Abstract
Current therapy for acute myeloid leukemia (AML) is largely hindered by the development of drug resistance of commonly used chemotherapy drugs, including cytarabine, daunorubicin, and idarubicin. In this study, we investigated the molecular mechanisms underlying the chemotherapy drug resistance and potential strategy to improve the efficacy of these drugs against AML. By analyzing data from ex vivo drug-response and multi-omics profiling public data for AML, we identified autophagy activation as a potential target in chemotherapy-resistant patients. In THP-1 and MV-4-11 cell lines, knockdown of autophagy-regulated genes ATG5 or MAP1LC3B significantly enhanced AML cell sensitivity to the chemotherapy drugs cytarabine, daunorubicin, and idarubicin. In silico screening, we found that chloroquine phosphate mimicked autophagy inactivation. We showed that chloroquine phosphate dose-dependently down-regulated the autophagy pathway in MV-4-11 cells. Furthermore, chloroquine phosphate exerted a synergistic antitumor effect with the chemotherapy drugs in vitro and in vivo. These results highlight autophagy activation as a drug resistance mechanism and the combination therapy of chloroquine phosphate and chemotherapy drugs can enhance anti-AML efficacy.
Similar content being viewed by others
Introduction
AML is a heterogenous malignancy of myeloid progenitor cells in the patients’ bone marrow and peripheral blood, and is characterized by the fast accumulation of clonal myeloid blasts, with immature differentiation. Increased knowledge about its molecular biology and genetic basis leads to tremendous advances of new treatments [1, 2]. The general treatment options for AML are induction therapy, consolidation therapy and Hematopoietic-Cell Transplantation [1, 3]. Most of the drugs used in these treatment regimens are cytarabine, daunorubicin and idarubicin. However, the complete remission (CR) rate of AML is not optimistic. The CR rate of first-line treatment in young and middle-aged adults is 60%–80%, and that of elderly people >65 years old is 40%–60% [4, 5]. At the same time, AML patients treated with current treatment strategies tend to have shorter remissions (less than 6 months), and most relapse within 3 years [1].
Drug resistance is considered as the leading cause of treatment failure, and much interest exists in elucidating the mechanisms of resistance to the drug [6]. Several mechanisms of resistance to cytarabine (Ara-C) and daunorubicin have been described [7]. For example, increasing the expression of the drug efflux transporter ATP-binding cassette (ABC) [8], decreasing the activity of DNA topoisomerase II [9], and the activating the apoptotic pathway [10] have been reported to be associated with daunorubicin resistance, deregulation of Ara-C metabolism [11,12,13], cell quiescence [14] and the DNA damage response [15,16,17,18] have been reported to be associated with Ara-C resistance. Based on public data mining [19] and experimental verification, we revealed that the activation of autophagy pathway may be the drug resistance mechanism of AML first-line treatment drugs, pharmacological downregulation of autophagy pathway resulted in enhanced sensitivity of AML cells to chemotherapy drugs. Our findings suggest a combination of chemotherapy agents and inactivation of autophagy pathways as a potential therapeutic strategy for AML.
Materials and methods
Nu/Nu mouse subcutaneous xenograft model evaluation
Female nu/nu mice (6–8 weeks old) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. Animal use procedures were approved by the Committee for Laboratory Animal Research Guidelines of the Shanghai Research Center for Model Organisms (IACUC approval 2023-01-LJ-140). MV-4-11 cells were resuspended in serum-free growth medium and mixed 1:1 with Matrigel (354248, CORNING, California, USA), and then 5 × 106 MV-4-11 cells were implanted subcutaneously on the right side of mice. Drug or vehicle (normal saline) was administered daily for 3 weeks after the tumor volume had grown to 100–300 mm3. Tumor volume was monitored twice weekly as an indicator of tumor growth by using digital calipers measurements. Body weight was measured twice a week. The formula for calculating tumor volume is VT = 1/2×a×b2 (Note: a and b represent length and width, respectively). If the animal loses weight due to diet reduction during the experiment, the fur of the animal is messy and dull, or the animal is injured due to fighting, etc., stop administering the drug to the animal. The maximum permitted weight loss is 15%. Animals were humanely killed when they became visibly ill, in accordance with Institutional Animal Care and Use Committee (IACUC) Ethical Protocols.
Western blotting
Cell lines were exposed to Ara-C and chloroquine phosphate (PCQ) or DMSO. Then protein lysates were made by 4× Laemmli Sample Buffer (161073, BIORAD, Virgina, USA). Protein samples were separated by SDS-polyacrylamide gel electrophoresis (1660013, BIO-RAD, Virgina, USA), and then transferred to Nitrocellulose (NC) membranes. After blocking with 5% skim milk for 50 min at room temperature, NC membranes were incubated with the primary antibody at 4 °C overnight before being washed three times with TBST. Then NC membranes were incubated with secondary antibodies for 1 h at room temperature. The signals were detected with ECL (KF8001&KF8003, Affinity, Jiangsu, China) using BIO-RAD ChemiDoc Touch Imaging system. The following antibodies were used for Western blotting analysis: β-actin (am1021b, Abcepta, Jiangsu, China), LC3B (#3868 S, CST, Massachusetts, USA) and peroxidase-conjugated AffiniPure goat anti-rabbit IgG (115-035-003) or goat anti-mouse IgG (111-035-003) from Jackson ImmunoResearch Laboratories (Pennsylvania, USA).
