Abstract
Background
We aimed to develop and validate a plasma extracellular vesicle circular RNA (circRNA)-based signature that can predict overall survival (OS) in first-line abiraterone therapy for metastatic castration-resistant prostate cancer (mCRPC) patients.
Methods
In total, 582 mCRPC patients undergoing first-line abiraterone therapy from four institutions were sorted by three phases. In the discovery phase, 30 plasma samples from 30 case-matched patients with or without early progression were obtained to generate circRNA expression profiles using RNA sequencing. In the training phase, differentially expressed circRNAs were examined using digital droplet PCR in a training cohort (n = 203). The circRNA signature was constructed using a least absolute shrinkage and selection operator Cox regression to predict OS. In the validation phase, the prognostic ability of this signature was prospectively validated in two external cohorts (Cohort I, n = 183; Cohort II, n = 166).
Results
We developed a five-circRNA signature, based on circCEP112, circFAM13A, circBRWD1, circVPS13C and circMACROD2, which successfully stratified patients into high-risk and low-risk groups. The prognostic ability of this signature was prospectively validated in two external cohorts (P < 0.0001, P < 0.0001). Patients with high-risk scores had shorter OS than patients with low-risk scores.
Conclusion
This five-circRNA signature is a reliable predictor of OS for mCRPC patients undergoing abiraterone.
Similar content being viewed by others
Background
Prostate cancer is the second most commonly diagnosed cancer in men and the fifth leading cause of cancer death worldwide. For the past 70 years, the standard of care for locally advanced or metastatic prostate cancer is androgen deprivation therapy. Although up to 85% of patients initially respond to androgen deprivation therapy, virtually all patients would progress to metastatic castration-resistant prostate cancer (mCRPC) [1]. Several available therapies could improve outcomes for mCRPC patients [2].
Abiraterone is an inhibitor of cytochrome P450 17A1 that potently blocks the production of androgens by the adrenal glands and testes and within the prostate tumour [3, 4]. Despite the significant advances in the treatment of mCRPC, ~20–40% of patients have no response to abiraterone with respect to serum prostate-specific antigen (PSA) values. Among patients who initially have a response, nearly all eventually exhibit acquired resistance [5, 6]. One potential explanation for the resistance to abiraterone is the emergence of constitutively active androgen receptor (AR) splice variants, of which AR splice variant-7 (AR-V7) is the most extensively characterised [7,8,9]. Recent studies have shown that noncoding RNAs, including circular RNAs (circRNAs), are strongly associated with AR/AR-V7 signalling in prostate cancer castration-resistant progression [10,11,12,13,14,15,16]. Compared with linear RNAs, circRNAs have covalently linked ends of a single RNA molecular and appear to have a higher stability [17]. This remarkable feature makes them to be more advantageous as potential molecular diagnostic and prognostic markers [18].
Extracellular vesicles (exosomes and microvesicles) are small membrane vesicles which could be released into circulatory fluids by cancer cells [19, 20]. Recently, plenty of plasma extracellular vesicle noncoding RNAs were reported as prognostic and predictive biomarkers in cancer patients [21]. However, few studies have been reported on plasma extracellular vesicle circRNAs in prostate cancer [22] and no data are available on their use in the identification of mCRPC patients receiving first-line abiraterone at greatest risk for overall survival. In this study, we aimed to analyse circRNA expression profiles from plasma-derived extracellular vesicles in mCRPC patients receiving first-line abiraterone to develop a multi-circRNA-based signature to predict overall survival. We prospectively evaluated the prognostic ability of this multi-circRNA-based signature in the training cohort and validated externally it in two independent cohorts. This prognostic circRNA signature may substantially improve the existing clinicopathological prognostic markers in mCRPC patients receiving first-line abiraterone therapy.
Methods
Patient population and study design
We designed a prospective biomarker discovery analysis according to the TRIPOD statement [23] to identify circRNA biomarkers associated with overall survival in mCRPC patients receiving first-line abiraterone. This study, which was conducted in accordance with the Declaration of Helsinki, was approved by the Institutional Ethical Review Board at The Third Affiliated Hospital of Sun Yat-sen University, and written informed consent was obtained from all study participants. The study is registered with www.chictr.org/cn/, number ChiCTR1800019529.
Patients in the discovery, training and validation phases, will be eligible for inclusion only if all of the following criteria apply: (1) age ≥18 years; (2) histologically confirmed diagnosis of adenocarcinoma of the prostate; (3) castrate levels of serum testosterone (<50 ng/dL) at most recent assessment and continued treatment with androgen deprivation therapy; (4) radiographic evidence of at least one metastatic lesion via MRI/CT scan or bone scan; (5) patients are about to begin treatment with first-line abiraterone acetate; (6) evidence of disease progression on or following most recent therapy as evidenced by at least one of the following: (a) Having ≥3 rising serum PSA values obtained 2 or more weeks apart, with the last value being 2.0 ng/mL or higher (Prostate Cancer Clinical Trials Working Group 2 guidelines) [24]; (b) having ≥20% increase in the sum of the diameters of soft-tissue lesions evaluated by MRI or CT using Response Evaluation Criteria in Solid Tumours [25] or ≥2 new bone lesions on technetium-99 bone scanning. Exclusion criteria were as follows: inadequate RNA (less than 1 ng/μL) available and loss of follow-up.
