Abstract
Oncolytic reovirus administration has been well tolerated by cancer patients in clinical trials. However, its anti-cancer efficacy as a monotherapy remains to be augmented. We and others have previously demonstrated the feasibility of producing replication-competent reoviruses expressing a heterologous transgene. Here, we describe the production of recombinant reoviruses expressing murine (mm) or human (hs) GM-CSF (rS1-mmGMCSF and rS1-hsGMCSF, respectively). The viruses could be propagated up to 10 passages while deletion mutants occurred only occasionally. In infected cell cultures, the secretion of GM-CSF protein (up to 481 ng/106 cells per day) was demonstrated by ELISA. The secreted mmGM-CSF protein was functional in cell culture, as demonstrated by the capacity to stimulate the survival and proliferation of the GM-CSF-dependent dendritic cell (DC) line D1, and by its ability to generate DCs from murine bone marrow cells. Importantly, in a murine model of pancreatic cancer we found a systemic increase in DC and T-cell activation upon intratumoral administration of rS1-mmGMCSF. These data demonstrate that reoviruses expressing functional GM-CSF can be generated and have the potential to enhance anti-tumor immune responses. The GM-CSF reoviruses represent a promising new agent for use in oncolytic virotherapy strategies.
Similar content being viewed by others
Introduction
Oncolytic viruses represent a promising tool in experimental anti-cancer treatment strategies. The features that determine the malignancy and outgrowth of cancer cells are often associated with an increased sensitivity to viral infections [1]. Many of the current strategies to improve the potential of oncolytic viruses are focused on the modulation and stimulation of a long-lasting adaptive anti-cancer immune response. T-VEC (Talimogene laherparepvec), the first oncolytic virus that was approved for treatment of a subset of melanoma patients, is a Herpes Simplex Virus that has been genetically modified to express GM-CSF [2, 3]. The approval of T-VEC may pave the way for clinical application of other oncolytic viruses.
Mammalian orthoreovirus T3D, hereafter called reovirus, is a non-enveloped virus carrying a segmented double-stranded RNA genome with a total size of 23.5kB. Reovirus has the natural preference to replicate in and kill tumor cells, and therefore is studied as an oncolytic agent for a variety of cancer types [4]. Treatment with the reovirus product Reolysin (Pelareorep) is well tolerated, but the efficacy and effects on progression-free and overall survival remain to be augmented.
One of the approaches to increase the anti-cancer potency of oncolytic viruses is through incorporating therapeutic transgenes in the virus backbone. Genetic modification of dsRNA viruses can be challenging but the development of a plasmid-based reverse genetics system has allowed researchers worldwide to make adaptations to the reovirus genome [5]. We have previously demonstrated that this system can be used to incorporate a small transgene in the reovirus backbone. We successfully replaced the S1 segment-encoded head domain of the reovirus spike protein σ1 that is responsible for binding the reovirus receptor JAM-A, for the small fluorescent protein iLOV [6], generating the recombinant reovirus rS1-iLOV. The mutation that causes the G196R substitution in the spike protein that was identified in the jin-3 reovirus mutant that is able to infect a variety of cancer cell lines that lack the JAM-A receptor [7], was incorporated in the same segment to allow for sialic-acid-dependent but JAM-A-independent viral entry. More recently, we published the generation and characterization of two additional recombinant reoviruses, rS1-E4orf4 and rS1-RFA [8]. In the rS1-E4orf4 virus the S1 head domain was replaced for a gene encoding E4orf4, an adenovirus-derived protein that is involved in the induction of tumor-cell selective non-classical apoptosis [9, 10]. This cell death induction partially depends on E4orf4 binding to phosphatase 2A (PP2A). The other reovirus, rS1-RFA, encodes a double mutant (RFA) of E4orf4 that is unable to bind PP2A. We showed that truncation of σ1 seems to result in improved tumor cell killing, but that the expression of E4orf4 does not enhance the oncolytic potential [8]. Based on these results, we speculated that it would be a more effective approach to generate a reovirus that leads to the synthesis of a secreted immune-modulating protein instead of a protein that acts in cis, i.e., only in the infected cell.
Here, we describe the generation and characterization of two reoviruses, rS1-mmGMCSF and rS1-hsGMCSF which harbor the codons for murine or human GM-CSF, respectively. GM-CSF is widely used to generate dendritic cells (DCs) in vitro. It promotes the survival and function of both monocyte-derived DCs [11] as well as CD8α+ DCs [12]. By enhancing dendritic cell (DC) maturation, and stimulating cross-presentation of tumor antigens, GM-CSF can boost subsequent anti-tumor immune responses. We demonstrate the secretion and the functionality of GM-CSF upon viral infection in vitro. Importantly, treatment of mice bearing pancreatic tumors with rS1-mmGMCSF systemically increased the percentage of activated DCs and effector T-cells compared to treatment with the control reovirus rS1-iLOV. These results show the successful production of a recombinant reovirus expressing a functional transgene that could be of clinical value.
Materials and methods
Cell lines
The human embryonic retinoblast-derived cell line “911” [13] was cultured in high-glucose Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen, Breda, the Netherlands), supplemented with penicillin, streptomycin (pen-strep) and 8% fetal bovine serum (FBS) (Invitrogen, Breda, the Netherlands). The polyclonal 911scFvHis cell line was established by lentiviral transduction of 911 cells with the pLV-scFvHis-IRES-Neo vector as described before [14]. The KPC3 cell line are low-passage cells derived from a primary tumor in KPC mice [15] and were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM) (Gibco, Life Technologies, Thermo Fisher Scientific) supplemented with 8% FCS (Gibco, Life Technologies, Thermo Fisher Scientific, cat. nr 10270-106), pen-strep and glutamine. The T7 RNA polymerase-expressing cell line BSR-T7 [16] was kindly provided by K. Conzelmann. The 911scFvHis and BSR-T7 cell lines were cultured in DMEM containing 8% FBS, pen-strep and 400 µg/ml G418. The growth factor-dependent dendritic cell (DC) line D1 was cultured in IMDM (Lonza) containing 50 µM β-mercaptoethanol (β-ME) (Sigma-Aldrich), 2 mM GlutaMAX (Thermo Fisher Scientific), 100IU/ml penicillin (Sandoz), and 10% FCS (Sigma-Aldrich), supplemented with 30% conditioned supernatant derived from murine GM-CSF producing NIH/3T3 cells (hereafter referred to as R1 sup) [17, 18]. All cell lines were cultured in a 5% CO2 atmosphere at 37 °C. All cell lines were routinely checked to be free of mycoplasma contamination.
