Abstract
The abnormal activation of the nuclear factor-kappa B (NF-κB) signaling pathway is an important precipitating factor for the inception and development of colorectal cancer (CRC), one of the most common tumors worldwide. As a pro-apoptotic transcription factor, monocyte chemotactic protein-induced protein 1 (MCPIP1) has been closely associated with many tumor types. In the present study, the expression of MCPIP1 was firstly discovered reduced in CRC tissues and correlated with poor patient prognosis. The decreased expression was caused by promoter hypermethylation. Overexpressed MCPIP1 was found to inhibit the proliferative and migratory abilities of CRC cells, whereas knockdown of MCPIP1 produced the opposite result. The subsequent investigation demonstrated that MCPIP1 exerted its “anti-cancer” effect by suppression of the NF-κB signaling pathway through negative regulation of K63-linked ubiquitylation of TNF receptor associated factor 6 (TRAF6). Therefore, our results indicate a prognostic marker for CRC and a theoretical basis for MCPIP1 as a treatment.
Similar content being viewed by others
Background
There are over a million new cases and more than 570,000 deaths from colorectal cancer (CRC), defining it as one of the most common malignancies worldwide [1]. In China, the trends in CRC incidence and mortality rates have risen during the past twenty years [2]. Effective treatment of CRC is urgently needed. However, due to tumor heterogeneity, patients with an identical disease stage can have significantly different prognoses. In recent years, newly discovered molecular markers of CRC have been identified with development of genome and proteomics projects. For example, patients with metastatic CRC carrying the wild-type RAS gene have a remarkably longer overall survival (OS) when treated with chemotherapy combined with cetuximab, compared to those treated with chemotherapy alone [3]. The prognosis of patients with MSI-H (high microsatellite instability) or dMMR (mismatch repair deficient) is dramatically improved following treatment with immunotherapy compared to those without MSI-H/dMMR [4]. These important advances inform the exploration of new molecular markers for CRC which may facilitate the development of novel diagnosis and treatment methods.
The nuclear factor-kappa B (NF-κB) signaling pathway has well-established involvement in regulating physiological and pathological events, including inflammation, the immune response and differentiation [5]. Controlling the expression of more than 400 genes, deregulated NF-κB signaling was demonstrated to enhance cancer cell proliferation and metastasis and also mediate radio-/chemo-resistance [6]. Ubiquitination and de-ubiquitination have been proven to play essential regulatory roles in the NF-κB pathway [7, 8]. Usually, K63-polyubiquitin (K63-Ub) regulates the target protein for non-proteolytic functions, such as signaling complex assembly, DNA damage response and kinase activation; and K48-polyubiquitin mediates protein for degradation by the 26S proteasome [9, 10]. Previous studies have shown that the stimulation of inflammation causes TNF receptor-associated factor 6 (TRAF6), a critical protein of the classical NF-κB signaling pathway, to activate ubiquitinase and synthesize a K63-Ub chain which recruits and binds TAK1- TABs complexes and IκB kinase (IKK) α/β/γ complexes. Later the phosphorylation of IκB ultimately releases the transcription factor, p50/RELA(p65), into the nuclei to initiate downstream signal transduction [11, 12]. The abnormal activation of the NF-κB signaling pathway is an important precipitating factor for inflammation-cancer transformation and for the occurrence and development of CRC [13, 14]. TRAF6 expression is reported to be elevated in CRC and correlates with poor patient prognosis [15]. Knockdown of TRAF6 inhibited the proliferation of CRC cells and enhanced the sensitivity to the chemotherapeutic regimen of 5-fluorouracil and etoposide [16]. In the present study, monocyte chemotactic protein-induced protein 1 (MCPIP1) was found to repress TRAF6 through inhibiting its K63-ubiquitynation, thus negatively regulating the NF-κB signaling pathway and suppressing the proliferation and migration of CRC cells; it was firstly identified as a CRC anti-oncogene. Moreover, decreased expression of MCPIP1 due to epigenetic abnormalities predicted advanced CRC prognosis.
Materials and methods
RT-qPCR assays
2 μg of total RNA were extracted from colorectal cell lines/ paired cancer tissues. After DNase I treatment, complementary DNA was reverse-transcribed with an oligo-dT primer (Promega, Madison, WI, USA) and real-time semi-quantitative polymerase chain reaction (RT-qPCR) was used to detect the mRNA level of ZC3H12A (encoding MCPIP1 protein). Gene expression was normalized to the GAPDH transcript level according to the △△Ct values and then compared with controls. Primers sequences as follows: ZC3H12A (coding MCPIP1) forward 5′ GTCTGACGGGATCGTGGTTT 3′, reverse 5′ TCAGGGGGCATAAACTTGTCA 3′; GAPDH forward 5′ AGAAGGCTGGGGCTCATTTG 3′, reverse 5′ AGGGGCCATCCACAGTCTTC 3′.
Immunohistochemical (IHC) staining assays and samples
Between September 2000 and January 2006, 172 sporadic CRC samples of single primary lesion without history of antitumor therapy were resected at Sun Yat-sen University Cancer Center and included in the IHC assay. Data on clinical and pathological characteristics were collected. The median follow-up conducted by phone was 70.9 months. Samples were collected with informed consent given in accordance with the institution’s ethical review board.
We performed IHC staining assays on paraffin-embedded sections with antibodies against MCPIP1 (#D222666, Sangon Biotech, Shanghai, China), COX-2 and p50 (#sc-376861 and #sc-8414, Santa Cruz Biotechnology, Santa Cruz, CA, USA), followed by incubation with the EnVision III Detection System (Dako, Kobenhavn, Denmark). We used 200× microscopy (Olympus, Tokyo, Japan) to obtain histological images. Then we assessed the proportion of cells with different intensities (0, negative; 1, weak; 2, moderate; 3, strong) by the inForm software (INFORM, Aachen, Nordrhein-Westfalen, Germany) and created a histoscore (H-score) by multiplying each staining intensity category with the percentage of positively stained cells. With the help of receiver operating characteristic (ROC) curve referencing to OS status, the H-scores were dichotomized into high versus low. The maximized sum of sensitivity and specificity was set as the boundary threshold.
DNA methylation analysis
The MethPrimer software (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi) was used to design specific primers for amplifying methylated or unmethylated CpG sites. Genomic DNA isolated from cell lines was converted with an EZ-DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, USA). Methylation-Specific PCR (MSP), followed by agarose gel electrophoresis to qualitatively analyze DNA methylation. Primer sequences were as follows: Methylated forward 5′ TTAAGGTGCGTGGATTAATTTTC 3′, reverse 5′ CAAAACTACGACGACGACGA 3′; unmethylated forward 5′ TTTTTAAGGTGTGTGGATTAATTTTT 3′, reverse 5′ ACCTCAAAACTACAACAACAACAAA 3′.
Immunoprecipitation and Western blotting assays
For immunoprecipitation assays, cells were harvested, lysed and centrifuged. Supernatants were incubated with antibodies and protein G beads (#sc-2002, Santa Cruz Biotechnology) overnight at 4 oC. Beads were washed and proteins precipitated with β-mercaptoethanol for immunoblot assays.