In the discovery phase, to generate circRNAs expression profiles, we obtained 30 plasma samples from 30 case-matched mCRPC patients with early progression (defined as the radiographic appearance of new documented metastases within 3 months from first-line abiraterone treatment) or without at The Third Affiliated Hospital of Sun Yat-sen University between March 2014 and April 2015. Cases were strictly matched on a 1:1 basis in an automated fashion using SPSS (version 25.0; SPSS Inc., Chicago, IL, USA) and the following rules: the similar age at the time of CRPC (within 5 years), the same Gleason score at initial diagnosis, the same clinical T staging at initial diagnosis, the similar number of positive biopsy cores at initial diagnosis, the similar serum PSA levels at initial diagnosis (within 5 ng/mL), and the similar duration time from initiation of androgen deprivation therapy for the hormone-sensitive stage to CRPC stage (within 3 months). No patients had previous treatment with radiotherapy or chemotherapy for castration-resistant diseases.
In the training and validation phases, we used three cohorts of plasma specimens from mCRPC patients: the training cohort, and two independent external validation cohorts. For the training cohort, data were prospectively acquired from 203 patients from The Third Affiliated Hospital of Sun Yat-sen University between May 2015 and December 2019. For the independent validation sets, 183 patients in Cohort I were prospectively obtained from Foshan First Municipal People’s Hospital between May 2015 and June 2020. Another 166 patients in Cohort II were included from The First Affiliated Hospital of the University of South China and The First Affiliated Hospital of Hainan Medical College between May 2015 and June 2020 (Fig. 1).
mCRPC metastatic castration-resistant prostate cancer, circRNAs circular RNAs, ddPCR digital droplet polymerase chain reaction, LASSO least absolute shrinkage and selection operator. *The early progression was defined as radiographic appearance of new documented metastases (per Response Evaluation Criteria in Solid Tumours 1.1 criteria) within 3 months from abiraterone treatment.
Abiraterone was given at a dose of 1000 mg daily, with prednisone at a dose of 5 mg twice daily. For follow-up, all patients were required to have PSA determination at least once every 1–2 months. The intervals of imaging (chest/abdomen/pelvis CT or MRI and technetium-99 bone scan evaluations) are baseline prior to abiraterone treatment initiation and every 2–3 months, or sooner if clinically indicated. Abiraterone therapy should continue until radiographic progression, or unmanageable drug-related toxicity.
Extracellular vesicle RNA extraction
For plasma specimens, whole blood (5 × 5 mL) was collected in ethylenediaminetetraacetic acid tubes and centrifuged at 1900×g for 10 min at 4 °C within 2 h after drawing. The supernatant was then fractioned into multiple aliquots, and stored at −80 °C until analysed centrally at The Core Lab Plat for Medical Science of Sun Yat-sen University.
The extracellular vesicle isolation and characterisation, RNA extraction, and quality control are described previously [26] (Supplementary Material and Supplementary Figs. S1–3).
Extracellular vesicle RNA sequencing library preparation and circRNAs sequencing
The total RNA was treated with RiboZero rRNA Removal Kit (Epicentre, WI, USA) to delete rRNA and digest linear RNA using RNase-R (Epicentre) according to the manufacturer’s instructions. Subsequently, strand-specific libraries were constructed using VAHTS Total RNA-seq (H/M/R) Library Prep Kit for Illumina (Vazyme, Nanjing, CHN) following the manufacturer’s instructions. In brief, the enriched circRNAs were fragmented using fragmentation buffer and then reverse transcribed into first-strand cDNA and second-strand cDNA in an orderly. Then, the cDNA fragments were purified, subjected to end-repair, and modified to add A at the 3’ end. The sequencing adapters were ligated to the cDNA fragments. The double-strand cDNA was digested by uracil-DNA glycosylase (Cwbio, Beijing, CHN) to remove the second-strand cDNA before sequencing. The purified ligation products were performed PCR amplification. The PCR amplification products of cDNA were purified with Agencourt AMPure XP Beads (Beckman Coulter, Inc. Kraemer Boulevard Brea, CA, USA) and then sequenced by Illumina Hiseq X Ten system (Illumina, San Diego, CA, USA) on paired-end mode with length 150 bases following the vendor’s recommended protocol.
