Abstract
Animal development and homeostasis require the programmed removal of cells. Autophagy-dependent cell deletion is a unique form of cell death often involved in bulk degradation of tissues. In Drosophila the steroid hormone ecdysone controls developmental transitions and triggers the autophagy-dependent removal of the obsolete larval midgut. The production of ecdysone is exquisitely coordinated with signals from numerous organ systems to mediate the correct timing of such developmental programs. Here we report an unexpected role for the Drosophila bone morphogenetic protein/transforming growth factor β ligand, Decapentaplegic (Dpp), in the regulation of ecdysone-mediated midgut degradation. We show that blocking Dpp signaling induces premature autophagy, rapid cell death, and midgut degradation, whereas sustained Dpp signaling inhibits autophagy induction. Furthermore, Dpp signaling in the midgut prevents the expression of ecdysone responsive genes and impairs ecdysone production in the prothoracic gland. We propose that Dpp has dual roles: one within the midgut to prevent improper tissue degradation, and one in interorgan communication to coordinate ecdysone biosynthesis and developmental timing.
Similar content being viewed by others
Introduction
In animal development tissue and organ morphogenesis is dependent on an intricate balance between cell division and programmed cell death (PCD) [1]. Although caspase-dependent apoptosis is the primary mode of developmental PCD, context specific modes of cell death that are dependent on autophagy, the catabolic process of cellular self-digestion through the action of lysosomal enzymes, also play a critical role [2,3,4,5]. Steroid hormones are important regulators of physiology and developmental transitions such as puberty in mammals and metamorphosis in insects [6]. The hormone-mediated juvenile-to-adult transition requires the remodeling or removal of juvenile structures by PCD. In Drosophila, the major developmental transitions are triggered by the steroid hormone 20-hydroxyecdysone (20E, ecdysone). The hormone binds its heterodimeric receptor, ecdysone receptor/ultraspiracle (EcR/Usp), in a spatiotemporal manner to regulate proliferation, differentiation, and PCD [7,8,9,10]. Multiple environmental and developmental signals coordinate hormone production to regulate developmental transitions and overall body size. However, the molecular links integrating hormone biosynthesis with other signals require further investigation.
A large ecdysone pulse just prior to the larval–pupal transition triggers metamorphosis accompanied by the degradation of the larval midgut characterized by the initial contraction of the gastric caeca which requires an autophagy-dependent cell death mechanism [11]. In addition to the hormonal cues, down-regulation of growth signaling through Ras and PI3K is critical for the appropriate induction of autophagy-dependent midgut PCD [12]. Here, we identify the bone morphogenetic protein/transforming growth factor β (BMP/TGF-β) ligand, decapentaplegic (Dpp), a morphogen required for developmental patterning [13], as an essential regulator of autophagy-dependent midgut degradation. The canonical Dpp signaling pathway in Drosophila is similar to that in mammals [14]. Signaling is initiated by Dpp binding to a type II receptor (Punt/Put), which recruits and phosphorylates a type I receptor (Thickveins/Tkv or Saxophone/Sax). The type I receptor phosphorylates and activates the Receptor-Smad (R-Smad) Mothers against dpp (Mad), which then binds to the common-Smad (co-Smad) Medea (Med). The active R-Smad/co-Smad (Mad-Med) complex translocates to the nucleus to regulate target gene expression. Inhibitory Smads (Daughters against Dpp/Dad) act as a negative feedback control by competing with R-Smads [14]. Several studies have implicated TGF-β in the regulation of autophagy [15,16,17]. In certain cancer cell lines, TGF-β has been shown to stimulate autophagy, with upregulation of autophagy genes Beclin-1, Atg5, and Atg7 [15, 17]. Similarly, TGF-β1 treatment of kidney cells resulted in increased reactive oxygen species and autophagy [18]. Sustained TGF-β can induce either autophagy or apoptosis depending on context [16], and TGF-β1 has also been implicated in down-regulating excessive autophagy [19]. Thus the physiological relationship between TGF-β signaling and autophagy-dependent cell death remains poorly understood. In this study we describe an unexpected and novel role of Dpp signaling in larval midgut growth and inhibition of autophagy. We found that an interaction between Dpp and ecdysone signaling plays a crucial function in regulating autophagy-dependent midgut degradation.
Results
Dpp prevents the premature removal of larval midgut cells
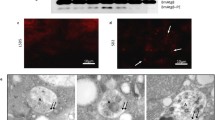
The ecdysone-triggered larval gut removal requires autophagy and cell size reduction. To identify regulators of this process we investigated clonal cells with loss of Dpp signaling and found that clones of Mad or tkv mutant cells underwent rapid elimination just prior to the initiation of the programmed larval midgut removal. Intact clones homozygous for Mad8–2 were difficult to find and image as the mutant cells in the midgut appeared to be rapidly removed, often showing degraded subcellular contents (Fig. 1a). This dramatic cell death was also observed in cell clones homozygous for tkv [5] (Fig. 1a), thus demonstrating an essential role for Dpp signaling in survival of midgut cells. Examination of autophagy using mChery-Atg8a revealed large puncta present in the rapidly dying cells, with abnormally diffuse DNA (Supplementary information, Figure S1a). The surrounding wild-type cells that have also initiated autophagy-dependent PCD remain intact as expected at this stage of PCD. This unique morphology of dying Mad and tkv deficient cells suggests that autophagy-dependent cell death may occur by the bulk degradation of cellular components. In addition, feeding the larvae chloroquine resulted in a partial rescue of the rapid removal of these cells observed by the presence of intact nuclei (Supplementary information, Figure S1b). Chloroquine inhibits lysosomal enzyme function and prevents autophagosomal fusion and degradation, highlighting the important role of autophagy in the removal of the midgut cells.
