Abstract
Hepatocellular carcinoma (HCC) is the second leading cause of cancer-related deaths worldwide, with more than 800,000 deaths each year, and its 5-year survival rate is less than 12%. The role of the HN1 gene in HCC has remained elusive, despite its upregulation in various cancer types. In our investigation, we identified HN1’s heightened expression in HCC tissues, which, upon overexpression, fosters cell proliferation, migration, and invasion, unveiling its role as an oncogene in HCC. In addition, silencing HN1 diminished the viability and metastasis of HCC cells, whereas HN1 overexpression stimulated their growth and invasion. Gene expression profiling revealed HN1 silencing downregulated 379 genes and upregulated 130 genes, and suppressive proteins associated with the lipogenic signaling pathway networks. Notably, suppressing HN1 markedly decreased the expression levels of SREBP1 and SREBP2, whereas elevating HN1 had the converse effect. This dual modulation of HN1 affected lipid formation, hindering it upon HN1 silencing and promoting it upon HN1 overexpression. Moreover, HN1 triggers the Akt pathway, fostering tumorigenesis via SREBP1-mediated lipogenesis and silencing HN1 effectively curbed HCC tumor growth in mouse xenograft models by deactivating SREBP-1, emphasizing the potential of HN1 as a therapeutic target, impacting both external and internal factors, it holds promise as an effective therapeutic strategy for HCC.

Similar content being viewed by others
Introduction
Cancer is a foremost cause of global morbidity and mortality [1, 2], with liver cancer ranking as the sixth most prevalent malignancy and the second leading contributor to cancer-related fatalities, accounting for more than 800,000 deaths annually worldwide [3]. Hepatocellular carcinoma (HCC) is the most prevalent form of liver cancer and is associated with a heightened mortality rate in individuals who also have cirrhosis [4,5,6]. HCC can develop progressively within a healthy liver, spurred by various risk factors, including hepatitis B and C, exposure to aflatoxins, cirrhosis, alcohol consumption, anabolic steroid use, and hemochromatosis [7, 8]. Despite the current efficacy of liver transplantation and surgical resection as the primary means of treating HCC patients [9, 10], the postoperative prognosis remains subject to individual patient factors and health status [11, 12]. Furthermore, the specter of recurrence following treatment looms large, posing a pervasive and detrimental impediment to the prospects of achieving a curative outcome.
Clinical staging systems currently provide a valuable and practical method for forecasting the prognosis of HCC patients and categorizing them according to observable physical signs and symptoms [13,14,15]. Nevertheless, their predictive accuracy remains constrained, and researchers are as yet unable to discern how the biological traits of HCC are linked to clinical diversity [16, 17]. In light of recent strides in gene-targeted prognostication and therapeutic advances, novel prediction models for HCC have emerged. Moreover, gene expression signatures could present more precise insights into HCC progression.
The hematopoietic- and neurologic-expressed sequence 1 (HN1) gene exhibits elevated expression levels in numerous cancer types, including melanoma [18], prostate cancer [19], breast cancer [20], and thyroid carcinoma [21]. It has been linked to tumor growth stimulation [22] and metastatic carcinoma progression [23], and it has been shown to play pivotal roles in neural development, nerve regeneration, and retina regeneration [24]. Notably, the depletion of HN1 in both melanoma and prostate cells leads to the suppression of cell cycle arrest [18, 21].
In prior investigations, a total of 1016 and 628 genes were identified as having associations with the survival of HCC patients following surgical resection [25,26,27]. Within those two studies, we pinpointed 65 genes characterized by a high-risk score and closely correlated with a heightened risk of liver cancer incidence and an adverse prognosis for liver cancer patients [28]. By examining the expression patterns of these 65 genes [28], we observed that HN1 exhibited significantly elevated expression levels, and HCC patients with heightened HN1 expression experienced unfavorable overall survival and recurrence-free survival durations. Another study demonstrated that a five-gene score, based on the combined expression levels of HN1, RAN, RAMP3, KRT19, and TAF9, was intricately linked to the biology and prognosis of HCC and showed that HCC patients whose five-gene scores indicated overexpression had the lowest survival rates [29].
Despite this compelling evidence pointing toward the pivotal role of HN1 in HCC, its specific biological function in this context remains poorly understood. Furthermore, the molecular mechanisms governing how HN1 regulates HCC cell proliferation and invasion and its interactions with signaling pathways remain elusive. Therefore, this study represents our initial endeavor to illuminate the underlying molecular mechanisms by which HN1 governs the proliferation, invasion, metastasis, and intracellular metabolism of HCC cells.
Results
HN1 exhibited pronounced overexpression in human HCC and was associated with poor overall survival times in individuals with HCC
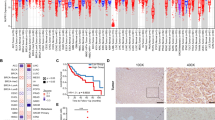
To investigate the role of HN1 in human HCC, we accessed patient survival data and gene expression profiles from the Cancer Genome Atlas (TCGA, CELL PMID: 28622513) and the National Cancer Institute database (NCI, Nature Med, PMID: 16532004). Patients were stratified into two groups based on their HN1 expression levels: the HN1 low-expression cohort and the HN1 high-expression cohort. Subsequently, we conducted a Kaplan–Meier survival analysis to scrutinize disparities in overall survival (OS) between those groups. Our findings unequivocally show that patients with diminished HN1 expression enjoyed a significantly prolonged OS compared with their counterparts with heightened HN1 expression (Fig. 1A, P = 7.0 × 10−7 (TCGA), P = 0.003 (NCI)). Moreover, in our quest to scrutinize the potential implications of HN1 in hepatocarcinogenesis, we explored the correlation between HN1 mRNA expression and HN1 DNA methylation levels within the TCGA HCC patient dataset. Those results unveiled a conspicuous negative correlation between HN1 mRNA expression and the methylation status of the HN1 promoter region in HCC (Fig. 1B, R = −0.405, P = 1.23 × 10−15). These findings strongly suggest that HN1 mRNA expression inversely correlates with HN1 promoter region methylation in HCC patients, hinting that HN1 is involved in hepatocarcinogenesis.
A Kaplan–Meier analysis for the overall survival times of hepatocellular carcinoma patients with high HN1 expression and low HN1 expression in TCGA (left) and the NCI (right) database. TCGA, The Cancer Genome Atlas. NCI, National Cancer Institute. OS, overall survival. B The correlation between HN1 mRNA expression and HN1 DNA methylation was analyzed in HCC patients. C The protein expression of HN1 in paired HCC tissues and adjacent non-tumoral liver tissues from seven patients. T liver tumor tissue. N adjacent non-tumoral liver tissue. D The protein expression of HN1 in unpaired HCC tissues from 7 patients. E The relative levels of HN1 mRNA were examined using qPCR in paired HCC and adjacent non-tumoral liver tissues from seven patients. F The protein levels of HN1 in seven hepatocellular carcinoma cell lines were detected. Data were expressed as the mean ± SEM of three independent experiments. Protein quantification was calculated by ImageJ software. *p < 0.05, **p < 0.01, and ***p < 0.001 compared with the corresponding control.
To further investigate the role of HN1 in human HCC, we conducted an analysis of HN1 expression in liver tumor tissues obtained from HCC patients. Those findings unequivocally demonstrate the significant overexpression of HN1 in liver tumor tissues compared with adjacent non-tumoral liver tissues (Fig. 1C, D). Furthermore, HN1 expression exhibited a noteworthy elevation in liver tumor tissues across different HCC patients when contrasted with adjacent non-tumoral liver tissues (Fig. 1E). In addition to tissue analysis, we explored the endogenous expression of HN1 in various HCC cell lines: Hep3B, HLE, Huh7, SNU638, SHJ-1, HepG2, and SNU449. Among them, SNU449 exhibited the highest HN1 expression levels, whereas Hep3B, HLE, SNU638, and HepG2 displayed comparatively low expression of HN1. For the sake of a representative investigation, we selected the poorly differentiated SNU449 cell line and the well-differentiated HepG2 cell line for further experiments.
HN1 suppression inhibited cell proliferation and induced apoptosis in HCC cells
To assess the effects of HN1 on cell proliferation in HCC cell lines, we used the WST-1 assay. Our results, depicted in Fig. 2A, demonstrate that HN1 knockdown led to a reduction in cell viability, whereas HN1 overexpression enhanced cell viability in both HepG2 and SNU449 cells. Furthermore, we conducted a colony formation assay to evaluate cell growth in those cell lines. Following transfection with either HN1 shRNA or HN1 overexpression plasmid for 7 days, we observed a significant decrease or increase, respectively, in the number of colonies, compared with the control group (Fig. 2B). These findings strongly indicate that HN1 knockdown inhibits cell proliferation and cell growth in the HepG2 and SNU449 cell lines.
A WST-1 assay to assess the viability of the HepG2 and SNU449 cell lines after HN1 shRNA knockdown or HN1 overexpression. B A colony staining assay showed the colony formation ability of HepG2 and SNU449 cells with HN1 shRNA knockdown or HN1 overexpression. Colonies were counted at least in five fields. C Apoptosis marker proteins (PARP/cleaved-PARP and caspase-9/cleaved-caspase-9) were detected in pairs by western blots of HepG2 and SNU449 cells after HN1 shRNA knockdown or HN1 overexpression. GAPDH was used as the internal control. Data were expressed as the mean ± SEM of three independent experiments. Protein quantification was calculated by ImageJ software. *p < 0.05, **p < 0.01, and ***p < 0.001 compared with the corresponding control. shCTRL, control shRNA (control). shHN1, HN1 shRNA knockdown. Vector, control vector plasmid (control). HN1 OE HN1 overexpression.