For the identification and annotation of circRNAs, the raw sequencing reads were subjected to trimming process with Trimmomatic (v.0.32). Briefly, the sequencing adapters, leading and trailing bases below Q30 were first trimmed. The reads were then scanned from both ends, using a 4 bp-wide sliding window, within which the low-quality (lower than Q20) bases were trimmed. Finally, the resulting reads of length at least 50 bases were selected for further analyses. High-quality reads were aligned to the Homo sapiens reference genome (GRCH37/hg19) that was obtained from UCSC genome browser (http://genome.ucsc.edu/) using the Burrows-Wheeler Alignment tool (v.0.7.12). The resulted sequence alignment files (bam formatted file) were then sorted by reference position as required by SAM tools (v.2.6.2).
Sequencing data analysis
CircRNAs were identified and quantified by CIRI2. The reads that partly aligned to the genome were considered as the candidate back-spliced junction reads and the unaligned part of reads were further spliced to multiple short seed reads, then aligned to the downstream and upstream region of aligned part and the key segments of both side were calculated with maximum likelihood estimation algorithm. The reads whose key segment of upstream region larger than that of downstream will be considered as the junction region reads of circRNAs. The identified circRNAs were then annotated with circBase database that contains data from studies of large-scale circRNAs identification published to date [27].
We compared circRNAs expression profiles in plasma extracellular vesicles across 30 case-matched mCRPC patients with or without early progression using the edgeR package of R (v.3.1.2) [28]. The expression fold change between each comparison group was calculated by spliced reads per billion mapping (SPRBM = number of circular reads/total mapped reads [units in billion]) and log-transformed. We defined the statistical criteria for selecting differentially expressed circRNAs using |fold changes| ≥2.0 with P values < 0.01. We have deposited the RNA sequencing data reported in this study into the National Center for Biotechnology Information’s Gene Expression Omnibus (GSE125256) (token: atutcmssvrczxmh).
Digital droplet PCR (ddPCR) analysis
We used ddPCR EvaGreen Supermix (Bio-Rad, Foster City, CA, USA) on the QX200 Droplet System as described in our previous study [29]. The primers used are listed in Supplementary Table S5. The reactions were assembled into each well according to the following protocol: 2 μl cDNA, 10 μl QX200 EvaGreen ddPCR Supermix (Bio-Rad), 4 μM of each primer and nuclease-free water up to 20 μl. A no template control, where nuclease-free water was added instead of cDNA samples, was set. The droplet generation procedure followed the manufacturer’s instructions. Each 70 μl QX200 Droplet generation oil and 20 μl ddPCR reaction, respectively, were added into the 8-channel droplet generation cartridge, and the cartridge was loaded in the QX200™ Droplet Generator. Subsequently, the 40 μl of resulting droplets solution was then transferred into a 96-well PCR plate. The cycling conditions were as follows: hot-start at 95 °C for 5 min; followed by 45 cycles of 95 °C for 30 s, 64 °C for 1 min; then, a signal stabilisation step at 4 °C for 5 min and 90 °C for 5 min, finally holding at 4 °C. Droplets were detected on the QX200 Droplet Reader (Bio-Rad) and analysed by QuantaSoft software (Bio-Rad). The resulting copies per microliter of reaction were the numbers exported by the software (Supplementary Figs. S4–6).
The laboratory investigators have no information regarding patients’ clinical and outcome data and researchers at all institutions were masked to circRNAs testing results. After all samples had been tested by the same central laboratory (The Core Lab Plat for Medical Science of Sun Yat-sen University), these blinded data were released to and compared with patient outcomes.
Outcomes
The primary endpoint was overall survival. Overall survival was defined as the time from plasma collection (the initiation of abiraterone) to the date of death of any cause. The secondary endpoint was (1) the proportion of patients with a serum PSA response (≥50% reduction in PSA from baseline, maintained for ≥4 weeks) at any time after the initiation of abiraterone therapy; The best PSA response (maximal percentage decrease in serum PSA from baseline) was also calculated; (2) radiographic progression was defined as ≥20% increase in the sum of the diameters of soft-tissue lesions evaluated by MRI or CT using Response Evaluation Criteria in Solid Tumours [25] or ≥2 new bone lesions on technetium-99 bone scanning. The intervals of imaging (chest/abdomen/pelvis CT or MRI and technetium-99 bone scan evaluations) are baseline prior to abiraterone treatment initiation and every 2–3 months, or sooner if clinically indicated. The presence or absence of metastatic lesions was confirmed by a central independent review.
Statistical analysis
We calculated the sample size of our study using the programme PASS (version 11, NCSS, Kaysville, UT, USA). The sample size was determined on the basis of the primary endpoint of overall survival. It was assumed that the 3-year overall survival rates for patients with mCRPC were 40% in the low-risk group and 20% in the high-risk group [30]. For statistical purposes, we will suppose that circRNAs with a high level of expression will be detectable from baseline samples in 50% of abiraterone-treated men. Under this assumption, a sample size of 133 patients per cohort will yield 80% power to detect a difference in overall survival from 20% (in the high-risk group) to 40% (in the low-risk group) in a median 3-year follow-up, using a two-sided log-rank test at a significance level of 0.05.
