Abstract
Citalopram is commonly prescribed to patients suffering from major depressive disorder. Some of them do not respond adequately to therapy with citalopram, while many of them experience type A adverse drug reactions. Current research revealed that CYP2C19 isoenzyme is involved in the biotransformation of citalopram. The objective of our study was to investigate the impact of 681G>A polymorphism of the CYP2C19 gene on the efficacy, safety and the concentration/dose indicator of citalopram. Our study enrolled 130 patients with major depressive disorder and comorbid alcohol use disorder (average age–38.7 ± 14.1 years). Therapy regimen included citalopram in an average daily dose of 31.1 ± 14.4 mg per week. Therapy efficacy and safety were evaluated using the international psychometric scales. For genotyping, we performed the real-time polymerase chain reaction. Our findings revealed the statistically significant results in terms of the treatment efficacy evaluation (HAMD scores at the end of the treatment course): (GG) 8.0 [8.0; 9.0] and (GA) 10.0 [9.0; 11.0], p < 0.001. In the safety profile (the UKU scores), the statistical significance was also obtained: (GG) 3.0 [3.0; 4.0] and (GA) 5.0 [4.0; 5.0], p < 0.001. We revealed a statistical significance for concentration/dose indicator of citalopram in patients with different genotypes: (GG) 2.543 [1.659; 4.239] and (GA) 4.196 [2.643; 5.753], p < 0.001). The effect of CYP2C19 genetic polymorphism on the efficacy and safety profiles of citalopram was demonstrated in a group of 130 patients with major depressive disorder.
Similar content being viewed by others
Introduction
Depressive disorders are among the most common comorbid psychiatric disorders in patients with alcohol use disorder [1]. Today antidepressants remain a cornerstone in the treatment of depression, and citalopram is a selective serotonin-reuptake inhibitor antidepressant, which is commonly used in the treatment of major depressive disorders [2]. Although clinical practice guidelines on the management of depressive disorders recommend antidepressant treatment, the proportion of resistant patients and those with dose-dependent adverse drug reactions remains high [3].
Most antidepressants are metabolized by the CYP2C19 isoenzyme [4]. Meanwhile, the CYP2C19 gene is highly polymorphic, which can affect the CYP2C19 isoenzyme activity [5]. Such changes may affect the metabolism of xenobiotic drug substrates of the enzyme, modifying the drug efficacy and safety profiles [6]. Research has found that people fall into one of four general metabolizer categories distinguished by their metabolic rates: extensive metabolizers considered a normal metabolic rate; poor metabolizers carrying mutations in the CYP2C19 gene, which may lead to a decreased functional metabolic activity resulting in an increased risk of dose-dependent adverse drug reactions; intermediate metabolizers having a mutation in only one of the homologous chromosomes, which reduces the CYP2C19 metabolic activity, but to a lesser degree than in poor metabolizers; and ultra-rapid metabolizers having the congenital increased CYP2C19 metabolic activity leading to the accelerated elimination of substrate drugs and the reduced treatment efficacy [7]. Hence, changes in the efficacy and safety profiles of citalopram may depend on, inter alia, changes in genes encoding pharmacodynamics and pharmacokinetics and, in particular, CYP2C19. Currently, a number of studies demonstrating the effect of CYP2C19 on the efficacy and safety of citalopram in patients with lucid depression have been conducted [8].
The objective of our study was to assess the impact of 681G>A polymorphism of the CYP2C19 gene on the efficacy, safety and the concentration/dose indicator of citalopram in patients suffering from major depressive disorder and comorbid alcohol use disorder.
Material and methods
Clinical characteristics of the study subjects
The study enrolled 130 male patients (average age–38.67 ± 14.08 years). The inclusion criteria were the dual diagnosis of “Depressive episode (F32.x, according to ICD-10)” and “Mental and behavioral disorders due to use of alcohol. Dependence syndrome. Currently abstinent but in a protected environment (F.10.212)”; signed informed consent, and 8-weeks citalopram monotherapy. Exclusion criteria were the diagnosis of any other mental disorder; severe somatic disorders (except alcoholic hepatitis and toxic encephalopathy); presence of any other psychotropic medications in treatment regimen; creatinine clearance values <50 mL/min, creatinine plasma concentration >1.5 mg/dL (133 mmol/L), bodyweight less than 60 kg or greater than 100 kg, age of 75 years or more and presence of any contraindications for citalopram use.
