Abstract
Triple-negative (ER-PR-HER2-) breast cancers (TNBC) are highly aggressive and difficult to treat. TNBC exhibit high genomic instability, which enables them to adapt and become resistant to chemo/radiation therapy, leading to rapid disease relapse and mortality. The pro-survival factors that safeguard genome integrity in TNBC cells are poorly understood. LBH is an essential mammary stem cell-specific transcription regulator in the WNT pathway that is aberrantly overexpressed in TNBC, correlating with poor prognosis. Herein, we demonstrate a novel role for LBH in promoting TNBC cell survival. Depletion of LBH in multiple TNBC cell models triggered apoptotic cell death both in vitro and in vivo and led to S-G2M cell cycle delays. Mechanistically, LBH loss causes replication stress due to DNA replication fork stalling, leading to ssDNA breaks, ɣH2AX and 53BP1 nuclear foci formation, and activation of the ATR/CHK1 DNA damage response. Notably, ATR inhibition in combination with LBH downmodulation had a synergistic effect, boosting TNBC cell killing and blocking in vivo tumor growth. Our findings demonstrate, for the first time, that LBH protects the genome integrity of cancer cells by preventing replicative stress. Importantly, they uncover new synthetic lethal vulnerabilities in TNBC that could be exploited for future multi-modal precision medicine.

Similar content being viewed by others
Introduction
Triple-negative breast cancer (TNBC) is a highly aggressive and treatment-resistant breast cancer subtype that disproportionally contributes to cancer deaths, especially in young women [1]. TNBC patients have few treatment options, as their cancers lack expression of targetable markers, i.e., estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) [1]. The mainstay of treatment remains chemotherapy [2] to which >50% of TNBC patients show either de novo or acquired resistance, leading to early relapse, distant metastasis, and often death within 3–5 years of diagnosis [3, 4]. Thus, novel molecular targets and treatments are needed.
TNBC tumors exhibit high genomic instability due to frequent loss-of-function mutations in genome caretaker genes, i.e., TP53, and DNA repair enzymes, BRCA1/2, RAD51C, and PALB2 [5,6,7,8,9], contributing to their heterogeneity and ability to rapidly adapt and develop resistance to therapy. Additionally, TNBC display gains/amplifications of oncogenes, i.e., MYC, PI3KCA, EGFR [8, 9], and high proliferation rates [1], which makes it further challenging to safeguard the genome. The pro-survival factors that protect TNBC cells from genomic stress remain poorly understood. Identification of these factors is an unmet medical need, as they may represent an ‘Archilles heel’ that could be exploited therapeutically for the development of more efficient treatments for TNBC, and/or for overcoming resistance to genotoxic chemo/radiation therapy.
Our prior work identified Limb-Bud and Heart (LBH), a vertebrate-specific transcription regulator in the WNT/ß-catenin signaling pathway [10,11,12,13]. LBH is implicated in normal adult breast stem cell regulation [13], progenitor cell proliferation, differentiation and cell migration during embryonic development [14,15,16,17], cell cycle control [18,19,20], angiogenesis [15, 21], autoimmunity [19, 20], and inner ear hair cell survival [22]. Deregulation of LBH is observed in many cancers [12, 18, 21, 23,24,25,26], and has context-specific function, acting as an oncogene in breast [12, 27], gastric [25] and brain cancer [21, 28], and as a tumor suppressor in nasopharyngeal [18] and lung cancer [24]. In breast cancer, LBH is predominantly expressed in high-grade, basal-like TNBC [12], correlating with early disease relapse and resistance to chemotherapy. In contrast, LBH is under-expressed in treatable, lower-grade ER+ luminal breast cancers compared to normal breast tissue and lacking in ER+ luminal breast cancer cell lines [12, 26, 29]. Notably, LBH is specifically expressed in CD44+CD24-/low breast cancer stem cells [30], which are enriched in TNBC [31, 32], and show increased resistance to chemo/radiation therapy [33, 34], suggesting a potential contribution of LBH to survival under genotoxic stress.
In this study, we discovered that depletion of LBH was sufficient to promote genomic instability and TNBC cell death due to increased replication stress, triggering an ATR DNA damage response. Importantly, LBH downmodulation sensitized TNBC cells to ATR inhibitor (ATRi) treatment and synergized with ATRi to enhance TNBC cell killing, blocking in vivo tumor growth. Our findings identify LBH as a potent pro-survival factor in TNBC that protects cancer genome integrity, as well as uncover a new synthetic lethal therapeutic opportunity for hard-to-treat TNBC.