To further probe the effects of HN1 on cell apoptosis, we assessed the levels of several apoptosis-related proteins (PARP, cleaved-PARP, caspase-9, and cleaved-caspase-9) using western blot analyses. As depicted in Fig. 2C, HN1 knockdown increased the expression of cleaved-PARP and cleaved-caspase-9 and decreased the levels of PARP and caspase-9, compared with cells treated with the control shRNA. Conversely, HN1 overexpression suppressed cleaved-PARP and cleaved-caspase-9 and elevated the expression of PARP and caspase-9, compared with cells treated with the vector control plasmid. These results provide compelling evidence that HN1 knockdown significantly induces cell apoptosis in the HepG2 and SNU449 cell lines.
HN1 suppression resulted in G1 phase arrest in HCC cells
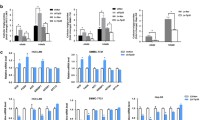
To investigate the role of HN1 in regulating the cell cycle, we used flow cytometry (FACS) for a PI staining analysis. The FACS results clearly demonstrate that HN1 knockdown in the HepG2 and SNU449 cell lines resulted in an accumulation of cells in the G1 phase (Fig. 3A). To corroborate those FACS findings, we conducted western blot analyses to assess the expression of G1 phase-related proteins in HepG2 and SNU449 cells. The outcomes of that analysis indicate that HN1 knockdown led to a reduction in the expression of cyclin D1, CDK4, and CDK6—proteins crucial for G1 phase progression—while also increasing the expression of the p53 protein (Fig. 3B). These results provide compelling evidence that HN1 knockdown facilitates cell cycle arrest in HCC cells, leading to an accumulation of cells in the G1 phase.
A FACs cell cycle analysis of the HepG2 and SNU449 cell lines with HN1 shRNA knockdown. B Western blot analysis of the G1 cycle-related proteins CDK4, CDK6, Cyclin D1, and p53 in the HepG2 and SNU449 cell lines with HN1 shRNA knockdown. GAPDH was used as the internal control. Data represent the mean ± SEM of three independent experiments. *p < 0.05 and **p < 0.01 compared with the corresponding control. shCTRL control shRNA (control), shHN1 HN1 shRNA knockdown.
HN1 suppression attenuated the migration and invasion of HCC cells
To assess the effects of HN1 on the migratory capacity of HCC cells, we conducted a wound-healing assay in the HepG2 and SNU449 cell lines. Our findings reveal a significant, time-dependent reduction in the migration rate following HN1 knockdown in both HepG2 and SNU449 cells. Conversely, HN1 overexpression led to an enhanced, time-dependent migration ability in both cell lines (Fig. 4A). These results strongly indicate that HN1 knockdown impedes the migration capacity of HCC cells. To further elucidate whether HN1 knockdown affects the invasiveness of HCC cells, we performed an invasion assay. As depicted in Fig. 4B, HN1 knockdown resulted in a substantial decrease in the number of invading cells in both the HepG2 and SNU449 cell lines. Conversely, overexpression of HN1 bolstered the invasiveness of both cell lines. Additionally, we examined metastasis-related markers, uPA and vimentin, to probe the underlying molecular mechanism. Our western blot analysis revealed that HN1 knockdown reduced the expression of uPA and vimentin in both HepG2 and SNU449 cells (Fig. 4C). The real-time PCR results further confirmed a significant reduction in the mRNA levels of uPA and vimentin in HepG2 cells (Fig. 4D). In contrast, HN1 overexpression induced the expression of uPA and vimentin at both the protein (Fig. 4C) and mRNA levels (Fig. 4D). Collectively, these findings strongly suggest that HN1 knockdown hinders the metastasis of HCC cells and concurrently suppresses the expression of metastasis-related markers at both the protein and mRNA levels. Taken together, our findings provide substantial evidence that HN1 knockdown curtails the migration and invasion of HCC cells.
A Wound-healing assay to assess the cell migration rate in the HepG2 and SNU449 cell lines after HN1 shRNA knockdown or HN1 overexpression. Representative images were obtained at 0, 24, and 48 h. Migration ability was quantified by measuring the gap distance. B Matrigel transwell assay to assess the cell invasion ability of the HepG2 and SNU449 cell lines after HN1 shRNA knockdown or HN1 overexpression for 72 h. The migration-related molecular uPA and vimentin protein levels (C) and mRNA levels (D) were detected by western blotting and rt-qPCR, respectively, in HepG2 and SNU449 cells after HN1 shRNA knockdown or HN1 overexpression. GAPDH was used as the internal control. Data are expressed as the mean ± SEM of three independent experiments. Protein quantification was calculated by ImageJ software. *p < 0.05, **p < 0.01, and ***p < 0.001 compared with the corresponding control. shCTRL control shRNA (control), shHN1 HN1 shRNA knockdown, Vector control vector plasmid (control), HN1 OE HN1 overexpression.
Gene expression levels were altered upon HN1 knockdown in HCC cells
When considering the effects of HN1 on the proliferation, migration, and invasion of HCC cells, the need to delve into the underlying molecular mechanisms governing HCC progression is pressing. In our effort to comprehend HN1’s regulatory role in HCC, we conducted a microarray analysis. This comprehensive analysis unveiled significant alterations in more than 1000 genes within HCC cells following HN1 knockdown. Several genes, including NQO1, GSTA5, GPR150, SPP1, HN1, UBASH3B, GSTA1, DLK1, HEPACAM, and GSTA5, experienced significant reductions in expression, whereas genes such as PAX6, UGT2B10, SLC16A9, DKK1, UGT2B11, WDR72, UGT2B28, and PRSS8 displayed substantial upregulation upon HN1 knockdown, compared with the control group (Fig. 5A). A gene ontology analysis revealed that the altered genes were associated with various diseases and disorders, molecular functions, cancer-related processes, lipid metabolism, and physiological systems (Fig. 5B).
A cDNA microarray heatmap showing the effects of HN1 shRNA knockdown on AKT signaling pathway-related genes in HCC cells. The data were expressed in matrix format, with rows representing individual genes and columns representing individual samples. Red and green indicate increased and decreased gene expression, respectively. B Gene ontology analysis showed that HN1 knockdown was related to a series of diseases and disorders. shCTRL control shRNA (control), shHN1 HN1 shRNA knockdown.
Inhibition of Akt led to the downregulation of HN1 expression in HCC cells
Within the gene analysis results derived from HCC cells following HN1 knockdown, we identified that SPP1, SCD, and NQO1 were downstream target genes of Akt, as revealed by the upstream gene networks analyzed through an ingenuity pathway analysis (Fig. 6A). Furthermore, the expression of those genes was effectively suppressed by the Akt inhibitor LY294002 (Fig. 6A). Those findings suggest a potential link between HN1’s functions in HCC cells and Akt activity. To substantiate that hypothesis, we conducted further experiments using the Akt inhibitor LY294002 and the Akt activator SC79 (Fig. 6B). Western blot results unequivocally indicate that LY294002 led to a decrease in HN1 expression, and SC79-induced HN1 expression in the HepG2 and SNU449 cell lines (Fig. 6B). In addition, reducing HN1 expression led to a decrease in p-Akt levels, whereas HN1 overexpression resulted in increased p-Akt levels in both the HepG2 and SNU449 cell lines (Fig. 6C). These results provide robust evidence supporting the notion that Akt can function as an upstream regulator of HN1 expression in HCC.
A Upstream gene networks from the ingenuity pathway analysis. The genes reduced by HN1 knockdown were downstream target genes of Akt, and the Akt inhibitor LY294002 also inhibited the expression of those genes. B The HN1 protein level was detected by western blotting after treatment with LY294002 (10 μM) and SC79 (10 μM) for 24 h. C HN1, Akt, and p-Akt levels were analyzed by western blotting in HepG2 and SNU449 cells following HN1 shRNA knockdown or overexpression. D Western blot analysis of HN1, TSC2, PRAS40, and PRAS40-S183 protein levels was conducted in HepG2 and SNU449 cells after transfection with HN1 siRNA. E The protein expression levels of HN1, Akt, and p-Akt in HepG2 and SNU449 cells were examined after transfection with HN1 siRNA, followed by treatment with SC79 (10 μM) as indicated. GAPDH was used as the internal control. Data were expressed as the mean ± SEM of three independent experiments. Protein quantification was calculated by ImageJ software. *p < 0.05, **p < 0.01, and ***p < 0.001 compared with the corresponding control. Ns not significant. Vehicle, DMSO-treated control group.
To further address the importance of HN1 for Akt’s oncogenic activity and its role in Akt activation, we examined how essential HN1 is for Akt activation by assessing the phosphorylation status of Akt downstream targets such as PRAS40 and TSC2 following HN1 knockdown (Fig. 6D, E). Our findings revealed that HN1 knockdown led to a significant decrease in the phosphorylation of these downstream targets, indicating that HN1 is vital for Akt activation and its signaling cascade (Fig. 6D). Additionally, to test the essential role of HN1 in Akt activation, we conducted experiments using the Akt activator SC79 in both control and siHN1-transfected HCC cells. As shown in Fig. 6E, HN1 knockdown significantly decreased the level of p-Akt. While SC79 treatment significantly increased p-Akt levels in control cells, the reduced p-Akt levels in HN1 knockdown cells were partially restored with SC79 treatment. This suggests that HN1 plays an important regulatory role in Akt activation, as the activation of Akt is limited in the absence of HN1. Therefore, these results emphasize that HN1 is essential for optimal Akt activation.