Equal amounts of denatured cell protein were separated by SDS-PAGE gels and transferred to a polyvinylidene difluoride membrane (Roche, Basel, Switzerland). Membranes were incubated with primary antibodies against: MCPIP1 (Sangon Biotech); COX-2, p50, TRAF6 and GAPDH (#sc-376861, #sc-8414, #sc-8409 and #sc-32233 respectively, Santa Cruz Biotechnology); HA-tag, Myc-tag (#3724, #2276, Cell Signaling Technology, Danvers, MA, USA); Flag-tag (#F1804, Sigma Aldrich, St. Louis, MO, USA); followed by appropriate secondary antibodies linked to HRP (anti-mouse, #7076; anti-rabbit #7074; Cell Signaling Technology). GAPDH served as loading control. An ECL Western blot detection kit (Thermo Fisher Scientific, Waltham, MA, USA) was used for detection.
Cell culture
The human non-tumor colon epithelial cell line, NCM460, was obtained from BLUEFBIO (Shanghai, China); the human embryonic kidney cell line 293T and CRC cell lines SW480, SW620, SW837, SW1116, SW1463, HCT-8, HT-29, HCT-15, HCT116, DLD-1 and RKO from the American Type Culture Collection (ATCC, Manassas, VA, USA). GP293 (human embryonic kidney cell line) was a gift from Prof. Anthony Chan (The Chinese University of Hong Kong). Cell lines were grown in DMEM/RPMI-1640 medium with 10% fetal bovine serum at 37 oC in a humidified chamber with 5% CO2. All experiments were performed in cultures which were in log-phase growth at 70–80% confluence. All cell lines were confirmed mycoplasma-free.
Construction of stable cell lines
The coding sequence of wild-type, ZC3H12A, confirmed by Sanger sequencing, was cloned and ligated into the pLNCX2 Retroviral Expression System (Clontech, Mountain View, CA, USA). A packaging cell line, GP293, was transfected with the retroviral vectors containing ZC3H12A or control vectors. Supernatants of GP293 cells were collected 48 h later and delivered to the HCT116 cell line. We applied G418 (Thermo Fisher Scientific) 1600μg/ml for 10 days to screen MCPIP1 stably over-expressing cells.
Three shRNA sequences (shRNA1: GCGTAAGAAGCCACTCACTTT, shRNA2: CCAGCGTGTATACTAAGCTGT, shRNA3: CCATCCTTTATTCAGAGTCAT) targeting ZC3H12A were used, as well as the negative control (shNC: TTCTCCGAACGTGTCACGTTT) and ligated into the pSUPER RNAi system (Oligoengine, Seattle, WA, USA). The pSUPER-shRNAs were transfected to a 293T packaging cell line. The supernatants of retrovirus containing shRNAs were collected 48 h later and delivered to the HT-29 cell line. 1.2 μg/ml puromycin (Thermo Fisher Scientific) was used for 2 weeks to screen MCPIP1 stably down-regulated HT-29 cells.
Colony formation assay and MTT assay
Cell proliferation was measured by seeding cells into 96-well plates (1.5 × 103 cells per well) for 1, 3, 5 and 7 days and adding 20 µl MTT (5 mg/ml) with incubation at 37 °C for 4 h. Cells were suspended in 100 µl DMSO (Sigma-Aldrich) and absorbance was measured at 490 nm by an ELISA reader. Colony formation was measured by seeding stable cell lines into 6-well plates with complete medium (500 cells per well). After 12 days, visible colonies were stained with 0.1% crystal violet and numbers calculated.
Apoptosis assays
Cells were seeded in 6-well plates (3 × 105 cells per well), allowed to adhere and 30 μM/L cisplatin was added to one of the wells as positive control. After 24h-incubation, apoptosis assays were performed using Annexin V-FITC and Propidium Iodide staining (Annexin-V-FLUOS Staining Kit, Roche) with flow-cytometry. Percentages of apoptotic HCT116 and HT-29 cells were calculated by the FlowJO software (Ashland, Oregon, USA).
Migration and invasion assays
200 µl of single-cell suspensions (105 cells) in serum-free DMEM were added to the 24-well upper chamber with or without Matrigel (8-μm pore, BD Falcon, San Jose, CA, USA) for migration or invasion assay, respectively, while the lower chambers were filled with 500 µl medium containing 10% FBS. After 24-hour of incubation, the cells that had migrated to the underside of the polycarbonate membrane were fixed and stained with 0.1% crystal violet. Migrated cells were counted in 5 independent fields for each sample (at 200× magnification).
Dual luciferase reporter assay
Cells were seeded into 24-well plates (4 × 104 per well) and incubated for 24 h. Reporter plasmids (pGL3 vector with the sequence of transcription factor binding sites, 0.2 μg per well) together with internal control (pRL-TK, the Renilla luciferase reporter, 0.01 μg per well) were transfected. After 24 h incubation, the firefly and Renilla luciferase activities were quantified with a luminometer using a Dual-Luciferase Reporter Kit (Promega).
Bioinformatics analysis
The expression matrices and corresponding clinical characteristics of the colorectal samples were obtained from the TCGA databases. Patients without survival information or with incomplete clinicopathological characteristics for further assessment were excluded. A total of 567 patients were enrolled. The expression matrix data of the TCGA-COADREAD cohort (FPKM format) were downloaded from the Genomic Data Commons platform and the FPKM value was converted to the million transcripts value per thousand bases (TPM). OS and RNA datasets were downloaded from TCGA or GEO data. Levels of ZC3H12A mRNA were dichotomized into high versus low based on the OS data and the boundary threshold with the strongest specificity and sensitivity on the ROC curve. Kaplan-Meier analysis of the TCGA data was performed by the survminer and survival package of R software (Version 3.3.3). Survival analysis of GEO data (GSE39582, GSE16125) was conducted via online database (http://www.progtools.net/). The methyltransferase list was identified by reviewing literature. By referring to the STRING database (https://string-db.org/, Version 11.5), items with corrected p value ≤ 0.05 were screened to reveal the network between proteins. R package ggplot2 was used to construct a co-expression network for MCPIP1, NF-κB regulators and NF-κB pathway associated proteins. Datasets, including biological processes, cellular components and molecular functions were downloaded from the DAVID database (https://david.ncifcrf.gov/) for Gene Ontology (GO) analysis. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was used to annotate the potential functions of MCPIP1. A significance level of p < 0.05 was set as the cut-off criterion and the plots were constructed by the gplots package in R software (Version 3.3.3). The DEmRNAs of MCPIP1 were enrolled in a PPI network and visualized with Cytoscape (Version 3.7.2).