Results
LBH depletion in TNBC cells induces apoptotic cell death
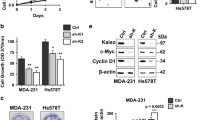
To study the effects of LBH on TNBC cell survival, we depleted LBH in three different TNBC lines (MDA-MB-231, HCC1806, and HCC1395) using RNAi. Transient knockdown (KD) of LBH with two independent siRNAs (KD1 and KD2) reduced LBH expression at the mRNA and protein levels by 65–90% relative to a non-targeting siRNA sequence (NT) used as control (Fig. 1A, B). MTS assays showed that LBH depletion with KD1 and KD2 significantly reduced cell viability in all three TNBC lines (Fig. 1C). Since KD2 outperformed KD1 in reducing cell viability and showed less residual LBH expression, KD2 siRNA (hereafter referred to as KD) was used in subsequent experiments.
Transient LBH knockdown (KD) in TNBC cell lines, MDA-MB-231, HCC1806, and HCC1395, with two individual siRNAs (KD1 and KD2). A non-targeting siRNA (NT) was used as control. Levels of LBH mRNA (A) and protein (B) were measured by RT-qPCR and Western blot analysis, respectively. Actin served as loading control. C MTS assays showing normalized cell viability. D Representative Annexin-V FACS plots (left), with quantification of Annexin-V+ cells (right), show increased apoptosis in LBH KD compared to NT control cells. Propidium Iodide (PI) was used as DNA stain. E Bar graphs showing normalized caspase 3/7 (Casp 3/7) activity. F Western blot analysis of pro-apoptotic (Cl-PARP, Cl-Caspase-3, and BAX) and anti-apoptotic (Bcl2, Bcl-XL) markers. Error bars represent the mean ± s.e.m. (n = 3 biological replicates). *P < 0.05; **P < 0.01; and ***P < 0.001 (two-tailed Student’s t-test in A, D, E; one-way ANOVA in C). All experiments were repeated three times.
To determine if reduced viability of LBH KD cells was due to increased cell death, we performed flow cytometry for the apoptosis marker, Annexin-V. LBH depletion significantly induced apoptosis in TNBC cells, as both early apoptotic (Annexin-V single positive/Q4) and late apoptotic (Annexin-V/PI double positive/Q2) cells were increased in LBH KD compared to NT control cells (Fig. 1D). Notably, LBH KD caused a significant increase in cleaved caspases 3/7 activity (Fig. 1E), which is a rate limiting step in the irreversible activation of programmed cell death. Moreover, Western blot analysis showed that hallmark pro-apoptotic markers, cleaved PARP, cleaved caspase-3, and BAX, were markedly increased, whereas anti-apoptotic markers, Bcl-2, and Bcl-XL, were decreased upon LBH loss (Fig. 1F). Thus, LBH deficiency causes apoptotic cell death.
Since LBH overexpression in other cancer types has been shown to activate survival pathways, i.e., AKT and MAPK signaling [21, 25], we next examined expression and phosphorylation status of survival proteins. However, the active, phosphorylated forms of these proteins, p-AKT, p-MAPK-38 (p38), and p-ERK, were up-regulated or unaffected rather than down-regulated in LBH KD cells (Fig. S1), indicating that the increased apoptosis upon LBH loss was not due to impaired survival signaling. Together these results demonstrate that depletion of LBH induces TNBC cell death, implying a critical function of LBH in maintaining TNBC cell survival.
LBH loss results in S-G2/M cell cycle delays and DNA replication stalling
To identify the causes of programmed cell death triggered by LBH downmodulation, we evaluated cell cycle progression in LBH KD MDA-MB-231 and HCC1806 cells. FACS analysis showed that LBH KD TNBC cells were attenuated in S and G2/M phase when compared to NT controls (Fig. 2A, B). EdU Click iT assays further revealed that upon LBH KD, DNA incorporation of EdU was significantly less during early-S and accumulated in late-S phase (Fig. 2C, D), suggestive of delayed DNA replication. Western Blot analysis of cell cycle markers also showed that LBH KD cells were halted in S-phase, as Cyclin A2, which promotes replication origin firing, and phospho-CDK2, which sustains the cell cycle in S phase to complete replication [35, 36], were increased (Fig. 2E, F). In contrast, expression of G1 and G2 cyclins, Cyclin D1 and Cyclin B1, respectively, was unchanged (Fig. 2E, F). Thus, depletion of LBH arrests cell cycle progression of TNBC cells specifically in S-phase due to under-replication of DNA.