HN1 suppression in HCC cells diminished the expression of SREBP-1 and hindered the nuclear translocation of SREBP-1
To gain a more profound understanding of the downstream target genes influenced by HN1, an in-depth analysis of the downstream gene networks (Ingenuity Pathway Analysis) was conducted. Those comprehensive results reveal that HN1 also plays a regulatory role over SREBP1 and SREBP2 (Fig. 7A), renowned downstream target genes of Akt [30, 31] that encode the SREBP1 and SREBP2 proteins, which are well-recognized for their involvement in lipid synthesis regulation [32]. We further assessed the protein levels of SREBP1 through a western blot analysis. As a pivotal component in lipogenesis, precursor SREBP-1c initially resides in the endoplasmic reticulum [33, 34]. Upon cleavage, mature SREBP-1c becomes activated and translocated into the nucleus [35]. Our western blot results unequivocally demonstrate that alterations in HN1 levels led to changes in both precursor SREBP-1c and mature SREBP-1c in HCC cells, signifying that HN1 not only governs SREBP-1 expression but also influences SREBP-1c activation (Fig. 7B). Additionally, our immunofluorescence findings directly show that HN1 knockdown inhibited the nuclear translocation of SREBP-1c, whereas HN1 overexpression induced it (Fig. 7C). These compelling findings substantiate that HN1 knockdown inhibits both the expression and nuclear translocation of SREBP-1. To further validate those observations, we conducted real-time qPCR, and those results show that HN1 knockdown resulted in a significant reduction in the mRNA levels of SREBP1 and SREBP2, whereas their levels were elevated following HN1 overexpression (Fig. 7D). These results conclusively establish that HN1 knockdown diminishes the expression of SREBP1 and SREBP2 and impedes the nuclear translocation of SREBP-1 in HCC cells.
A Downstream gene networks from the ingenuity pathway analysis. HN1 knockdown inhibited the SREBF-1 and SREBF-2 genes and their downstream genes. B Precursor and mature SREBP-1c protein expression was examined by western blotting in HepG2 and SNU449 cells after HN1 shRNA knockdown or HN1 overexpression. GAPDH was used as the internal control. C SREBP-1 cell distribution was observed directly by immunofluorescence against the SREBP-1 antibody. Green, SREBP-1. Blue, DAPI (nucleus). The distribution of fluorescence density in the nucleus and protein quantification were measured by ImageJ software. D The mRNA levels of precursor and mature SREBP-1 were measured by rt-qPCR after HN1 shRNA knockdown in the HepG2 and SNU449 cell lines. Data were expressed as the mean ± SEM of three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001 compared with the corresponding control. shCTRL control shRNA (control). shHN1 HN1 shRNA knockdown, Vector control vector plasmid (control), HN1 OE HN1 overexpression.
HN1 suppression in HCC cells hindered the process of lipogenesis
Given SREBP-1’s established role in governing glucose metabolism [36], fatty acid synthesis, and lipid production within cells [37], the interaction between HN1 and SREBP-1 impelled us to explore HN1’s effects on lipogenesis in HCC cells. To evaluate the effects of varying HN1 expression levels on lipogenesis under diverse conditions (normal, low glucose, high glucose, and high insulin medium), we conducted an oil red O staining assay. Two analytical methods were used to analyze the stained images: one measured the percentage of the positively stained area, and the other assessed the absorbance of the solution after the stained area had been dissolved with isopropanol. As illustrated in Fig. 8A, compared with the control group, the HN1 knockdown group exhibited a reduction in both the percentage of positively stained lipid droplets and the absorbance of the dissolving solution. Conversely, HN1 overexpression expanded the area of positively stained lipid droplets and increased the absorbance of the dissolving solution in both the HepG2 and SNU449 cell lines. These findings compellingly indicate that HN1 knockdown significantly impedes intracellular lipid accumulation in HCC cells. Furthermore, we quantified the levels of cholesterol and triglycerides in both the HN1 knockdown and HN1 overexpression groups. HN1 knockdown markedly lowered the levels of cholesterol and triglycerides in both HepG2 and SNU449 cells (Fig. 8B, C). Conversely, HN1 overexpression significantly elevated the levels of cholesterol and triglycerides in those cell lines (Fig. 8B, C). Additionally, we assessed the expression of FAS and ACC, lipogenesis-related proteins downstream of SREBP-1, through western blot analyses. HN1 knockdown suppressed the protein expression of FAS and ACC, and HN1 overexpression enhanced their expression in the HepG2 and SNU449 cell lines (Fig. 8D). These results further validate that HN1 knockdown curtails lipogenesis in HCC cells.
A Lipid droplet formation was detected by the oil red O staining assay after HN1 siRNA knockdown or HN1 overexpression in HepG2 and SNU449 cells. The percentage of positive stained area was analyzed by ImageJ software. After dissolving the stained area with isopropanol, the absorbance level of the stained area was detected at 510 nm. B Cholesterol levels were detected by a cholesterol fluorometric assay after HN1 siRNA knockdown or HN1 overexpression in HepG2 and SNU449 cells. C Triglyceride levels were detected by a triglyceride fluorometric assay after HN1 siRNA knockdown or HN1 overexpression in HepG2 and SNU449 cells. D The expression of the lipogenesis-related SREBP-1 downstream proteins FAS and ACC were detected by western blotting after HN1 siRNA knockdown or HN1 overexpression. GAPDH was used as the internal control. Protein quantification was calculated by ImageJ software. Data were expressed as the mean ± SEM of three independent experiments. *p < 0.05, **p < 0.01, and ***p < 0.001 compared with the corresponding control. siCTRL control siRNA (control), siHN1 HN1 siRNA knockdown, shCTRL control shRNA (control), shHN1 HN1 shRNA knockdown, Vector control vector plasmid (control), HN1 OE HN1 overexpression.
SREBP-1 played a pivotal role in mediating HN1-induced regulation of cell proliferation and lipogenesis in HCC cells
To further elucidate the role of SREBP-1 in HN1-regulated cell proliferation and lipogenesis, we conducted transfections involving SREBP-1 siRNA, HN1 overexpression plasmid, and both the SREBP-1 siRNA and HN1 overexpression plasmid together in HepG2 and SNU449 cells. As demonstrated in Fig. 9A, B, SREBP-1 knockdown significantly attenuated the cell viability and colony formation induced by HN1 overexpression in the HepG2 and SNU449 cell lines. In addition, we have investigated the effects of HN1-dependent SREBP expression on apoptosis, cell cycle, and migration in HCC cells. As demonstrated in Fig. 9C, silencing SREBP increased cell apoptosis, while overexpression of HN1 inhibited apoptosis in SREBP-silenced HCC cells. The combination treatment reversed the G2 cell cycle arrest induced by overexpressing HN1 or silencing SREBP (Fig. 9D). Wound-healing assays showed that overexpression of HN1 increased the migration rate, whereas silencing SREBP inhibited the migration rate of HCC cells (Fig. 9E). The inhibited migration rates due to SREBP silencing were recovered by overexpression of HN1 in HCC cells. These data suggest that HN1 regulates SREBP expression and its associated effects on apoptosis and migration in HCC cells. Furthermore, we investigated the function of SREBP-1 in HN1-mediated lipogenesis. Even though HN1 overexpression led to an increase in FAS and ACC protein levels, SREBP-1 knockdown abrogated that induction (Fig. 9F). Additionally, HN1 overexpression heightened cholesterol and triglyceride levels (Fig. 9G, H), and those increases were subsequently reversed by SREBP-1 knockdown in the HepG2 and SNU449 cell lines. These results provide compelling evidence supporting the notion that SREBP-1 is a key downstream target of HN1 in regulating cell proliferation and lipogenesis in HCC cells.
HepG2 and SNU449 cells were transfected with an HN1 overexpression plasmid, SREBP-1 siRNA, or both the HN1 overexpression plasmid and SREBP-1 siRNA for 48 h. A The WST-1 assay was performed to assess the viability of the HepG2 and SNU449 cell lines after transfection. B A colony staining assay was performed to assess the colony formation ability of HepG2 and SNU449 cells after transfection. Colonies were counted in at least in five fields. C Flow cytometry was used to determine cell apoptosis in HepG2 and SNU449 cells. D Flow cytometry was performed to assess the cell cycle distributions and the relationship between HN1 and SREBP. E Wound healing assays were conducted to evaluate the migration rate of HepG2 and SNU449 cells. F The protein levels of HN1, precursor, and mature SREBP-1C, FAS, and ACC were detected by western blotting. GAPDH was used as the internal control. Protein quantification was calculated by ImageJ software. G Cholesterol levels in HepG2 and SNU449 cells were detected by a cholesterol fluorometric assay after transfection. H Triglyceride levels in HepG2 and SNU449 cells were detected by a triglyceride fluorometric assay after transfection. *, compared with the vector plasmid plus control siRNA; #, compared with HN1 overexpression plus SREBP-1 siRNA knockdown. * or # p < 0.05; ** or ## p < 0.01. N.s not significant, siCTRL control siRNA, siSREBP-1 SREBP-1 siRNA knockdown, Vector control vector plasmid, HN1 OE HN1 overexpression.