Reoviruses
The wild-type T3D strain R124 was isolated by plaque purification from a heterogeneous reovirus T3D stock and propagated as described previously [7]. The jin-3 mutant reovirus was isolated from U118MG cells upon infection with wild-type T3D [7]. The genomes of the R124 and jin-3 viruses were fully sequenced [7]. Infectious virus titers were quantified by plaque assays read-out on 911 cells. All experiments were performed using CsCl-purified virus stocks, unless indicated otherwise. For purification, a freeze-thaw lysate containing virus particles was incubated with 0,1% Triton (Sigma-Aldrich, Zwijndrecht, the Netherlands) and 25 units/ml Benzonase (Santa Cruz, Bio-Connect B.V. Huissen, the Netherlands) for 15 min on ice followed by 15 min at 37 °C. After two extractions with Halotec CL10 (FenS B.V. Goes, the Netherlands), the cleared lysate was loaded onto a discontinuous CsCl gradient (1.45 and 1.2 g/cm3 in phosphate-buffered saline (PBS)). After centrifugation in a SW28 rotor (Beckman Coulter, Woerden, the Netherlands) at 95,000 × g for 4 h at 16 °C, the lower band containing the infectious particles was harvested and desalted in an Amicon Ultra 100 K device according to the manufacturer’s protocol (Millipore, Merck Chemicals BV, Amsterdam, the Netherlands). The CsCl-purified reoviruses were recovered in reovirus storage buffer (RSB: 10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2 • 6 H2O, 5% sucrose), aliquoted and stored at 4 °C until use. The amount of particles was calculated based on the OD260 values. The recombinant reoviruses expressing transgenes are described in a separate Materials and Methods section.
Plasmid constructs
The S1 segments encoding the murine or (codon-optimized) human GM-CSF gene were designed in silico and the DNA was synthesized by Eurofins Genomics (Ebersberg, Germany). Prior to the GM-CSF sequences, a porcine teschovirus-1 2A (P2A) sequence was incorporated. Behind the GM-CSF sequences, the S1 3′ untranslated region (UTR) consisted of the C-box and the remaining 3′ end sequence of S1. The P2A-GMCSF sequences were flanked by a 5′ NcoI site and inserted in the pEX-A2 plasmid by Eurofins Genomics. This NcoI site allowed the exchange of the NcoI-SacII 614 bp fragment (containing sequences P2A-GM-CSF-3′UTR S1) from the pEX-GMCSF constructs with the NcoI-SacII 584 bp fragment (removing the P2A-iLOV-3′UTR sequences) of plasmid pBT7-S1His-2A-iLOV [6], using standard cloning techniques. This results in the plasmids: pBacT7-S1His-2A-mmGMCSF, pBacT7-S1His-2A-hsGMCSF and pBacT7-S1His-2A-hsGMCSFco.
The structure of the S1-His-2A-mmGMCSF (1407 bp) and S1His-2A-hsGMCSF(co) (1416 bp) constructs is shown in Fig. 1: they contain (1) nt 1-768 of the reovirus jin-3 S1 segment, including the 5′ UTR, the entire σ1 s ORF, and the codons for the first 252 amino acids of the jin-3 σ1 protein which harbors the G196R change near the sialic-acid binding region; (2) the codons (18 bp) for a 6x His-tag that are in frame with the σ1 ORF; (3) the codons (66 + 3 additional bp) for the P2A sequence; (4) the sequence encoding the murine (423 bp) or (codon-optimized) human (432 bp) GM-CSF protein; (5) a stop codon; and (6) the 3′ UTR (nt 1219–1416) of the reovirus T3D S1 segment, including the C-box but not the A- and B-boxes. The A-, B-, and C-boxes have been implicated in packaging of the positive RNA strand in the viral capsid [19].
Structure of the S1 segments in the various recombinant reoviruses. The S1 segments of the recombinant reoviruses contain nt 1-768 of the reovirus jin-3 S1 segment, including the mutation underlying the G196R change near the sialic-acid binding region; the codons for the 6x His-tag in frame with the σ1 ORF; the codons for the porcine teschovirus-1 2A (P2A) sequence; the transgene encoding the heterologous protein—iLOV, E4orf4, RFA, murine GM-CSF (mmGM-CSF), human GM-CSF (hsGM-CSF), or codon-optimized hsGM-CSF (hsGM-CSFco)—including a stop codon; and the 3′ UTR (nt 1219–1416) of the reovirus T3D S1 segment. In contrast to S1His-2A-iLOV, -E4orf4, and -RFA, the 3′ UTR region in the GM-CSF encoding S1 segments contains the C-box but not the A- and B-boxes. The occurrence or absence of deletion mutants is indicated by Y (yes) or N (no), respectively. The use of the P2A sequence results in the separate synthesis of a σ1-His-tagged spike protein and the transgene-encoded protein
Recombinant reoviruses
The reovirus expressing iLOV (rS1His-2A-iLOV, hereafter referred to as rS1-iLOV) was previously generated as described [6]. Reoviruses expressing murine (rS1His-2A-mmGMCSF) or human GM-CSF (rS1His-2A-hsGMCSF) are here referred to as rS1-mmGMCSF and rS1-hsGMCSF. The reovirus expressing the codon-optimized variant of hsGM-CSF (rS1His-2A-hsGMCSFco) is referred to as rS1-hsGMCSFco. All three recombinant viruses were constructed by transfection of 2.5 × 105 BSR-T7 cells with the following plasmids: pT7-M2-S3-S4T3D (Addgene, plasmid #33302), pT7-L3-M1T3D (Addgene, plasmid #33301), pT7-L2-M3T3D (Addgene, plasmid #33300), pT7-L1T1L (Addgene, plasmid #33286), and pBacT7-S1His-2A-mm/hsGMCSF (2 µg per plasmid, total of 10 µg DNA). TransIT-LT1 (Mirus, Sopachem BV, Ochten, the Netherlands) was used as a transfection reagent. The cells were harvested 72 h post transfection, lyzed by three cycles of freezing and thawing, and subsequently added to 911scFvHis cells to promote amplification of the produced viruses. The 911scFvHis cells were harvested two weeks post infection, and lyzed by three cycles of freezing and thawing. Individual virus clones were obtained by limiting dilution of the reoviruses originated from the first passage on 911scFvHis cells. RNA was isolated 48 h post infection from cells infected with the individual clones using the Absolutely RNA miniprep kit (Stratagene, Agilent Technologies, Amstelveen, the Netherlands). An RT-PCR was performed to analyze the stability of the S1 segments with the following primers: S1EndR (GATGAAATGCCCCAGTGC) for S1 cDNA synthesis, and S1For (GCTATTGGTCGGATGGATCCTCG) and S1EndR for the S1 PCR, using Pfu polymerase (Fermentas, Thermo Fisher Scientific, Landsmeer, the Netherlands). The resulting PCR product was analyzed by gel electrophoresis, and further purified for analysis by Sanger Sequencing at the Leiden Genome Technology Center (Leiden University Medical Center, Leiden, the Netherlands). The clones with an S1 segment containing GM-CSF were further propagated for up to 10 passages to study the stability of the clones. Infectious virus titers were determined by TCID50 assays on 911 cells. All experiments were performed with CsCl-purified virus stocks, unless indicated otherwise. Virus batches were stored in RSB supplemented with 0.1% BSA (RSB/0.1%BSA).