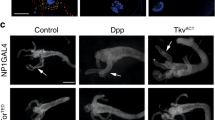
Dpp signaling prevents cell size contraction and autophagy during larval midgut degradation. a Loss-of-function clones of Mad and tkv die prematurely. Mad8-2 (hsFLP; Mad8-2 FRT-40A/Ubi-GFP FRT-40A) or tkv5 (hsFLP tkv5 FRT-40A/Ubi-GFP FRT-40A) homozygous cells detected by the absence of GFP are rapidly removed compared to the neighboring control cells marked by GFP (green) at −4 h RPF. LysoTracker red (red) and DNA stained by Hoechst (blue). Scale bar represents 25 μm. Live imaging reveals the rapid degradation of the Mad8-2 and tkv5 mutant cells (outlined), and show different stages of degradation. Due to the rapid degradation of the mutant cells, they are difficult to image. b Blocking Dpp signaling induces premature autophagy with small cell size while activating Dpp signaling reduces autophagy with larger cell size. Dad-expressing cells (hsFLP; pmCherry-Atg8a/UAS-Dad; Act > CD2 > GAL4, UAS-nlsGFP/+) marked by GFP (green) have increased levels of mCherry-Atg8a puncta (red, outlined) compared to the neighboring control cells and smaller cell size at −8 h RPF. TkvACT-expressing cell (hsFLP; pmCherry-Atg8a/+; Act > CD2 > GAL4, UAS-nlsGFP/UAS-tkvACT) marked by GFP (green) has decreased levels of mCherry-Atg8a puncta (red, outlined) compared to the neighboring control cells and larger cell size from +1 h RPF gastric caeca. DNA is stained by Hoechst (blue). Scale bar represents 25 μm. c Quantitation of cell size from (b) measured using ImageJ (average ± SD) (**p < 0.0001, compared to control). d Quantitation of puncta per cell from (b) measured using ImageJ (average ± SD.) (**p < 0.0001, compared to control)
To further characterize the role of Dpp in autophagy-dependent cell death we generated clones in the midgut that give rise to cells-expressing Dad (an inhibitor of Dpp signaling) adjacent to control cells. The Dad clone cells had a less severe phenotype compared to the Mad or tkv mutant cells, enabling a more detailed analysis of the timing of autophagy induction and cell size contraction. The Dad-expressing cells were smaller in size with increased levels of autophagy at an early stage (−8 h RPF) compared to the neighboring control cells indicating the premature onset of autophagy-dependent cell removal (Fig. 1b–d). Conversely, to determine the effect of sustained Dpp signaling we examined the expression of the constitutively active mutant form of the receptor Thickveins, TkvQ253D (referred to as TkvACT) [20]. Clone cells-expressing TkvACT in the midgut were larger with reduced autophagy compared to the neighboring control cells following the onset of midgut PCD (+1 h RPF) indicating a delay in their removal (Fig. 1b–d). At an earlier stage (−8 h RPF) prior to the initiation of PCD the TkvACT cells were similar in size to the adjacent wild-type cells indicating that TkvACT does not influence midgut cell size in third instar larvae, but rather prevents the cell size contraction during cell death (Supplementary information, Figure S1c). These findings reveal that Dpp signaling blocks autophagy-dependent PCD in vivo, and that down-regulation of Dpp signaling is required for the correct timing of midgut removal.
Dpp signaling in the larval midgut blocks degradation
Although Dpp activity is known to be present in the larval midgut [21,22,23,24], the function of Dpp in this tissue remains unknown. To examine the spatial localization of Dpp we used dpp-GAL4 to drive expression of UAS-GFP (dpp > GFP), which revealed distinct GFP-positive cells in the midgut (Figs. 2a, b). The larval midgut comprises a number of cell types including the intestinal stem cells (ISCs), which give rise to a new ISC and an enteroblast (EB) that differentiates into the absorptive enterocyte (EC) or enteroendocrine cell (EE) (Fig. 2c). The dpp > GFP (green) was detected in distinct GFP-positive ISC and EB, but EE cells that are identified by Prospero expression lacked GFP (Fig. 2b). Based on nuclear size and location, GFP was not detected in the ECs or the undifferentiated adult midgut progenitors identified as clusters of small cells [25]. Additionally, using reporters for Dpp activity, dpp-lacZ [26] and omb-lacZ, β-GAL activity could be detected along the midgut, consistently expressed at high levels in the middle part of the anterior midgut (Figure 2d). β-GAL immunostaining of dpp-lacZ confirmed this localization and identified specific cells in the midgut with strong staining consistent with the dpp > GFP cell localization (Fig. 2e). Nuclear phospho-Smad (p-Smad) staining, a marker of Dpp signaling activity, was detected in feeding third instar larval midguts (−16 h RPF) that decreased in wandering (−8 h RPF), and late third instar larval midguts (−4 h RPF) (Fig. 2f). This suggests that Dpp signaling is present in midguts prior to the onset of removal, which then decreases coincident with midgut degradation. Thus, it appears that Dpp signaling plays an important role in communication between the cells in the larval gut to prevent the premature degradation of this tissue.
Dpp expression and activity in the midgut. a Dpp-GAL4 driving UAS-GFP as a marker for Dpp expressing midgut cells (Dpp > GFP). GFP-positive cells (green) can be found along the anterior midgut (arrows). Inset shows higher magnification. Scale bar represents 100 μm. b Dpp > GFP (green) is expressed in the larval intestinal stem cells (ISC)/enteroblasts (EB) and not in the Prospero-labeled enteroendocrine (EE) cells. Higher magnification inset shows the GFP and Pros cells do not colocalize. DNA is stained by Hoechst (blue). Scale bar represents 50 μm. c Schematic representation of larval midgut cells. The larval midgut cells include intestinal stem cells (ISC), enteroblasts (EB), enteroendocrine (EE), and enterocytes (EC) cells. Other cell types include adult midgut progenitors (AMP), and the basement membrane (BM) and visceral muscle (VM). d In control, dpp-lacZ and omb-LacZ expression of LacZ reporter for Dpp signaling can be detected by β-GAL activity in the midgut at −4 h RPF. X-gal activity is present in distinct cells along the gastric caeca (arrow) and along the midgut. Scale bar represents 100 μm. e β-GAL immunolocalization in dpp-LacZ midguts at −4 h RPF marks distinct cells along the gastric caeca and along the midgut body. Scale bar represents 20 μm. f Nuclear localization of phospho-Smad (red) and DNA (blue) in midgut ECs is high in −16 h RFP, with reduced levels in −8 h RPF and low-nuclear levels in −4 h RPF. Scale bar represents 10 μm
Together, this suggests there is a complex role of Dpp signaling in the larval midgut that may function to coordinate midgut degradation. We propose that Dpp is expressed in the ISC/EB and the Dpp signal is received by the ECs to promote activation and nuclear localization of Mad. A similar role for Dpp has also been identified during spermatogenesis, where dpp expression in the somatic cells provides the Dpp ligand signal to the neighboring germ cells [27]. Activation of the Dpp pathway in ECs in the adult gut inhibits Dpp ligand expression [28], which is consistent with our findings of low levels of Dpp in larval ECs. Given the role of Dpp in communication between different cell/tissue types, it appears that Dpp signaling between the cells in the larval gut plays an important role in the correct timing of midgut removal.
Dpp signaling prevents autophagy and larval midgut degradation
Having uncovered a critical role of Dpp signaling for the survival of midgut cells we examined the effect of persistent Dpp signaling on the whole tissue. We found that Dpp signaling blocked larval midgut removal (Figs. 3a, d). Expression of Dpp or TkvACT using the highly specific midgut driver, mex-GAL4, or an alternative midgut driver, NP1-GAL4, [29] resulted in larger midguts that failed to show gastric caeca contraction compared to the control that had begun to contract (Fig. 3a, d; Supplementary information, Fig. 2a). While the drivers are expressed during earlier gut development, the phenotype was not apparent until late third instar larval stage as there was no significant difference in the size of Dpp and TkvACT midguts compared to controls from early third instar larvae (−16 to −8 h RPF) (Figure S2b). In addition, animals that expressed Dpp in the midgut did not undergo metamorphosis and died as late third instar larvae, while the mex-tkvACT animals died at a later stage as early pupae.