Depleting HN1 in xenograft mouse models markedly restrained the growth of human HCC tumors and triggered apoptosis
Building upon those in vitro findings, we evaluated the functional effects of HN1 in vivo using xenograft nude mouse models. The HepG2 negative control and HN1 shRNA cell lines were transplanted into mice, and the results, depicted in Fig. 10A, B, reveal that the tumor size in the HN1 shRNA group was noticeably smaller than in the control group. Additionally, both the tumor weight (Fig. 10D) and volume (Fig. 10A) in the HN1 shRNA group were significantly reduced compared with the control group, though no significant difference in body weight was observed (Fig. 10C). To further examine the state of tumor growth, staining assays were performed on the mouse tumor tissues. Remarkably, mice that received control cells exhibited a greater number of dark-blue-stained nuclei, indicating vigorous tumor growth (Fig. 10E). Conversely, mice that received HN1 shRNA cells displayed more apoptotic cells and DNA fragmentation, suggesting that HN1 knockdown increased cell apoptosis in the mouse tumor tissues (Fig. 10E). These results strongly support the notion that HN1 knockdown significantly inhibited the growth of HCC tumors and induced apoptosis in the xenograft mouse models. To investigate whether HN1 regulates lipogenesis-related proteins such as SREBP-1, FAS, and ACC, we performed immunostaining on the mouse tumor tissue sections using anti-SREBP-1 and HN1 antibodies. The HN1 knockdown group exhibited reduced brown-positive staining for both SREBP-1 and HN1, compared with the control group (Fig. 10G). Consistent with those findings, western blot results showed decreased expression of both the precursor and mature forms of SREBP-1, FAS, and ACC proteins in the HN1 knockdown group. These results indicate that HN1 knockdown suppressed the expression of SREBP-1 and its downstream targets FAS and ACC in the xenograft mouse models. We have further conducted additional in vivo experiments to examine the effect of HN1 overexpression and HN1 overexpression combined with SREBP knockdown. In this experiment, we used Fatostatin, a SREBP inhibitor, as a replacement for SREBP knockdown. As shown in Fig. 11, the increased tumor size in HN1-overexpressed mice was significantly reduced in HN1-overexpressed mice treated with Fatostain (Fig. 11A, B, D). These results indicate that HN1 and SREBP-1 are interconnected in the progression of HCC, and that HN1 cannot function properly without SREBP-1. This highlights that SREBP-1 is an important mediator in the progression of HCC.
The xenograft nude mouse models were established with HepG2 cells transfected with control shRNA or HN1 shRNA. The mice were separated into two groups: (i) control shRNA group and (ii) HN1 shRNA group. A, B Images of mice and tumors. Body weight (C) and tumor volume (E) were measured every 3 days. Tumor weight (D) and images of tumor size (B) were evaluated after euthanasia. F A histopathological analysis of H&E stained tissues and the TUNEL assay were performed to determine the histological characteristics of the mouse tumor tissues. G Tumor tissues were immunostained with HN1 and SREBP-1 antibodies. H Western blot analysis of HN1, precursor and mature SREBP-1C, FAS, and ACC in mouse tumor tissues. GAPDH was used as the cytoplasm internal control. Protein quantification was calculated by ImageJ software. *p < 0.05, **p < 0.01, and ***p < 0.001 compared with the control. shCTRL control shRNA (control), shHN1 HN1 shRNA knockdown.
Xenograft nude mouse models were established using HepG2 cells transfected with either a control or HN1 overexpression vector. The mice were divided into three groups: (i) control group, (ii) HN1 overexpression group, and (iii) HN1 overexpression combined with Fatostatin. A Tumor volume was measured every 3 days. After euthanasia, images of the tumors (B), body weight (C), and tumor weight (D) were evaluated. *p < 0.05, **p < 0.01, and ***p < 0.001: vector vs. HN1 OE. ## p < 0.01 and ###p < 0.001: HN1 OE vs. HN1 OE plus Fatostatin. “Vector” refers to the control vector plasmid, “HN1 OE” to HN1 overexpression, and “Fatostatin” to the SREBP inhibitor.
Discussion
In this study, we revealed heightened HN1 expression in HCC tissues, demonstrating its oncogenic role through the promotion of cell proliferation, migration, and invasion. Silencing HN1 inhibited those tumorigenic processes, strongly suggesting that HN1 is a key driver of HCC development. Additionally, HN1 activated the Akt pathway, driving tumorigenesis through SREBP1-mediated lipogenesis. Silencing HN1 effectively restrained HCC tumor growth in mouse xenograft models by deactivating SREBP-1, highlighting the potential of HN1 as a therapeutic target that affects both external and internal factors.
In recent decades, HCC’s mortality rate has outpaced that of other cancer types [15]. Surgical procedures and liver transplantation are the primary curative approaches for HCC patients [38], but the recurrence and clinical heterogeneity of HCC pose challenges to effective treatment because tumor biology can vary based on each patient’s physical condition [39,40,41]. Therefore, understanding the complexity of tumorigenesis is paramount. Accurate cancer prediction and characterization of its biological attributes are critical steps toward targeted treatment, the alleviation of patient suffering, and the extension of patient lifespans.
HN1 is a gene that is overexpressed in various carcinomas, compared with benign tumors [29]. It plays a critical and causative role in the aggressive malignant phenotype of head and neck cancer [21], and it acts as an interaction marker for the inactive GSK3/β-catenin /APC complex and promotes the ubiquitin-dependent proteasomal degradation of β-catenin, contributing to metastasis in prostate cancer [42, 43]. In addition, HN1 closely correlates with tumor biology and prognosis in HCC [28], serving as a significant risk predictor for a clinical diagnosis [28]. Our findings confirm HN1’s overexpression in HCC tumor tissues, compared with adjacent non-tumoral tissues, and its presence in seven HCC cell lines. Knocking down HN1 suppressed HCC cell proliferation, migration, and invasion and induced apoptosis, whereas HN1 overexpression in HCC cells promoted tumorigenesis, signifying its pivotal role in HCC pathogenesis.
Increased understanding of the intricate cross-talk among multiple signaling transduction pathways, as well as cooperation among various target factors and the ongoing emergence of genetic diversity, continually expands the horizons of research and treatment during the complex process of tumorigenesis [44, 45]. Recent investigations using gene expression profiling technology have produced remarkable enhancements in risk assessment models for HCC [46,47,48]. Our microarray analysis unveiled the profound influence of HN1 knockdown on 406 genes that were also suppressed by the Akt signaling pathway. In addition, our gene ontology analysis revealed these genes’ close ties to cancer, lipid metabolism, cell survival, and apoptosis. Moreover, our analysis identified HN1 as an intermediate regulator within the Akt-SREBP signaling pathway. Previous studies have established the prominent role of the Akt signaling pathway in various cancers, often stemming from genetic mutations [49]. Akt, a serine/threonine kinase, has a central and pivotal role in governing human biological processes and various diseases [50]. In collaboration with PI3K, Akt contributes significantly to the regulation of cellular homeostasis [51]. The PI3K-Akt signaling pathway is known for its involvement in cell proliferation and viability [52]. Akt activation is prompted by extracellular stimuli that initiate a cascade of downstream events encompassing cell growth, metastasis, and angiogenesis [53]. Cumulatively, those findings substantiate the crucial role of Akt in cellular development. During our current investigation, we confirmed that an Akt inhibitor downregulated HN1 expression, and an Akt activator induced its expression, suggesting that HN1 expression in HCC can be regulated by the upstream Akt gene.
HN1 has been implicated in modulating Akt activity [54]. Research has shown that Akt is a crucial node in cancer signaling pathways, and its activity is essential for many oncogenic processes [55]. Specifically, studies have demonstrated that inhibiting Akt phosphorylation and downstream signaling components like PRAS40 and TSC2 can significantly impair cancer cell growth and survival, highlighting the importance of Akt signaling in oncogenesis [56]. Our findings indicate that HN1 knockdown significantly reduces the phosphorylation of downstream targets, underscoring HN1’s critical role in Akt activation and its subsequent signaling cascade. Although HN1 silencing markedly decreased p-Akt levels, treatment with SC79 notably restored p-Akt levels in HN1 knockdown cells. This restoration suggests that HN1 is a key regulator of Akt activation, thus, our results highlight the necessity of HN1 for optimal Akt activation within this signaling pathway. To delve deeper into the molecular mechanism of HN1 during tumorigenesis, we conducted an Ingenuity Pathway Analysis using gene expression profiling of HCC cells. That analysis revealed that HN1 regulates SREBP1 and SREBP2, along with their downstream genes. The SREBPs belong to the basic-helix-loop-helix leucine zipper category of transcription factors and reside initially in an inactive form within nuclear membranes and endoplasmic reticulum envelopes [57]. Upon activation, SREBPs bind to specific sterol regulatory element gene sequences, thereby governing the expression of genes and enzymes crucial for lipogenesis [58]. SREBP1, composed of two isoforms, SREBP-1a and SREBP-1c, is primarily responsible for fatty acid synthesis and cholesterol uptake, whereas SREBP2 plays a pivotal role in cholesterol metabolism [59]. Recent studies have indicated that the PI3K/Akt signaling pathway stimulates the expression of SREBP1 and SREBP2, which are encoded by the SREBF-1 and SREBF-2 genes, respectively [60,61,62]. The regulatory roles of SREBP1 and SREBP2 in lipogenesis and cholesterol metabolism contribute significantly to cancer proliferation, growth, and the different stages of cancer progression [63,64,65]. Reprogrammed energy metabolism is a new hallmark of cancer that supports the energy demands of tumor malignancy throughout disease progression [66,67,68]. Lipids play a vital role in forming cellular membrane structures, act as signaling molecules, and provide energy resources for cell proliferation and growth [69, 70]. Enhanced lipid accumulation, excess energy storage, and heightened lipogenesis are evident in many cancers and promote rapid tumor growth [71, 72]. Those findings underscore the potential effects of targeting lipid synthesis pathways in cancer treatment. Given the association between HN1 and SREBP1, we investigated HN1’s role in lipid production. Our findings indicate that HN1 suppression led to a reduction in lipid-stained droplets, demonstrating lipogenesis inhibition. Conversely, HN1 overexpression stimulated lipogenesis. Further exploration of HN1’s downstream target genes revealed its interaction with SREBP1 and SREBP2, pivotal regulators of lipogenesis and cholesterol metabolism. Silencing HN1 significantly decreased the protein and mRNA levels of both SREBP1 and SREBP2, which was accompanied by reduced protein expression of FAS and ACC. These collective results suggest that HN1 might promote tumor proliferation and growth in HCC by influencing lipid metabolism via SREBP-1.