RT‒qPCR
Cell lines were exposed to different concentrations of PCQ, then RNA was extracted from cell lines. After reverse-engineering the RNA of the sample into cDNA, the primers of the target gene (ATG4A, ATG5, ATG12, PIK3C3, MAP1L3CB) were designed to perform PCR amplification reaction on the sample. Then fluorescent chemical materials (AceQ qPCR SYBR Green Master Mix, Q121-02, Vazyme, Nanjing, China) were added to the reaction system so that the fluorescent signal emitted by each cycle product could be monitored in realtime. A fluorescent signal amplification curve was obtained with the accumulation of reaction products to quantify the template. The expression of genes was normalized to the geometric mean of the housekeeping gene ACTB to control the variability in expression levels and was analyzed using the 2-ΔΔCt method. All samples within an experiment were reverse-transcribed at the same time. n = 3 biologically independent samples.
Primer sequence:
Gene | Primer sequence |
|---|---|
ATG4A-F | 5′-TGCTGGTTGGGGATGTATGC-3′ |
ATG4A-R | 5′-GCGTTGGTATTCTTTGGGTTGT-3′ |
ATG5-F | 5′-AAAGATGTGCTTCGAGATGTGT-3′ |
ATG5-R | 5′-CACTTTGTCAGTTACCAACGTCA-3′ |
ATG12-F | 5′-CTGCTGGCGACACCAAGAAA-3′ |
ATG12-R | 5′-CGTGTTCGCTCTACTGCCC-3′ |
PIK3C3-F | 5′-TAGGAGGAACAACGGTTTCGC-3′ |
PIK3C3-R | 5′-GCTTCTACATTAGGCCAGACTTT-3′ |
MAP1L3CB-F | 5′-GATGTCCGACTTATTCGAGAGC-3′ |
MAP1L3CB-R | 5′-TTGAGCTGTAAGCGCCTTCTA-3′ |
In vitro drug sensitivity assay
AML cell lines (1×104 cells/well) were seeded into 96-well plates containing dose gradients of PCQ or chemotherapy drugs combinations in triplicate and cultured for 3 days. Cell viability was measured by CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS, G3581, Promega, Wisconsin, USA). Cell viability was determined by comparing the 490 nm absorbance amount of drug-treated cells to that of untreated controls, which was set as 100%. n = 3 biologically independent samples.
Cell proliferation assay
Invitrogen CellTrace Cell Proliferation Kits (C34572, Invitrogen, California, USA) were used to monitor the generation of proliferating cells by FACS. Briefly, cell proliferation was tracked for four days using CellTraceTM Far Red reagent. Cells were stained with 1 μM CellTraceTM Far Red reagent on day 0, cultured with or without drug treatment at the indicated concentrations for 4 days, and then analyzed using flow cytometry (CytoFLEX Flow Cytometer, Beckman Coulter, Inc, California, USA) by 630-nm excitation and 660-nm emission filters (APC).
Lentivirus packaging and production
The endotoxin-free lentiviral vector and its packaging original vector plasmids were co-transfected into 293 T cells with HG transgene reagent(GMLCP-10, Genomeditech, Shanghai, China). After 8 h, fresh medium was added to the cells, and the cells were incubated for 48 h. Particle virus-enriched cell supernatant was concentrated to yield high-titer lentivirus. The knockdown target sequence of ATG5 is 5′-CCTGAACAGAATCATCCTTAA-3′ [20]. The knockdown target sequence of MAP1LC3B is 5′-GTGCATGTCAGTTGTGGAGAA-3′ [21].
Lentivirus infection of AML cell lines
Cells (4×104 in 250 μL of medium) were inoculated in a 24-well plate, and 10 μL of quantitative virus (MOI = 100) stock solution was added, suspended cells were centrifuged at 2000 rpm for 90 min at a flat angle and incubated for 2.5 h, and then, 250 μL of fresh medium was added per well. After 24 h, the cells were centrifuged, washed, and cultured with fresh medium. After 48 h, puromycin was added to the cells, and the cells were screened for expansion and cultivation.
Apoptosis assay
Apoptosis was detected by an Annexin V-APC/7AAD Apoptosis Detection Kit (GA1023-KGA1026, Keygentec, Nanjing, China) followed by flow cytometry analysis. Briefly, cells were treated with the PCQ or chemotherapy drugs combinations, washed with PBS, stained with Annexin V-APC/7AAD and detected using flow cytometry (CytoFLEX Flow Cytometer, Beckman Coulter, Inc, California, USA). The apoptosis rate of stained cells was counted in APC/PC5.5.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed using GSEA 4.1.0 software (https://www.gsea-msigdb.org/gsea/datasets.jsp). Significance of GSEA results was determined by the two-sided permutation test, and P-value was adjusted for multiple comparisons.