Dpp signaling blocks midgut removal and autophagy. a Morphology of control (NP1-GAL4/+), Dpp (NP1-GAL4/+; UAS-dpp/+), and TkvACT (NP1-GAL4/+; UAS-tkvACT/+) midguts from late third instar animals (−4 h RPF) shows enlarged midgut and gastric caeca (arrows). Scale bar represents 200 μm. b Autophagy induction detected by Atg8a puncta (red) and LysoTracker Red (red) in control midgut cells with low basal levels in Dpp and TkvACT midguts at −4 h RPF. DNA is stained by Hoechst (blue). Scale bar represents 100 μm. c Representative TEM images from sections of midgut at −4 h PRF. Control cells possess autolysosomal structures (arrows), while Dpp and TkvACT midgut cells lack structures. Scale bar represent 1 μm. d Quantitation of the gastric caeca size from (a) (average pixels ± SD) (**p < 0.0001, compared to control), autophagy puncta from (b) measured using ImageJ (average ± SD) (**p < 0.0001), and number of autophagic vesicles (solid shape indicates autophagosomes and autolysosomes) and lysosomes (outlined shape indicates lysosomes) from c (average ± SD) (**p < 0.0001)
To further establish the critical period required for Dpp expression we used the temperature-sensitive Gal4 inhibitor, Gal80ts, to regulate Dpp expression from the NP1-GAL4 driver (referred to as NP1ts). At the permissive temperature (18 or 25 °C), Gal4 activity is blocked by Gal80ts, thus preventing Dpp or TkvACT expression. Transfer of NP1ts > dpp animals to the restrictive temperature (29 °C) inactivating Gal80ts following the third instar larval molt resulted in enlarged midguts and late larval or early pupal lethality (Supplementary information, Figure S2c). This indicates that the critical time for Dpp signaling in the midgut is just prior to the critical weight developmental checkpoint prior to nutrient independent growth. These findings support a new role for Dpp signaling in regulation of autophagy-dependent midgut degradation during metamorphosis.
The ecdysone-triggered midgut removal is dependent on autophagy induction [11, 30]. To determine if Dpp mediated blockade of midgut degradation was due to a loss of autophagy, we examined markers of autophagy. At the onset of midgut degradation the control midguts had induced autophagy with high levels of autophagy (Atg8a puncta) whereas the midguts-expressing Dpp and TkvACT showed low basal levels of Atg8a puncta (Fig. 3b; Supplementary information, Figure S2d). LysoTracker staining has also been used in Drosophila to detect autophagy-associated lysosomal activity in the fat body and midgut [30, 31]. Consistent with low-Atg8a puncta, the Dpp and TkvACT midgut cells also had reduced LysoTracker staining (Fig. 3b, d). Ultrastructural analysis showed that the Dpp and TkvACT midguts lacked autolysosomal structures and contained more mitochondria (Fig. 3c, d), similar to the effect of blocking autophagy [32]. Together, these data indicate that autophagy induction is blocked by Dpp and TkvACT expression in the midgut.
Blocking Dpp signaling induces autophagy and midgut removal
To examine the effect of blocking Dpp signaling we expressed the inhibitory Smad, Dad, in the midgut. This resulted in both premature midgut degradation, as evidenced by shorter gastric caeca (+1 h RPF) and premature induction of autophagy (−4 h RPF) (Fig. 4a–d). Similarly, the knockdown of dpp in the midgut resulted in premature gastric caeca contraction and early autophagy induction (Fig. 4a–d). Similar results were seen using another independent dpp knockdown line. The phenotype observed by knockdown of dpp was less pronounced compared to the phenotype observed with the expression of Dad, and this may be due to the incomplete knockdown of dpp (Supplementary information, Figure S3a).
Blocking Dpp pathway induces premature autophagy and midgut removal. a Morphology of control (NP1-GAL4/+; pmCherry-Atg8a/+), Dad-expressing (NP1-GAL4/UAS-Dad; pmCherry-Atg8a/+) and dppi (NP1-GAL4/UAS-dppRNAi; pmCherry-Atg8a/+) midguts from +1 h shows smaller midgut and more contracted gastric caeca (arrows). Scale bar represents 200 μm. b Autophagy monitored by mCherry-Atg8a puncta (red) shows that expression of Dad and knockdown of dpp promotes premature induction of autophagy in the larval midguts at −4 h RPF (arrows). Scale bar represents 50 μm. c Quantitation of gastric caeca size at +1 h RPF from a (average pixels ± SD) (**p < 0.0001). d Quantitation of puncta at −4 h RPF from b (average ± SD) (*p = 0.003, **p = 0.0003). e Morphology at +1 h RPF shows contracted gastric caeca (arrows) from knockdown of Madi (NP1 > MadRNAi) and Medi (NP1 > MedRNAi) with the co-expression of Dpp or TkvACT. Scale bar represents 200 μm. f Quantitation of gastric caeca size from (E) (average pixels ± SD). (ns, not significant). g Number of pupae from combined Mad or Med knockdown with either expression of Dpp or TkvACT
To determine if the role of Dpp signaling on autophagy-dependent midgut removal was due to downstream Dpp signaling components, we examined the effects of combining the knockdown of Smads, Mad and Med, (Supplementary information, Figure S3b) with the expression of Dpp or TkvACT. The knockdown of Mad and Med combined with Dpp or TkvACT showed strong suppression following induction of midgut PCD (+1 h RPF), while the Dpp or TkvACT animals developmentally arrest prior to this stage (Fig. 4e, f). This rescue was not observed by knockdown of independent pathway genes (e.g., Figs 7c–e). Thus, Mad and Med knockdown was sufficient to rescue both midgut morphology and the larval lethality caused by NP1 > dpp or NP1 > tkvACT (Fig. 4g). These results are consistent with Dpp signaling through the canonical signaling pathway blocking midgut degradation.
Dpp modulates the expression of genes involved in autophagy and ecdysone response
The multistep autophagy pathway is regulated by a number of distinct Atg genes that are upregulated during autophagy-dependent midgut removal [11, 30]. To determine if the reduced autophagy and block in midgut removal by Dpp signaling was due to lower expression of genes essential for autophagy we examined the expression of several Atg genes in both Dpp or TkvACT midguts (Fig. 5a). The expression of several key Atg genes required for midgut degradation was significantly reduced in both Dpp and TkvACT expressing midguts (Fig. 5a). These observations further support the role of Dpp in preventing autophagy and Atg gene expression.
Dpp signaling impairs ecdysone dependent signaling. a Atg gene expression is reduced in larval midguts-expressing Dpp or TkvACT. Transcript levels were measured by qRT-PCR in control, Dpp and TkvACT larval midguts at −4 h RPF. Data are from 3 experiments, with 20 midguts per sample (average ± SEM) (*p < 0.05). b EcR knockdown blocks autophagy and midgut histolysis. Morphology of control (NP1-GAL4/+) and EcRi (NP1-GAL4/+; UAS-EcRRNAi/+) midguts at +4 h RPF show enlarged midgut and gastric caeca (arrows). Scale bar represents 200 μm. Quantitation of the gastric caeca size (average pixels ± SD) (**p < 0.0001). Autophagy monitored by mCherry-Atg8a puncta (red) from control and EcRi midguts from +2 h RPF. DNA is stained by Hoechst (blue). Scale bar represents 50 μm. Quantitation of puncta at +2 h RPF (average ± SD) (*p = 0.003). c Atg gene expression is reduced in larval midguts in two independent EcR RNAi lines. Transcript levels were measured by qRT-PCR in contro1 (NP1/+), NP1 > EcRi #1 and NP1 > EcRi #2 larval midguts at −4 h RPF (average ± SEM) (*p < 0.05)
The requirement for ecdysone in the removal of the obsolete larval tissue during metamorphosis is well established [33, 34]. Ecdysone signaling, mediated by the heterodimeric receptor EcR/Usp, regulates the transcription of target genes directly or through early (or primary) regulatory genes encoding transcription factors [7, 9, 10]. During salivary gland histolysis, the expression of several autophagy and apoptosis genes increases in response to ecdysone, in part due to the direct binding of EcR/Usp to the promoters [35, 36]. To establish the requirement of ecdysone signaling in regulation of autophagy during midgut degradation, the effect of EcR knockdown in the midgut was examined. The knockdown of EcR in the midgut blocked autophagy, delayed midgut removal (Fig. 5b; Supplementary information, Figure S4a), and resulted in decreased expression of all Atg genes examined (Fig. 5c).