Targeting cancer energy metabolism, including the regulation of glucose and lipids, has emerged as a promising therapeutic approach [73, 74]. The “Warburg effect,” also known as aerobic glycolysis, which was discovered in the last century, provides significant evidence of reprogrammed energy metabolism in cancer cells [75,76,77]. In this study, we found that SREBP-1 knockdown significantly reduced the cell growth induced by HN1 overexpression in the HepG2 and SNU449 cell lines, supporting the notion that HN1 cooperates with SREBP1 to drive HCC tumorigenesis. Because SREBP1 plays crucial roles as a regulator of cholesterol and triglyceride formation [63], we transfected human cells (with or without HN1 overexpression) with SREBP1 siRNA and measured the levels of cholesterol and triglycerides. Those measurements confirmed that SREBP-1 knockdown led to a reduction in the elevated cholesterol and triglyceride levels we had previously confirmed with HN1 overexpression in the HepG2 and SNU449 cell lines. The regulatory network between HN1 and SREBP1/SREBP2 in HCC tumorigenesis presents a novel focal point for further investigation, as demonstrated in our research. Consistent with our in vitro results, our in vivo results demonstrate that mice that received a transplant of cells transfected with HN1 shRNA exhibited significantly lower tumor weights and volumes than those that received a transplant of control cells. Furthermore, immunohistochemistry and western blot assays revealed decreased SREBP1 protein expression levels in xenograft tissue from the HN1 shRNA–treated animals. Those findings confirm that inhibiting HN1 suppressed HCC tumorigenesis in vivo by inhibiting SREBP1-mediated lipogenesis. In contrast, overexpression of HN1 led to increases in both tumor weight and volume. Treatment with the SREBP inhibitor fatostatin significantly mitigated the HN1-induced augmentation of tumor growth. These findings highlight a critical interaction between HN1 and SREBP-1 in the progression of hepatocellular carcinoma (HCC), suggesting that the oncogenic effects of HN1 are contingent upon SREBP-1 activity. This underscores the integral role of SREBP-1 as a mediator with HN1 in the pathogenesis of HCC.
In summary, HN1, potentially regulated by the Akt signaling pathway, led to the activation of SREBP1, thereby inducing lipogenesis, including triglyceride and cholesterol synthesis. That excessive lipid formation provided ample energy for cell proliferation and tumor growth in HCC. Significantly, HN1 knockdown elicited substantial tumor regression in xenograft mouse models by suppressing SREBP1-mediated lipogenesis. Collectively, our results highlight the therapeutic potential of HN1 inhibition in a preclinical model of HCC. Therefore, HN1 might promote the proliferation, migration, and invasion of HCC cells, partially through activation of the Akt-HN1-SREBP signaling pathway (Fig. 12). Given the genetic diversity of tumorigenesis [78] and the complex biological characteristics of cancer development, challenges persist in treatment and research approaches. Although further investigations are required to fully elucidate the intricate mechanisms of this gene, our current findings suggest that targeting HN1 could be a promising therapeutic strategy for HCC.
Materials and methods
Cell culture and patient tissue
The HCC cell lines used in this study, Hep3B, HLE, Huh7, SHJ-1, and HepG2, were obtained from ATCC, while SNU368 and SNU449 were sourced from Korean Cell Line Bank. The cells were cultured as monolayers in DMEM (Gibco by Life Technologies, Thermo Fisher Scientific Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, Welgene Gold Serum, Gyeongsan-si, Korea) and 1% penicillin (Sigma, St. Louis, MO, USA) in 100-nm dishes under standard conditions at 37 °C and 5% CO2 humidity Liver tissue samples from patients were sourced from the fresh-frozen tissue bank at the University of Texas MD Anderson Cancer Center (MDACC), and this study received approval from the Institutional Review Board at MDACC (LAB09-0687) in Houston, TX.
Plasmids and reagents
Control siRNA-A (SC-37007), HN1 siRNA (sc-93940), SREBP-1 siRNA (sc-36557), control shRNA plasmid-A (sc-108060), HN1 shRNA plasmid (sc-93940-SH), control shRNA lentiviral particles (sc-108080), and HN1 shRNA lentiviral particles (sc-93904-V) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Human HN1 c-flag tag (HG14335-CF) and pCMV3-C-FLAG vector were purchased from Sino Biological Inc. (Beijing, China). Primary antibodies against caspase-9 (#9502), c-caspase-9 (#9501), PARP (#9542), c-PARP (#9541), CDK4 (#2906), CDK6 (#3136 s), cyclin D1 (#2922), vimentin (#5741), Akt (#9272), p-Akt (#2775), ACC (#8578 s), and GAPDH (#2118) and secondary anti-rabbit (#7074) and anti-mouse (#7076) antibodies were all purchased from Cell Signaling Technology (Beverly, MA, USA). HN1 (14914-1-AP) antibody was bought from Proteintech Group Inc. (Rosemont, IL, USA). SREBP-1 (sc-13551), uPA (sc-14019), and GAPDH (sc-47724) antibodies were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA). HN1 (ab126705), SREBP-1 (ab235177), and FAS (ab96863) antibodies were purchased from Abcam (Cambridge, UK). LY294002 was purchased from Sigma-Aldrich (St. Louis, MO, USA) and diluted in dimethyl sulfoxide (DMSO; Sigma, Saint Louis, MO, USA). SC79 was purchased from Tocris Bioscience (Bristol, United Kingdom) and diluted in DMSO (Sigma, Saint Louis, MO, USA).
siRNA transduction
HepG2 and SNU449 cells were seeded in 6-well plates. After overnight incubation, transfection was conducted with a 100 pmol concentration of siRNA in Opti-MEM (Gibco by Life Technologies, Thermo Fisher Scientific Inc., Waltham, MA, USA) and lipofectamine 3000 (Invitrogen, Thermo Fisher Scientific Inc., Waltham, MA, USA). The cells were incubated with transfection mixture for 6 h that was then changed to complete medium for 48 h, according to the manufacturer’s protocol. Those cells were then used for further experiments.
Plasmid transduction
After cells were seeded overnight, they were rinsed with Opti-MEM. The transfection reagents were prepared as follows: Solution A: plasmid, p-3000, and Opti-MEM, and Solution B: lipo3000 and Opti-MEM. The mixture of Solution A and Solution B was incubated for 30 min at room temperature and then added to the cells. The cells were incubated for an additional 6 h. After incubation, DMEM containing 10% FBS and 1% antibiotics was added to the cells, and they were incubated for an additional 48 h. For long-term plasmid transfection, the transfected cells were cultured and selected in DMEM supplemented with 10% FBS and 3.5 µg/mL puromycin for one week prior to use in subsequent experiments. Transfection efficacy was confirmed by western blot analyses.
shRNA lentiviral particle transduction
HepG2 and SNU449 cells were seeded in 12-well plates and incubated for 24 h until the cell confluence was around 50%. After incubation, the complete medium was replaced with polybrene (sc-134220, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA). The lentiviral particles were thawed and added to the cultured cells. The volume of the particles was determined based on the cell density (8 μl of particles were used for 0.8 × 105 cells; 10 μl of particles were used for 1.0 × 105 cells). After overnight incubation, the complete medium was changed, and the cells were incubated again overnight. On the following day, a complete medium with a specific concentration of puromycin was added and maintained for at least 2 weeks to select the stably transfected cell line. Western blotting was performed to detect the efficacy of transfection.
WST-1 assay
HepG2 and SNU449 cells were seeded in 96-well plates. After transfection, the cells were incubated for 48 to 72 h. Cell viability was determined using an EZ-CYTOX kit (DoGen Bio, Seoul, South Korea) according to the manufacturer’s instructions. The absorbance was measured at 450 nm using a VICTOR3 multilabel reader (Perkin Elmer, MA, USA). Three independent experiments were performed with each cell line.
Colony staining assay
HepG2 and SNU449 cells (104/well) were seeded in a 6- or 12-well plate overnight. After transfection, the cells were incubated for 1 week. The cells were stained with 0.5% crystal violet (Sigma-Aldrich, St. Louis, MO, USA) and 25% methanol (SAMCHUN CHEMICALS Co., Ltd, Seoul, South Korea) in DPBS for 1 h. The stained cells were imaged, and colonies were counted using ImageJ software.
Cell cycle analysis
HepG2 and SNU449 cells were seeded in six-well plates and transfected for 48 h. The cells were then harvested using EDTA-trypsin (Welgene Gold Serum, Gyeongsan-si, Korea) and re-suspended in DPBS (Welgene Gold Serum, Gyeongsan-si, Korea). The cells were centrifuged at 13,200 rpm for 5 min and fixed in 75% ethanol in DPBS at −20 °C for 2 h. After fixation, the cell pellets were rinsed twice with DPBS and re-suspended in 500 µL of DPBS. RNase was added, and the cells were incubated for 15 min at 37 °C. Propidium Iodide (PI) stain was added, and the cells were incubated for 30 min at room temperature in the dark. The cell cycle was then analyzed using a FACStar flow cytometer (Becton-Dickinson, San Jose, CA, USA).