We prepared RNK files (Pearson correlation value between gene expression and drug sensitivity) and GMT files (pathway database, Reactome pathways, and GObp pathways, https://www.gsea-msigdb.org/gsea/msigdb/collections.jsp) and input the series of documents into GSEA software. The pathways were enriched according to running sum statistics and weighted Kolmogorov–Smirnov-like statistics. Finally, we ranked the pathways according to the normalized enrichment score and false discovery rate to find the significant changed Reactome pathways and GObp pathways correlated with drug resistance.
Protein interaction network analysis
We input the gene set into the STRING [22] database, selected the species as human, selected the “as tabular text output” format on the “exports” section to output the network file, then imported the network file into cytoscape [23], and used the cyroHubba [24] app to find the core protein in the protein interaction network by Maximal Clique Centrality algorithm.
The mining flowchart of repurposed drugs via connectivity map (cMAP)
The cMAP was queried with the common genes that were upregulated in chemotherapy drug-resistant AML cells (Supplementary Table S8). By choosing a “reverse mode” configuration, we searched for small molecule signatures that could reverse the input signature. Our final query result was displayed as the tau (τ) score (connectivity score) based on the weighted Kolmogorov‒Smirnov statistic. We obtained 3 lists of drugs (Ara-C, daunorubicin, and idarubicin) in the range of -100 to 100 that were pooled together by calculating a “connectivity score,” with reversing (low score) or mimicking (high score) expression signatures with input genes. Finally, the summary drug scores were recalculated with the average statistical analysis. We found that chloroquine scored below -95. Generally, we considered that τ of +90 or higher and -90 or lower was a strong score, which could be used as a hypothesis for further research. In this way, our results suggested that chloroquine can downregulate highly expressed genes in chemotherapy drug-resistant AML cells.
Statistics & Reproducibility
In cell experiments, the experimenter randomly took different replicates of the same cells and assigned them to control or experimental groups. In the animal experiment, mice were randomly assigned to the vehicle or administration groups. Comparisons of two groups were performed equal variance two-tailed Student’s t test. Significance was set at *P < 0.05, **P < 0.01, ***P < 0.001. The results were depicted as the mean ± SEM. Pearson tests were performed to calculate the correlation between samples.
Results
Autophagy is an important target in chemotherapy drug resistance
To identify targets involved in chemotherapy drug resistance, we analyzed AML patient samples from the ex vivo drug-response and multi-omics profiling public data [19], which had been subjected to 347 emerging and 168 approved drugs (included Ara-C, daunorubicin and idarubicin) screening and had detailed clinical annotations, whole exome sequencing (WES), RNA-seq data and in vitro sensitivity data (Fig. 1a). We also analyzed AML cell lines data from Depmap database [25], GDSC database [26] and past research [27] which had proteomic data, Crispr activation screening data and in vitro sensitivity data (Fig. 1a). We compared the selective drug-sensitivity scores (sDSS, the larger the value indicated the higher sensitivity to the drug) to the clinical characteristics and found a significant positive association between chemotherapy drug resistance and the activity of lactate dehydrogenase in peripheral blood plasma (p-ld), indicating that AML samples with high p-ld were more sensitive to chemotherapy drug (Fig. 1b, Supplementary Table S1). We also evaluated the relationship between the sDSS of drug sensitivity and common somatic mutations in AML (Fig. 1c) and found that no common somatic mutation was significantly associated with drug sensitivity. Based on the RNA-seq data, we calculated the Pearson correlation between global gene expression levels and sDSS. Then we performed gene set enrichment analysis (GSEA) according to the correlation score, and we found that the genes negatively correlated with sDSS were significantly enriched in the autophagy pathway (Fig. 1d), which indicated that the autophagy pathway was significantly activated in chemotherapy drug-resistant AML primary cells. In addition, previous studies have reported that the reduced activity of lactate dehydrogenase is associated with the activation of the autophagy pathway [28, 29]. Then we calculated the correlation of all drug sDSSs to global gene expression in the public database. And the upregulation degree of all autophagy pathways in drug-resistant samples was calculated according to the Pearson correlation value and GSEA (Supplementary Table S2. The lower the NES indicated the higher upregulated degree of autophagy in the drug-resistant sample). Autophagy pathways were significantly upregulated in samples resistant to Ara-C, daunorubicin, and idarubicin, whereas there were no significant changes in autophagy pathways in samples resistant to other approved anti-AML targeted inhibitors such as Midostaurin and Venetoclax (Supplementary Table S2).
a Schematic diagram of identifying biomarkers associated with chemotherapy resistance based on in vitro screening data of chemotherapy drugs at the level of AML primary cells and cell lines (Clinical, genomic, transcriptomic, proteomic and CRISPR screening data). b Venn analysis of clinical indicators significantly associated with drug sensitivity (sDSS) of three chemotherapeutic drugs (cytarabine, daunorubicin, idarubicin). c The sDSS of three chemotherapy drugs in AML common somatic mutation groups. sDSS in per group are presented as mean ± SEM. d GSEA enrichment profile of genes associated with resistance to three chemotherapy drugs.