Dpp signaling in the midgut blocks the developmental ecdysone response
With the essential role of ecdysone in midgut removal and the requirement of EcR for the expression of Atg genes, it is possible that Dpp signaling represses EcR and, thus its downstream primary response genes to alter Atg gene expression during midgut removal. Examination of the expression of EcR and primary response genes, BrC and E74 by qRT-PCR revealed a dramatic decrease in the transcript levels of these genes in Dpp and TkvACT-expressing midguts (Fig. 6a). The levels of EcR and primary response genes were also decreased in whole animals expressing Dpp and TkvACT in the midgut (Fig. 6a). Furthermore, the levels of EcR in the nucleus of midgut cells was dramatically reduced in response to Dpp or TkvACT expression in the midgut (Fig. 6b; Supplementary information, Figure S4b). Interestingly, the transcript levels of EcR in mex > tvkACT were similar to the control (Supplementary information, Figure S4c), yet the expression of EcR target genes was reduced, consistent with the weaker phenotype of this line. However, in clones of cells expressing TkvACT in the midgut there was no decrease in EcR staining (Fig. 6c). This suggests that the reduced EcR in the midguts is not simply due to Dpp expression in this tissue, but that a signal is produced from the midguts with maintained Dpp signaling that blocks the EcR response. It also implies that the developmental arrest due to Dpp expression in the midgut is not a consequence of the enlarged tissue. This is in agreement with the knockdown of other genes (e.g., Atg1 [11] or EcR that block autophagy) that result in enlarged midguts due to delayed degradation yet these animals undergo metamorphosis with no developmental block observed.
Dpp signaling blocks ecdysone production. a The levels of EcR, Br-C, and E74 are reduced in larval midgut and in whole larvae expressing Dpp or TkvACT in the midgut. Transcript levels were measured by qRT-PCR from control, NP1 > dpp and NP1 > tkvACT larval midguts at −4 h RPF. Data are from 3 experiments, with 20 midguts per sample (average ± SEM) (*p < 0.01). b Control, NP1 > dpp and NP1 > tkvACT midguts from larvae at −4 h RPF stained with EcR antibody (red) show dramatically reduced EcR. DNA is stained by Hoechst (blue). Scale bar represents 100 μm. c TkvACT-expressing cell (hsFLP; pmCherry-Atg8a/+; Act > CD2 > GAL4, UAS-nlsGFP/UAS-tkvACT) marked by GFP (green) has similar levels of EcR (red) compared to the neighboring control cells from −4 h RPF gastric caeca. DNA is stained by Hoechst (blue). Scale bar represents 25 μm. d Transcript levels of ptth, spo, phm, dib, sad, shd, and ecd were measured by qRT-PCR from control (NP1-GAL4/+), NP1 > dpp and NP1 > tkvACT late third instar larvae (−4 h RPF). Data are from three experiments, with three larvae per sample (average ± SEM) (*p < 0.001). e The ecdysone titer from whole late third instar larvae (−4 h RPF) expressing Dpp (NP1 > dpp) or TkvACT (NP1 > tkvACT) in the midgut. Data are from 3 experiments, with a minimum of 15 larvae per sample (average ± SD) (*p < 0.05, compared to control). f Survival of NP1 > dpp or NP1 > tkvACT to pupal stage following feeding of food supplemented with 20E or vehicle (ethanol)
The transcriptional activity of EcR/Usp requires ecdysone binding and EcR expression is directly induced by ecdysone [37]. Given the reduced EcR signaling in animals expressing Dpp and TkvACT in the midgut it was possible that this was due to a reduction in ecdysone levels. The production of ecdysone occurs in the prothoracic glands (PG), and requires a number of biosynthetic enzymes. Following its release from the PG ecdysone is taken up by target tissues where it is converted into the active form 20E [38]. Prothoracicotropic hormone stimulates ecdysone production in the PG and requires the function of several biosynthesis genes including spook (spo), phantom (phm), disembodied (dib), shadow (sad), and shade (shd). To determine if ecdysone production was disrupted we examined the levels of ecdysone biosynthesis genes in whole larvae that expressed Dpp and TkvACT in the midgut. This showed significantly reduced expression of several ecdysone biosynthesis genes (Fig. 6d, Supplementary information Figure S4d). Consistent with this, late third instar larvae expressing Dpp and TkvACT in the midgut had significantly reduced ecdysone titer (Fig. 6e). These results indicate that maintaining Dpp signaling in the midgut blocks the developmental ecdysone response. Thus, the transcriptional activation of genes required for ecdysone production is blocked in response to activation of the Dpp pathway in the midgut impeding ecdysone production. In support of this we found the block in metamorphosis in NP1 > dpp and NP1 > tkvACT animals could be partially rescued by feeding larvae with 20E (Fig. 6f). Thus, reduced ecdysone production contributes to the developmental arrest, and Dpp produced by the midgut plays a key role in regulating the correct timing of ecdysone biosynthesis. This is consistent with the down-regulation of Dpp activity indicated by the decrease in nuclear pSmad in the midgut prior to metamorphosis (Fig. 2f).
Dpp blocks the production of ecdysone
As the animals with maintained Dpp signaling in the midgut resulted in a block in ecdysone production we investigated the potential signal from the Dpp-expressing midgut that prevents ecdysone production in the PG. The insulin/relaxin-like peptide Dilp8 acts to ensure the coordination of organ growth with animal maturation [39, 40]. During larval development abnormally growing imaginal discs secrete Dilp8 to delay metamorphosis by inhibiting ecdysone biosynthesis. To examine if upregulation of Dilp8 is contributing to the block in ecdysone production from NP1 > dpp or NP1 > tkvACT, we examined a GFP reporter of Dilp8. Expression of GFP could be detected in the wing and eye imaginal disc, but could not be detected in the midgut or the brain and ring gland complex (Fig. 7a). The low levels of Dilp8 detected in the NP1 > dpp and NP1 > tkvACT animals that fail to pupate are not consistent with the ectopic expression of Dilp8 in imaginal discs that has been shown to delay pupariation by 2–3 days [39, 40], suggesting that Dilp8 alone is unlikely to be responsible for blocking ecdysone production preventing metamorphosis.