Therapy efficacy and safety evaluation
To evaluate citalopram efficacy, two international psychometric scales were used: Hospital Anxiety and Depression Scale (HADS) [9] and Hamilton Depression Rating Scale (HAMD) [10]. The safety profile was evaluated using the UKU Side-Effect Rating Scale (UKU) [11]. Patients were examined on weeks 1, 4, and 8 of citalopram therapy.
Genotyping
For genotyping, venous blood samples were collected into VACUETTE® (Greiner Bio-One, Austria) vacuum tubes on the 8 week of citalopram therapy. The DNA amplifiers “Dtlite” by DNA Technology (Moscow, Russia), CFX96 Touch Real-Time System with CFX Manager software by Bio-Rad Laboratories Inc. (Hercules, CA, USA) and the “SNP-screen” sets by “Syntol” (Moscow, Russia) were used to perform the real-time polymerase chain reaction in order to determine the single-nucleotide polymorphisms (SNPs) 681G>A of the CYP2C19 (rs4244285) gene. We used two allele-specific hybridizations in every “SNP-screen” set, which have enabled us to determine separately two alleles of the studied SNP on two fluorescence channels.
Local ethical committee
The local ethical committee of the Russian Medical Academy of Continuous Professional Education of the Ministry of Health of the Russian Federation approved the research (The protocol No. 6 from 5/16/2017).
Statistical analysis
Statistical analysis of the results was performed with nonparametric methods using the “Statsoft Statistica v. 10.0” (Dell Statistica, Tulsa, OK, USA). The normality of sample distribution was evaluated using the W-Shapiro–Wilk test and taken into account when choosing a method. The differences were considered statistically significant at р < 0.05 (power above 80%). Two samples of continuous independent data were compared using the Mann–Whitney U-test, which takes into account the abnormal nature of data distribution, with further correction of the obtained p value using the Benjamin–Hochberg test, due to the multiple comparison procedure. Several samples of continuous data were analyzed using the Kruskal–Wallis H-test. Correlation analysis was performed using the Spearman nonparametric test, taking into account the abnormal nature of sample distribution. Pearson’s Chi2 test for evaluation of the sampling distribution of the alleles (Hardy–Weinberg equilibrium) has been used. Research data are presented in the form of the median and interquartile range (Me [Q1; Q3]), or in case of their normal distribution, as the arithmetic mean and standard deviation (Mean ± SD).
Study results
The CYP2C19 genotyping by polymorphic marker 681G>A (rs4244285) performed in 130 subjects have revealed the following:
-
The amount of patients with the GG genotype (extensive metabolizers) was 84 (64.6%);
-
The number of patients with the GA genotype (intermediate metabolizers) was 46 (35.4%);
-
There were no subjects with the AA genotype (poor metabolizers).
Genotype distributions didn’t follow a Hardy–Weinberg equilibrium (Chi2 = 6.01, p value = 0.01).
The results of data analysis performed for psychometric assessments (HADS, HAMD) and side-effect rating scale (UKU) on weeks 1, 4, and 8 in patients who received citalopram can be found in Table 1.
Dynamics of changes in HAMD scale scores among the patients with different genotypes are demonstrated in Fig. 1. At the baseline, there were no statistically significant differences across patients with different genotypes: (GG) 22.0 [21.0; 23.0], (GA) 22.0 [21.0; 23.0], p = 0.868. By week 4, statistically significant differences were not revealed, as well: (GG) 13.5 [11.0; 15.2], (GA) 14.0 [14.0; 15.0], p = 0.033. On the 8th week of the study, statistically significant differences were found: (GG) 8.0 [8.0; 9.0], (GA) 10.0 [9.0; 11.0], p < 0.001. For the HADS scale, the same dynamics of changes in scores was demonstrated.