To confirm the differential expression of candidate circRNAs from the RNA sequencing results, we further determined their expression in 64 plasma samples from the training cohort using ddPCR. Using X-tile plots (X-tile software version 3.6.1, Yale University School of Medicine, New Haven, CT, USA), we chose the optimal cutoff score for the expression of every significantly differentially expressed circRNA based on the association with the patient’s overall survival. We employed LASSO Cox regression model [31] to select the most useful prognostic circRNAs identified with the training cohort, and constructed a multi-circRNA-based signature for predicting overall survival.
The overall survival and progression-free survival were estimated by the Kaplan–Meier method and assessed by the log-rank test. HRs were calculated by the use of univariate Cox regression analyses. We used multivariable Cox regression analyses to test the independent significance of different variables. Covariates included multi-circRNA-based signature, and the traditional risk factors (including age, ECOG PS, disease site, opioid analgesic use, PSA, LDH, haemoglobin, alkaline phosphatase and albumin), which were reported to be associated with overall survival for patients with mCRPC [32].
We investigated the prognostic accuracy of existing clinicopathological prognostic factors and multi-circRNA-based signature using receiver operating characteristic (ROC) analysis. AUC was used to assess model performance. Decision curve analysis was used as described in our previous study [33] to compare the net benefit associated with overall survival prediction using multi-circRNA-based signature. Calibration was assessed to compare the observed rates with the multi-circRNA-based signature predicted probabilities.
All statistical analyses were done with SPSS (version 25.0; SPSS Inc, Chicago, IL, USA) or R software version 3.4.3 (R Foundation for Statistical Computing, Vienna, Austria), and a two-tailed P value of less than 0.05 was regarded as statistically significant.
The TRIPOD statement was followed throughout this study [23].
Results
Patient characteristics
In the discovery phase, between March 2014 and April 2015, we obtained 30 plasma samples from 30 case-matched mCRPC patients at The Third Affiliated Hospital of Sun Yat-sen University to generate circRNA expression profiles. The clinicopathological characteristics of these patients were shown in Supplementary Table S1. Between May 2015 and December 2020, we enrolled 552 men with mCRPC from four academic medical centres for the training and validation phases (Fig. 1). Baseline characteristics of these cohorts are listed in Table 1. The median follow-up time for the entire cohort was 32.0 months (interquartile range [IQR]: 30.5–33.5 months); 33.0 months (IQR: 31.3–34.7 months) for the training cohort, 32.0 months (IQR: 29.8–34.2 months) for the validation Cohort I, and 32.0 months (IQR: 28.8–35.2 months) for the validation Cohort II. During follow-up, 85.1% of the patients (470 of 552) developed progression, and 78.8% of the patients (435 of 552) died.
Selection of candidate circRNAs
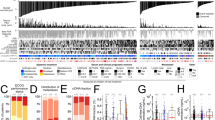
In RNA sequencing analysis, we identified 96 plasma extracellular vesicle circRNAs that had significantly different levels between case-matched mCRPC patients with or without early progression (Fig. 2). We further confirmed the RNA sequencing results of 96 circRNAs in the training cohort by ddPCR analysis. Twenty-nine circRNAs were identified to be significantly differentially expressed between mCRPC patients with progression and those without (P < 0.01). Of these 29 circRNAs, 16 circRNAs were upregulated circRNAs and 13 downregulated circRNAs (Supplementary Table S2).
a Clustered heatmap of the differentially expressed plasma-derived extracellular vesicle circRNAs from 15 paired mCRPC patients receiving abiraterone with early progression (Group 2, n = 15) or without (Group 1, n = 15). The early progression was defined as radiographic appearance of new documented metastases within 3 months from abiraterone treatment. Each row represents an individual sample, and each column represents an individual circRNA. The expression values were log2 transformed. b LASSO coefficient profiles of the 29 progressive mCRPC-associated circRNAs. The dotted vertical line is drawn at the minimum lambda value chosen by tenfold cross-validation. circRNAs circular RNAs, mCPRC metastatic castration-resistant prostate cancer, LASSO least absolute shrinkage and selection operator.
Subsequently, we employed X-tile plots to generate the optimal cutoff score for these 29 significantly differentially expressed circRNAs in the training cohort. The univariate analysis between each of the 29 circRNAs and overall survival was shown in Supplementary Table S3. We used a least absolute shrinkage and selection operator (LASSO) Cox regression model to build a prognostic signature, which chose five circRNAs from the 29 circRNAs identified in the training cohort: (circCEP112, circFAM13A, circBRWD1, circVPS13C, and circMACROD2, Fig. 2b). In the five circRNAs, expression levels of three circRNAs (circCEP112, circFAM13A, and circBRWD1) were positively associated with overall survival, and the expression levels of the other two (circVPS13C and circMACROD2) were inversely associated with overall survival (Supplementary Fig. S7).