Wound healing assay
HepG2 and SNU449 cells were seeded in a six-well plate. After transfection, a wound was created in the monolayer using a 200 µl pipette tip. Images were taken after 0, 24, and 48 h, and the wound area was measured using AxioVision Rel 4.8 software (Carl Zeiss, Berlin, Germany).
Matrigel invasion assay
In vitro cell invasion was tested using BD BioCoatTM MatrigelTM invasion chambers (BD Biosciences, San Jose, CA, USA). The chambers were rehydrated in a humidified tissue culture incubator at 37 °C and 5% CO2. Cells were seeded in each Matrigel-coated transwell insert in 500 µl of medium. The lower chamber of the transwell was filled with 500 µl of medium. After incubation, the inserts were washed and stained with a Diff-Quik kit (Sysmex Corp., Kobe, Japan). The stained inserts were sequentially transferred through solutions of increasing concentrations of ethanol to visualize the invading cells and observed and photographed under a light microscope. The number of cells was counted in five fields, and the invasion rates were calculated as the percentage of cells that had invaded through the Matrigel transwell.
Western blotting analysis
Cells were harvested and suspended in RIPA buffer (Thermo Fisher Scientific Inc., Waltham, MA, USA) containing a phosphatase and protease inhibitor cocktail (Pierce, Rockford, IL, USA). The extracts were incubated on ice for 10 min and then centrifuged at 13,200 rpm for 20 min at 4 °C. The supernatant was collected, and the protein concentration was determined using a BCA protein assay kit (Pierce Biotechnology, Inc., Rockford, IL, USA). The whole lysate was loaded onto an SDS-PAGE gel and transferred to PVDF membranes (Bio-Rad, Hercules, CA, USA). The membranes were probed with specific primary antibodies and peroxidase-conjugated secondary antibodies. The bands were visualized using an enhanced chemiluminescence kit (Amersham, Arlington Heights, IL, USA).
RNA extraction and cDNA synthesis
RNA was extracted from cells using TRIzol reagent (Ambion by Life Technologies, Thermo Fisher Scientific Inc., Waltham, MA, USA) according to the manufacturer’s protocol. Briefly, the cells were harvested and incubated with TRIzol at room temperature for 10 min. Chloroform (Sigma, St. Louis, MO, USA) was added, and the mixture was incubated at room temperature for 3 min. The mixture was then centrifuged at 13,200 × g for 10 min at 4 °C. The supernatant was transferred to a new tube, and isopropanol (Sigma, St. Louis, MO, USA) was added at a 1:1 ratio. The mixture was incubated at room temperature for 10 min and then centrifuged at 13,200 × g for 20 min at 4 °C. The supernatant was removed, and the RNA pellet was washed twice with 75% ethanol. The RNA pellet was then dissolved in RNase-free water (Welgene Gold Serum, Gyeongsan-si, Korea). After RNA quantification, 1 µg of total RNA from each sample was subjected to reverse transcription using a PrimeScript RT reagent kit (Takara Bio Inc., Otsu, Shiga, Japan) according to the manufacturer’s protocol to synthesize stable cDNA for further use.
Real-time qPCR
A real-time quantitative PCR reaction mixture was added to a PCR plate and analyzed using an Applied Biosystems 7900HT fast real-time PCR system (Applied Biosystems, Foster City, CA, USA) with TaKaRa SYBR Premix EX TaqTM (Takara Bio Inc., Otsu, Shiga, Japan). Data were analyzed according to the ΔΔCt method and normalized to GAPDH expression in each sample. The primer sequences used were as follows: uPA sense: 5′-AGAATTCACCACCATCGAGA-3′ and 5′-ATCAGCTTCACAACAGTCAT-3′, vimentin sense: 5′-CCCTCACCTGTGAAGTGGAT-3′ and 5′-TCCAGCAGCTTCCTGTAGGT-3′, SREBP-1 sense: 5′-CCTGGTGGTGGGCACTGA-3′ and 5′-GTGCTGTAAGAAGCGGATGTAGTC-3′, SREBP-2 sense: 5′-GCAACCAGCTTTCAAGTCCT-3′ and 5′-TACCGTCTGCACCTGCTG-3′, HN1 sense: 5′-TGACTGTCCTGAACGTCT-3′ and 5′-AGATCCTCTAACACATCTGG-3′, GAPDH sense: 5′-GTCTCCTC TGACTTCAACAGCG-3′ and 5′-ACCACCCTGTT GCTGTAGCCAA-3′.
Microarray analysis
A microarray analysis was performed using a mirVana miRNA isolation labeling kit (Ambion Inc., TX, USA) according to the manufacturer’s protocol. Biotin-labeled cRNA was prepared using an Illumina Total Prep RNA amplification kit (Ambion Inc) for hybridization. Gene expression data were extracted from Genome Studio (Illumina, San Diego, CA). Data were normalized using the quantile normalization method in the Linear Models for Microarray data package in the R program. A heatmap of gene expression was generated using the Cluster and Tree view programs. Microarray data can be accessed in NCBI’s Gene Expression Omnibus public database (GSE271950).
Immunofluorescence staining
Immunofluorescent staining was performed on HepG2 and SNU449 cells seeded on cover glasses (Paul Marienfeld GmbH & Co. KG, Lauda-Königshofen, Germany) in a 6-well plate overnight. After transfection, the cover glasses were removed to a new plate and washed twice with DPBS (Gibco, Grand Island, NY, USA). The cells were fixed with 4% methanol in DPBS for 40 min at room temperature. Permeabilization was performed with 0.1% triton X-100 (Sigma-Aldrich, St. Louis, MO, USA) in DBPS for 30 min. The cells were blocked with 5% bovine serum albumin (BSA, Gendepot, Katy, TX, USA) in 0.1% Triton X-100 in DBPS for 30 min. The primary antibody, SREBP-1 (sc-13551, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), was diluted 1:100 in 5% BSA in 0.1% Triton X-100 in DBPS and added to the cover glasses, which were were rinsed and incubated with a mouse secondary antibody (1:100 diluted in 5% BSA in 0.1% Triton X-100 in DBPS) at room temperature for 1 h in the dark. The cells were then stained with DAPI (1:500 diluted in 5% BSA in 0.1% Triton X-100 in DBPS) for 5 min at room temperature in the dark. The cover glasses were then mounted on microscope slides (Paul Marienfeld GmbH & Co. KG, Lauda-Königshofen, Germany) with mounting solution (Vector Laboratories, Inc., Burlingame, CA, USA) and sealed with Canada balsam (Junsei Chemical Co., Chuo-ku, Tokyo, Japan). Images were taken under a fluorescence microscope (Zeiss Model Axio Imager M1, Jena, Germany). Fluorescence values were detected and merged using ImageJ (NIH, Bethesda, MD, USA).
Oil red O staining
Oil red O staining was performed on cells cultured in a six-well plate. After transfection, the cells were washed twice with DPBS (Gibco, Grand Island, NY, USA) and fixed with 10% formalin at room temperature for 2 h. The cells were then rinsed with 60% isopropanol (Sigma-Aldrich, St. Louis, MO, USA) for 30 s and stained with Oil red O solution (Sigma, St. Louis, MO, USA) for 1 h. The cells were rinsed again with 60% isopropanol for 5 seconds and then with distilled water for 2–3 min. The cells were then observed and photographed under a light microscope. The percentage of the positive stained area was analyzed using ImageJ software. The stained area was then dissolved with isopropanol, and the absorbance at 510 nm was detected.
Cholesterol fluorometric assay
Cholesterol was extracted from cells using a cell scraper and 200 µl of a mixed solution of chloroform: isopropanol: NP-40 (7: 11: 0.1) in a micro-homogenizer. The extracts were centrifuged at 15,000 × g for 10 min. The liquid organic phase was transferred to a new eppendorf tube, and the sample was air-dried at 50 °C. Then, 200 µl of cholesterol assay buffer was added to the extracts, and the extracts were analyzed using a cholesterol assay kit (Cayman Chemical, Ann Arbor, MI, USA). These experiments were performed more than three times.
Triglyceride colorimetric assay
Triglycerides were extracted from transfected cells using a cell scraper and 1 ml of a 5% NP- 40 / distilled water solution. The samples were heated to 80–100 °C in a water bath for 2–5 min or until the NP-40 became cloudy and then cooled to room temperature. This step was repeated until all triglyceride was solubilized. The samples were then centrifuged at 13,200 × g for 2 min to remove any insoluble material. The samples were diluted tenfold with distilled water before proceeding with the assay. The extracts were then analyzed using a triglyceride assay kit (Cayman Chemical, Ann Arbor, MI, USA). These experiments were performed more than three times.