We further detected the drug sensitivity data (IC50) of 10 AML cell lines to Ara-C (Fig. 2a, Supplementary Table S3) and collected the drug sensitivity data (IC50) of 27 AML cell lines to Ara-C in the GDSC database (Fig. 2a, Supplementary Table S4). Based on the common cell line data of the 2 types of data, our drug sensitivity test results have a significant positive correlation with public databases (Fig. 2a). We then collected the proteomic data of all AML cell lines in the Depmap database and calculated the Pearson correlation between the IC50 of Ara-C and the global protein expression level of the local (Fig. 2b, Supplementary Table S5) and GDSC databases (Fig. 2c, Supplementary Table S6). GSEA was performed according to the correlation score, and we found that the proteins positively correlated with IC50 were significantly enriched in the autophagy pathway (Fig. 2b, c). At the same time, the CRISPR activation (CRISPRa) screening data of MV-4-11 cell line showed that Ara-C resistance-related activated transcripts were mainly enriched in the autophagy pathway (Fig. 2d, Supplementary Table S7). The above data of AML cell lines indicated that the autophagy pathway was significantly activated in chemotherapy drug-resistant AML cell lines.
a Based on the cytotoxicity test data (IC50) of Ara-C in 10 AML cell lines and the IC50 data of 27 AML cell lines in GDSC data, Pearson correlation analysis was performed between the two types of data. GSEA enrichment profile of Ara-C resistance-related proteins based on the correlation analysis between the AML cell line proteome and the IC50 of Ara-C in local experiments (b) and GDSC database (c). d GSEA enrichment profile of Ara-C resistance-related transcripts based on the change of sgRNA count in CRISPRa screening data of MV-4-11. e An overlap analysis of the core genes upregulated in the autophagy pathway in three chemotherapy drug-resistant samples from public databases (left). The interaction network of proteins (from the result of overlap analysis) based on the String database and analyzed by Maximal Clique Centrality algorithm (right).
Therefore, clinical indicators, transcriptome data, proteomic data, and CRISPRa screening data suggested autophagy pathway activation as a potential target in chemotherapy-resistant patients.
We performed an overlap analysis of the core genes upregulated in the autophagy pathway in three chemotherapy drug-resistant samples (cytarabine, daunorubicin and idarubicin) in public databases by GSEA (Fig. 2e). We found 21 core genes that were upregulated in common. Based on the String database [22], then we explored the interaction network of the proteins encoded by these 21 genes. We further used the Cytoscape [23] software package cytoHubba [24] to determine the most important node proteins in the protein interaction network based on the Maximal Clique Centrality algorithm, and finally found the most important core protein (the highest score of interaction in the network): LC3B (gene symbol: MAP1LC3B), which is also closely related to autophagy pathway. LC3B is one of the members of the LC3 protein family [30]. The LC3 family is involved in the early stage of autophagosome membrane elongation and is indispensable for the autophagy process of mammalian cells [31].
The knockdown of autophagy-related genes enhanced chemotherapy drug efficacy
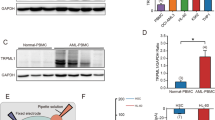
Since autophagy-related gene 5 (ATG5) is also a core autophagy gene upregulated in cytarabine-resistant samples, and ATG5 is an essential gene for autophagosome formation [32]. To determine whether the knockdown of autophagy-regulated genes could enhance the response to chemotherapy drug, we generated stable ATG5 and MAP1LC3B knockdown (KD) MV-4-11 and THP-1 cell lines by lentivirus-mediated shRNA, validated at the protein and mRNA levels (Fig. 3a, b). Accordingly, in the in vitro drug sensitivity assay of MV-4-11 and THP-1, the knockdown of ATG5 and MAP1LC3B could significantly reduce the IC50 of Ara-C, daunorubicin and idarubicin (Fig. 3c, d), but not obviously affect the IC50 of Venetoclax (Fig. 3c, d), which is consistent with the results of Supplementary Table S2. In the cell proliferation assay, the knockdown of ATG5 or MAP1LC3B could enhance Ara-C, daunorubicin, and idarubicin-induced proliferation inhibition (Fig. 3e, f). The results proved that the knockdown of ATG5 or MAP1LC3B could specifically increase the sensitivity of MV-4-11 and THP-1 to three chemotherapy drugs (Ara-C, daunorubicin, and idarubicin).
ATG5 or MAP1LC3B knockdown in THP-1 and MV-4-11 cell lines, as confirmed by Western blotting (a) and qPCR (b). In vitro drug sensitivity assay of Ara-C, daunorubicin and idarubicin with or without knockdown of ATG5 and MAP1LC3B in THP-1 (c) and MV-4-11 (d) cell lines. Cell viability was measured by CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS). Significance was analyzed by equal variance two-tailed t test. CellTrace™ Far Red staining of THP-1 (e) and MV-4-11 (f) cells with or without knockdown of ATG5 and MAP1LC3B treated with DMSO, Ara-C, daunorubicin and idarubicin for 4 days.