GM-CSF ELISAs
911scFvHis cells were infected with rS1-mmGMCSF, rS1-hsGMCSFco or jin-3 at an MOI of 1. After several time points, the levels of murine GM-CSF (mmGM-CSF) or human GM-CSF (hsGM-CSF) were determined using Quantikine® ELISA immunoassay kits (R&D Systems, Abingdon, United Kingdom) according to the manufacturer’s instructions. The experiments were repeated twice to validate the results obtained. For the in vivo experiments, levels of mmGM-CSF in serum and tumor tissue from treated C57BL/6 mice were determined using the same kit.
mmGM-CSF functionality in vitro
To study the functionality of rS1-mmGMCSF-induced secreted murine GM-CSF in vitro, we used bone marrow cells from C57BL/6 mice. Supernatants from rS1-mmGMCSF or rS1-iLOV (negative control) infected 911 cells were pre-exposed to short-wave UV irradiation for 15 min to inactivate any virus that may be present. GM-CSF concentrations were determined by ELISA. Freshly isolated bone marrow cells were cultured in IMDM containing 50 µM β-ME, 2 mM GlutaMAX, 100IU/ml penicillin, and 10% FCS, supplemented with rS1-mmGMCSF sup at 5 ng/ml GM-CSF or an equal volume of negative control rS1-iLOV sup. As a positive control, R1 sup was used at the same concentration of GM-CSF [17, 18]. Medium was refreshed at days 3 and 6. Finally, after 9 days of culturing, the cells were analyzed by flow cytometry. Used antibodies were anti-MHCII (clone M5/114.15.2, eBioscience, cat. nr. 12-5321-82), anti-CD11b (clone M1/70, eBioscience, cat. nr. 48-0112-82), anti-CD11c (clone N418, eBioscience, cat. nr. 47-0114-82), and anti-CD86 (clone GL1, eBioscience, cat. nr. 17-0862-82). The experiment was repeated twice to confirm the results.
Alternatively, the DC line D1 [17] was exposed to various concentrations of GM-CSF. Survival and proliferation of this cell line is dependent on the presence of growth factors such as GM-CSF. As for the experiment with bone marrow cells, supernatants from rS1-mmGMCSF or rS1-iLOV (negative control) infected 911 cells were pre-treated with short-wave UV irradiation for 15 min to inactivate any reovirus that may be present. The absence of infectious virus was confirmed by TCID50 assay on 911 cells (≤10pfu/ml). GM-CSF concentrations were determined by ELISA, and D1 cells were exposed to culture medium containing rS1-mmGMCSF at 1 or 5 ng/ml GM-CSF. As a positive control, D1 cells were treated with medium containing R1 sup at the same concentrations. Cells were monitored daily for viability by light microscopy, and a WST-1 cell proliferation assay (Roche, Woerden, the Netherlands) was performed after 3 days according to the manufacturer’s protocol. The experiment was repeated twice to validate the results obtained.
Animal experiments
The mouse experiments were controlled by the animal welfare committee (IvD) of the Leiden University Medical Center and approved by the national central committee of animal experiments (CCD) under the permit number AVD116002015271, in accordance with the Dutch Act on Animal Experimentation and EU Directive 2010/63/EU. Appropriate group sizes were determined based on variation observed in previous experiments. Mice were divided in groups with equal tumor volumes. Mice were not randomized. Treatment was not blinded. The 8–10 weeks aged male C57BL/6 mice (Charles River, L’Arbresle Cedex, France) were inoculated subcutaneously with 1 × 105 low-passage KPC3 cells in PBS/0.1%BSA in a total volume of 100 µl per injection. Tumor volume was measured twice weekly using a caliper and calculated (tumor volume = width × length × height). At tumor volume of 50–200 mm3 the tumors were treated intratumorally with 1 × 107 pfu of rS1-iLOV or rS1-mmGMCSF, or as control, reovirus storage buffer (RSB), all in a total volume of 30 µl per injection. All treatments were repeated on day 2 and day 3 after the first treatment. Six days after the first treatment, mice were sacrificed and tumors, tumor-draining lymph nodes (TDLNs), and spleens were collected for analyses.
RT-qPCR analysis of tumors
To check for the presence of viral genome product S4, and virus-induced CXCL10 and/or mmGM-CSF expression in the tumors, reverse transcription-quantitative PCR (RT-qPCR) analysis was performed. A proportion of the tumors was snap-frozen in liquid nitrogen, and stored at −80 °C until use. Of each tumor, 10–30 mg was taken up in 500 µl LBP lysis buffer (NucleoSpin Plus kit, Macherey-Nagel™) in an RNAse-free Eppendorf tube. The tissue was disrupted by addition of a sterile 5 mm stainless steel bead and placing the tubes in a TissueLyser system for 5 min at high-speed 30 Hz. Subsequently, RNA isolation was performed according to the NucleoSpin Plus protocol with the minor modification that all centrifugation steps were performed at 10,000 rpm. For S4 analysis, 150 ng of RNA was used to generate cDNA with primer S4EndR (GATGAATGAAGCCTGTCCCACGTCA). For CXCL10 and mmGM-CSF analysis, 500 ng of RNA was used to generate cDNA using the High-Capacity RNA-to-cDNATM Kit (Thermo Fisher Scientific, cat. nr. 4387406) according to the manufacturer’s protocol. The qPCRs were performed with Biorad IQTM SYBR ® Green Supermix (Cat. nr. 170-8886) and the following primer sets: S4Q Forw (CGCTTTTGAAGGTCGTGTATCA) and S4Q Rev (CTGGCTGTGCTGAGATTGTTTT), CXCL10 Forw (ACGAACTTAACCACCATCT) and CXCL10 Rev (TAAACTTTAACTACCCATTGATACATA), GM-CSF Forw (CTAACATGTGTGCAGACCCG) and GM-CSF Rev (GGCTGTCTATGAAATCCGCA), household gene MZT2 For (TCGGTGCCCATATCTCTGTC) and MZT2 Rev (CTGCTTCGGGAGTTGCTTTT), and household gene PTP4A2 For (AGCCCCTGTGGAGATCTCTT) and PTP4A2 Rev (AGCATCACAAACTCGAACCA). The used PCR program contained the following steps: (1) 3 min at 95 °C; (2) 40× a) 10 s at 96 °C, b) 30 s at 60 °C, and c) plate read; (3) 10 s at 95 °C; (4) Melt curve 65–95 °C with increment of 0.2 °C every 10 s, and plate read. Expression of CXCL10 and mmGM-CSF genes was normalized to reference genes MZT2 and PTP4A2 in Bio-rad CFX Manager 3.1 software. Reovirus S4 10log copy numbers were determined based on a standard curve, generated with serial dilutions of plasmid pcDNA_S4. Calculations of copy numbers were performed according to a described formula for rotavirus NSP3 quantification [20]. All samples were measured in duplo.