Xenograft mouse model
All experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Jeonbuk National University and conducted in accordance with university and NIH guidelines and regulations (CBNU 2020-072). Four-week-old CAnN.Cg-Foxn1nu/CrlOri (SPF/VAF immunodeficient) male mice were purchased from Orient Bio Inc (Deajeon, South Korea). The mice were acclimated to their new surroundings for one week and used at 5 weeks of age. All mice were maintained under specific pathogen-free conditions in filter-topped cages, with autoclaved food and water. Mice were randomly assigned to experimental groups (n = 5 per group) using a simple random sampling method. They were then injected subcutaneously in their two flanks with a mixture of 50 µl of DPBS and 50 µl of Matrigel (Corning® Matrigel® Basement Membrane Matrix, Bedford, MA, USA) containing 5 × 106 HepG2 cells that had been transfected with either an HN1 shRNA plasmid or a control shRNA plasmid and HN1 overexpression plasmid or a control plasmid. No animals were excluded from the study. Tumor volume and body weight were measured every 3 days. Tumor volume was measured using a caliper and calculated as (width)2 x length/2. Blinding was implemented during the data measurement and analysis phases. The tumor-bearing mice were sacrificed after the tumor volume reached 2000 mm3, and the tumors were extracted for further study.
H&E staining assay
Tumor sections were deparaffinized and rehydrated. Then, they were stained with hematoxylin, rinsed with tap water, stained with eosin, and rinsed with tap water again. Finally, the sections were dehydrated in alcohol solutions, a coverslip was applied, and the samples were left to dry overnight. Images were taken under a light microscope.
TUNEL assay
Mouse tumor tissue sections were deparaffinized, rehydrated, and treated with protease. The TUNEL assay was conducted using a cell death detection kit and following the manufacturer’s instructions. TUNEL-positive cells were analyzed using imaging software. Images were taken under a light microscope.
Immunohistochemistry staining
Immunohistochemistry staining was performed on tumor tissue sections that were incubated overnight with anti-HN1 (14914-1-AP, Proteintech Group Inc.), and SREBP-1 (sc-13551, Santa Cruz Biotechnology, Inc.) antibodies at 4 °C. The sections were then probed with an anti-mouse HRP/DAB IHC kit (Abcam, Cambridge, UK) for 2 h at room temperature. Images were taken under a light microscope.
Statistical analysis
All the experiments were repeated more than three times. Statistical analysis was performed using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA). Data were presented as the mean ± SEM. Differences between the two groups were analyzed using the unpaired Student’s t‐test. One‐way ANOVA followed by Tukey’s post hoc test was used to assess differences among multiple groups. A p value <0.05 was considered to indicate a statistically significant result.
Data availability
All the data generated or analyzed during this study are included in this published article.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J. 2021;134:783–91.
Liu Y, Zheng J, Hao J, Wang RR, Liu X, Gu P, et al. Global burden of primary liver cancer by five etiologies and global prediction by 2035 based on global burden of disease study 2019. Cancer Med. 2022;11:1310–23.
Chidambaranathan-Reghupaty S, Fisher PB, Sarkar D. Hepatocellular carcinoma (HCC): epidemiology, etiology and molecular classification. Adv Cancer Res. 2021;149:1–61.
Farah M, Anugwom C, Ferrer JD, Baca EL, Mattos AZ, Possebon JPP, et al. Changing epidemiology of hepatocellular carcinoma in South America: a report from the South American liver research network. Ann Hepatol. 2023;28:100876.
Tran NH. Shifting epidemiology of hepatocellular carcinoma in far eastern and southeast Asian patients: explanations and implications. Curr Oncol Rep. 2022;24:187–93.
Lliovet, J. M. et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6.
Konyn P, Ahmed A, Kim D. Current epidemiology in hepatocellular carcinoma. Expert Rev Gastroenterol Hepatol. 2021;15:1295–307.
Mehta N, Bhangui P, Yao FY, Mazzaferro V, Toso C, Akamatsu N, et al. Liver transplantation for hepatocellular carcinoma. working group report from the ILTS transplant oncology consensus conference. Transplantation. 2020;104:1136–42.
Berenguer M, Burra P, Ghobrial M, Hibi T, Metselaar H, Sapisochin G, et al. Posttransplant management of recipients undergoing liver transplantation for hepatocellular carcinoma. Working group report from the ILTS transplant oncology consensus conference. Transplantation. 2020;104:1143–9.
Bednarsch J, Czigany Z, Heise D, Joechle K, Luedde T, Heij L, et al. Prognostic evaluation of HCC patients undergoing surgical resection: an analysis of 8 different staging systems. Langenbecks Arch Surg. 2021;406:75–86.
Zhang W, Zhang B, Chen XP. Adjuvant treatment strategy after curative resection for hepatocellular carcinoma. Front Med. 2021;15:155–69.
Tellapuri S, Sutphin PD, Beg MS, Singal AG, Kalva SP. Staging systems of hepatocellular carcinoma: a review. Ind J Gastroenterol. 2018;37:481–91.
Tumen D, Heumann P, Gulow K, Demirci CN, Cosma LS, Muller M, et al. Pathogenesis and current treatment strategies of hepatocellular carcinoma. Biomedicines. 2022;10:3202.
Bruix J, Reig M, Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology. 2016;150:835–53.
Ahn JC, Teng PC, Chen PJ, Posadas E, Tseng HR, Lu SC, et al. Detection of circulating tumor cells and their implications as a biomarker for diagnosis, prognostication, and therapeutic monitoring in hepatocellular carcinoma. Hepatology. 2021;73:422–36.
Salazar J, Le A. The heterogeneity of liver cancer metabolism. Adv Exp Med Biol. 2021;1311:127–36.
Laughlin KM, Luo D, Liu C, Shaw G, Warrington KH Jr, Law BK, et al. Hematopoietic- and neurologic-expressed sequence 1 (Hn1) depletion in B16.F10 melanoma cells promotes a differentiated phenotype that includes increased melanogenesis and cell cycle arrest. Differentiation. 2009;78:35–44.
Varisli L. Decreased expression of HN1 sensitizes prostate cancer cells to apoptosis induced by docetaxel and 2-methoxyestradiol. Ann Clin Lab Sci. 2022;52:196–201.
Zhang C, Xu B, Lu S, Zhao Y, Liu P. HN1 contributes to migration, invasion, and tumorigenesis of breast cancer by enhancing MYC activity. Mol Cancer. 2017;16:90.
Pan Z, Fang Q, Li L, Zhang Y, Xu T, Liu Y, et al. HN1 promotes tumor growth and metastasis of anaplastic thyroid carcinoma by interacting with STMN1. Cancer Lett. 2021;501:31–42.
Wang R, Fu Y, Yao M, Cui X, Zhao Y, Lu X, et al. The HN1/HMGB1 axis promotes the proliferation and metastasis of hepatocellular carcinoma and attenuates the chemosensitivity to oxaliplatin. FEBS J. 2022;289:6400–19.
Chen J, Qiu J, Li F, Jiang X, Sun X, Zheng L, et al. HN1 promotes tumor associated lymphangiogenesis and lymph node metastasis via NF-kappaB signaling activation in cervical carcinoma. Biochem Biophys Res Commun. 2020;530:87–94.
Goto T, Tokunaga F, Hisatomi O. Hematological- and neurological-expressed sequence 1 gene products in progenitor cells during newt retinal development. Stem Cells Int. 2012;2012:436042.
Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–76.
Lee JS, Thorgeirsson SS. Genome-scale profiling of gene expression in hepatocellular carcinoma: classification, survival prediction, and identification of therapeutic targets. Gastroenterology. 2004;127:S51–55.
Woo HG, Park ES, Cheon JH, Kim JH, Lee JS, Park BJ, et al. Gene expression-based recurrence prediction of hepatitis B virus-related human hepatocellular carcinoma. Clin Cancer Res. 2008;14:2056–64.
Kim SM, Leem SH, Chu IS, Park YY, Kim SC, Kim SB, et al. Sixty-five gene-based risk score classifier predicts overall survival in hepatocellular carcinoma. Hepatology. 2012;55:1443–52.
Nault JC, De Reynies A, Villanueva A, Calderaro J, Rebouissou S, Couchy G, et al. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology. 2013;145:176–87.
Ricoult SJ, Yecies JL, Ben-Sahra I, Manning BD. Oncogenic PI3K and K-Ras stimulate de novo lipid synthesis through mTORC1 and SREBP. Oncogene. 2016;35:1250–60.
Yecies JL, Zhang HH, Menon S, Liu S, Yecies D, Lipovsky AI, et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 2011;14:21–32.
Zhao Q, Lin X, Wang G. Targeting SREBP-1-mediated lipogenesis as potential strategies for cancer. Front Oncol. 2022;12:952371.
Du X, Pham YH, Brown AJ. Effects of 25-hydroxycholesterol on cholesterol esterification and sterol regulatory element-binding protein processing are dissociable: implications for cholesterol movement to the regulatory pool in the endoplasmic reticulum. J Biol Chem. 2004;279:47010–6.
Ferre P, Phan F, Foufelle F. SREBP-1c and lipogenesis in the liver: an update1. Biochem J. 2021;478:3723–39.
Wu X, Romero D, Swiatek WI, Dorweiler I, Kikani CK, Sabic H, et al. PAS kinase drives lipogenesis through SREBP-1 maturation. Cell Rep. 2014;8:242–55.
Cheng C, Ru P, Geng F, Liu J, Yoo JY, Wu X, et al. Glucose-mediated N-glycosylation of SCAP is essential for SREBP-1 activation and tumor growth. Cancer Cell. 2015;28:569–81.
Cheng C, Geng F, Li Z, Zhong Y, Wang H, Cheng X, et al. Ammonia stimulates SCAP/Insig dissociation and SREBP-1 activation to promote lipogenesis and tumour growth. Nat. Metab. 2022;4:575–88.