Dpp dependent block in ecdysone is not mediated by Dilp8 or Impl2. a Dilp8-GFP expression is undetectable in the midgut and brain complexes with modest GFP detected in eye and wing disc from NP1 > dpp, NP1 > tkvACT. Scale bars, 100 µm. b The level of Impl2 transcripts measured by qRT-PCR from NP1 > dpp and NP1 > tkvACT larvae was increased compared to the control at −4 h RPF. Data are from three experiments, with three larvae per sample (average ± SEM) (**p < 0.01, *p < 0.05 compared to control). c Knockdown of Impl2i (NP1-GAL4/UAS-Impl2RNAi), did not suppress the phenotype of NP1 > dpp and NP1 > tkvACT midguts at −4 h RPF by comparing contraction of the gastric caeca (arrows). Scale bar represents 200 μm. d Quantitation of gastric caeca size in (B) (average pixels ± SD). (**p < 0.001, *p < 0.01 and ns, not significant). e Quantitation by qRT-PCR of the level of RNAi mediated knockdown of Impl2 (average ± SEM)
The insulin/IGF antagonist, Imaginal morphogenesis protein-Late 2 (ImpL2) acts as a sensor of the nutritional state of larvae and coordinates dietary information and ecdysone production to modulate developmental transitions [41]. In the adult, ImpL2 is secreted from intestinal tumors that mediates organ wasting [42, 43]. We detected increased levels of Impl2 in animals expressing Dpp and TkvACT in the midgut (Fig. 7b). To investigate if the block in ecdysone production in the PG was due to the expression and secretion of ImpL2 from the midgut we ablated Impl2 in the Dpp or TkvACT larval midgut (Fig. 7c–e). However, this did not rescue the midgut morphology or the developmental arrest, suggesting that Impl2 is not secreted from the midgut (Fig. 7c, d).
An alternative possibility was that Dpp from midgut directly contributed to block ecdysone production by activating Dpp pathway in the PG. A recent study has shown that Dpp expressed in the fat body is secreted into the hemolymph resulting in perturbed Dpp signaling in the wing disc [44]. We investigated the potential for Dpp expressed in the midgut to promote Dpp signaling in peripheral tissue. Compared to the distinct pattern of activated Mad localization in the control wing disc, in animals expressing Dpp in the midgut homogeneous Dpp signaling in the wing disc was detected by the altered pattern of activated Mad staining (pSmad, Fig. 8a). This was in contrast to animals expressing TkvACT in the midgut, where the pattern of Mad localization was similar to control discs (Fig. 8a). More importantly, strong nuclear Mad localization was detected in the prothoracic gland of animals expressing Dpp in the midgut but the localization remained cytoplasmic in response to TkvACT expression in the midgut (Fig. 8b). This suggests that there is a signal produced from midguts that express Dpp that can signal to other peripheral tissue including the PG.
Block in ecdysone production due to Dpp expressed in the midgut. a Spatial localization of phospho-Smad (red) and DNA (blue) in wing discs from control, mex > dpp and mex > tkvACT larvae at −4 h RPF showing loss of the distinct pattern of pMad in mex > dpp. Scale bar represents 50 μm. b Nuclear localization of phospho-Smad (red) and DNA (blue) in ring glands from control, mex > dpp and mex > tkvACT larvae at −4 h RPF. Scale bar represents 50 μm. c Model of the proposed crosstalk between Dpp and ecdysone signaling during metamorphosis
Discussion
Our data identifies a previously unreported role for Dpp signaling as a critical regulator of autophagy-dependent midgut degradation. We show that canonical Dpp signaling prevents midgut degradation, while blocking Dpp signaling prematurely induces autophagy and midgut removal. In mosaic clones, cells expressing TkvACT have delayed cell size contraction and reduced autophagy during midgut PCD, whereas cells expressing Dad show premature cell size reduction and increased autophagy. Importantly, Mad8–2 and tkv5 mutant cells undergo rapid removal from the larval midgut. The unique morphology of dying Mad8–2 and tkv5 mutant cells also provides the first visual glimpses into the mechanism of autophagy-mediated cell death, as the cell contents appear to be rapidly degraded by lysosomal enzymes leaving behind cell corpses which appear as holes in the midgut tissue.
The spatial and temporal control of midgut growth and PCD involves the integration of several major inputs, including growth and hormonal signals. Down-regulation of growth signaling is a prerequisite for autophagy-dependent midgut PCD as is the input from hormonal cues. Interestingly, in the PG signaling pathways InR and Torso, acting via PI3K and Ras, respectively, are important for the upregulation of ecdysone biosynthesis genes and require BMP/activin signaling to ensure their expression [45]. Our findings now reveal that Dpp signaling prevents autophagy and ecdysone production, and it will be important to examine the coordination between Dpp, InR, and Ras signaling in the midgut.
The removal of the larval midgut occurs when the larvae stop feeding, and our data suggest that the midgut plays an important role in the coordination of growth and hormonal cues to establish a developmental checkpoint. Our studies also indicate that Dpp has dual roles, locally in the midgut during ecdysone-mediated PCD at the onset of metamorphosis and upstream as a regulator of ecdysone production from PG (Fig. 8c). Our data suggest that Dpp expressed in the midgut produces a signal responsible for the developmental arrest and block in ecdysone production. One possible model is that Dpp acts a sensor; if developmental cues are compromised Dpp activity results in a feedback to block ecdysone production, preventing midgut histolysis and metamorphosis thereby establishing a developmental transition checkpoint. Future studies to dissect out this important function of Dpp activity in the midgut will need to be undertaken to establish how the long-range signal is established and how this leads to the developmental checkpoint.
It is interesting that Dpp has distinct roles in the gut at various developmental stages. During embryonic gut formation Dpp is required in a complex feedback, both within and between tissue layers, to maintain the correct patterning of homeotic gene expression during midgut morphogenesis [46, 47]. In the Drosophila adult gut, Dpp has multiple complex roles in the regulation of ISCs in both an autocrine and paracrine mode [48,49,50,51,52,53]. Additionally, the TGF-β/activin pathway acts as a carbohydrate-sensing mechanism in the adult midgut to regulate digestive enzyme expression [54]. After more than 20 years exploring the role of Dpp as a tissue organizer, our studies have now uncovered another key role for Dpp, required for hormone-mediated autophagy-dependent midgut removal. The involvement of TGF-β signaling as a regulator of autophagy has also been reported in Caenorhabditis elegans [55, 56], thus suggesting that the function of Dpp may be conserved.
Materials and methods
Fly stocks
The midgut driver P{GawB}Myo31DFNP0001 referred to as NP1-GAL4, was obtained from the Drosophila Genetic Resource Center (Kyoto, Japan), and mex-GAL4 from R. Burke (Monash University, Vic., Australia). The following stocks were from the Bloomington Drosophila Stock Center (Bloomington, IN, USA) including RNAi lines from the Transgenic RNAi Project (http://www.flyrnai.org): w1118, UAS-dpp (w*; UAS-dpp.S 42B), UAS-tkvACT (w*; UAS-tkv.Q253D/TM3, Sb1 Ser1), EcRi #2 (w1118; UAS-EcR-RNAi s104), Mad8-2 (w*; Mad8-2 FRT-40A/CyO) dpp-GAL4 (w*; wgSp-1/CyO; P{GAL4-dpp.blk1}40C.6/TM6B, Tb1), omb-lacZ, dpp-lacZ (cn1; P{dppshv-lacZ.RD2}RD2, ry506), dppi (y1 v1; P{TRiP.JF01371}attP2), Madi (y1 v1; P{TRiP.JF01263}attP2), Medi (y1 v1; P{TRiP.JF02218}attP2), impl2i y1 w*; {MiMIC}Ilp8MI00727 and UAS-GFP (w1118; UAS-EGFP 5a.2). The ImpL2i (P{KK112218}VIE-260B) line was from Vienna Drosophila RNAi Center, EcRi #1 (UAS-EcR-RNAi) line was from NIG-Fly, ywhsFlp; pmCherry-Atg8a; Act > CD2 > GAL4, UAS-nlsGFP/TM6B from E. Baehrecke, hsFLP; Ubi-GFP FRT-40A, tkv5 FRT-40A /CyO, and UAS-Dad from H. Richardson (La Trobe University, Vic., Australia), and tub-Gal80ts from G. Hime (Melbourne University, Vic., Australia. The control was w1118 crossed to the driver line. Verification and quantitation of the RNAi lines was determined by qRT-PCR from late third instar larvae (−4 h RFP) from 20 midguts/sample in triplicate. All flies were maintained and crossed performed at 25 °C on cornmeal, molasses, and yeast medium.