Flow cytometric analysis of tissues
To examine the effect of treatment on the immune composition of the tumors, TDLNs, and spleens, these tissues were analyzed by flow cytometry. The tumor tissue was cut into pieces and Liberase TL (Roche, 05401020001) was added. After 15 min incubation at 37 °C, the reaction was stopped by the addition of medium. The mixture was transferred to a cell strainer and gently dissociated until a single cell suspension was obtained. Cells were incubated with Zombie AquaTM Fixable Viability Dye (Biolegend, cat. nr. 423101) in PBS at room temperature and 2.4G2 FcR blocking (BD Biosciences, cat. nr. 553141) in FACS buffer prior to antibody stainings for flow cytometry analyses. Used antibodies are anti-CD3 (clone 145-2C11, BD Biosciences, cat. nr. 562286), anti-CD4 (clone RM4-5, Biolegend, cat. nr. 100547), anti-CD8 (clone 53-6.7, eBioscience, cat. nr. 56-0081-82), anti-CD45.2 (clone 104, eBioscience, cat. nr. 47-0454-82), anti-CD62L (clone MEL−14, Biolegend, cat. nr. 104436), anti-CD69 (clone H1.2F3, Biolegend, cat. nr. 104507), anti-CD25 (clone PC61, Biolegend, cat. nr. 102012), anti-KLRG1 (clone 2F1, eBioscience, cat. nr. 25-5893-82), anti-CD8α (clone S3-6.7, Biolegend, cat. nr. 100734), anti-CD86 (clone GL−1, eBioscience, cat. nr. 12-0862-82), anti-CD11c (clone N418, Biolegend, cat. nr. 117310), and anti-CD11b (clone M1/70, Invitrogen, cat. nr. 48-0112-82). Samples were fixed in 1% PFA (Pharmacy Leiden University Medical Center, the Netherlands) and acquired on a BD Fortessa X20 Flow Cytometer. Flow cytometry data were analyzed by FlowJo software (version 10).
Statistical analysis
All statistical analyses were performed using GraphPad Prism software (version 7.02). Data represent means and standard deviations. For the flow cytometry data, the means of the groups were compared with analysis of variance (ANOVA) including Holm–Sidak’s multiple comparisons test. For the RT-qPCR data, the means of the groups were compared by multiple Mann–Whitney tests. Significant differences are indicated by asterisks, with p values < 0.05 shown as *, <0.01 as **, <0.001 as ***, and <0.0001 as ****. Non-significant differences are indicated by n.s.
Results
Generation of the rS1-mmGMCSF and rS1-hsGMCSF reoviruses
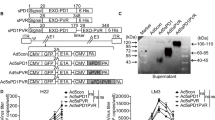
Previously, we described a method for the generation and characterization of recombinant reoviruses. The strategy was based on the replacement of the JAM-A binding head domain of the S1 segment by a sequence containing the codons for a His tag, a 2A peptide-bond skipping sequence, and a small transgene [6, 8]. This modification results in reoviruses with truncated σ1 spikes in their capsids. We utilized the mutation leading to the G196R substitution in the S1 segment to allow the virus to enter cells independent from JAM-A, through facilitating enhanced sialic-acid binding. The first recombinant virus we produced harbors a transgene encoding the small plant-derived green fluorescent protein iLOV [6]; subsequently, we generated a reovirus with S1 carrying a transgene coding for the adenovirus-derived apoptotic E4orf4 protein [8]. Based on the results of experiments with these viruses we decided to generate a recombinant reovirus encoding an immunostimulatory rather than a direct cytolytic product. We chose GM-CSF, which nicely fits within the limited space available in the S1 genome segment of reovirus (max. 522 bp). Fig. 1 shows the structure of the previously and newly constructed S1 segments: they contain nt 1–768 of the reovirus jin-3 S1 segment harboring the G196R change near the sialic-acid binding region, a 6x His-tag, the porcine teschovirus-1 2A (P2A) sequence, the transgene sequence encoding the heterologous protein – iLOV, E4orf4, RFA, human (432 bp) GM-CSF (hsGM-CSF) or murine (425 bp) GM-CSF (mmGM-CSF)—including a stop codon, and the 3′ UTR of the reovirus T3D S1 segment. In contrast to S1His-2A-iLOV, -E4orf4, and -RFA, the 3′ UTR region in GM-CSF encoding S1 segments contains the C-box but lacks the A- and B-boxes. The A-, B-, and C-boxes have been implicated in packaging of the positive RNA strand in the viral capsid [19].
Using a plasmid-based reverse genetics system, we attempted to generate reoviruses armed with these S1 segments encoding mmGM-CSF or hsGM-CSF. To this end, BSR-T7 cells were transfected with plasmids encoding the different reovirus segments. After 72 h, the cells were harvested and the lysates were added to 911scFvHis cells for further viral replication. Two weeks later, clear CPE was seen (Fig. 2), indicating that infectious virus was produced.
Reoviruses rS1-mmGMCSF and rS1-hsGMCSF retrieved from transfected BSR-T7 cells induce CPE in infected 911scFvHis cells. BSR-T7 cells were transfected with plasmids encoding the different reovirus segments. The S1 segment was replaced for the S1His-2A-mmGMCSF or -hsGM-CSF segment. After 72 h, the cells were harvested, and 911scFvHis cells were exposed to the cleared lysates for further viral replication. Two weeks later, clear CPE was seen, indicating that infectious virus was produced. Scale bars represent 100 µm
Stability of S1-mmGMCSF and S1-hsGMCSF
The 911scFvHis cells were harvested and a limiting dilution assay was performed to obtain individual virus clones. Then, 911scFvHis cells were infected with the clones, and RNA was isolated for use in an RT-PCR procedure to amplify the S1 segment. The S1 PCR product was analyzed to detect the presence of the S1 segment carrying the GM-CSF transgene. For rS1-mmGMCSF, the migration of the PCR product in agarose electrophoresis confirmed the expected size of the recombinant segment. Sequencing validated the presence of complete S1-His-2A-mmGMCSF. Upon further propagation up to passage #10, the clone continued to express the full-length segment, showing that we obtained a clone that stably expresses the full-length S1His-2A-mmGMCSF transgene (Fig. 3a). Further experiments were performed with a passage #10 virus batch of rS1-mmGMCSF.