Statistical analysis
All experiments were performed in unique triplicates. Investigators were blinded to the group allocation during the procedure and data analysis. We used the PASW Statistics 18.0 software package (SPSS Inc., Chicago, IL, USA) to perform statistical analysis. All CRC patients were included in Kaplan-Meier survival analysis and survival curves were compared by a log-rank test. We applied a Pearson’s chi-squared test (2-sided) to assess associations between clinical-pathological parameters and MCPIP1 expression. We applied multivariate Cox proportional hazards models to explore the prognostic effects of MCPIP1 expression on OS and progression-free survival (PFS). Data are presented as mean ± SD. The Student’s t test (two-tailed) was used for evaluating statistical difference among two groups, while one-way ANOVA was used for more than two groups of differences. A value of p < 0.05 was considered statistically significant.
Results
Low expression of MCPIP1 indicated poor prognosis of CRC patients
mRNAs were isolated from 71 CRC tissue specimens with matched normal intestinal epithelial tissues and ZC3H12A (encoding MCPIP1 protein) levels were detected using semi-qPCR after reverse transcription. The expression of ZC3H12A mRNA was demonstrated to be markedly reduced in 84.5% cases (60/71) of CRC tissue specimens in contrast to normal controls (p = 0.014, Fig. 1A). There were similar ZC3H12A levels when stage I-IV cases were compared (p = 0.397, Fig. 1B). We further analyzed the ZC3H12A mRNA level of 567 cases in the TCGA dataset for validation. It showed that the ZC3H12A level was significantly higher in stage I than in stage II-IV (p = 0.005, Fig. 1C). The difference between our data and TCGA data may be caused by the sample size of stage I in ours being small (n = 14), still, both of the data suggest that MCPIP1 expression was probably silenced in early-stage of CRC.
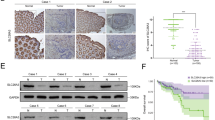
A ZC3H12A mRNA level of 71 paired CRC tissues via semi-quantitate PCR (n = 3). B The ZC3H12A mRNA level of the 71 cases was similar when stage I-IV cases were compared (n = 3). GAPDH was used as a control. *p < 0.05. Data are shown as median ± quartile. C The mRNA levels of ZC3H12A in different stages of 567 CRC cases obtained from the TCGA- COADREAD project. TPM, transcripts per million. D Representative images of IHC staining for MCPIP1 in colorectal tumor tissues. E MCPIP1 expression was down-regulated in cancer tissues (black arrows) compared with paratumor tissues (red arrows). Representative immunohistochemistry (IHC) staining images are shown. Scale bar = 50 μm. F Kaplan-Meier survival analysis of our CRC dataset with high or low MCPIP1 expression in tumors. G Kaplan-Meier survival analysis of CRC patients from TCGA OS and ZC3H12A mRNA level data (n = 567). HR hazard ratio, CI confidence interval.
IHC assay was utilized to determine MCPIP1 protein in 172 CRC tissue sections. The content of MCPIP1 was reduced in tumor tissues compared to non-malignant paratumor tissues, consistent with the qPCR results. The positive expression rate of MCPIP1 was 82.7% and the histochemistry (H)-score was 1.60 in tumor tissues, whereas the comparison positive expression rate and H-score were 92.3% and 2.29, respectively, in paratumor tissues (p < 0.001, p < 0.001, Fig. 1D, E).
The correlation between MCPIP1 and the prognosis of the 172 CRC patients was analyzed. The H-score values of MCPIP1 were dichotomized into low versus high based on the OS data and the boundary threshold with the strongest specificity and sensitivity on the ROC curve. According to the analysis of the correlations of MCPIP1 with clinicopathological factors, a low expression of MCPIP1 in tumor tissues was associated with malignant features, such as with distant metastasis in our 172 cases dataset (p = 0.024, Supplementary Table S1), and with deeper lymph node infiltration (p = 0.003), late TNM stage (p = 0.010), and elevated CEA level (p = 0.011) in 567 cases of TCGA dataset (Supplementary Table S2). Furthermore, multivariate regression analysis demonstrated that a low expression of MCPIP1 in tumor tissues proved to be an independent factor for poor prognosis of CRC patients (PFS: p = 0.005, hazard ratio [HR] = 2.899, 95%CI 1.377–6.098; OS: p = 0.002, HR = 4.065, 95%CI 1.656-10.00, Table 1), similar to the commonly used factors for poor prognosis such as TNM stage, CEA level and CA19-9 level. We further investigated whether MCPIP1 also acted as an independent prognostic factor in TCGA data. Results proved that the predictive power of MCPIP1 for OS remained significant in the multivariate analysis after controlling for age, gender and tumor stage (p = 0.038, HR = 1.494, Supplementary Table S3).
Kaplan-Meier survival analysis showed that patients with high tumor tissue MCPIP1 (H-score > 2.1) had distinctly increased PFS and OS compared to patients with low MCPIP1 (PFS: p = 0.026, HR = 0.49; OS: p = 0.014, HR = 0.40, Fig. 1F). Survival analysis with TCGA and GEO data also revealed that a high level of ZC3H12A in CRC tissue correlated with better OS (TCGA: n = 567, p = 0.008, HR = 0.61, Fig. 1G; GEO39582: n = 533, p = 0.013, HR = 0.69, Supplementary Fig. S1A; GEO16125: n = 32, p = 0.001, HR = 0.28, Supplementary Fig. S1B). Therefore, low expression of MCPIP1 was statistically associated with poor prognosis of CRC patients.
ZC3H12A promoter was generally hypermethylated
The specific cause of reduced MCPIP1 expression in CRC was investigated. A PubMed search, sequence analysis and online web prediction of the promoter region of ZC3H12A revealed a CpG island containing raised GC content with a span above 450 bp (Fig. 2A). Methylation-specific PCR (MSP) primers were designed to qualitatively detect the methylation site of the ZC3H12A promoter. Using MSP electrophoresis, the normal human colonic epithelial cell line, NCM460, without methylation in this region was compared to 6 CRC cell lines in which partial hypermethylation was demonstrated (Fig. 2B).
A Percentage of C + G nucleotides is presented for a region spanning 2 kbp upstream of ZC3H12A. Specific primers for CpG island are indicated by convergent arrows. B Qualitative methylation-specific PCR of the unmethylated (USP) or methylated (MSP) CpG island DNA in CRC cell lines. C ZC3H12A mRNA level (Top, by semi-qPCR) and promoter methylation level (Bottom, by qualitative methylation-specific PCR) in CRC cells pre- and post-treatment with 5-Aza-dC (n = 3), ***p < 0.001 and *p < 0.05. Data are shown as mean ± s.d.
To clarify the relationship between the abnormal hypermethylation of the ZC3H12A promoter region and its transcription level, CRC cells were treated with a DNA methyltransferase inhibitor, 5-Aza-2′-deoxycytidine (5-Aza-dC). Real-time fluorescence qPCR assay showed that the mRNA expression level was elevated when the methylation of the ZC3H12A promoter region of CRC cells treated with 5-Aza-dC was attenuated (Fig. 2C). Therefore, we concluded that abnormal hypermethylation of the ZC3H12A promoter region significantly inhibited mRNA transcription, thereby reducing MCPIP1 expression.