Larval staging, midgut morphology analysis, and 20E feeding
Larvae were staged by feeding on food containing 0.05% bromophenol blue and transferred as wandering third instar larvae into a petri dish lined with moist Whatmann paper to monitor for gut clearance as visualized by loss of blue in the gut [57]. For temperature shift experiments, crosses were allowed to lay eggs in 4 h batches and aged at 18 °C prior to shifting to 29 °C. For morphological analysis, a minimum of 10 midguts from appropriately staged animals were dissected in PBS, fixed in 4% formaldehyde/PBS and imaged using a stereozoom microscope (Olympus, Tokyo, Japan). Measurements of the gastric caeca size from images was done using Photoshop (Adobe, San Jose, CA, USA) magnetic lasso tool and the histogram function used to determine pixels included in the area as previously described [12]. For ecdysone feeding larvae were collected as first instar and placed in groups of 10 in 1.5 ml tubes containing 200 μl standard food supplemented with 20E (Sigma-Aldrich, St. Louis, MO, USA) dissolved in ethanol (0.5 mg/ml), capped with cotton wool. Control larvae were fed with standard food mixed with the same volume of ethanol. A total of 300 larvae were collected from each genotype.
Live mCherry and LysoTracker imaging
To assay for mCherry-Atg8a, a minimum of 10 midguts were dissected from appropriately staged animals in PBS with Hoechst 33342 (Sigma-Aldrich) to stain DNA and imaged immediately without fixation using a Zeiss LSM 700 confocal (Detmold Imaging Core Facility, SA Pathology, Adelaide, SA, Australia). The images were quantitated using ImageJ to count puncta with a size larger than two pixels and represented as the average puncta per cell. To assay for LysoTracker, a minimum of 10 midguts were dissected in PBS with 1 µM LysoTracker Red and Hoechst 33342. The images were quantitated using ImageJ to count puncta with a size larger than 30 pixels, and represented as the average puncta per cell.
Induction of cell clones and chloroquine feeding
To induce TkvACT or Dad clones, ywhsFlp; pmCherry-Atg8a; Act > CD2 > GAL4, UAS-nlsGFP/TM6B was crossed to UAS-tkvACT/TM6B or UAS-Dad and 1-day-old embryos were heat shocked at 37 °C for 5 min. Cell size was quantitated using ImageJ [58]. To generate mutant clones hsFlp; ubi-GFP FRT-40A was crossed to Mad8–2 FRT-40A/CyO actGFP and tkv5 FRT-40A/CyO actGFP mutant allele lines on the same chromosome as the FRT. The cross was allowed to lay for 8 h then embryos were heat shock at 37 °C for 60 min. For chloroquine feeding of larvae, chloroquine diphosphate salt (CQ, Sigma) was dissolved in water (25 mg/ml), and added to standard food to a final concentration of 2.5 mg/ml. Control larvae were fed with standard food mixed with the same volume of PBS.
Immunohistochemistry
Midguts of the required genotype were dissected from appropriately staged animals in PBS and fixed in 4% paraformaldehyde in PBS with phosphatase inhibitors (1:100) (Sigma, P5726) for 20 min at room temperature as described [31]. Primary antibodies used were rabbit anti-Phospho-Smad 1/5 (Ser463/465) (1:500) (Cell Signaling, 9516, Danvers, MA, USA), rabbit anti-GABARAP1, referred to as ATG8a, (1:200) (Abcam, Cambridge, MA, USA), mouse anti-EcR B1(1:400) (Development Studies Hybridoma Bank, DSHB), anti-prospero (1:10) (DSHB), rabbit anti-β-galactosidase (1:2000) (MP Biomedicals, Solon, OH, USA), and rabbit anti-GFP (1:500) (Abcam). Secondary antibodies were anti-rabbit Alexa-FLUOR 568 and anti-mouse Alexa-FLUOR 488 (Molecular Probes, Eugene, CA, USA), and Hoechst 33342 (Sigma-Aldrich) was used to detect DNA. The samples were imaged using a Zeiss LSM 700 confocal microscope.
X-gal staining
The lacZ activity was detected using standard X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining procedure. Briefly, midguts from larvae were dissected in PBS, fixed in 0.8% glutaraldehyde in PBS for 15 min, washed in PBS and stained in 0.2% X-gal in staining buffer (10 mM NaH2PO4/Na2HPO4 pH7.2, 1 mM MgCl2, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 0.4% SDS) at room temperature.
Transmission electron microscopy
Midguts from appropriately staged larvae were dissected in 1×PBS then fixed in 1.25% glutaraldehyde, 4% sucrose, 4% paraformaldehyde in PBS for 30 min at room temperature. Samples were then washed with 4% sucrose in PBS, postfixed in 1% osmium tetroxide for 1 h, dehydrated, treated with propylene oxide, and infiltrated for embedding in resin as described [31]. Ultrathin sections were cut on grids, stained for 2 min with 4% uranyl acetate in 25% ethanol and 2 min in Reynold’s lead citrate before examination using Tecnai G2 Spirit TEM (Adelaide Microscopy).
Confocal imaging
Confocal images were obtained using Zeiss LSM 700 or Zeiss LSM 800 inverted confocal microscope with Argon ion 488 nm (14 mw) and Green HeNe 543 nm (1.5 mw) lasers and a 40 × UPLAPO (NA = 1.2 water) objective. The dual labeled samples were imaged with two separate channels (PMT tubes) in a sequential setting. Green fluorescence was excited with an Ar 488 nm laser line and the emission viewed through a HQ515/30 nm narrow band barrier filter in PMT1. Red fluorescence was excited with a HeNe 543 nm laser line and the emission viewed through a long pass barrier filter (E570LP) in PMT2 (Detmold Facility). On LSM 700, Zen grey was used to capture the images. On LSM 800, Airyscan detector was used, pictures were captured and Airyscan processed using Zen blue. Images were then processed using Photoshop (Adobe).