Dynamics of changes in the UKU scores among the patients are presented in Fig. 1. At the baseline, there were no statistically significant differences: (GG) 0.0 [0.0; 0.0], (GA) 0.0 [0.0; 0.0], p = 0.673. By week 4, statistically significant differences were not revealed, as well: (GG) 3.0 [2.0; 3.0], (GA) 3.0 [3.0; 4.0], p = 0.033. A statistically significant difference was revealed on week 8 of therapy: (GG) 3.0 [3.0; 4.0], (GA) 5.0 [4.0; 5.0], p < 0.001.
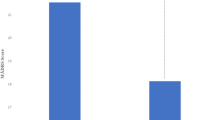
Table 2 summarizes the data on concentration/dose ratio (C/D) values obtained for citalopram through pharmacokinetic studies in terms of quantitative and nominal units. We revealed a statistical significance for C/D indicator in patients with different genotypes: (GG) 2.543 [1.659; 4.239], (GA) 4.196 [2.643; 5.753], p < 0.001 (Fig. 2).
Discussion
The findings of this study revealed a statistically significant difference between the values of citalopram equilibrium concentration in patients with different CYP2C19 genotypes by polymorphic marker 681G>A (rs4244285): patients carrying the A allele have a lower level of drug equilibrium concentration than those with the G allele (p < 0.001). This appears to be due to the reduced biotransformation and elimination rates of citalopram in patients carrying the A allele, which in turn leads to a cumulation of the drug in plasma. It may lead to an increased risk of adverse drug reactions and pharmacoresistance. Our results are consistent with the results of previous studies, which also demonstrate a decrease in the metabolic rate of citalopram in the A allele carriers [8].
Statistical analysis of data on the clinical efficacy profile of citalopram in patients carrying different genotypes by the polymorphic marker 681G>A of the CYP2C19 gene (rs4244285) revealed the statistically significant differences (p < 0.001). The analysis of citalopram safety data also demonstrated the statistically significant difference (p < 0.001). Values of these parameters were significantly higher in the carriers of the minor allele in comparison with individuals with the dominant allele. This may indicate that carriage of the polymorphic marker may lead to an increased risk of the adverse drug reactions of citalopram, which is probably due to a reduction of CYP2C19 activity in these patients, a decrease in its biotransformation and elimination rates, and accumulation of the drug in plasma.
Discussing the study limitations, it should be noted that all patients had comorbid alcohol use disorder, but the severity of liver damage did not differ in patients with different genotypes. We included only male patients in our study to exclude the influence of gender on the effectiveness of therapy.
Thus, based on the results showing that the genetic polymorphism affects the efficacy and safety rates of citalopram in patients with major depressive disorder, we can assume that it is reasonable to regard the genotyping results before prescribing citalopram to such cohort of patients. According to the results of our earlier studies, it was also shown that polymorphism of the CYP2C19 gene should be taken into account when administering citalopram, as this may have an impact on the efficacy and safety profiles of citalopram [12,13,14].
Conclusion
The effect of CYP2C19 genetic polymorphism on the efficacy and safety profiles of citalopram was demonstrated in a group of 130 patients with major depressive disorder.
References
Boschloo L, Vogelzangs N, Smit JH, van den Brink W, Veltman DJ, Beekman AT, et al. Comorbidity and risk indicators for alcohol use disorders among persons with anxiety and/or depressive disorders: Findings from the Netherlands Study of Depression and Anxiety (NESDA). J Affect Disord. 2011;131:233–42. https://doi.org/10.1016/j.jad.2010.12.014.
Shiv G, Akhilesh J, Manaswi G. Guidelines for the pharmacological management of depression. Indian J Psychiatry. 2017;59:34–50. https://doi.org/10.4103/0019-5545.196973.