To identify the methyltransferase that targets the ZC3H12A promoter region, a Spearman correlation analysis of TCGA data was applied in Supplementary Fig. S2. It showed that among the DNA methyltransferases (DNMT1, DNMT3A and DNMT3B), DNMT3A was the most negatively correlated with ZC3H12A.
MCPIP1 repressed proliferation, migration, invasion and enhanced apoptosis of CRC cells
The mRNA and protein levels of MCPIP1 in normal colonic epithelial cell line, NCM460, and 10 CRC cell lines were measured using real-time fluorescence qPCR and Western blotting. Results showed that expression of MCPIP1 was down-regulated in 6 of 10 CRC cell lines to varying degrees (Fig. 3A).
Protein and mRNA level of MCPIP1 in normal (NCM460) and 10 CRC cell lines (A) and in stably MCPIP1-overexpressing HCT116 cells and stably MCPIP1-knocked down HT-29 cells (B). Ctrl, negative control. Effects of MCPIP1 over-expression and knockdown on oncogenic growth of CRC cells in vitro assayed by MTT (C) and colony formation (D). E Apoptosis assays of CRC cells were performed after cisplatin 30 µM/L incubation for 24 h. F CRC cell migration and invasion evaluated by transwell assay using MCPIP1 over-expressing or knocked down CRC cells. ***p < 0.001, **p < 0.01, and *p < 0.05. Data are shown as mean ± s.d, n = 3.
To investigate the function of MCPIP1 in CRC cells, the HCT116 cell line with low endogenous expression was selected for stable overexpression (HCT116-Ctrl/MCPIP1) and HT-29 with high endogenous expression was used for stable knockdown (HT-29-Ctrl/shRNA, Fig. 3B).
Using the MTT assay, MCPIP1 was demonstrated to inhibit proliferation of HCT116 cells so that the number of HCT116-MCPIP1 cells was only 59.2% of the control group on Day 7 (p = 0.006, Fig. 3C). When MCPIP1 was knocked down by shRNA, the proliferation of HT-29 cells was accelerated in contrast to the control group (1.20 times on Day 7, p = 0.02, Fig. 3C). According to the colony formation assay, less colonies were formed by HCT116 cells overexpressing MCPIP1 (94 ± 13) after 12 days of culture compared with that in the control group (185 ± 10, p = 0.001). Moreover, the opposite experimental result was obtained in HT-29 cells with stable knockdown of MCPIP1, that is, the HT-29-shRNA group formed significant more colonies (164 ± 8) than the HT-29-Ctrl group (77 ± 5, p < 0.001, Fig. 3D).
Following treatment with 30 µM/L cisplatin for 24 hours, overexpressed MCPIP1 enhanced apoptosis of HCT116 cells (15.52% in the control group and 20.97% in the overexpression group, p = 0.022). In HT-29 cells, the percentage of apoptotic cells was decreased with the knockdown of MCPIP1 (average 16.32% in the control group and 10.61% in the shRNA group, p = 0.037, Fig. 3E). STRING functional enrichment analysis was performed to reveal the network between proteins from literature data and it showed that MCPIP1 expression level was associated with AEN, caspase3, caspase 8 and BCL2, intrinsic apoptotic signaling pathway proteins (Supplementary Fig. S3). These results indicated that MCPIP1 induced apoptosis of CRC cells.
A transwell system was employed to detect changes in the migratory/invasive ability of HCT116/HT-29 cells following overexpression or knock down of MCPIP1. The number of HCT116 cells overexpressing MCPIP1 that migrated through the chambers was reduced (39.4% of the control group, p < 0.001) and a similar result was observed using the invasion assay plated with Matrigel (31.8% of the control group, p < 0.001). After knocking down MCPIP1 with shRNA, the migratory ability of HT-29 cells was enhanced in comparison to the control group (1.54 times of the control group, p = 0.001) and invasion was 2.35 times that of the control group when plated with Matrigel (p < 0.001, Fig. 3F).
In summary, in-vitro functional experiments showed that overexpressed MCPIP1 inhibited colony formation, promoted apoptosis and repressed both migration and invasion of CRC cells. Therefore, MCPIP1 functioned as an anti-oncogene in CRC.
MCPIP1 suppressed the NF-κB signaling pathway via K63-linked deubiquitylation of TRAF6
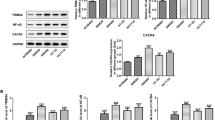
To investigate the underlying mechanism of MCPIP1, we used the protein‑protein interaction (PPI) network visualization, the Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of TCGA data to reveal that MCPIP1 was engaged in ubiquitin-like protein ligase binding, the NF-κB signaling pathway and cellular response to interlukin-1 (an activator of the NF-κB signaling pathway, Supplementary Fig. S4A, S4B, and S4C). Next we applied a dual-luciferase reporter gene assay to analyze the MAPK, p53, NF-κB and Wnt signaling pathways which are known to be involved in CRC cell proliferation, apoptosis and migration [17]. Results demonstrated that 293T cells transiently overexpressing MCPIP1 had the greatest degree of inhibition to the NF-κB signaling pathway among the four pathways by 36% (p = 0.027, Fig. 4A). The RNA-Sequencing dataset of TCGA also showed that MCPIP1 was positively associated with NF-κB inhibitor family members (Fig. 4B). These results were further corroborated using HCT116/HT-29 cell lines in which the NF-κB signaling pathway activity was decreased by 50% through stably overexpressing MCPIP1 in HCT116 cells (p = 0.01) and increased by 32% via stable knockdown of MCPIP1 in HT-29 cells (p = 0.001, Fig. 4C).
A Dual luciferase reporter assay of NF-κB, MAPK (AP-1, SRE), P53 (P21) and Wnt (Top1-flash) pathway activation in 293T cells in the presence of transient MCPIP1 over-expression. The pGL3-control vector was used as positive control and the pRL-TK vector as negative control (n = 3). B The correlation between ZC3H12A and NF-κB pathway inhibitors in CRC of RNA data obtained from TCGA with Spearman’s assay. Red grids represent a positive correlation while blue grids represent a negative correlation. C Dual luciferase reporter assay of NF-κB pathway activation and real-time qPCR of the mRNA level of NF-κB target genes in control (Ctrl) or HCT116 cells with over-expressed MCPIP1 (D) and HT-29 cells stably targeted by shRNA against ZC3H12A (E) (n = 3). F The correlation of MCPIP1 with COX-2 and p50 in CRC tissues. Left: Images of representative CRC cases. Right: Linear regression analysis of MCPIP1 and COX-2 or p50 H-scores of 20 CRC tumor tissues. ***p < 0.001, **p < 0.01, *p < 0.05, and n.s. no significance. Data represents mean + s.d.