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from 20 midguts/sample or 3 whole larvae/sample in triplicate for each sample using TRIzol reagent (Invitrogen). cDNA was synthesized using High Capacity cDNA Reverse Transcription Kit (Applied Biosciences, Life Technologies, Carlsbad, CA, USA) and random or oligo dT primers (Geneworks, Thebarton, SA, Australia), with 1 μg of total RNA. qRT-PCR was performed on a Qiagen Rotor-Gene Q using RT [2] SYBR Green qPCR MasterMix (Qiagen, Valencia, CA, USA) as per the manufacturer’s instructions. Reactions were performed in triplicate using three independent biological samples and the transcript levels were normalized using rp49 as the reference gene. Data were analyzed using either the comparative CT method with optimized efficiencies for Target and Reference genes or using the Q-Gene software with standard curves [59], and samples on the same graph were run simultaneously. Primers for Atg genes are described [11, 30], and primers used are as follows:
dpp F 5′-gccaacacagtgcgaagtt; R 5′-cgcgtgatgtcgtagacaag Mad F 5′-ctgagcaacgtgaacaggaa; R 5′-gatggaatccgtggtggtag Med F 5′-accctcacctacacgcagtc; R 5′-tatgcgatggagcaccagtaBr-C F 5′-acaacaacagccccgactt; R 5′-gcttgtcgctgatggagatt E74 F 5′-ccacaatctgcttagcggc; R 5′-ctgggcggaaatgaacctgt EcR F 5′-ctcagctgcaaggtcaactg; R 5′-ccatgtattcgctgctcgta ptth F 5′-ggctgcgactgcaaagttac; R 5′-ccacgaataggggtgatcgg spo F 5′-tatctcttgggcacactcgctg; R 5′-gccgagctaaatttctccgctt phm F 5′-ggatttctttcggcgcgatgtg; R 5′-tgcctcagtatcgaaaagccgt dib F 5′-tgccctcaatccctatctggtc; R 5′-acagggtcttcacacccatctc sad F 5′-ccgcattcagcagtcagtgg; R 5′-acctgccgtgtacaaggagag shd F 5′-cgggctactcgcttaatgcag; R 5′-agcagcaccacctccatttc Impl2 F 5′-tacaagtgcatagcccgcaa; R 5′-ttcacgcatcttcgaaggca
Measurement of ecdysone titer
Late third instar larvae were collected from control, Dpp and TkvACT (minimum of 15 larvae/sample in triplicate) snap frozen in liquid nitrogen and stored at −80 °C. The weighed samples were homogenized in 300 µl of methanol, centrifuged at 16,000×g for 20 min twice with the combined methanol supernatants dried prior to resuspending in 50 µl enzyme immunoassay buffer (Cayman Chemicals, Inc., USA). The ecdysteroid levels were measured as 20E equivalents and the assay was performed using 20E antiserum, 20E-acetylcholinesterase Tracer and Ellman reagent (Cayman Chemicals) according to the manufacture’s protocol. Briefly, 50 µl of the standards and diluted samples were incubated with 50 µl of tracer and antiserum in antirabbit IgG coated ELISA plates overnight, washed five times and developed by adding 200 µl of Ellman’s reagent. The absorbance was read at 405 nm on Spectrophotometer (Fluostar) and all assays were performed in triplicate.
Statistical analysis of data
Student’s t test, two-tailed equal variance was used for statistical analysis to compare two groups and data are expressed as mean ± standard deviation (SD) or mean ± standard error of the mean (SEM), as appropriate. P < 0.05 was considered significant. For multiple analyses an ordinary one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test was performed using Prism (GraphPad Software) and data are expressed as mean ± SD. There was no statistical method used to predetermine sample size. The experiments were not randomized and no samples were excluded. There was no blind allocations of the experiments or interpretation of results.
References
Fuchs Y, Steller H. Live to die another way: modes of programmed cell death and the signals emanating from dying cells. Nat Rev Mol Cell Biol. 2015;16:329–44.
Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19:107–20.
Denton D, Xu T, Kumar S. Autophagy as a pro-death pathway. Immunol Cell Biol. 2015;93:35–42.
Yuan J, Kroemer G. Alternative cell death mechanisms in development and beyond. Genes Dev. 2010;24:2592–602.
Arakawa S, Tsujioka M, Yoshida T, Tajima-Sakurai H, Nishida Y, Matsuoka Y, et al. Role of Atg5-dependent cell death in the embryonic development of Bax/Bak double-knockout mice. Cell Death Differ. 2017;24:1598–1608.
King-Jones K, Thummel CS. Nuclear receptors-a perspective from Drosophila. Nat Rev Genet. 2005;6:311–23.
Baehrecke EH. Steroid regulation of programmed cell death during Drosophila development. Cell Death Differ. 2000;7:1057–62.
Kumar S, Cakouros D. Transcriptional control of the core cell-death machinery. Trends Biochem Sci. 2004;9:193–9.
Denton D, Aung-Htut MT, Kumar S. Developmentally programmed cell death in Drosophila. Biochim Biophys Acta. 2013;833:3499–506.
Nicolson S, Denton D, Kumar S. Ecdysone-mediated programmed cell death in Drosophila. Int J Dev Biol. 2015;59:23–32.
Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, et al. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol. 2009;19:1741–6.
Denton D, Chang T-K, Nicolson S, Shravage B, Simin R, Baehrecke EH, et al. Relationship between growth arrest & autophagy in midgut programmed cell death in Drosophila. Cell Death Differ. 2012;19:1299–307.
Bier E, De Robertis EM. BMP gradients: a paradigm for morphogen-mediated developmental patterning. Science. 2015;348:aaa5838.
Hamaratoglu F, Affolter M, Pyrowolakis G. Dpp/BMP signaling in flies: from molecules to biology. Semin Cell Dev Biol. 2014;32:128–36.
Suzuki HI, Kiyono K, Miyazono K. Regulation of autophagy by transforming growth factor-b (TGF-b) signaling. Autophagy. 2010;6:645–7.
Jiang Y, Woosley AN, Sivalingam N, Natarajan S, Howe PH. Cathepsin-B-mediated cleavage of Disabled-2 regulates TGF-beta-induced autophagy. Nat Cell Biol. 2016;18:851–63.
Kiyono K, Suzuki HI, Matsuyama H, Morishita Y, Komuro A, Kano MR, et al. Autophagy is activated by TGF-beta and potentiates TGF-beta-mediated growth inhibition in human hepatocellular carcinoma cells. Cancer Res. 2009;69:8844–52.
Xu Y, Yang S, Huang J, Ruan S, Zheng Z, Lin J. Tgf-beta1 induces autophagy and promotes apoptosis in renal tubular epithelial cells. Int J Mol Med. 2012;29:781–90.
Ni BB, Li B, Yang YH, Chen JW, Chen K, Jiang SD, et al. The effect of transforming growth factor beta1 on the crosstalk between autophagy and apoptosis in the annulus fibrosus cells under serum deprivation. Cytokine. 2014;70:87–96.
Nellen D, Burke R, Struhl G, Basler K. Direct and long-range action of a DPP morphogen gradient. Cell . 1996;85:357–68.
Cherbas L, Hu X, Zhimulev I, Belyaeva E, Cherbas P. EcR isoforms in Drosophila: testing tissue-specific requirements by targeted blockade and rescue. Development. 2003;130:271–84.
Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–20.
Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–87.
Wilk R, Hu J, Blotsky D, Krause HM. Diverse and pervasive subcellular distributions for both coding and long noncoding RNAs. Genes Dev. 2016;30:594–609.
Micchelli CA, Sudmeier L, Perrimon N, Tang S, Beehler-Evans R. Identification of adult midgut precursors in Drosophila. Gene Expr Patterns. 2011;11:12–21.
Hursh DA, Padgett RW, Gelbart WM. Cross regulation of decapentaplegic and Ultrabithorax transcription in the embryonic visceral mesoderm of Drosophila. Development. 1993;117:1211–22.
Tang Y, Geng Q, Chen D, Zhao S, Liu X, Wang Z. Germline proliferation is regulated by somatic endocytic genes via JNK and BMP signaling in Drosophila. Genetics. 2017;206:189–97.