Spear BB, Heath-Chiozzi M, Huff J. Clinical application of pharmacogenetics. Trends Mol Med. 2001;7:201–4. https://doi.org/10.1016/s1471-4914(01)01986-4.
Aldrich SL, Poweleit EA, Prows CA, Martin LJ, Strawn JR, Ramsey LB. Influence of CYP2C19 metabolizer status on escitalopram/citalopram tolerability and response in youth with anxiety and depressive disorders. Front Pharmacol 2019;10:99. https://doi.org/10.3389/fphar.2019.00099.
Dorji PW, Tshering G. Na-Bangchang KCYP2C9, CYP2C19, CYP2D6 and CYP3A5 polymorphisms in South-East and East Asian populations: a systematic review. J Clin Pharm Ther. 2019;44:508–24. https://doi.org/10.1111/jcpt.12835.
Huang B, Cui DJ, Ren Y, Han B, Yang DP, Zhao X. Effect of cytochrome P450 2C19*17 allelic variant on cardiovascular and cerebrovascular outcomes in clopidogrel-treated patients: a systematic review and meta-analysis. J Res Med Sci. 2017;22:109. https://doi.org/10.4103/jrms.JRMS_590_16. Published 2017 Sep 26.
Moriyama B, Obeng AO, Barbarino J, Penzak SR, Henning SA, Scott SA, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP2C19 and Voriconazole Therapy [published correction appears in Clin Pharmacol Ther. Clin Pharmacol Ther. 2017;102:45–51. https://doi.org/10.1002/cpt.583.
Mrazek DA, Biernacka JM, O’Kane DJ, Black JL, Cunningham JM, Drews MS, et al. CYP2C19 variation and citalopram response. Pharmacogenet Genomics. 2011;21:1–9. https://doi.org/10.1097/fpc.0b013e328340bc5a.
Zigmond AS, Snaith RP. The hospital anxiety and depression scale. Acta Psychiatr Scand. 1983;67:361–70.
Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry. 1960;23:56–62.
Lingjaerde O, Ahlfors UG, Bech P, Dencker SJ, Elgen K. The UKU side effect rating scale. A new comprehensive rating scale for psychotropic drugs and a cross-sectional study of side effects in neuroleptic-treated patients. Acta Psychiatr Scand Suppl. 1987;334:1–100.
Zastrozhin MS, Skryabin VY, Torrado M, Petrovna A, Sorokin AS, Grishina EA, et al. Effects of CYP2C19*2 polymorphisms on the efficacy and safety of phenazepam in patients with anxiety disorder and comorbid alcohol use disorder. Pharmacogenomics. 2020;21:111–23. https://doi.org/10.2217/pgs-2019-0019.
Zastrozhin MS, Sorokin AS, Agibalova TV, Grishina EA, Antonenko AР, Rozochkin IN, et al. Using a personalized clinical decision support system for bromdihydrochlorphenylbenzodiazepine dosing in patients with anxiety disorders based on the pharmacogenomic markers. Hum Psychopharmacol. 2018;33:e2677. https://doi.org/10.1002/hup.2677. Epub 2018 Oct 25.
Skryabin VY, Zastrozhin MS, Torrado MV, Grishina EA, Ryzhikova KA, Shipitsyn VV, et al. How do CYP2C19*2 and CYP2C19*17 genetic polymorphisms affect the efficacy and safety of diazepam in patients with alcohol withdrawal syndrome? Drug Metab Pers Ther. 2020. https://doi.org/10.1515/dmpt-2019-0026.
Funding
This work was financially supported by the project 16-15-00227 entitled “Fundamental research and exploratory research in priority areas of research” of the Russian Science Foundation. This work was supported by the grant of the Russian Science Foundation (project No. 18-75-10073).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zastrozhin, M.S., Skryabin, V.Y., Petukhov, A.E. et al. Effects of CYP2C19 genetic polymorphism on the steady-state concentration of citalopram in patients with major depressive disorder. Pharmacogenomics J 21, 435–439 (2021). https://doi.org/10.1038/s41397-021-00219-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41397-021-00219-7
- Springer Nature Limited