Repurposed drugs targeting autophagy enhance chemotherapy drug efficacy
We further speculate that targeting autophagy pathways and drug resistance-related genes may enhance chemotherapy drug sensitivity and overcome drug resistance. Connectivity Map (CMAP) is a collection of genome-wide gene expression reads from cell lines treated with more than 2000 chemicals to find compounds of interest based on expression profile similarities [33, 34]. We input three lists of top 100 genes significantly associated with resistance to three chemotherapy drugs (Ara-C, daunorubicin, and idarubicin) into the CMAP database (Fig. 4a, Supplementary Table S8). The connectivity scores obtained (see Methods for details) represent either reversed (low scores) or mimicked (high scores) expression signatures of the input genes (Fig. 4a). Total drug scores were calculated using average statistical analysis (Supplementary Table S9). Chloroquine appeared in the top 10 drugs that could downregulate the input genes (Supplementary Table S9). We found that chloroquine phosphate (PCQ) could significantly downregulate the autophagy pathway of MV-4-11 (Fig. 4b). RT‒qPCR results showed that the concentration gradient of PCQ could significantly reduce the expression of autophagy-related genes in the MV-4-11 cell line.
a The drug repurposing discovery process of targeting top 100 chemotherapy resistance-associated genes via CMAP analysis. A score ≤ -90 indicated significant downregulation. The score of chloroquine is -95 and ranked in the top ten. b Effects of doses gradient PCQ on mRNA levels of autophagy-related genes in MV-4-11 (24 h). c Cytotoxicity of PCQ alone in MV-4-11 and THP-1 cell lines. Cytotoxicity of combination of PCQ with Ara-C, daunorubicin and idarubicin in THP-1 (d) and MV-4-11 (e) cell lines, Cell viability was measured by CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS). Significance was analyzed by equal variance two-tailed t test. **P < 0.01, *** P < 0.001. CellTrace™ Far Red staining of THP-1 (f) and MV-4-11 (g) cells treated with DMSO, Ara-C, daunorubicin and idarubicin in the absence or presence of PCQ (10 μM) for 4 days.
Meanwhile, PCQ alone did not significantly inhibit cell viability in AML cell lines MV-4-11 and THP-1 (Fig. 4c), but PCQ can significantly synergistically increase the activity of cytarabine, daunorubicin and idarubicin in MV-4-11 and THP-1 in cell growth and cell proliferation assays (Fig. 4d–g). We further found that PCQ significantly enhanced the apoptosis induced by Ara-C, daunorubicin and idarubicin in MV-4-11 and THP-1 cells (Fig. 5a, b).
a Cell apoptosis of MV-4-11 and THP-1 cells treated with DMSO, Ara-C (1000 nM) or daunorubicin (5 nM or 15 nM) or idarubicin (1 nM or 5 nM) in the absence or presence of PCQ (10 μM) for 48 h. b Quantification of the apoptosis changes in (a) with three independent experiments. Significance was analyzed by equal variance two-tailed t test. *** P < 0.001. c The tumor growth of MV-4-11 cells treated with vehicle (normal saline, n = 10 mice, s.c), Ara-C (20 mg/kg, n = 4 mice, s.c), PCQ (50 mg/kg, n = 4 mice, i.p), or the combination (n = 4 mice) in the subcutaneous transplantation tumor model (nu/nu mice). Data are means ± SEM. Significance was analyzed by equal variance two-tailed t test. *P < 0.05.
To determine whether PCQ could enhance Ara-C sensitivity in vivo, we established a xenograft tumor model by subcutaneous inoculation of MV-4-11 cells into nu/nu mice and treated the animals with vehicle (normal saline, NS), the single agent PCQ, Ara-C, or the combination when the tumors reached 100–300 mm3 (Fig. 5c, Supplementary Table S10). PCQ (50 mg/kg, daily) alone caused no significant changes in tumor growth (P = 0.349), while Ara-C (20 mg/kg, daily) alone slowed tumor growth compared to the vehicle group but not significantly (P = 0.099). Importantly, the combination of PCQ and Ara-C significantly reduced the tumor burden compared with vehicle treatment (P = 0.043).
Collectively, the findings that PCQ could enhance chemotherapy drugs sensitivity in vitro and in vivo highlight that the autophagy pathway could be successfully targeted by chloroquine with apparent efficacy.
Discussion
Autophagy captures, degrades, and recycles intracellular proteins and organelles in lysosomes, which is a survival-enhancing pathway. Although in some cases autophagy suppresses tumorigenesis, in most cases autophagy promotes tumorigenesis [35]. In the environment of hypoxia and nutrient deficiency, tumor cells upregulate the autophagy pathway to maintain their survival [36]. Autophagy supplies metabolic substrates by recycling intracellular components to meet the high metabolic and energy demands of proliferative tumors [37, 38]. Autophagy can be inhibited by deleting Beclin 1 to increase cell death [36, 39]. In addition, RAS-driven cancers often have an activated autophagy pathway, RAS-activating mutations increase autophagy, thereby promoting tumor growth, survival, and tumorigenesis, and are associated with the development of some fatal cancers [40,41,42].