Tian A, Wang B, Jiang J. Injury-stimulated and self-restrained BMP signaling dynamically regulates stem cell pool size during Drosophila midgut regeneration. Proc Natl Acad Sci USA. 2017;114:E2699–E708.
Phillips MD, Thomas GH. Brush border spectrin is required for early endosome recycling in Drosophila. J Cell Sci. 2006;119:1361–70.
Xu T, Nicolson S, Denton D, Kumar S. Distinct requirements of autophagy-related genes in programmed cell death. Cell Death Differ. 2015;22:1792–802.
Xu T, Kumar S, Denton D. Characterization of autophagic responses in Drosophila melanogaster. Methods Enzymol. 2017;588:445–65.
Chang T-K, Shravage BV, Hayes SD, Powers CM, Simin RT, Wade Harper J, et al. Uba1 functions in Atg7- and Atg3-independent autophagy. Nat Cell Biol. 2013;15:1067–78.
Lee CY, Cooksey BA, Baehrecke EH. Steroid regulation of midgut cell death during Drosophila development. Dev Biol. 2002;250:101–11.
Lee CY, Simon CR, Woodard CT, Baehrecke EH. Genetic mechanism for the stage- and tissue-specific regulation of steroid triggered programmed cell death in Drosophila. Dev Biol. 2002a;252:138–48.
Cakouros D, Daish TJ, Kumar S. Ecdysone receptor directly binds the promoter of the Drosophila caspase dronc// regulating its expression in specific tissues. J Cell Biol. 2004;165:631–40.
Denton D, Aung-Htut MT, Lorensuhewa N, Nicolson S, Zhu W, Mills K, et al. UTX coordinates steroid hormone-mediated autophagy and cell death. Nat Commun. 2013a;4:2916.
Karim FD, Thummel CS. Temporal coordination of regulatory gene expression by the steroid hormone ecdysone. EMBO J. 1992;11:4083–93.
Gilbert LI, Rybczynski R, Warren JT. Control and biochemical nature of the ecdysteroidogenic pathway. Annu Rev Entomol. 2002;47:883–916.
Colombani J, Andersen DS, Leopold P. Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science. 2012;336:582–5.
Garelli A, Gontijo AM, Miguela V, Caparros E, Dominguez M. Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science. 2012;336:579–82.
Sarraf-Zadeh L, Christen S, Sauer U, Cognigni P, Miguel-Aliaga I, Stocker H, et al. Local requirement of the Drosophila insulin binding protein imp-L2 in coordinating developmental progression with nutritional conditions. Dev Biol. 2013;381:97–106.
Kwon Y, Song W, Droujinine IA, Hu Y, Asara JM, Perrimon N. Systemic organ wasting induced by localized expression of the secreted insulin/IGF antagonist ImpL2. Dev Cell. 2015;33:36–46.
Figueroa-Clarevega A, Bilder D. Malignant Drosophila tumors interrupt insulin signaling to induce cachexia-like wasting. Dev Cell. 2015;33:47–55.
Ma M, Cao X, Dai J, Pastor-Pareja JC. Basement membrane manipulation in Drosophila wing discs affects Dpp Retention but not growth mechanoregulation. Dev Cell. 2017;42:97–106 e4.
Gibbens YY, Warren JT, Gilbert LI, O’Connor MB. Neuroendocrine regulation of Drosophila metamorphosis requires TGF-beta/activin signaling. Development. 2011;138:2693–703.
Bienz M. Endoderm induction in Drosophila: the nuclear targets of the inducing signals. Curr Opin Genet Dev. 1997;7:683–8.
Nakagoshi H. Functional specification in the Drosophila endoderm. Dev Growth Differ. 2005;47:383–92.
Li H, Qi Y, Jasper H. Dpp signaling determines regional stem cell identity in the regenerating adult Drosophila gastrointestinal tract. Cell Rep. 2013;4:10–8.
Li Z, Zhang Y, Han L, Shi L, Lin X. Trachea-derived dpp controls adult midgut homeostasis in Drosophila. Dev Cell. 2013;24:133–43.
Guo Z, Driver I, Ohlstein B. Injury-induced BMP signaling negatively regulates Drosophila midgut homeostasis. J Cell Biol. 2013;201:945–61.
Tian A, Jiang J. Intestinal epithelium-derived BMP controls stem cell self-renewal in Drosophila adult midgut. eLife. 2014;3:e01857.
Zhou J, Florescu S, Boettcher AL, Luo L, Dutta D, Kerr G, et al. Dpp/Gbb signaling is required for normal intestinal regeneration during infection. Dev Biol. 2014;399:89–203.
Ayyaz A, Li H, Jasper H. Haemocytes control stem cell activity in the Drosophila intestine. Nat Cell Biol. 2015;17:736–48.
Chng WB, Bou Sleiman MS, Schupfer F, Lemaitre B. Transforming growth factor beta/activin signaling functions as a sugar-sensing feedback loop to regulate digestive enzyme expression. Cell Rep. 2014;9:336–48.
Guo B, Huang X, Zhang P, Qi L, Liang Q, Zhang X, et al. Genome-wide screen identifies signaling pathways that regulate autophagy during Caenorhabditis elegans development. EMBO Rep. 2014;15:705–13.
Aladzsity I, Toth ML, Sigmond T, Szabo E, Bicsak B, Barna J, et al. Autophagy genes unc-51 and bec-1 are required for normal cell size in Caenorhabditis elegans. Genetics. 2007;177:655–60.
Denton D, Mills K, Kumar S. Methods and protocols for studying cell death in Drosophila. Methods Enzymol. 2008;446:17–37.
Osman OS, Selway JL, Kepczynska MA, Stocker CJ, O’Dowd JF, Cawthorne MA, et al. A novel automated image analysis method for accurate adipocyte quantification. Adipocyte. 2013;2:160–4.
Muller PY, Janovjak H, Miserez AR, Dobbie Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques. 2002;32:1372–9.
Acknowledgments
This work was supported by the National Health and Medical Research Council of Australia Project Grant (1124490), an Australian Research Council Grant (DP170100623), and a NHMRC Senior Principal Research Fellowship (1103006). We thank the Australian Drosophila Research Support Facility (Monash University, Melbourne), Drosophila Genetic Resource Center (Kyoto, Japan), Vienna Drosophila Resource Center, and Bloomington Drosophila Stock Center for Drosophila stocks, Developmental Studies Hybridoma Bank (University of Iowa) for antibodies, Amanda Rogers for H&E tissue processing and Ruth Williams at Adelaide Microscopy for sample preparation for TEM. We thank the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing transgenic RNAi fly stocks.
Author contributions
S.K. and D.D. conceptualized and designed the project, D.D., T.X., S.D., and S.N. performed experimental work; D.D., S.K., T.X., S.D., and S.N. analyzed data; D.D. and S.K. prepared the manuscript. All authors discussed the results and commented on the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Edited by E. Baehrecke
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Denton, D., Xu, T., Dayan, S. et al. Dpp regulates autophagy-dependent midgut removal and signals to block ecdysone production. Cell Death Differ 26, 763–778 (2019). https://doi.org/10.1038/s41418-018-0154-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-018-0154-z
- Springer Nature Limited
We’re sorry, something doesn't seem to be working properly.
Please try refreshing the page. If that doesn't work, please contact support so we can address the problem.