Reovirus rS1-mmGMCSF but not rS1-hsGMCSF(co) S1 segments are stable during prolonged propagation. 911scFvHis cells were infected with individual clones of rS1-mmGMCSF, rS1-hsGMCSF, or rS1-hsGMCSFco for analysis of the S1 segment by RT-PCR. a One rS1-mmGMCSF clone was propagated for 10 passages in total and RNA was extracted for RT-PCR analysis of the S1 segment. A band at the expected size of S1His-2A-mmGMCSF was detected, and confirmed to be full-length S1His-2A-mmGMCSF by sequencing. In the PCR, plasmid pBacT7S1His-2A-mmGMCSF was used as a positive control and Milli-Q water as a negative control. b All the rS1-hsGMCSF PCR products consisted of shorter fragments than that of the full-length S1His-2A-hsGMCSF size. In the PCR, plasmid pBacT7S1His-2A-hsGMCSF was used as a positive control and Milli-Q water as a negative control. c In the PCR products of rS1-hsGMCSFco clones, a predominant full-length size fragment and a shorter fragment are detected. In the PCR, plasmid pBacT7S1His-2A-hsGMCSFco was used as a positive control and Milli-Q water as a negative control. d Upon prolonged propagation of the rS1-hsGMCSFco clones, only the shorter PCR products could be detected in the S1 RT-PCR
For rS1-hsGMCSF, all virus clones showed a S1 PCR product smaller than the expected size of S1His-2A-hsGMCSF (Fig. 3b), indicating the generation of deletion mutants. Three deletion mutants were sequenced and found to have lost the hsGM-CSF sequences. Similar results were obtained in a number of repeat experiments (data not shown). In an attempt to improve transgene preservation, we constructed an alternative S1 plasmid harboring a codon-optimized hsGM-CSF sequence, S1His-2A-hsGMCSFco. We repeated the procedure by transfection of BSR-T7 cells and subsequent infection of 911scFvHis cells, and analyzed the S1His-2A-hsGMCSFco RT-PCR product after RNA isolation of infected 911scFvHis cells. As before, deletion mutants seemed to be present in all clones (Fig. 3c). However, a predominant band at the expected full-length size was evident, indicating that the complete S1His-2A-hsGMCSFco RNA sequence is encoded by the virus. Sequencing of this PCR product confirmed the integrity of the fragment and the presence of an intact S1 segment carrying His, p2A and the complete human GM-CSF gene.
For rS1-hsGMCSFco, also three S1 PCR products of deletion mutants were sequenced. The deletion mutants were found to have lost hsGM-CSFco sequences. These deletion mutants overgrew the virus population upon further propagation (Fig. 3d). Therefore, we continued with low passage (#2) rS1-hsGMCSFco clones for further experiments. An overview of all deletion mutants is depicted in Fig. 4.
Secretion of mmGM-CSF and hsGM-CSF from infected 911 cells
To see whether the reovirus clones induce the secretion of GM-CSF from infected cell cultures, 911 cells were infected with rS1-mmGMCSF, rS1-hsGMCSFco, or jin-3. Supernatants were collected at several time points post infection, and analyzed for the presence of mmGM-CSF or hsGM-CSF by ELISA. A clear induction of GM-CSF secretion was observed, increasing in time (Fig. 5a, b). This indicates that the mmGM-CSF or hsGM-CSFco sequences present in the S1 segments are effectively expressed, stimulating the production and secretion of GM-CSF protein.
Infection of 911 cells with rS1-mmGMCSF/rS1-hsGMCSF leads to secretion of detectable GM-CSF in the supernatant. 911 cells were infected with a a passage#10 (p#10) CsCl-purified rS1-mmGMCSF batch at an MOI of 1, or b a p#2 freeze-thaw (FT) batch of rS1-hsGMCSFco at unknown MOI (20 µl of a 5 ml T25 harvest). As a control, cells were infected with a) a CsCl-purified batch of jin-3 or b) freeze-thaw (FT) batch of jin-3 at an MOI of 1. At 16, 24, 40, and 48 h post infection (hpi), the supernatant was collected for each condition and analyzed for the presence of mmGM-CSF/hsGM-CSF by ELISA. A clear induction of GM-CSF secretion is apparent, increasing in time
Functionality of secreted mmGM-CSF in vitro
GM-CSF is known to stimulate the generation of bone marrow-derived dendritic cells (BMDCs) from murine bone marrow cells [2]. To address the question whether the secreted mmGM-CSF from infected cells is functional, murine bone marrow leukocytes isolated from C57BL/6 mice were exposed to supernatant of rS1-mmGMCSF infected 911scFvHis cells (referred to as rS1-mmGMCSF sup) at a concentration of 5 ng/ml mmGM-CSF. As a positive control, we used conditioned medium from GM-CSF producing NIH/3T3 cells [17] (referred to as R1 sup) at the same concentration of mmGM-CSF. To check for any effects of virus-related factors in the supernatants, the same volume of supernatant from rS1-iLOV-infected 911scFvHis cells (referred to as rS1-iLOV sup) was added as a negative control. The replication efficiencies of rS1-mmGMCSF and rS1-iLOV in 911scFvHis cells are comparable (data not shown), so the supernatants should contain similar levels of virus-related products. Prior to the treatment, the rS1-iLOV and rS1-mmGMCSF supernatants had been irradiated with UV light to inactivate any reovirus that may be present. After 9 days of culture, the bone marrow leukocytes were analyzed for DC markers by flow cytometry. We found multiple sub-populations of CD11c+ cells, differing in their expression level of CD11b and MHC-II. The results are shown in Fig. 6a, b, demonstrating that mmGM-CSF is very efficient at generating CD11bINT, MHC-IIINT cells, indicative for immature DCs, and of CD11bINT, MHC-IIHIGH cells, indicative for mature DCs [21]. The control rS1-iLOV supernatant did not induce the generation of CD11c+ cells. The secreted mmGM-CSF even outcompetes the control R1 supernatant at generating CD11c+ sub-populations, perhaps due to additional factors present in the rS1-mmGMCSF sup. Although the rS1-iLOV supernatant should control for effects of virus-related products, the secreted mmGM-CSF could synergize with virus-related products to induce higher amounts of CD11c+ cells. Both DC populations clearly expressed the co-stimulatory marker CD86, the mature DCs (Fig. 6d) to a higher level than the immature DCs (Fig. 6c), implying they are activated DCs.
Secreted mmGM-CSF induces the maturation and activation of dendritic cells (DCs) from bone marrow cells. Bone marrow cells were exposed to D1 medium supplemented with UV-irradiated supernatants from rS1-mmGMCSF or rS1-iLOV (negative control) infected 911 cells at 5 ng/ml GM-CSF or an equal volume of negative control rS1-iLOV sup. As a positive control, R1 sup was used at 5 ng/ml of GM-CSF. After 3 and 6 days, the treatments on the cells were refreshed. The cells were analyzed by flow cytometry after 9 days of treatment. Shown are a the percentages of CD11bINT MHC-IIINT cells in the CD11c+ population, b the percentages of CD11bINT MHC-IIHIGH cells in the CD11c+ population, c the CD86 expression in the CD11bINT MHC-IIINT CD11c+ population, and d the CD86 expression in the CD11bINT MHC-IIHIGH CD11c+ population. In c and d, gray shaded areas represent the CD86 expression upon R1 treatment, and the black-boxed areas the CD86 expression upon rS1-mmGMCSF treatment. The experiment was repeated twice to confirm the results. Error bars indicate standard deviations (SD)
Alternatively, the growth factor-dependent murine DC line D1 was used [17]. The cells were treated with rS1-mmGMCSF sup at concentrations of 1 and 5 ng/ml mmGM-CSF. As a positive control, R1 sup was taken along at the same concentrations of mmGM-CSF. As a negative control, cells were treated with the same volume of rS1-iLOV sup. After 3 days, the proliferative capacity of the cells was measured by a WST-1 assay. The proliferative capacity of the D1 cells was increased in the presence of both concentrations of rS1-mmGMCSF and R1 sup, but not rS1-iLOV sup (Fig. 7). This implies that there is functional mmGM-CSF present in the rS1-mmGMCSF sup, which stimulates the survival and proliferation of D1 cells.