Several studies have shown that the resistance of cancer cells to multiple anticancer drugs can be increased through the upregulation of autophagy [43, 44]. Autophagy is a protective mechanism of cancer cells undergoing anticancer therapy. The efficacy of chemotherapy drugs in many cancers is limited by the unintended induction of protective autophagy, for example, as a mechanism of cisplatin-mediated drug resistance, autophagy promotes drug resistance in ovarian cancer through regulation of the ERK pathway and overexpression of Beclin 1 [45, 46]. Therefore, targeting autophagy is an important strategy for cancer treatment. Cisplatin therapy combined with autophagy inhibition significantly increased the cytotoxicity of esophageal cancer [47]. Another study showed that the inhibition of autophagy leads to the promotion of apoptosis and the therapeutic effect of anticancer therapy [48]. Preclinical studies show that chloroquine or hydroxychloroquine can inhibit cancer cell growth by inhibiting autophagy in bladder and pancreatic cancer [49, 50].
However, there are still few studies on the relationship between autophagy pathway and the mechanism of first-line chemotherapy resistance in AML. The drug resistance mechanism of Ara-C and daunorubicin is currently considered to be mainly related to drug metabolism, DNA damage and the activation of DNA topoisomerase II [9, 11,12,13, 15, 17, 18]. Combined with in vitro drug sensitivity data, we found that the autophagy pathway is the important target in first-line chemotherapy drug resistance of AML by analyzing the transcriptome and clinical data of the public database of AML patients, the proteome and CRISPRa screening data of the public database of AML cell lines [19, 25,26,27] (Figs. 1 and 2). Autophagy pathway is significantly activated in AML primary cells and AML cell lines resistant to chemotherapy drugs (Ara-C, daunorubicin, idarubicin). At the same time, consistent with previous studies, the activity of lactate dehydrogenase in drug-resistant primary cells is also decreased when the autophagy pathway is activated [28, 29].
In additiion, based on protein interaction network analysis (Fig. 2e), we found that MAP1LC3B may be the key gene upregulated in chemotherapy-resistant samples. Since ATG5 is also a core autophagy gene upregulated in cytarabine-resistant samples, we showed that the knockdown of ATG5 and MAP1LC3B (Fig. 3) can increase the sensitivity of MV-4-11 and THP-1 to three chemotherapy drugs (cytarabine, daunorubicin, and idarubicin) but not BCL2 inhibitor Venetoclax by in vitro drug sensitivity assay (MTS) and cell proliferation assay (CellTraceTM Far Red), which proved activation of autophagy-related pathways may be a specific feature of chemotherapy drug resistance (Fig. 3, Supplementary Table S2). ATG5 and MAP1LC3B are important genes in autophagy [30,31,32], targeting ATG5 has been reported to increase the sensitivity of pancreatic and lung cancers to chemotherapy drugs [51, 52], and it has been reported that inhibition of LC3B can increase the sensitivity of chemotherapy drug in ovarian cancer cells [53], and LC3B is also a marker of poor prognosis in triple-negative breast cancer [54]. Our findings further expanded the understanding of the relationship between ATG5, LC3B and chemotherapy drug resistance.
Currently, Ara-C and daunorubicin are still the mainstays of first-line chemotherapy for AML. To develop more advanced treatment options, drug repurposing is an attractive strategy that can be used to develop drugs beyond their original use [55], and CMAP containing integrated cellular signatures based on network pharmacology has been used to address drug repurposing in AML [56]. Our CMAP analysis showed that chloroquine, a known autophagy inhibitor, could significantly downregulate the expression of genes significantly and positively associated with chemotherapy drug resistance (Fig. 4a). Chloroquine, as an approved antimalarial drug, has had good antitumor therapeutic effects in clinical trials [57, 58]. Impressively, chloroquine phosphate (PCQ) enhanced the sensitivity of cytarabine, daunorubicin and idarubicin, exhibiting a significant synergy with chemotherapy drugs in vitro and in vivo (Figs. 4d–g and 5). Mechanistically, chloroquine directly inhibits autophagy by changing lysosomal pH, inhibiting autophagic degradation, and autophagosome accumulation [59, 60]. We have experimentally demonstrated that chloroquine can inhibit the autophagy pathway in AML cell lines (Fig. 4b). As the results of public data analysis show that autophagy is an important target of chemotherapy drug resistance in AML, this may be the reason why chloroquine can have a synergistic effect with chemotherapy drugs.
In summary, we have identified autophagy activation as a key resistance mechanism to first-line chemotherapeutics in AML, and further determined that chloroquine, an existing drug targeting the autophagy pathway, combined with chemotherapeutics can overcome drug resistance and improve the efficacy of anti-AML. We recommend future clinical studies to evaluate combination therapy of chemotherapeutic agents and inhibition of autophagy as a next-generation AML treatment strategy.
References
Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–52.
Estey EH. Treatment of acute myeloid leukemia. Haematologica. 2009;94:10–16.
Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–47.
Luppi M, Fabbiano F, Visani G, Martinelli G, Venditti A. Novel agents for acute myeloid leukemia. Cancers (Basel). 2018;10:429.