Building a predictive signature
To investigate the effectiveness of these five circRNAs as a circRNA-based signature for overall survival prediction, we assigned each patient a risk score using a formula derived from their individual five circRNAs expression levels (Supplementary Table S4).
Risk score = (0.182 × expression level of circCEP112) + (0.094 × expression level of circFAM13A) + (0.028 × expression level of circBRWD1) − (0.076 × expression level of circVPS13C) − (0.336 × expression level of circMACROD2).
Using X-tile plots to generate an optimal cutoff score (0.84) (Supplementary Fig. S7), we divided patients into high-risk and low-risk groups (Figs. 3–5). This assay categorised 113 (55.6%) of 203 patients in the training cohort to the low-risk group and 90 (44.3%) to the high-risk group. The PSA response rate among patients with high-risk scores was 17.8% (16 of 90 men), and the rate among patients with low-risk scores was 31.9% (36 of 113 men; P = 0.034) (Fig. 6a). Patients with high-risk scores had shorter overall survival (hazard ratio [HR] 2.69, 95% confidence interval [CI] 1.96–3.71; P < 0.0001; Fig. 6a) than patients with low-risk scores. Similarly, patients with high-risk scores had shorter progression-free survival (HR 2.41, 1.76–3.29; P < 0.0001; Fig. 6a) than those with low-risk scores. The circRNA risk-score distributions and patients’ overall survival status in each risk group are shown in Fig. 3.
Upper panel: circRNA risk-score distribution. Middle panel: patients’ survival status. Bottom panel: colour-gram of circRNA expression profiles of mCRPC patients; rows represent high-risk and protective circRNAs, columns represent patients. The black dotted line represents the optimal circRNA signature cutoff dividing patients into low-risk and high-risk groups. mCPRC metastatic castration-resistant prostate cancer.
Upper panel: circRNA risk-score distribution. Middle panel: patients’ survival status. Bottom panel: colour-gram of circRNA expression profiles of mCRPC patients; rows represent high-risk and protective circRNAs, columns represent patients. The black dotted line represents the optimal circRNA signature cutoff dividing patients into low-risk and high-risk groups. mCPRC metastatic castration-resistant prostate cancer.
Upper panel: circRNA risk-score distribution. Middle panel: patients’ survival status. Bottom panel: colour-gram of circRNA expression profiles of mCRPC patients; rows represent high-risk and protective circRNAs, columns represent patients. The black dotted line represents the optimal circRNA signature cutoff dividing patients into low-risk and high-risk groups. mCPRC metastatic castration-resistant prostate cancer.
a Training cohort (n = 203). b Validation Cohort I (n = 183). c Validation Cohort II (n = 166). The dotted line shows the threshold for defining a PSA response (≥50% reduction in PSA level from baseline). Hazard ratios were calculated using a univariate Cox regression analysis and P values were calculated using the log-rank test. PSA prostate-specific antigen, circRNA circular RNA, HR hazard ratio, CI confidence interval.
Validating the signature
We used the same risk score formula as in the training cohort and calculated the five-circRNA signature risk score for each patient in two independent external validation cohorts (Cohort I, n = 183; Cohort II, n = 166). We then assigned them into the high-risk group or low-risk group, respectively, based on the same cutoff value obtained from the training cohort.
In the independent external validation Cohort I, 93 (50.8%) of 183 patients were classified into the low-risk group and 90 (49.2%) patients were classified into the high-risk group. The PSA response rate among patients with high-risk scores was 13.3% (12 of 90 men), and the rate among patients with low-risk scores was 30.1% (28 of 93 men; P = 0.010) (Fig. 6b). Patients with high-risk scores had shorter overall survival (HR 2.06, 95% CI 1.49–2.85; P < 0.0001; Fig. 6b) and progression-free survival (HR 2.16, 1.58–2.96; P < 0.0001; Fig. 6b) than patients with low-risk scores. Similarly, in the independent external validation Cohort II, the circRNA signature stratified 77 (46.4%) of 166 patients into the low-risk group and 89 (53.6%) patients into the high-risk group. The PSA response rate among patients with high-risk scores was 16.9% (15 of 89 men), and the rate among patients with low-risk scores was 40.3% (31 of 77 men; P = 0.001) (Fig. 6c). Patients with high-risk scores had shorter overall survival (HR 2.16, 95% CI 1.50–3.11; P < 0.0001; Fig. 6c) and progression-free survival (HR 1.96, 1.40–2.75; P < 0.0001; Fig. 6c). The circRNA risk-score distributions and patients’ overall survival status in each risk group are shown in Figs. 4 and 5.