Secreted mmGM-CSF facilitates the proliferation of murine DC line D1. D1 cells were cultured in the presence of rS1-mmGMCSF sup or R1 sup at 1 or 5 ng/ml mmGM-CSF. As negative controls, they were left untreated or the same volume of rS1-iLOV sup was added. After 3 days, the proliferative capacity of the cells was examined by a WST-1 assay. Error bars indicate SD. Both rS1-mmGMCSF sup and R1 sup caused an increase in D1 proliferative capacity
Altogether, these data clearly show that the produced and secreted mmGM-CSF from infected cells is functional in vitro, i.e., it efficiently triggers the generation of immature and mature DCs from murine bone marrow cells, and it stimulates the survival and proliferation of the murine DC line D1.
Activity of rS1-mmGMCSF in KPC3 mouse pancreatic tumor model
To study the functionality of the mmGM-CSF gene in vivo, we made use of a murine subcutaneous pancreatic ductal adenocarcinoma (PDAC) implantation model in C57BL/6 mice using a low-passage KPC-derived tumor cell line [15]. Mice were treated three times (day 0, day 2, and day 3) with intratumoral injections containing 1 × 107 pfu of rS1-mmGMCSF or control reovirus rS1-iLOV, or sham treatment (reovirus storage buffer). Six days after the first injection (day 6), the mice were sacrificed and the tumors, tumor-draining lymph nodes (TDLNs), and spleens were collected for analysis.
To examine the expression of viral genome products and of virally induced CXCL10 and/or mmGM-CSF in the tumors, a RT-qPCR analysis was performed for the viral S4 genome segment, CXCL10, and for mmGM-CSF mRNA expression. CXCL10 is an IFN-induced chemokine that is often expressed in virus-infected tissues, and therefore an indirect measure of viral infection. Compared to the sham-treated mice, the majority of mice treated with reoviruses had a detectable increase in S4 RNA and CXCL10 mRNA expression in the tumors (Fig. 8a, b). Compared to control and rS1-iLOV-treated mice, the majority of rS1-mmGMCSF-treated mice showed a clear increase in mmGM-CSF mRNA expression (Fig. 8c). Some virus-treated mice showed a (nearly) undetectable increase in S4 or mmGM-CSF expression (data points not shown). To study the effects of mmGM-CSF, mice were only included in subsequent analyses when S4 (for rS1-iLOV and rS1-mmGMCSF treatments) or mmGM-CSF (for rS1-mmGMCSF treatment) expression deviated more than 2 standard deviations (SD) above the mean of the SHAM-treated group. In total, 4 mice (1 rS1-iLOV treated mouse and 3 rS1-mmGMCSF treated mice) were excluded (non-responders), leaving a total of 5 mice per treatment group for further analysis. These results indicate that in the majority of the mice the reoviruses successfully replicated in the KPC3 tumors, and that rS1-mmGMCSF induced the expression of mmGM-CSF inside the tumor tissue.
Gene expression of S4, CXCL10, and mmGM-CSF in tumor-bearing mice treated with rS1-mmGMCSF or rS1-iLOV. Mice were treated at day 0, day 2, and day 3 with intratumoral injections of 107 pfu rS1-mmGMCSF or control reovirus rS1-iLOV, or sham treatment (reovirus storage buffer). At day-6, the mice were sacrificed, and the tumors were collected for (RT-)qPCR analysis of a reovirus segment S4 RNA, b CXCL10 mRNA, and c mmGM-CSF mRNA expression. For S4, 10log copy numbers are shown, calculated based on a standard curve of S4 plasmid. For CXCL10 and mmGM-CSF, normalized expression to reference genes MZT2 and PTP4A2 is shown. All samples were measured in duplo. Data are represented as box plots with whiskers indicating minimum and maximum values. Both rS1-mmGMCSF and rS1-iLOV induced S4 RNA and CXCL10 mRNA expression, and rS1-mmGMCSF specifically increased mmGM-CSF mRNA expression. Significant differences are illustrated by asterisks, with p values < 0.05 shown as *, and <0.01 as **. n.s. indicates non-significant differences
To specifically address the functionality of the mmGM-CSF gene, we analyzed the immune composition and activation of immune cells in the tumors, TDLNs, and spleens by flow cytometry. No significant differences were observed in the immune infiltrate of tumor tissue between rS1-iLOV and rS1-mmGMCSF. GM-CSF is known to enhance the recruitment, maturation and activation of DCs. In doing so, it can trigger a specific anti-tumor immune response by priming CD4+ and/or CD8+ T-cells. Our analyses of the tumors revealed no significant differences in T-cell activation markers (CD25, KLRG1, CD69, and loss of CD62L) for both CD4+ and CD8+ cells. Notably, we observed an increase in co-stimulatory marker CD86 and cross-presentation marker CD8α on CD11c+ cells (Fig. 9a, b) upon both virus treatments. The increase in CD86 expression was even more pronounced on the CD8α+ subset of CD11c+ cells, indicating the activation of cross-presenting DCs (Fig. 9c).
Effects of rS1-mmGMCSF on the immune composition in treated mice. Mice were treated at day 0, day 2, and day 3 with intratumoral injections of 107 pfu rS1-mmGMCSF or control reovirus rS1-iLOV, or sham treatment (reovirus storage buffer). At day 6, the mice were sacrificed and the tumors, tumor-draining lymph nodes (TDLNs), and spleens were collected for flow cytometric analysis. Shown are the percentages of a CD86+ of CD11c+ cells in the tumor, b CD8α+ of CD11c+ cells in the tumor, c CD86+ of CD8α+CD11c+ cells in the tumor, d CD25+ of CD4+ cells in the TDLN, e KLRG1+ CD62L− of CD8+ cells in the TDLN, f KLRG1+ CD62L− of CD8+ cells in the spleen, g CD86+ of CD11c+ cells in the TDLN, h CD86+ of CD11c+ cells in the spleen, i CD86+ of CD8α+ CD11c+ cells in the TDLN, j CD86+ of CD8α+ CD11c+ cells in the spleen, k KLRG1+ CD62L− of CD4+ cells in the TDLN, l CD69+ of CD4+ cells in the spleen, m CD62L− of CD4+ cells in the spleen, and n CD62L− of CD8+ cells in the spleen. Error bars indicate standard error of the mean (SEM). Significant differences are indicated by asterisks, with p values < 0.05 shown as *, <0.01 as **, <0.001 as ***, and <0.0001 as ****. n.s. indicates non-significant differences
Interestingly, we noted several effects on the immune profile at distant locations, i.e., the tumor-draining lymph nodes (TDLNs) and the spleens. Both viruses triggered an increase in activation marker CD25 on CD4+ T-cells in the TDLNs (Fig. 9d). In both TDLNs and spleens, activation marker KLRG1 was increased and naive T-cell marker CD62L was decreased on CD8+ cells (Fig. 9e, f). These findings indicate the systemic activation of CD4+ and CD8+ T-cells, suggesting the formation of effector T-cell responses. Like in the tumors, CD86 expression was increased on CD11c+ cells in the TDLNs and spleens (Fig. 9g, h), and even more pronounced on the CD8α+ subset of CD11c+ cells (Fig. 9i, j), indicating the activation of cross-presenting DCs.
