultured on glass coverslips in 12-well plates and transfected with the indicated plasmids or treated with viral infection. Then, the cells were collected and fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and blocked with PBS containing 5% BSA. The fixation, permeabilization, and blocking buffer reagents were purchased from Beyotime Biotechnology. The cells were reacted with the indicated primary antibodies at 4 °C overnight, rinsed, and then incubated with the corresponding secondary antibodies (Invitrogen). The nuclei were stained with DAPI (Abcam). Images were acquired with a Zeiss LSM780 confocal microscope, analyzed by ZEN imaging software (2012 SP1,8.1), and quantified by ImageJ software.
Statistical analysis
All the data are presented as the mean values based on three independent experiments. Statistical significance was determined by two-tailed Student’s t test or the log-rank Mantel‒Cox test. Statistical significance is indicated as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., no significance.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information files or are available from the corresponding author upon reasonable request.
References
Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell 2006;124:783–801.
Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–7.
Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010;140:805–20.
Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol. 2020;20:537–51.
Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev. 2009;227:54–65.
Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell 2011;147:409–22.
Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 2011;147:423–35.
Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M Jr., Patel SS, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 2011;479:423–7.
Peisley A, Wu B, Xu H, Chen ZJ, Hur S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature 2014;509:110–4.
Wu B, Peisley A, Tetrault D, Li Z, Egelman EH, Magor KE, et al. Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol Cell. 2014;55:511–23.
Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 2011;146:448–61.
Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 2010;141:315–30.
Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, et al. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 2012;36:959–73.
Okamoto M, Kouwaki T, Fukushima Y, Oshiumi H. Regulation of RIG-I Activation by K63-Linked Polyubiquitination. Front Immunol. 2017;8:1942.
Shi Y, Yuan B, Zhu W, Zhang R, Li L, Hao X, et al. Ube2D3 and Ube2N are essential for RIG-I-mediated MAVS aggregation in antiviral innate immunity. Nat Commun. 2017;8:15138.
Cadena C, Ahmad S, Xavier A, Willemsen J, Park S, Park JW, et al. Ubiquitin-Dependent and -Independent Roles of E3 Ligase RIPLET in Innate Immunity. Cell 2019;177:1187–200 e16.
Song B, Chen Y, Liu X, Yuan F, Tan EYJ, Lei Y, et al. Ordered assembly of the cytosolic RNA-sensing MDA5-MAVS signaling complex via binding to unanchored K63-linked poly-ubiquitin chains. Immunity 2021;54:2218–30 e5.
Liu B, Zhang M, Chu H, Zhang H, Wu H, Song G, et al. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat Immunol. 2017;18:214–24.
Hou J, Han L, Zhao Z, Liu H, Zhang L, Ma C, et al. USP18 positively regulates innate antiviral immunity by promoting K63-linked polyubiquitination of MAVS. Nat Commun. 2021;12:2970.
Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–7.
Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 2005;122:669–82.
Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–40.
He L, Bardiaux B, Ahmed M, Spehr J, Konig R, Lunsdorf H, et al. Structure determination of helical filaments by solid-state NMR spectroscopy. Proc Natl Acad Sci USA. 2016;113:E272–81.
Xu H, He X, Zheng H, Huang LJ, Hou F, Yu Z, et al. Structural basis for the prion-like MAVS filaments in antiviral innate immunity. Elife 2014;3:e01489.
Cai X, Xu H, Chen ZJ. Prion-Like Polymerization in Immunity and Inflammation. Cold Spring Harb Perspect Biol. 2017;9:a023580.
Frias-Staheli N, Giannakopoulos NV, Kikkert M, Taylor SL, Bridgen A, Paragas J, et al. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe. 2007;2:404–16.
Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 2009;461:114–9.
Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell 2006;124:1197–208.
Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 2007;446:916–20.
Li S, Zheng H, Mao AP, Zhong B, Li Y, Liu Y, et al. Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J Biol Chem. 2010;285:4291–7.
Peng Y, Guo J, Sun T, Fu Y, Zheng H, Dong C, et al. USP39 Serves as a Deubiquitinase to Stabilize STAT1 and Sustains Type I IFN-Induced Antiviral Immunity. J Immunol. 2020;205:3167–78.
Xu S, Han L, Wei Y, Zhang B, Wang Q, Liu J, et al. MicroRNA-200c-targeted contactin 1 facilitates the replication of influenza A virus by accelerating the degradation of MAVS. PLoS Pathog. 2022;18:e1010299.
Meyer C, Garzia A, Morozov P, Molina H, Tuschl T. The G3BP1-Family-USP10 Deubiquitinase Complex Rescues Ubiquitinated 40S Subunits of Ribosomes Stalled in Translation from Lysosomal Degradation. Mol Cell. 2020;77:1193–205 e5.
Higuchi M, Kawamura H, Matsuki H, Hara T, Takahashi M, Saito S, et al. USP10 Is an Essential Deubiquitinase for Hematopoiesis and Inhibits Apoptosis of Long-Term Hematopoietic Stem Cells. Stem Cell Rep. 2016;7:1116–29.
Blount JR, Johnson SL, Todi SV. Unanchored Ubiquitin Chains, Revisited. Front Cell Dev Biol. 2020;8:582361.
Hodge CD, Spyracopoulos L, Glover JN. Ubc13: the Lys63 ubiquitin chain building machine. Oncotarget 2016;7:64471–504.
Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 2010;140:384–96.
Niu J, Shi Y, Xue J, Miao R, Huang S, Wang T, et al. USP10 inhibits genotoxic NF-kappaB activation by MCPIP1-facilitated deubiquitination of NEMO. EMBO J. 2013;32:3206–19.
Acknowledgements
We thank Dr FaJian Hou (Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences) for the DDX58−/−, UBE2N−/− and MAVS−/− HEK293T cells. This work was supported by grants from the National Natural Science Foundation of China (31730026, 81930039, 32000633), National Key Research and Development Program (2021YFC2300603), Natural Science Foundation of Shandong Province (ZR2020QH136), China Postdoctoral Science Foundation (2020M682187), and Postdoctoral Innovation Project of Shandong Province (202002012).
Author information
Authors and Affiliations
Contributions
CG conceived the research; CG and FL designed the experiments; FL performed the research; WZ, BS, YY, and JL provided reagents and participated in the experiments; JZ revised the paper and participated in discussions; YZ, BL and WZ participated in discussions; CG and FL analyzed the data; CG and FL wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, F., Zhuang, W., Song, B. et al. MAVS-loaded unanchored Lys63-linked polyubiquitin chains activate the RIG-I-MAVS signaling cascade. Cell Mol Immunol 20, 1186–1202 (2023). https://doi.org/10.1038/s41423-023-01065-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41423-023-01065-2
- Springer Nature Limited
Keywords
This article is cited by
-
The E3 ligase ASB3 downregulates antiviral innate immunity by targeting MAVS for ubiquitin-proteasomal degradation
Cell Death & Differentiation (2024)
-
Unconventional posttranslational modification in innate immunity
Cellular and Molecular Life Sciences (2024)