The influence of MCPIP1 on the NF-κB signaling pathway was further confirmed by qPCR assay. Overexpression of MCPIP1 inhibited the transcription of several target genes [Bcl-2, Bcl-xL, IL-6, MMP-9, Vimentin, cyclooxygenase-2 (COX2), c-Myc and p50] related to tumor development, metastasis and inflammatory responses downstream of the NF-κB signaling pathway. Moreover, the transcription of these genes was strengthened by knocking down MCPIP1 (Fig. 4D, E). Correlations of MCPIP1 level with those of COX2 and p50 were investigated using IHC assay on 20 CRC tissues. Analyses showed that the protein level of MCPIP1 was inversely related to the expressions of both COX2 and p50 (Fig. 4F). That is, the higher the level of MCPIP1, the lower the expression of NF-κB signaling pathway transcription products, COX2 and p50. Correlation analysis of the TCGA data also showed that MCPIP1 was negatively associated with most of the downstream target genes in the NF-κB pathway (Supplementary Fig. S5). These results proved that MCPIP1 inhibited the transcriptional activity of the NF-κB signaling pathway.
To determine specific target sites of MCPIP1 in the NF-κB signaling pathway, the activators at different action nodes of the pathway (from extracellular stimulating factor TNF-α to intra-cellular signal transduction factors, TRAF2, TRAF6, TAK1, TAB1, IKKβ, to transcription factor p65/p50) were co-transfected with MCPIP1 or control plasmid into 293T cells and luciferase reporter gene activity of the NF-κB signaling pathway was examined. MCPIP1 repressed the activation of the NF-κB signaling pathway induced by TRAF6 and its upstream signaling molecules (Fig. 5A). Moreover, MCPIP1was significant negatively correlated with TRAF6 and TAB2 according to TCGA data (Supplementary Fig. S6A, S6B).
A Dual luciferase reporter assay of the NF-κB pathway activation in the presence or absence of MCPIP1 over-expression and different activators (n = 3). B Effect of MCPIP1 over-expression on TRAF6 ubiquitination with wildtype or mutant HA-tagged ubiquitin. C Protein and mRNA level of TRAF6 transfected with siTRAF6 (n = 3). Ctrl blank control; NC negative control. D Effect of TRAF6 over-expression with or without siTRAF6 on NF-κB dual luciferase reporter activity (n = 3). Effect of MCPIP1 over-expression with or without siTRAF6 on K63-ubiquitination of Flag-tagged TRAF6 (E) and activation of NF-κB luciferase reporter (F) (n = 3). ***p < 0.001, **p < 0.01, *p < 0.05, and n.s. no significance. Data represents mean + s.d.
As the bioinformatic analysis of TCGA data (Supplementary Fig. S4) revealed that MCPIP1 was involved in ubiquitin-like protein ligase binding, the impact of MCPIP1 on TRAF6 ubiquitylation was investigated and MCPIP1 was found to inhibit the K63-linked ubiquitylation of TRAF6 but had no significant influence on K48 ubiquitylation (Fig. 5B).
Fluorescence qPCR and Western blotting assays showed that TRAF6 was knocked down by small interfering RNA (siTRAF6) transfected into 293T cells on both mRNA and protein levels (Fig. 5C). Additionally, using the dual-luciferase reporter gene assay and the IP assay, induction of siTRAF6 dramatically attenuated the NF-κB signaling pathway activity (Fig. 5D) and TRAF6 K63-ubiquitylation level compared with negative control (Fig. 5E). In the presence of siTRAF6, cells were resistant to the inhibitory effect of MCPIP1 on both TRAF6 ubiquitylation and the pathway activity (Fig. 5E, F). These results demonstrated that MCPIP1 suppressed the NF-κB signaling pathway by inhibiting the K63-linked ubiquitylation of TRAF6.
MCPIP1 affected the proliferation and migration of CRC cells by restraining the activity of the NF-κB signaling pathway
Experimental results showed that the proliferative and migratory abilities of CRC cells were weakened following TRAF6 knockdown (HT-29/HCT116-Ctrl + siTRAF6) compared to controls (HT-29/HCT116-Ctrl, Fig. 6). This indicated that the TRAF6-activated NF-κB signaling pathway stimulated the malignant transformation of tumor cells. As shown in Fig. 6A, in contrast to cells with normal MCPIP1 expression (HT-29-Ctrl + siTRAF6), the proliferative and migratory abilities of CRC cells were partially restored when MCPIP1 and TRAF6 were simultaneously knocked down in HT-29 cells (HT-29-shRNA + siTRAF6). Overexpressing MCPIP1 from TRAF6 down-regulated HCT116 cells (HCT116-MCPIP1 + siTRAF6) further decreased the migratory and proliferative abilities of CRC cells (Fig. 6B). Therefore, it would be reasonable to postulate that MCPIP1 would be capable of influencing the migration and proliferation of CRC cells through K63-linked deubiquitylation of TRAF6 but this may not be the sole influencing factor.
In conclusion, the following hypotheses were proposed for the action of MCPIP1 in the NF-κB signaling pathway: The K63-linked ubiquitin chains of TRAF6 recruit downstream factors and activate the downstream NF-κB signaling pathway. In the cells with normal MCPIP1 expression, MCPIP1 disabled the recruitment of downstream signaling factors by repressing K63-linked ubiquitylation of TRAF6, ultimately suppressing the NF-κB signaling pathway (Supplementary Fig. S7). In CRC cells with a low MCPIP1 expression due to promoter methylation, the K63-linked ubiquitylation of TRAF6 would not be impacted, thus the transcription of the NF-κB signaling pathway would be initiated to mediate downstream activities related to cell proliferation, cell apoptosis and immune response.
Discussion
MCPIP1, the expression of which is found to be raised in monocytes treated with the pro-inflammatory cytokine MCP-1, has 599 amino acid residues and consists of an endoribonuclease domain, deubiquitylation-related domains and domains interacting with ZC3H12D [18, 19]. In the early stage of inflammation, MCPIP1 has been reported to promote the degradation of a series of inflammation-related mRNAs with translational activity (such as IL-2, IL-6, and IL-12B) or resect the terminal of miRNA precursor capable of regulating inflammatory responses to prevent the generation of mature miRNAs, thereby restraining inflammatory responses [20,21,22,23,24]. Moreover, MCPIP1 facilitates the dissociation of K48- or K63-linked ubiquitin chain in TRAFs to disrupt the conduction of the JNK or NF-κB signaling pathway, ultimately negatively regulating the inflammatory response and internal immune environment [25]. Regarding tumor-related studies, Marona et al. reported that MCPIP1 expression was reduced in renal clear cell carcinoma, consequently increasing the secretion of precursors of angiogenic factors, such as VEGF, IL-8 and CXCL2, which are prominently associated with tumor cell proliferation and tumor angiogenesis [26]. On this basis, Gorka et al. discovered that MCPIP1 overexpression increased the expression levels of cancer suppressive factors, PTEN, RECK and TIMP3 [27]. Ren et al. discovered that down-regulated MCPIP1 enhanced the proliferation, migration and invasion of osteosarcoma cells as well as promoting the pro-inflammatory cytokine IL-6 [28]. According to the report of Lu et al., MCPIP1 expression declined in breast cancer and correlated with poor patient prognosis [29].