Importantly, mice treated with rS1-mmGMCSF showed a significantly higher increase in CD86+ CD8α+ CD11c+ cells in the TDLNs compared to rS1-iLOV treated mice (Fig. 9g), highlighting a systemic immune-modulating effect specific for rS1-mmGMCSF. We also noticed an increase in KLRG1+ CD62L− CD4+ effector T-cells in the TDLNs specifically upon rS1-mmGMCSF treatment (Fig. 9k), implying an increase in activation of CD4+ T-cells in the TDLNs. Furthermore, elevated levels of activation marker CD69 on CD4+ T-cells, and loss of naive T-cell marker CD62L on CD4+ and CD8+ cells were observed in the spleens of rS1-mmGMCSF treated mice (Fig. 9l, m, n), suggesting that effector T-cell responses are formed in the spleen as well. These findings suggest that rS1-mmGMCSF administration activates T-cell responses in TDLNs and spleens.
Altogether, our findings demonstrate that rS1-mmGMCSF treatment induces the expression of functional mmGM-CSF in this murine model for pancreatic cancer which systemically influences the maturation and activation status of immune cells.
Discussion
Oncolytic viruses represent promising candidates for anti-cancer therapy, inducing both cancer cell death as well as triggering an immune response that could be directed against the tumor as well as the virus [1]. In 2015, the first oncolytic virus therapy was approved for cancer treatment in the US, namely T-VEC, a Herpes Simplex Virus carrying copies of the GM-CSF gene [3]. The U.S. Food and Drug Administration (FDA) approved the usage of T-VEC for patients with advanced melanoma that cannot be surgically removed (completely). This approval may pave the way for other oncolytic viruses to reach the clinic.
Oncolytic virotherapy using reovirus has entered a variety of clinical trials, which demonstrated both the safety and feasibility of the approach. However, its clinical efficacy has been limited in stand-alone therapies [4]. Genetic modification of reovirus can be challenging, due to its segmented dsRNA genomic nature and a relatively limited packaging capacity. We and others have previously demonstrated the feasibility of generating recombinant reoviruses expressing small transgenes such as iLOV [6], UnaG [22], E4orf4, and RFA [8]. We showed that the JAM-A binding region of the viral S1 segment can be replaced by a small transgene. A mutation in the sialic-acid binding area of S1 offered the viruses an alternative entry route through enhanced sialic-acid binding. We showed that the viruses with a truncated σ1 spike all seemed potent cancer-killing viruses, and that encoding the cell death inducing E4orf4 protein did not further increase oncolysis [8]. We postulated that encoding an oncolytic transgene may be obsolete. Combined with the current dogma that anti-tumor immunity is crucial for an oncolytic virotherapy to be clinically potent, it may be a better choice to incorporate a transgene encoding an immunostimulatory protein to enhance anti-tumor immune responses. Interestingly, during the previous production of the iLOV-encoding reovirus (rS1-iLOV), we encountered a deletion mutant virus that had deleted the A- and B-boxes [6]. The A-, B-, and C-boxes are thought to be important for packaging of the positive RNA strand in the viral capsid [19]. Therefore, we hypothesized that the A- and B-boxes are dispensable for efficient packaging of S1 into reovirus particles. As a result of this, the available space for transgene insertion increases from 451 to 522 bp.
Here, we describe the successful construction of recombinant reoviruses carrying a gene encoding murine (mm) or human (hs) GM-CSF. We demonstrate that the viruses trigger GM-CSF secretion in vitro upon infection of cell lines. This shows that encoding a secretory protein after a 2A sequence in the reovirus S1 segment can lead to efficient intracellular routing of the protein and release from the cell. Furthermore, we show that the produced mmGM-CSF is functional in vitro, as it facilitates the proliferation of cells that depend on GM-CSF for their survival, and it efficiently triggers the generation of immature and mature dendritic cells (DCs) from murine bone marrow cells. Importantly, our data reveal the functionality of the mmGM-CSF gene in vivo as well, evident from specific increases in DC activation and the generation of effector T-cells at distant locations in a KPC3 mouse tumor model.
During the production of rS1-hsGMCSF, we encountered the appearance of deletion mutants. Expressing a codon-optimized copy of the hsGM-CSF gene (hsGM-CSFco) allowed us to rescue reoviruses expressing full-length S1-hsGMCSFco. Nevertheless, limiting dilutions of the virus stock revealed that the S1 deletions were present from the first passage on in all of the virus clones. In the batches with the early passage number, both full-length S1-hsGMCSFco and deleted variants were detected by S1 RT-PCR. Upon further propagation, we noticed that the deletion mutants rapidly overgrew the reovirus cultures, thereby losing the viruses harboring full-length S1-hsGMCSFco. This implies that the deletion mutant has a growth advantage over the intact rS1-hsGMCSFco. Therefore, we postulated that either the hsGM-CSFco gene or the hsGM-CSF protein has negative effects on viral replication. As we grow our viruses on 911 cells, which are human cells, it could be that they are sensitive to the produced hsGM-CSF, and therefore better facilitate the replication of the deletion mutant that does not lead to production of hsGM-CSF. However, attempts to generate a genetically stable rS1-hsGMCSFco virus on a non-human cell line were unsuccessful, suggesting that the generation of deletion mutants is not due to sensitivity of the producer cell line to the hsGM-CSF. Further inspection of the sequences surrounding the deletions did not provide evidence for involvement of either splicing or homology-directed mechanisms (data not shown). It seems most likely that the cause of the deletions lies at the RNA level, possibly in the RNA structure of the segment. For our experiments with rS1-hsGMCSFco, we used a passage#2 virus in which predominantly the full-length S1His-2A-hsGMCSFco is still present.