Zane KE, Nagib PB, Jalil S, Mumtaz K, Makary MS. Emerging curative-intent minimally-invasive therapies for hepatocellular carcinoma. World J Hepatol. 2022;14:885–95.
Chang KV, Chen JD, Wu WT, Huang KC, Hsu CT, Han DS. Association between loss of skeletal muscle mass and mortality and tumor recurrence in hepatocellular carcinoma: a systematic review and meta-analysis. Liver Cancer. 2018;7:90–103.
Chen S, Ji R, Shi X, Wang Z, Zhu D. Retrospective analysis of efficacy, safety, and prognostic factors in a cohort of Chinese hepatocellular carcinoma patients treated with drug-eluting bead transarterial chemoembolization. Braz J Med Biol Res. 2019;52:e8467.
Sparchez Z, Mocan T, Radu P, Mocan LP, Sparchez M, Leucuta DC, et al. Prognostic factors after percutaneous radiofrequency ablation in the treatment of hepatocellular carcinoma. Impact of incomplete ablation on recurrence and overall survival rates. J Gastrointestin Liver Dis. 2018;27:399–407.
Hart MJ, de los Santos R, Albert IN, Rubinfeld B, Polakis P. Downregulation of beta-catenin by human Axin and its association with the APC tumor suppressor, beta-catenin and GSK3 beta. Curr Biol. 1998;8:573–81.
Varisli L, Ozturk BE, Akyuz GK, Korkmaz KS. HN1 negatively influences the beta-catenin/E-cadherin interaction, and contributes to migration in prostate cells. J Cell Biochem. 2015;116:170–8.
Park JH, Pyun WY, Park HW. Cancer metabolism: phenotype, signaling and therapeutic targets. Cells. 2020;9:2308.
Koundouros N, Poulogiannis G. Reprogramming of fatty acid metabolism in cancer. Br J Cancer. 2020;122:4–22.
Sun Y, Wu L, Zhong Y, Zhou K, Hou Y, Wang Z, et al. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell. 2021;184:404–421.e416.
Deng M, Sun S, Zhao R, Guan R, Zhang Z, Li S, et al. The pyroptosis-related gene signature predicts prognosis and indicates immune activity in hepatocellular carcinoma. Mol Med. 2022;28:16.
Shimada S, Mogushi K, Akiyama Y, Furuyama T, Watanabe S, Ogura T, et al. Comprehensive molecular and immunological characterization of hepatocellular carcinoma. EBioMedicine. 2019;40:457–70.
Fortin J, Mak TW. Targeting PI3K signaling in cancer: a cautionary tale of two AKTs. Cancer Cell. 2016;29:429–31.
Revathidevi S, Munirajan AK. Akt in cancer: mediator and more. Semin Cancer Biol. 2019;59:80–91.
Altomare DA, Khaled AR. Homeostasis and the importance for a balance between AKT/mTOR activity and intracellular signaling. Curr Med Chem. 2012;19:3748–62.
Yu JS, Cui W. Proliferation, survival and metabolism: the role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development. 2016;143:3050–60.
He Y, Sun MM, Zhang GG, Yang J, Chen KS, Xu WW, et al. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct Target Ther. 2021;6:425.
Varisli L, Gonen-Korkmaz C, Debelec-Butuner B, Erbaykent-Tepedelen B, Muhammed HS, Bogurcu N, et al. Ubiquitously expressed hematological and neurological expressed 1 downregulates Akt-mediated GSK3beta signaling, and its knockdown results in deregulated G2/M transition in prostate cells. DNA Cell Biol. 2011;30:419–29.
Tian LY, Smit DJ, Jucker M. The role of PI3K/AKT/mTOR signaling in hepatocellular carcinoma metabolism. Int J Mol Sci. 2023;24:2652.
Davies BR, Greenwood H, Dudley P, Crafter C, Yu DH, Zhang J, et al. Preclinical pharmacology of AZD5363, an inhibitor of AKT: pharmacodynamics, antitumor activity, and correlation of monotherapy activity with genetic background. Mol Cancer Ther. 2012;11:873–87.
Rishi V, Gal J, Krylov D, Fridriksson J, Boysen MS, Mandrup S, et al. SREBP-1 dimerization specificity maps to both the helix-loop-helix and leucine zipper domains: use of a dominant negative. J Biol Chem. 2004;279:11863–74.
Cheng C, Geng F, Cheng X, Guo D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun. 2018;38:27.
Zhu T, Wang Z, Zou T, Xu L, Zhang S, Chen Y, et al. SOAT1 promotes gastric cancer lymph node metastasis through lipid synthesis. Front Pharmacol. 2021;12:769647.
Chen H, Qi Q, Wu N, Wang Y, Feng Q, Jin R, et al. Aspirin promotes RSL3-induced ferroptosis by suppressing mTOR/SREBP-1/SCD1-mediated lipogenesis in PIK3CA-mutatnt colorectal cancer. Redox Biol. 2022;55:102426.
Zhao S, Cheng L, Shi Y, Li J, Yun Q, Yang H. MIEF2 reprograms lipid metabolism to drive progression of ovarian cancer through ROS/AKT/mTOR signaling pathway. Cell Death Dis. 2021;12:18.
Du F, Zhang HJ, Shao W, Tu YY, Yang KJ, Piao LS. Adenosine diphosphate-ribosylation factor-like 15 can regulate glycolysis and lipogenesis related genes in colon cancer. J Physiol Pharmacol. 2022;73:403–11.
Jin Y, Chen Z, Dong J, Wang B, Fan S, Yang X, et al. SREBP1/FASN/cholesterol axis facilitates radioresistance in colorectal cancer. FEBS Open Bio. 2021;11:1343–52.
Zhou C, Qian W, Li J, Ma J, Chen X, Jiang Z, et al. High glucose microenvironment accelerates tumor growth via SREBP1-autophagy axis in pancreatic cancer. J Exp Clin Cancer Res. 2019;38:302.
Li C, Zhang L, Qiu Z, Deng W, Wang W. Key molecules of fatty acid metabolism in gastric cancer. Biomolecules. 2022;12:706.
Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun. 2021;41:109–20.
Pavlova NN, Zhu J, Thompson CB. The hallmarks of cancer metabolism: still emerging. Cell Metab. 2022;34:355–77.
Zanotelli MR, Zhang J, Reinhart-King CA. Mechanoresponsive metabolism in cancer cell migration and metastasis. Cell Metab. 2021;33:1307–21.
Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17:351–9.
Liu W, Chakraborty B, Safi R, Kazmin D, Chang CY, McDonnell DP. Dysregulated cholesterol homeostasis results in resistance to ferroptosis increasing tumorigenicity and metastasis in cancer. Nat Commun. 2021;12:5103.
Holthuis JC, Menon AK. Lipid landscapes and pipelines in membrane homeostasis. Nature. 2014;510:48–57.
Lien EC, Westermark AM, Zhang Y, Yuan C, Li Z, Lau AN, et al. Low glycaemic diets alter lipid metabolism to influence tumour growth. Nature. 2021;599:302–7.
Stine ZE, Schug ZT, Salvino JM, Dang CV. Targeting cancer metabolism in the era of precision oncology. Nat Rev Drug Discov. 2022;21:141–62.
Kubik J, Humeniuk E, Adamczuk G, Madej-Czerwonka B, Korga-Plewko A. Targeting energy metabolism in cancer treatment. Int J Mol Sci. 2022;23:5572.
Zdralevic M, Vucetic M, Daher B, Marchiq I, Parks SK, Pouyssegur J. Disrupting the ‘Warburg effect’ re-routes cancer cells to OXPHOS offering a vulnerability point via ‘ferroptosis’-induced cell death. Adv Biol Regul. 2018;68:55–63.
Gwangwa MV, Joubert AM, Visagie MH. Crosstalk between the Warburg effect, redox regulation and autophagy induction in tumourigenesis. Cell Mol Biol Lett. 2018;23:20.
Jing ZL, Liu QM, He XY, Jia ZR, Xu ZZ, Yang BL, et al. NCAPD3 enhances Warburg effect through c-myc and E2F1 and promotes the occurrence and progression of colorectal cancer. J Exp Clin Canc Res. 2022;41:198.
Maley CC, Galipeau PC, Finley JC, Wongsurawat VJ, Li X, Sanchez CA, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–73.
Acknowledgements
The National Research Foundation (NRF), which is sponsored by the Korean government, provided funding for this study through the Basic Science Research Program (2021R1A2C2013505) and the Medical Research Center Program (NRF-2017R1A5A2015061) (MSIP).
Author information
Authors and Affiliations
Contributions
HJ and RM wrote the draft and performed biological experiments. CSL conducted the experimental studies and analytical assistance from S-HK, OHC, and B-HP on the various cellular experiments. Y-HL and J-SL conducted the histology study and statistical analyses. SMK designed the experiments and described the paper. The final draft of the manuscript was reviewed and approved by all the authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The human study was conducted following approval from the Ethics Committee of the University of Texas MD Anderson Cancer Center (LAB09-0687) in accordance with the Helsinki Declaration of 1975. Written informed consent was obtained from all patients prior to their inclusion in the study. All animal care procedures and sacrifices were carried out in strict accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) and the Center for Animal Experiments at Jeonbuk National University (CBNU 2020-072).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jin, H., Meng, R., Li, C.S. et al. HN1-mediated activation of lipogenesis through Akt-SREBP signaling promotes hepatocellular carcinoma cell proliferation and metastasis. Cancer Gene Ther (2024). https://doi.org/10.1038/s41417-024-00827-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41417-024-00827-y
- Springer Nature America, Inc.