Therefore, studies have corroborated that MCPIP1 exerts “anti-cancer” effects on a number of tumors but the mechanism of MCPIP1 action in CRC remained unknown. The present study demonstrated that the expression level of MCPIP1 was similarly lower in CRC in comparison with normal tissues and correlated with poor prognosis. The decrease was attributable to the promoter hypermethylation of MCPIP1. Furthermore, MCPIP1 was able to repress the proliferative and migratory abilities of CRC cells in vitro, promoting apoptosis and exhibiting a tumor suppressor function. Other studies also observed that MCPIP1 induced apoptosis and indicated that its proapoptotic properties are triggered by selectively enhancing the decay of antiapoptotic RNA transcripts and its involvement in NF-κB signaling inhibition. Lu et al. found by PCR Array that 25 proapoptotic genes were upregulated and 6 antiapoptotic genes were downregulated in the breast cancer cell line, MDA-MB-231, overexpressing MCPIP1. RNA immunoprecipitation further demonstrated that MCPIP1 directly binds and cleaves antiapoptotic gene transcripts, including Bcl2L1, Bcl2A1, RelB, Birc3 and Bcl3 [29]. In the Caki-1 cells of clear cell renal cell carcinoma, Ligeza et al. demonstrated that overexpression of MCPIP1 induced nuclear morphology characteristic of late apoptosis and enhanced caspase 3/7 activity [30]. Their further investigation indicated that MCPIP1 decreased the antiapoptotic BCL-2 transcript level and increased the proapoptotic Bax and Bak1 levels [27]. Suk et al. proved that MCPIP1 in T-REx-293 cells dramatically enhances TNF-α induced apoptosis by activating the caspase cascade (caspase-3, -7, -8 and -9 activation) after TNF-α treatment and that pan-caspase inhibitor and the caspase-1/−4 inhibitor could reverse this [31]. Our STRING functional enrichment analysis showed that the MCPIP1 level were associated with proapoptotic factors. We will validate their interaction in a following study.
Unresolved inflammation has been identified as a vital factor leading to the malignant transformation of cells and the occurrence of CRC. Clinical studies have revealed an increased risk of CRC in patients with inflammatory bowel disease [17], and aspirin, an anti-inflammatory drug, can prevent the occurrence and reduce postoperative CRC metastasis [32]. The mouse model of colitis associated cancer induced by the combination of mutagen azoxymethane (AOM) and the inflammatory agent dextran sodium sulphate (DSS) has been widely applied to investigate the pathogenesis of CRC [33]. As a crucial signaling pathway mediating inflammatory responses, the NF-κB signaling pathway is abnormally activated in CRC. Sakamoto et al. detected NF-κB activation in 40% CRC tissues and 67% of CRC cell lines [34]. Lin et al. reported that patients with KRAS mutations (detected in 35–40% of CRC cases [35]) had a higher activity of NF-κB signaling than those with wildtype KRAS and these patients had lower survival and poorer response to first-line treatment [36]. The downstream genes targeted by the NF-κB signaling pathway have been related to many tumor processes, including apoptosis, proliferation, invasion, angiogenesis and inflammatory responses (tumor microenvironment) [14, 37]. Moreover, activated NF-κB contributes to the resistance to 5-FU and camptothecin, two of the most commonly used chemotherapeutic agents in CRC treatment [34, 38]. Thus, finding a strategy for the NF-κB inhibition is of great importance in CRC treatment.
The p50/RELA(p65) nuclei translocation is a crucial step for NF-κB signaling activation [39]. Suk et al. have proved that the wildtype MCPIP1 but not the MCPIP1-D141N mutant (with deficient RNase and deubiquitinase activity [25, 40]) suppressed NF-κB activation, evidenced by inhibition of IκB kinase (IKK) phosphorylation, IKK activity, IκB degradation and p65 translocation to nuclei [31]. The reduction of p65 nuclei translocation by MCPIP1 expression has also been shown by other studies [25, 41, 42]. The luciferase reporter assay shown in Fig. 5F validated that MCPIP1 greatly inhibited the NF-κB pathway activity but when siTRAF6 was applied (which caused resistance to the inhibitory effect of MCPIP1 on TRAF6 K63-ubiquitylation, confirmed in Fig. 5E), the inhibition effect was diminished. We concluded that MCPIP1 suppressed the NF-κB signaling pathway by inhibiting the K63-linked ubiquitylation of TRAF6. In the present study, we showed that the proliferative and migratory abilities of HT-29 and HCT116 cells were weakened following TRAF6 knockdown, indicating that the TRAF6-activated NF-κB signaling pathway could stimulate the malignant transformation of tumor cells. In contrast to HT-29 and HCT116 cells with normal MCPIP1 and knockdown of TRAF6, the proliferative and migratory abilities of cells were partially restored in HT-29 cells with repressed MCPIP1 while the abilities of HCT116 cells were further decreased with overexpressed MCPIP1. Therefore, we postulate that MCPIP1 inhibits the migration and proliferation of CRC cells through K63-linked deubiquitylation of TRAF6 but this de-TRAF6-K63Ub may not be the sole influencing factor.
The accumulation of cytosine-phosphate-guanine (CpG) dinucleotides, also known as CpG islands, occurs in a great majority of human genes at the promoter regions. Changes in CpG island methylation affect the transcription of multiple genes through inhibiting specific transcription factors binding to DNA or recruiting mediators of chromatin remodeling (e.g., histone-modifying enzymes) [43,44,45]. DNA methylation have displayed great potential in diagnosis, prognosis and treatment of cancer. For instance, methylation levels of MGMT promoter regions are associated with the sensitivity to temozolomide in glioma [46]. Investigating the methylation status of SEPT9 presents high sensitivity and specificity for CRC detection and prognosis prediction [47]. In mammals, DNA methylation patterns are routinely established and maintained by three DNA methyltransferases (DNMT), including DNMT1, DNMT3A and DNMT3B [48]. In our study, we found that the decreased expression of MCPIP1 was caused by its promoter hypermethylation, furthermore, correlation analysis showed that DNMT3A was the most negatively correlated with ZC3H12A. Various DNA methyltransferase inhibitors have been developed to reactivate silenced genes caused by aberrant hypermethylated promoter regions. The nucleoside inhibitors of DNMT, azacytidine, decitabine and Zebularine, are cytosine analogs that can be incorporated into the DNA in place of cytosine and trap DNMTs, resulting in interference with the methylation and inducing proteosomal degradation of DNMTs [49, 50]. Azacytidine and decitabine have been approved for the treatment of hematologic malignancies, particularly acute myelogenous leukemia and myelodysplastic syndrome [51, 52]. A phase I/II study of decitabine in combination with panitumumab in 20 patients with wild-type KRAS metastatic CRC showed that two patients (10 %) had a partial response and ten patients had stable disease with a tolerable safety profile [53]. Other clinical trials of DNA methyltransferase inhibitors have been explored in CRC: azacytidine plus pembrolizumab [54]/entinostat [55]/capecitabine and oxaliplatin [56], an oral hypomethylating agent CC-486 plus durvalumab [57]. Thus, the development of novel agents targeting methyltransferase which is responsible for ZC3H12A promoter hypermethylation may yield novel therapeutic strategies for the treatment of CRC. The lack of detailed mechanisms of ZC3H12A promoter hypermethylation, such as the exact methyltransferase, and a xenograft model in vivo, are limitations of our study which will be explored in the following work.