The scope of this study was to generate recombinant reoviruses encoding GM-CSF and determine whether a reovirus-encoded GM-CSF gene has functionality both in vitro and in vivo. While the short duration of our performed animal study did not allow to examine any potential effects on tumor volume or survival, future studies are recommended to determine whether the GM-CSF expressing reovirus can further stimulate (local and distant) tumor reduction and/or improve survival of tumor-bearing mice. Despite distant effects on T-cell activation, we did not observe local effects on T-cell activation markers in the KPC3 tumors upon both rS1-mmGMCSF and rS1-iLOV treatments. Notably, the KPC3 tumor model has a very low abundance of T-cells [15], which may impede (the detection of) immune-mediated intratumoral effects. A higher immunogenic model could be a better choice to look at virus- and GM-CSF-induced effects on tumor reduction.
References
Keller BA, Bell JC. Oncolytic viruses-immunotherapeutics on the rise. J Mol Med. 2016;94:979–91.
Hu JC, Coffin RS, Davis CJ, Graham NJ, Groves N, Guest PJ, et al. A phase I study of OncoVEXGM-CSF, a second-generation oncolytic herpes simplex virus expressing granulocyte macrophage colony-stimulating factor. Clin Cancer Res. 2006;12:6737–47.
Pol J, Kroemer G, Galluzzi L. First oncolytic virus approved for melanoma immunotherapy. Oncoimmunology. 2016;5:e1115641.
Zhao X, Chester C, Rajasekaran N, He Z, Kohrt HE. Strategic combinations: the future of oncolytic virotherapy with reovirus. Mol Cancer Ther. 2016;15:767–73.
Komoto S, Kawagishi T, Kobayashi T, Ikizler M, Iskarpatyoti J, Dermody TS, et al. A plasmid-based reverse genetics system for mammalian orthoreoviruses driven by a plasmid-encoded T7 RNA polymerase. J Virol Methods. 2014;196:36–9.
van den Wollenberg DJ, Dautzenberg IJ, Ros W, Lipinska AD, van den Hengel SK, Hoeben RC. Replicating reoviruses with a transgene replacing the codons for the head domain of the viral spike. Gene Ther. 2015;22:267–79.
van den Wollenberg DJ, Dautzenberg IJ, van den Hengel SK, Cramer SJ, de Groot RJ, Hoeben RC. Isolation of reovirus T3D mutants capable of infecting human tumor cells independent of junction adhesion molecule-A. PLoS ONE 2012;7:e48064.
Kemp V, Dautzenberg IJC, Cramer SJ, Hoeben RC, van den Wollenberg DJM. Characterization of a replicating expanded tropism oncolytic reovirus carrying the adenovirus E4orf4 gene. Gene Ther. 2018;25:331–34.
Branton PE, Roopchand DE. The role of adenovirus E4orf4 protein in viral replication and cell killing. Oncogene. 2001;20:7855–65.
Marcellus RC, Chan H, Paquette D, Thirlwell S, Boivin D, Branton PE. Induction of p53-independent apoptosis by the adenovirus E4orf4 protein requires binding to the Balpha subunit of PP2A. J Virol. 2000;74:7869–77.
Xu Y, Zhan Y, Lew AM, Naik SH, Kershaw MH. Differential development of murine dendritic cells by GM-CSF versus Flt3 ligand has implications for inflammation and trafficking. J Immunol. 2007;179:7577–84.
Zhan Y, Carrington EM, van Nieuwenhuijze A, Bedoui S, Seah S, Xu Y, et al. GM-CSF increases cross-presentation and CD103 expression by mouse CD8(+) spleen dendritic cells. Eur J Immunol. 2011;41:2585–95.
Fallaux FJ, Kranenburg O, Cramer SJ, Houweling A, Van Ormondt H, Hoeben RC, et al. Characterization of 911: a new helper cell line for the titration and propagation of early region 1-deleted adenoviral vectors. Hum Gene Ther. 1996;7:215–22.
van den Wollenberg DJ, van den Hengel SK, Dautzenberg IJ, Cramer SJ, Kranenburg O, Hoeben RC. A strategy for genetic modification of the spike-encoding segment of human reovirus T3D for reovirus targeting. Gene Ther. 2008;15:1567–78.
Lee JW, Komar CA, Bengsch F, Graham K, Beatty GL. Genetically engineered mouse models of pancreatic cancer: the KPC model (LSL−Kras(G12D/+);LSL−Trp53(R172H/+);Pdx-1-Cre), its variants, and their application in immuno-oncology drug discovery. Curr Protoc Pharmacol. 2016;73:14.39.1–14.39.20.
Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73:251–9.
Winzler C, Rovere P, Rescigno M, Granucci F, Penna G, Adorini L, et al. Maturation stages of mouse dendritic cells in growth factor–dependent long-term cultures. J Exp Med. 1997;185:317–28.
Boross P, van Montfoort N, Stapels DA, van der Poel CE, Bertens C, Meeldijk J, et al. FcRgamma-chain ITAM signaling is critically required for cross-presentation of soluble antibody-antigen complexes by dendritic cells. J Immunol. 2014;193:5506–14.
Roner MR, Roehr J. The 3’ sequences required for incorporation of an engineered ssRNA into the Reovirus genome. Virol J. 2006;3:1.
Mijatovic-Rustempasic S, Tam KI, Kerin TK, Lewis JM, Gautam R, Quaye O, et al. Sensitive and specific quantitative detection of rotavirus A by one-step real-time reverse transcription-PCR assay without antecedent double-stranded-RNA denaturation. J Clin Microbiol. 2013;51:3047–54.
Helft J, Bottcher J, Chakravarty P, Zelenay S, Huotari J, Schraml BU, et al. GM-CSF mouse bone marrow cultures comprise a heterogeneous population of CD11c(+)MHCII(+) macrophages and dendritic cells. Immunity. 2015;42:1197–211.
Eaton HE, Kobayashi T, Dermody TS, Johnston RN, Jais PH, Shmulevitz M. African swine fever virus NP868R capping enzyme promotes reovirus rescue during reverse genetics by promoting reovirus protein expression, virion assembly, and RNA incorporation into infectious virions. J Virol. 2017;91:e02416–16.
Author information
Authors and Affiliations
Contributions
VK, DJMvdW, MGMC, TvH, PK, NPvM, and RCH all compiled data and jointly wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Kemp, V., van den Wollenberg, D.J.M., Camps, M.G.M. et al. Arming oncolytic reovirus with GM-CSF gene to enhance immunity. Cancer Gene Ther 26, 268–281 (2019). https://doi.org/10.1038/s41417-018-0063-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41417-018-0063-9
- Springer Nature America, Inc.
This article is cited by
-
Engineering strategies to enhance oncolytic viruses in cancer immunotherapy
Signal Transduction and Targeted Therapy (2022)
-
Gastrointestinal cancer-associated fibroblasts expressing Junctional Adhesion Molecule-A are amenable to infection by oncolytic reovirus
Cancer Gene Therapy (2022)
-
Oncolytic Viral Therapy for Malignant Glioma and Their Application in Clinical Practice
Neurotherapeutics (2022)
-
Immunotherapy for pancreatic cancer: chasing the light at the end of the tunnel
Cellular Oncology (2021)