Representative cell cycle histograms (left) and stacked percentage bar graphs (right) showing quantitative changes in cell cycle phase distribution upon LBH KD in MDA-MB-231 (A) and HCC1806 (B) TNBC cells. C, D Representative EdU Click-iT FACS plots showing active DNA replication by modified EdU nucleotide incorporation in the indicated TNBC lines (left). Quantification of total EdU-positive cells during early, mid-, and late S phase (right). Western blot analysis of cell cycle markers in LBH-deficient MDA-MB-231 (E) and HCC1806 (F) cells. G DNA fiber assays. Top: schematic representation of experiment with timeline for pulse labeling with two different modified nucleotides (CldU and IdU). Bottom: Representative confocal immunofluorescence images of DNA fiber tracks (left) displaying fork speed differences in LBH KD compared to NT control cells. Scatter dot plots (right) quantifying the fork speed in µm/min (N = 50 random tracks from n = 3 samples/group were scored). H Representative confocal immunofluorescence images of RPA32 expression. Single nucleus zoomed inserts displaying RPA32 nuclear foci formation in LBH KD compared to NT control cells. DAPI was used as nuclear counter stain. Scale bars, 20 µm. I Western blot showing RPA32 expression and its activated phosphorylated form (red arrows). Error bars represent the mean ± s.e.m. (n = 3 biological replicates). *P < 0.05; **P < 0.01; and ***P < 0.001; ns not significant (two-tailed Student’s t-test). All data are representative of experiments repeated three times.
To determine if cell death induced by LBH loss was dependent on the observed cell cycle arrest, we evaluated cell cycle profiles, in parallel, with cleaved caspase apoptosis assays at different time points after LBH knockdown (Fig. S2). In both MDA-MB-231, and HCC1806, LBH KD cells started to accumulate in S and G2/M phase as early as 24 h after transient transfection with LBH siRNA compared to NT transfected cells (Fig. S2A, B, D, E). However, cell death increases became noticeable only 60–72 h after LBH KD (Fig. S2C, F). Hence, cell death upon LBH loss occurs after cell cycle arrest.
Since DNA replication is the rate limiting step in S/G2 progression, we performed DNA fiber assays to measure replication fork progression. Control and LBH-depleted MDA-231 or HCC1806 cells were sequentially pulsed for 20 min each with CldU and IdU, followed by immunofluorescence detection. The fork speed was calculated as shown in Fig. 2G. The average length of red CldU and green IdU-labeled DNA fibers was visibly shorter in LBH KD TNBC cells (Fig. 2G, left images). Notably, the average fork progression speed in LBH KD cells was 70% lower than in NT control cells (Fig. 2G, right), indicative of DNA replication fork stalling.
During DNA replication, the exposed single strand DNA (ssDNA) at replication forks is coated with single strand binding protein RPA32 to protect from degradation and to facilitate gap filling during the discontinuous synthesis of the lagging strand [37]. Replication fork stalling causes RPA32 phosphorylation at Ser4/8, which acts as a sensor of replication stress [38,39,40]. We examined RPA32 expression by confocal immunofluorescence and its phosphorylation status by Western Blot analysis. Both RPA32 genome occupancy (Fig. 2H), and RPA32 phosphorylation (Fig. 2I) were markedly increased in LBH KD cells compared to NT control cells, consistent with increased replication stress.
Overall, these results indicate that LBH is required for normal DNA replication progression, and its depletion causes replication fork stalling and replicative stress, leading to S-G2/M cell cycle delays and increased cell death.
LBH loss induces DNA damage
Accumulation of ssDNA due to fork stalling and replicative stress can cause ssDNA breaks, leading to DNA damage [40, 41]. We used the alkaline comet assay to visualize ssDNA breaks and found that LBH KD increased the number of comet tail-positive nuclei and olive tail movement in both MDA-MB-231 and HCC1806 TNBC cells (Fig. 3A, B). Sustained ssDNA damage triggers the phosphorylation and recruitment of DNA damage markers, ɣH2AX and 53BP1, to sites of DNA damage within minutes. We found that LBH depletion resulted in a drastic increase in both ɣH2AX, and p-53BP1 nuclear foci formation (Fig. 3C). In addition, ɣH2AX protein levels were increased in LBH KD compared to NT control cells (Fig. 3D). We next performed FACS analysis to quantify ɣH2AX-positive cells during cell cycle progression (Fig. 3E–H). The total number of ɣH2AX+ cells was significantly increased in LBH KD compared to control cells (Fig. 3G, H - left panels). Overlay with EdU cell cycle profiles revealed that this increase occurred mostly in S-phase (Fig. 3E, F, G, H - right panels). From this data it is evident that LBH loss triggers ssDNA damage in TNBC cells leading to active recruitment of DNA damage markers.