In this study, we demonstrated that MCPIP1 suppressed the activity of the NF-κB signaling pathway but generally has low expression in CRC. In the future, a prognostic prediction tool based on the MCPIP1 level may be developed. A method for up-regulating the expression of MCPIP1, such as drugs targeting the methyltransferase responsible for the ZC3H12A promoter hypermethylation, or other treatments that can demethylate the promoter region to restore gene transcription to increase endogenous MCPIP1 level can be considered. In addition, treatments for overexpressing exogenous MCPIP1, for example, the introduction of the MCPIP1 protein (similar to Endostar, a recombinant human endostatin [58]), or an adenoviral vector loading the ZC3H12A coding DNA sequence into the tumor (similar to Gendicine and Advexin, replication-defective recombinant human p53 adenoviral vectors [59, 60]), would seem to have potential as individualized CRC treatments. The above strategies may guide clinical treatments for CRC in the future, especially where a decreased MCPIP1and activated NF-κB signaling pathway is present.
Conclusions
In the present study, MCPIP1 repressed the NF-κB signaling pathway by negatively regulating the K63-linked ubiquitylation of TRAF6, thereby inhibiting the proliferation and migration of CRC cells. The decreased expression of MCPIP1 in CRC tumor tissues, which was related to poor prognosis, was caused by promoter hypermethylation.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32.
Qin S, Li J, Wang L, Xu J, Cheng Y, Bai Y, et al. Efficacy and tolerability of first-line Cetuximab Plus Leucovorin, Fluorouracil, and Oxaliplatin (FOLFOX-4) Versus FOLFOX-4 in patients with RAS wild-type metastatic colorectal cancer: the open-label, randomized, Phase III TAILOR Trial. J Clin Oncol. 2018;36:3031–9.
Andre T, Shiu KK, Kim TW, Jensen BV, Jensen LH, Punt C, et al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N. Engl J Med. 2020;383:2207–18.
Singh V, Gupta D, Arora R. NF-kB as a key player in regulation of cellular radiation responses and identification of radiation countermeasures. Discoveries. 2015;3:e35.
Mirzaei S, Zarrabi A, Hashemi F, Zabolian A, Saleki H, Ranjbar A, et al. Regulation of Nuclear Factor-KappaB (NF-kappaB) signaling pathway by non-coding RNAs in cancer: Inhibiting or promoting carcinogenesis? Cancer Lett. 2021;509:63–80.
Harhaj EW, Dixit VM. Regulation of NF-κB by deubiquitinases. Immunol Rev. 2012;246:107–24.
Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem. 2009;78:769–96.
Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–86.
Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 2007;315:201–5.
Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-κB regulatory pathways. Annu Rev Biochem. 2009;78:769–96.
Martinez-Forero I, Rouzaut A, Palazon A, Dubrot J, Melero I. Lysine 63 polyubiquitination in immunotherapy and in cancer-promoting inflammation. Clin Cancer Res. 2009;15:6751–7.
Clevers H. At the crossroads of inflammation and cancer. Cell. 2004;118:671–4.
Maeda S, Omata M. Inflammation and cancer: role of nuclear factor-kappaB activation. Cancer Sci. 2008;99:836–42.
Zhang T, Wang H, Han L. Expression and clinical significance of tumor necrosis factor receptor-associated Factor 6 in patients with colon cancer. Iran Red Crescent Med J 2016;18:e23931.
Sun H, Li X, Fan L, Wu G, Li M, Fang J. TRAF6 is upregulated in colon cancer and promotes proliferation of colon cancer cells. Int J Biochem Cell Biol. 2014;53:195–201.
Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507.
Bidzhekov K, Zernecke A, Weber C. MCP-1 induces a novel transcription factor with proapoptotic activity. Circ Res. 2006;98:1107–9.
Zhou L, Azfer A, Niu J, Graham S, Choudhury M, Adamski FM, et al. Monocyte chemoattractant protein-1 induces a novel transcription factor that causes cardiac myocyte apoptosis and ventricular dysfunction. Circ Res. 2006;98:1177–85.
Garg AV, Amatya N, Chen K, Cruz JA, Grover P, Whibley N, et al. MCPIP1 Endoribonuclease activity negatively regulates Interleukin-17-mediated signaling and inflammation. Immunity 2015;43:475–87.
Mizgalska D, Wegrzyn P, Murzyn K, Kasza A, Koj A, Jura J, et al. Interleukin-1-inducible MCPIP protein has structural and functional properties of RNase and participates in degradation of IL-1beta mRNA. Febs j. 2009;276:7386–99.
Huang S, Liu S, Fu JJ, Tony Wang T, Yao X, Kumar A, et al. Monocyte chemotactic protein-induced Protein 1 and 4 form a complex but act independently in regulation of Interleukin-6 mRNA degradation. J Biol Chem. 2015;290:20782–92.
Xu J, Peng W, Sun Y, Wang X, Xu Y, Li X, et al. Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase. Nucleic Acids Res. 2012;40:6957–65.
Suzuki HI, Arase M, Matsuyama H, Choi YL, Ueno T, Mano H, et al. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol Cell. 2011;44:424–36.
Liang J, Saad Y, Lei T, Wang J, Qi D, Yang Q, et al. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-kappaB signaling. J Exp Med. 2010;207:2959–73.
Marona P, Gorka J, Mazurek Z, Wilk W, Rys J, Majka M, et al. MCPIP1 downregulation in clear cell renal cell carcinoma promotes vascularization and metastatic progression. Cancer Res. 2017;77:4905–20.
Gorka J, Marona P, Kwapisz O, Rys J, Jura J, Miekus K. The anti-inflammatory protein MCPIP1 inhibits the development of ccRCC by maintaining high levels of tumour suppressors. Eur J Pharm. 2020;888:173591.
Ren Z, He M, Shen T, Wang K, Meng Q, Chen X, et al. MiR-421 promotes the development of osteosarcoma by regulating MCPIP1 expression. Cancer Biol Ther. 2020;21:231–40.
Lu W, Ning H, Gu L, Peng H, Wang Q, Hou R, et al. MCPIP1 selectively destabilizes transcripts associated with an antiapoptotic gene expression program in breast cancer cells that can elicit complete tumor regression. Cancer Res. 2016;76:1429–40.
Ligeza J, Marona P, Gach N, Lipert B, Miekus K, Wilk W, et al. MCPIP1 contributes to clear cell renal cell carcinomas development. Angiogenesis 2017;20:325–40.