Megías-Vericat J, Rojas L, Herrero M, Bosó V, Montesinos P, Moscardó F, et al. Influence of ABCB1 polymorphisms upon the effectiveness of standard treatment for acute myeloid leukemia: a systematic review and meta-analysis of observational studies. Pharmacogenomics J. 2015;15:109–18.
Zhang J, Gu Y, Chen B. Mechanisms of drug resistance in acute myeloid leukemia. OncoTargets Ther. 2019;12:1937–45.
Arwanih EY, Louisa M, Rinaldi I, Wanandi SI. Resistance mechanism of acute myeloid leukemia cells against daunorubicin and cytarabine: a literature review. Cureus. 2022;14:e33165.
Shustik C, Dalton W, Gros P. P-glycoprotein-mediated multidrug resistance in tumor cells: biochemistry, clinical relevance and modulation. Mol Asp Med. 1995;16:1–78.
Ganapathi RN, Ganapathi MK. Mechanisms regulating resistance to inhibitors of topoisomerase II. Front Pharmacol. 2013;4:89–89.
Xu J, Patel NH, Gewirtz DA. Triangular relationship between p53, autophagy, and chemotherapy resistance. Int J Mol Sci. 2020;21:8991.
Cai J, Damaraju VL, Groulx N, Mowles D, Peng Y, Robins MJ, et al. Two distinct molecular mechanisms underlying cytarabine resistance in human leukemic cells. Cancer Res. 2008;68:2349–57.
Veuger MJ, Heemskerk MH, Honders MW, Willemze R, Barge RM. Functional role of alternatively spliced deoxycytidine kinase in sensitivity to cytarabine of acute myeloid leukemic cells. Blood J Am Soc Hematol. 2002;99:1373–80.
Veuger MJ, Honders MW, Willemze R, Barge RM. Deoxycytidine kinase expression and activity in patients with resistant versus sensitive acute myeloid leukemia. Eur J Haematol. 2002;69:171–8.
Di Tullio A, Rouault-Pierre K, Abarrategi A, Mian S, Grey W, Gribben J, et al. The combination of CHK1 inhibitor with G-CSF overrides cytarabine resistance in human acute myeloid leukemia. Nat Commun. 2017;8:1–12.
Cho SH, Toouli CD, Fujii GH, Crain C, Parry D. Chk1 is essential for tumor cell viability following activation of the replication checkpoint. Cell Cycle. 2005;4:131–9.
Loegering D, Arlander SJ, Hackbarth J, Vroman BT, Roos-Mattjus P, Hopkins KM, et al. Rad9 protects cells from topoisomerase poison-induced cell death. J Biol Chem. 2004;279:18641–7.
Mesa RA, Loegering D, Powell HL, Flatten K, Arlander SJ, Dai NT, et al. Heat shock protein 90 inhibition sensitizes acute myelogenous leukemia cells to cytarabine. Blood. 2005;106:318–27.
Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–39.
Malani D, Kumar A, Brück O, Kontro M, Yadav B, Hellesøy M, et al. Implementing a functional precision medicine tumor board for acute myeloid leukemia. Cancer Discov. 2022;12:388–401.
Lenain C, Gusyatiner O, Douma S, van den Broek B, Peeper DS. Autophagy-mediated degradation of nuclear envelope proteins during oncogene-induced senescence. Carcinogenesis. 2015;36:1263–74.
Zhao X, Liu L, Jiang Y, Silva M, Zhen X, Zheng W. Protective effect of metformin against hydrogen peroxide-induced oxidative damage in human retinal pigment epithelial (RPE) cells by enhancing autophagy through activation of AMPK pathway. Oxid Med Cell Longev. 2020;2020:1–14.
Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–D646.
Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–2.
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. CytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8:1–7.
Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER, et al. Next-generation characterization of the cancer cell line encyclopedia. Nature. 2019;569:503–8.
Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, et al. Genomics of drug sensitivity in cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2012;41:D955–D961.
Bester AC, Lee JD, Chavez A, Lee YR, Nachmani D, Vora S, et al. An integrated genome-wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell. 2018;173:649–64.e20.
Yang Y, Su D, Zhao L, Zhang D, Xu J, Wan J, et al. Different effects of LDH-A inhibition by oxamate in non-small cell lung cancer cells. Oncotarget. 2014;5:11886–96.
Zhao Z, Han F, Yang S, Wu J, Zhan W. Oxamate-mediated inhibition of lactate dehydrogenase induces protective autophagy in gastric cancer cells: involvement of the Akt–mTOR signaling pathway. Cancer Lett. 2015;358:17–26.
Mikhaylova O, Stratton Y, Hall D, Kellner E, Ehmer B, Drew AF, et al. VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell. 2012;21:532–46.
Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE‐16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–802.
Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–68.
Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–35.
Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–52.e17.
White E. The role for autophagy in cancer. J Clin Invest. 2015;125:42–6.
Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64.
Liu EY, Ryan KM. Autophagy and cancer–issues we need to digest. J Cell Sci. 2012;125:2349–58.
Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–8.