Using the multivariable Cox proportional hazard regression analysis, we found that the five-circRNA signature remained a strong independent predictor for overall survival (HR 2.62, 1.86–3.69; P < 0.0001 in training cohort; HR 2.19, 1.53–3.14; P < 0.0001 in Cohort I; HR 2.17, 1.47–3.19; P < 0.0001 in Cohort II) regardless of age, Eastern Cooperative Oncology Group performance status [ECOG PS], disease sites [lymph node-only metastasis; bone/bone+lymph node metastasis; any visceral metastasis], opioid analgesic use, PSA, lactate dehydrogenase [LDH], haemoglobin, alkaline phosphatase and albumin (Table 2). This five-circRNA signature also showed significantly higher prognostic accuracy than any clinicopathological variable, or single circRNA alone. We then integrated this five-circRNA signature to combined clinicopathological variables for predicting overall survival. The addition of this five-circRNA signature achieved superior performance than did combined clinicopathological variables alone, shown by a larger area under the receiver operating characteristic curve (AUC) (0.729 95% CI 0.679–0.779] versus 0.634 [0.579–0.688], P = 0.0004, Fig. 7). Thus, the five-circRNA signature could add significant prognostic value to existing clinicopathological prognostic factors.
a Comparisons of the prognostic accuracy by the five-circRNA signature and clinicopathological variables. b Comparisons of the prognostic accuracy by the five-circRNA signature, and circCEP112, circFAM13A, circBRWD1, circVPS13C, circMACROD2. P values show the AUC for the five-circRNA signature vs the AUC for other features. AUC area under the receiver operating characteristic curve, CI confidence interval, circRNA circular RNA, PSA prostate-specific antigen, LDH lactate dehydrogenase, ECOG Eastern Cooperative Oncology Group performance status.
In both the training and validation cohorts (Cohort I and Cohort II), this five-circRNA signature showed near-perfect calibration, with the predicted probabilities of overall survival at 24 months accurately, describing the true risk observed in all three cohorts (Supplementary Fig. S8). Compared with conditions where no prediction model would be used (i.e., all or none), the five-circRNA signature provided a high net benefit across a wide range of decision threshold probabilities (Fig. 8).
Compared with conditions where no prediction model would be used (i.e., all or none), the model with the five-circRNA signature provided a large net benefit in the training cohort (a) and the validation Cohort I (b) and Cohort II (c). “All” implied the assumption that all are at risk for death (all positive); “none” implied the assumption that no one is at risk for death (all negative). circRNA circular RNA.
Discussion
In this multi-centre, prospective cohort study, we developed and externally validated a novel prognostic tool based on five circRNAs that improves the ability to predict overall survival in mCRPC patients receiving first-line abiraterone. To our knowledge, this is the first study that has prospectively evaluated the global circRNA expression profiles from plasma-derived extracellular vesicles in mCRPC patients receiving first-line abiraterone. Our results showed that the five-circRNA signature could successfully categorise patients into high-risk and low-risk groups with significant differences in overall survival. Furthermore, we found that this five-circRNA signature was able to improve on existing clinicopathological prognostic factors for the risk stratification of mCRPC.
The difficulty in obtaining tumour sample from men with CRPC, as well as the molecular heterogeneity of tumour makes liquid biopsies-based biomarker assays crucially important to individualise management [34]. Cell-free RNA profiling, such as plasma extracellular vesicle circRNAs might offer an attractive and easy-to-use potential for prognostic biomarker development [17, 21, 35]. Recent studies have demonstrated that circRNAs may be associated with tumorigenesis [29], microRNA inhibition [36], and epithelial–mesenchymal transition [37]. Although the mechanisms of circRNAs underlying abiraterone resistance remain elusive, some noncoding RNAs are critical for the production of AR-Vs, specifically AR-V7 [10,11,12,13,14,15,16]. Thus, we hypothesised that detection of selective circRNAs in plasma extracellular vesicles from men with mCRPC would be associated with resistance to abiraterone. Interestingly, among these identified circRNAs in our study, circVPS13C and circBRWD1 were recently suggested to affect prostate cancer cell proliferation independently of their linear counterparts [22]. Although the biological roles of many candidate circRNAs on the mCRPC progression remain unknown, a lot of circRNAs have the ability to function as oncogenes or tumour suppressors in prostate cancer [22]. Our findings raise the possibility that these identified circRNAs could be crucial in the more lethal behaviour of mCRPC (e.g., drug resistance).