Suk FM, Chang CC, Sun PC, Ke WT, Chung CC, Lee KL, et al. MCPIP1 Enhances TNF-alpha-mediated apoptosis through downregulation of the NF-kappaB/cFLIP Axis. Biology. 2021;10:655.
Drew DA, Chan AT. Aspirin in the prevention of colorectal neoplasia. Annu Rev Med. 2021;72:415–30.
De Robertis M, Massi E, Poeta ML, Carotti S, Morini S, Cecchetelli L, et al. The AOM/DSS murine model for the study of colon carcinogenesis: From pathways to diagnosis and therapy studies. J Carcinog. 2011;10:9.
Sakamoto K, Maeda S, Hikiba Y, Nakagawa H, Hayakawa Y, Shibata W, et al. Constitutive NF-kappaB activation in colorectal carcinoma plays a key role in angiogenesis, promoting tumor growth. Clin Cancer Res. 2009;15:2248–58.
Bos JL, Fearon ER, Hamilton SR, Verlaan-de Vries M, van Boom JH, van der Eb AJ, et al. Prevalence of ras gene mutations in human colorectal cancers. Nature 1987;327:293–7.
Lin G, Zheng XW, Li C, Chen Q, Ye YB. KRAS mutation and NF-κB activation indicates tolerance of chemotherapy and poor prognosis in colorectal cancer. Dig Dis Sci. 2012;57:2325–33.
Soleimani A, Rahmani F, Ferns GA, Ryzhikov M, Avan A, Hassanian SM. Role of the NF-kappaB signaling pathway in the pathogenesis of colorectal cancer. Gene 2020;726:144132.
Wang CY, Cusack JC Jr., Liu R, Baldwin AS Jr. Control of inducible chemoresistance: enhanced anti-tumor therapy through increased apoptosis by inhibition of NF-kappaB. Nat Med. 1999;5:412–7.
Zhang Q, Lenardo MJ, Baltimore D. 30 years of NF-kappaB: a blossoming of relevance to human pathobiology. Cell 2017;168:37–57.
Wilamowski M, Gorecki A, Dziedzicka-Wasylewska M, Jura J. Substrate specificity of human MCPIP1 endoribonuclease. Sci Rep. 2018;8:7381.
Roy A, Zhang M, Saad Y, Kolattukudy PE. Antidicer RNAse activity of monocyte chemotactic protein-induced protein-1 is critical for inducing angiogenesis. Am J Physiol Cell Physiol. 2013;305:C1021–32.
Han S, Li Z, Ji P, Jia Y, Bai X, Cai W, et al. MCPIP1 alleviated lipopolysaccharide-induced liver injury by regulating SIRT1 via modulation of microRNA-9. J Cell Physiol. 2019;234:22450–62.
Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–34.
Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–91.
Cao J, Yan Q. Cancer epigenetics, tumor immunity, and immunotherapy. Trends Cancer. 2020;6:580–92.
Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl J Med. 2005;352:997–1003.
Song L, Yu H, Jia J, Li Y. A systematic review of the performance of the SEPT9 gene methylation assay in colorectal cancer screening, monitoring, diagnosis and prognosis. Cancer Biomark. 2017;18:425–32.
Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–20.
Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst. 2005;97:1498–506.
Hu C, Liu X, Zeng Y, Liu J, Wu F. DNA methyltransferase inhibitors combination therapy for the treatment of solid tumor: mechanism and clinical application. Clin Epigenetics. 2021;13:166.
Kantarjian H, Issa JP, Rosenfeld CS, Bennett JM, Albitar M, DiPersio J, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer 2006;106:1794–803.
Silverman LR, Demakos EP, Peterson BL, Kornblith AB, Holland JC, Odchimar-Reissig R, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002;20:2429–40.
Garrido-Laguna I, McGregor KA, Wade M, Weis J, Gilcrease W, Burr L, et al. A phase I/II study of decitabine in combination with panitumumab in patients with wild-type (wt) KRAS metastatic colorectal cancer. Invest N. Drugs. 2013;31:1257–64.
Kuang C, Park Y, Augustin RC, Lin Y, Hartman DJ, Seigh L, et al. Pembrolizumab plus azacitidine in patients with chemotherapy refractory metastatic colorectal cancer: a single-arm phase 2 trial and correlative biomarker analysis. Clin Epigenetics. 2022;14:3.
Azad NS, El-Khoueiry A, Yin J, Oberg AL, Flynn P, Adkins D, et al. Combination epigenetic therapy in metastatic colorectal cancer (mCRC) with subcutaneous 5-azacitidine and entinostat: a phase 2 consortium/stand up 2 cancer study. Oncotarget 2017;8:35326–38.
Overman MJ, Morris V, Moinova H, Manyam G, Ensor J, Lee MS, et al. Phase I/II study of azacitidine and capecitabine/oxaliplatin (CAPOX) in refractory CIMP-high metastatic colorectal cancer: evaluation of circulating methylated vimentin. Oncotarget 2016;7:67495–506.
Taylor K, Loo Yau H, Chakravarthy A, Wang B, Shen SY, Ettayebi I, et al. An open-label, phase II multicohort study of an oral hypomethylating agent CC-486 and durvalumab in advanced solid tumors. J Immunother Cancer. 2020;8:e000883.
Ling Y, Yang Y, Lu N, You QD, Wang S, Gao Y, et al. Endostar, a novel recombinant human endostatin, exerts antiangiogenic effect via blocking VEGF-induced tyrosine phosphorylation of KDR/Flk-1 of endothelial cells. Biochem Biophys Res Commun. 2007;361:79–84.
Senzer N, Nemunaitis J. A review of contusugene ladenovec (Advexin) p53 therapy. Curr Opin Mol Ther. 2009;11:54–61.
Zhang WW, Li L, Li D, Liu J, Li X, Li W, et al. The first approved gene therapy product for cancer Ad-p53 (Gendicine): 12 years in the clinic. Hum Gene Ther. 2018;29:160–79.
Funding
This work is supported by the grants from the National Natural Science Foundation of China (No. 81702312) and the Natural Science Foundation of Guangdong Province (No. 2017A030310489).
Author information
Authors and Affiliations
Contributions
WY, WH, and YL contributed to the conceptualization and design of the experiment. WY, JR, ZZ, and AL contributed to the methodology, investigation, and data curation. YC and YL contributed to the software, validation, and visualization. WY also contributed to the supervision and funding acquisition. All authors contributed to the data analysis, paper writing, and approved submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ye, W., Cui, Y., Rong, J. et al. MCPIP1 Suppresses the NF-κB Signaling Pathway Through Negative Regulation of K63-Linked Ubiquitylation of TRAF6 in Colorectal Cancer. Cancer Gene Ther 30, 96–107 (2023). https://doi.org/10.1038/s41417-022-00528-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-022-00528-4
- Springer Nature America, Inc.
This article is cited by
-
Network pharmacology and experimental verification study on the mechanism of Hedyotis diffusa Willd in treating colorectal cancer
Naunyn-Schmiedeberg's Archives of Pharmacology (2024)