White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer autophagy in cancer therapy. Clin Cancer Res. 2009;15:5308–16.
Goel S, Huang J, Klampfer L. K-Ras, intestinal homeostasis and colon cancer. Curr Clin Pharmacol. 2015;10:73–81.
Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–70.
Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–31.
Chen Z, Jiang Q, Zhu P, Chen Y, Xie X, Du Z, et al. NPRL2 enhances autophagy and the resistance to Everolimus in castration‐resistant prostate cancer. Prostate. 2019;79:44–53.
Xiao X, Wang W, Li Y, Yang D, Li X, Shen C, et al. HSP90AA1-mediated autophagy promotes drug resistance in osteosarcoma. J Exp Clin Cancer Res. 2018;37:1–13.
Bao L, Jaramillo MC, Zhang Z, Zheng Y, Yao M, Zhang DD, et al. Induction of autophagy contributes to cisplatin resistance in human ovarian cancer cells. Mol Med Rep. 2015;11:91–8.
Wang J, Wu GS. Role of autophagy in cisplatin resistance in ovarian cancer cells. J Biol Chem. 2014;289:17163–73.
Cheng C, Liu J, Wang J, Li Y, Pan J, Zhang Y. Autophagy inhibition increased the anti-tumor effect of cisplatin on drug-resistant esophageal cancer cells. J Biol Regul Homeost Agents. 2017;31:645–52.
Jin F, Wang Y, Li M, Zhu Y, Liang H, Wang C, et al. MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis. 2018;8:e2540.
Frieboes HB, Huang JS, Yin WC, McNally LR. Chloroquine-mediated cell death in metastatic pancreatic adenocarcinoma through inhibition of autophagy. JOP. 2014;15:189–97.
Lin YC, Lin JF, Wen SI, Yang SC, Tsai TF, Chen HE, et al. Chloroquine and hydroxychloroquine inhibit bladder cancer cell growth by targeting basal autophagy and enhancing apoptosis. Kaohsiung J Med Sci. 2017;33:215–23.
Wang ZC, Huang FZ, Xu HB, Sun JC, Wang CF. MicroRNA-137 inhibits autophagy and chemosensitizes pancreatic cancer cells by targeting ATG5. Int J Biochem Cell Biol. 2019;111:63–71.
Chen J, Zhang L, Zhou H, Wang W, Luo Y, Yang H, et al. Inhibition of autophagy promotes cisplatin-induced apoptotic cell death through Atg5 and Beclin 1 in A549 human lung cancer cells. Mol Med Rep. 2018;17:6859–65.
Tang J, Zhu J, Ye Y, Liu Y, He Y, Zhang L, et al. Inhibition LC3B can increase chemosensitivity of ovarian cancer cells. Cancer Cell Int. 2019;19:1–15.
Lefort S, Joffre C, Kieffer Y, Givel AM, Bourachot B, Zago G, et al. Inhibition of autophagy as a new means of improving chemotherapy efficiency in high-LC3B triple-negative breast cancers. Autophagy. 2014;10:2122–42.
Chong CR, Sullivan DJ. New uses for old drugs. Nature. 2007;448:645–6.
Valli D, Gruszka AM, Alcalay M. Has drug repurposing fulfilled its promise in acute myeloid leukaemia? J Clin Med. 2020;9:1892.
Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–66.
Sotelo J, Briceño E, López-González MA. Adding chloroquine to conventional treatment for glioblastoma multiforme: a randomized, double-blind, placebo-controlled trial. Ann Intern Med. 2006;144:337–43.
Manic G, Obrist F, Kroemer G, Vitale I, Galluzzi L. Chloroquine and hydroxychloroquine for cancer therapy. Mol Cell Oncol. 2014;1:e29911.
Njaria PM, Okombo J, Njuguna NM, Chibale K. Chloroquine-containing compounds: a patent review (2010–2014). Expert Opin Ther Pat. 2015;25:1003–24.
Acknowledgements
This work was supported by National Natural Science Foundation of China (81821005), Guangdong High-level New R&D Institute (2019B090904008), Guangdong High-level Innovative Research Institute (2021B0909050003), Science and Technology Commission of Shanghai Municipality (18431907100 and 19430750100).
Author information
Authors and Affiliations
Contributions
These authors participated in conception and design: YBZ, JL, HLW. These authors participated in bioinformatics data analysis: HLW, GHL. These authors participated in development of methodology: HLW, WJK, JNL, GYX, NS, GHL, WBW, BF, JFF, YTT, MML, RX. These authors participated in analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): YBZ, JL, HLW, GHL, JNL. These authors participated in writing, review, and/or revision of the manuscript: YBZ, HLW, JL, GW, JNL. These authors participated in study supervision: YBZ, JL, GW.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Hl., Li, Jn., Kan, Wj. et al. Chloroquine enhances the efficacy of chemotherapy drugs against acute myeloid leukemia by inactivating the autophagy pathway. Acta Pharmacol Sin 44, 2296–2306 (2023). https://doi.org/10.1038/s41401-023-01112-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41401-023-01112-8
- Springer Nature Singapore Pte Ltd.