There are several strengths of our study. First, we employed rigorous statistical methodology in accordance with TRIPOD statement to prospectively validate that five-circRNA signature could be used as an independent biomarker to predict overall survival in mCRPC. We selected overall survival as the primary endpoint, which is considered as less subject to interpretation bias [32]. Second, blood measurements are repeatable, minimally invasive, and easily implemented during the course of treatment. Third, compared with expensive and technically challenging circulating tumour cell counts [38], the extraction and processing of plasma-derived extracellular vesicles is considerably cheap and less labour-intensive. Last, ddPCR shows many potential advantages over traditional real-time PCR for plasma nucleic acid quantification analysis, including greater precision and improved day-to-day reproducibility [39].
This study has several limitations. First, all patients are Chinese, and the effects of race on the value of the five-circRNA signature is unclear. Second, the biological roles by which many candidate circRNAs in the five-circRNA signature, such as circCEP112, circFAM13A and circMACROD2, contribute to mCRPC progression remain unknown. Third, we did not evaluate the five-circRNA signature in patients receiving other novel hormonal therapy agents (e.g., enzalutamide, apalutamide). It is for the reason that enzalutamide and apalutamide were just officially approved for clinical use in China in 2019. Herein, further investigations are warranted to elucidate the biological mechanisms behind the prognostic value of this five-circRNA signature in mCRPC patients receiving novel hormonal therapy.
In summary, our findings show that the five-circRNA signature is a potential prognostic tool for predicting overall survival in mCRPC patients receiving first-line abiraterone. It could improve existing clinicopathological prognostic factors to provide a more accurate prognosis. Thus, our findings could have therapeutic implications in directing personalised regimen selection for patients with mCRPC.
Data availability
All data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Kozminsky M, Fouladdel S, Chung JS, Wang Y, Smith DC, Alva A, et al. Detection of CTC clusters and a dedifferentiated RNA-expression survival signature in prostate cancer. Adv Sci. 2019;6:1801254.
Posadas EM, Chi KN, de Wit R, de Jonge MJA, Attard G, Friedlander TW, et al. Pharmacokinetics, safety, and antitumor effect of apalutamide with abiraterone acetate plus prednisone in metastatic castration-resistant prostate cancer: phase Ib study. Clin Cancer Res. 2020;26:3517–24.
O’Donnell A, Judson I, Dowsett M, Raynaud F, Dearnaley D, Mason M, et al. Hormonal impact of the 17alpha-hydroxylase/C(17,20)-lyase inhibitor abiraterone acetate (CB7630) in patients with prostate cancer. Br J Cancer. 2004;90:2317–25.
Attard G, Reid AH, Yap TA, Raynaud F, Dowsett M, Settatree S, et al. Phase I clinical trial of a selective inhibitor of CYP17, abiraterone acetate, confirms that castration-resistant prostate cancer commonly remains hormone driven. J Clin Oncol. 2008;26:4563–71.
Romanel A, Gasi Tandefelt D, Conteduca V, Jayaram A, Casiraghi N, Wetterskog D, et al. Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med. 2015;7:312re10.
Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013;368:138–48.
De Laere B, van Dam PJ, Whitington T, Mayrhofer M, Diaz EH, Van den Eynden G, et al. Comprehensive profiling of the androgen receptor in liquid biopsies from castration-resistant prostate cancer reveals novel intra-AR structural variation and splice variant expression patterns. Eur Urol. 2017;72:192–200.
Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–38.
Armstrong AJ, Halabi S, Luo J, Nanus DM, Giannakakou P, Szmulewitz RZ, et al. Prospective multicenter validation of androgen receptor splice variant 7 and hormone therapy resistance in high-risk castration-resistant prostate cancer: the PROPHECY study. J Clin Oncol. 2019;37:1120–9.
Wang R, Sun Y, Li L, Niu Y, Lin W, Lin C, et al. Preclinical study using Malat1 small interfering RNA or androgen receptor splicing variant 7 degradation enhancer ASC-J9((R)) to suppress enzalutamide-resistant prostate cancer progression. Eur Urol. 2017;72:835–44.
Greene J, Baird AM, Casey O, Brady L, Blackshields G, Lim M, et al. Circular RNAs are differentially expressed in prostate cancer and are potentially associated with resistance to enzalutamide. Sci Rep. 2019;9:10739.
Cao S, Ma T, Ungerleider N, Roberts C, Kobelski M, Jin L, et al. Circular RNAs add diversity to androgen receptor isoform repertoire in castration-resistant prostate cancer. Oncogene. 2019;38:7060–72.
Wu G, Sun Y, Xiang Z, Wang K, Liu B, Xiao G, et al. Preclinical study using circular RNA 17 and micro RNA 181c-5p to suppress the enzalutamide-resistant prostate cancer progression. Cell Death Dis. 2019;10:37.
Sharp A, Coleman I, Yuan W, Sprenger C, Dolling D, Rodrigues DN, et al. Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. J Clin Invest. 2019;129:192–208.
Zhang Y, Pitchiaya S, Cieslik M, Niknafs YS, Tien JC, Hosono Y, et al. Analysis of the androgen receptor-regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat Genet. 2018;50:814–24.
Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602.
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–4.
Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, et al. Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23:3918–28.
Del ReM, Biasco E, Crucitta S, Derosa L, Rofi E, Orlandini C, et al. The detection of androgen receptor splice variant 7 in plasma-derived exosomal RNA strongly predicts resistance to hormonal therapy in metastatic prostate cancer patients. Eur Urol. 2017;71:680–7.
Krug AK, Enderle D, Karlovich C, Priewasser T, Bentink S, Spiel A, et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann Oncol. 2018;29:700–6.
Huang X, Yuan T, Liang M, Du M, Xia S, Dittmar R, et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. 2015;67:33–41.
Chen S, Huang V, Xu X, Livingstone J, Soares F, Jeon J, et al. Widespread and functional RNA circularization in localized prostate. Cancer Cell. 2019;176:831–43. e22
Moons KG, Altman DG, Reitsma JB, Ioannidis JP, Macaskill P, Steyerberg EW, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162:W1–73.
Scher HI, Halabi S, Tannock I, Morris M, Sternberg CN, Carducci MA, et al. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: recommendations of the Prostate Cancer Clinical Trials Working Group. J Clin Oncol. 2008;26:1148–59.
Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228–47.
He YD, Tao W, He T, Wang BY, Tang XM, Zhang LM, et al. A urine extracellular vesicle circRNA classifier for detection of high-grade prostate cancer in patients with prostate-specific antigen 2-10 ng/mL at initial biopsy. Mol Cancer. 2021;20:96.
Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–70.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165:289–302.
Halabi S, Lin CY, Kelly WK, Fizazi KS, Moul JW, Kaplan EB, et al. Updated prognostic model for predicting overall survival in first-line chemotherapy for patients with metastatic castration-resistant prostate cancer. J Clin Oncol. 2014;32:671–7.
Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16:385–95.
Black WC, Haggstrom DA, Welch HG. All-cause mortality in randomized trials of cancer screening. J Natl Cancer Inst. 2002;94:167–73.
Gao X, Li LY, Rassler J, Pang J, Chen MK, Liu WP, et al. Prospective study of CRMP4 promoter methylation in prostate biopsies as a predictor for lymph node metastases. J Natl Cancer Inst. 2017;109:djw282.
Olmos D, Brewer D, Clark J, Danila DC, Parker C, Attard G, et al. Prognostic value of blood mRNA expression signatures in castration-resistant prostate cancer: a prospective, two-stage study. Lancet Oncol. 2012;13:1114–24.
Li Y, Zhao J, Yu S, Wang Z, He X, Su Y, et al. Extracellular vesicles long RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human blood as potential biomarkers for cancer diagnosis. Clin Chem. 2019;65:798–808.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8.
Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–34.
Gold B, Cankovic M, Furtado LV, Meier F, Gocke CD. Do circulating tumor cells, exosomes, and circulating tumor nucleic acids have clinical utility? A report of the association for molecular pathology. J Mol Diagn. 2015;17:209–24.
Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10:1003–5.
Acknowledgements
We thank all patients involved in this study.
Funding
This study was funded by National Natural Science Foundation of China (81472383, 81502206, 81772753, 81972419), Guangdong Provincial Natural Science Foundation of China (2014A030313088, 2017A030313478), Guangdong Provincial Science and Technology Provincial Science and Technology Foundation of China (2016A020215073), Guangzhou Science and Technology Foundation of China (201804010324), and the Fundamental Research Funds for the Central Universities (17ykzd21).
Author information
Authors and Affiliations
Contributions
XG and L-YL: conceptualisation. WT, Z-HL, Y-DH and B-YW: methodology and writing manuscript original draft. WT, Z-HL, Y-DH and B-YW: formal analysis. WT, T-LX, W-MD, L-XZ, X-MT and Z-AM: data collection and curation. X-MT and Z-AM: biological sample analysis and interpretation of these data. All authors: reviewing and editing the manuscript. XG and L-YL: supervision. XG and L-YL: project administration. L-YL and Y-DH: funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was conducted in compliance and in accordance with the Declaration of Helsinki. This study was approved by the Institutional Ethical Review Board at The Third Affiliated Hospital of Sun Yat-sen University, and signed informed consent was obtained for all subjects included in the study.
Consent to publish
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tao, W., Luo, ZH., He, YD. et al. Plasma extracellular vesicle circRNA signature and resistance to abiraterone in metastatic castration-resistant prostate cancer. Br J Cancer 128, 1320–1332 (2023). https://doi.org/10.1038/s41416-023-02147-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-023-02147-8
- Springer Nature Limited
This article is cited by
-
Therapeutic importance and diagnostic function of circRNAs in urological cancers: from metastasis to drug resistance
Cancer and Metastasis Reviews (2024)
-
Abiraterone
Reactions Weekly (2023)