Alkaline comet assay showing increased ssDNA strand breaks upon LBH KD in MDA-MB-231 (A) and HCC1806 (B) cells compared to NT control cells. Representative images showing DNA comet tail formation (white arrows; left panels), with quantification of olive tail movement (right) (N = 50 random nuclei from n = 3 samples/group were scored). C Representative immunofluorescence images, with single nuclei close-ups (insets), showing expression and nuclear foci formation of DNA damage markers, ɣH2AX and p-53BP1. DAPI served as nuclear counter stain. Scale bars, 20 µm. D Western blot showing increased ɣH2AX protein levels upon LBH KD. E, F Representative FACS plots of ɣH2AX-positive cells (red) overlayed with EdU cell cycle profiles. G, H Quantification of total ɣH2AX+ cells (left) and their distribution during the cell cycle (right). Error bars represent the mean ± s.e.m. (n = 3 biological replicates). **P < 0.01; ***P < 0.001; ns not significant (two-tailed Student’s t-test).
LBH loss activates the ATR/CHK1 DNA damage response
It has been established that replication stress and ssDNA breaks mainly activate the ATR/CHK1 DNA repair pathway, while dsDNA breaks activate the ATM/CHK2 pathway [42, 43]. To investigate the pathways that are activated to resolve the DNA damage caused by LBH loss in TNBC cells, we examined both the ATR and ATM DNA damage response (DDR). Confocal (Fig. 4A, B) and Western blot analysis (Fig. 4C) showed that LBH loss in MDA-MB-231 and HCC1806 cells increased genome occupancy and levels of phospho-ATR, and its substrate, phospho-CHK1. Moreover, phosphorylated ATRIP, which recruits ATR to DNA damage sites [42,43,44,45], was increased relative to total ATRIP protein. In contrast, phospho-ATM, and its substrate, phospho-CHK2, over their total forms did not change, and no nuclear foci formation by these proteins was observed (Fig. 4D, F).
Representative immunofluorescence images showing expression of p-ATR (A) and p-CHK1 (B) relative to DAPI in MDA-MB-231 and HCC1806 cells with LBH KD compared to NT control cells. Single nucleus zoomed inserts display p-ATR and p-CHK1 nuclear foci formation. Scale bars, 20 µm. C Western blot showing expression levels of phosphorylated versus total ATR, CHK1, and ATRIP proteins. Representative immunofluorescence images showing expression of p-ATM (D) and p-CHK2 (E) relative to DAPI in TNBC cells. Scale bars, 20 µm. F Western blot showing the levels of phospho- vs. total ATM and CHK2 proteins in LBH KD compared to NT control cells. G, H Dose dependent sigmoidal graphs and IC50 values display the effects of ATR (AZD6738) and ATM (KU55933) specific inhibitors in LBH KD compared to NT control MDA-MB-231 cells. Error bars represent the mean ± s.e.m. (n = 5 biological replicates).
To investigate the functional significance of these findings, we performed ATR and ATM inhibition studies in the MDA-MB-231 TNBC model with LBH KD (Fig. 4G, H). Addition of the ATR inhibitor AZD6738 reduced the IC50 of LBH KD cells by over 60-fold compared to control cells (Fig. 4G), suggesting that LBH KD sensitizes TNBC cells to ATR inhibition. In contrast, we observed only moderate shifts in IC50 concentrations when LBH KD MDA-MB-231 cells were treated with the ATM specific inhibitor KU55933 (Fig. 4H). Together these data demonstrate that LBH loss specifically activates the ATR/CHK1 ssDNA damage response and renders TNBC cells responsive to ATR inhibition, uncovering a potential novel targetable approach.
ATR inhibition synergizes with LBH knockdown to induce TNBC cell lethality
Based on the foregoing, we hypothesized that targeting the ATR pathway with clinically established inhibitors might act synergistically with LBH downmodulation to reduce TNBC cell survival. The ATR inhibitor AZD6738 (ceralasertib) is used in phase II clinical trials for solid tumor metastatic cancers [45], including BRCA1/2-mutated breast cancer and metastatic TNBC (https://www.clinicaltrials.gov/NCT04090567; NCT05582538; NCT03801369), with little toxicity and side effects [46, 47]. Hence, we used AZD6738 in all subsequent studies to block ATR activation triggered by LBH loss.
While ATR inhibition (ATRi) alone had little effect on viability of control NT-transfected MDA-MB-231 and HCC1806 cells, AD6738 treatment in LBH KD cells drastically reduced viability, and to a greater extent than LBH KD alone (Fig. 5A, B). Moreover, ATRi in combination with LBH KD increased caspase 3/7 activity (Fig. 5C, D), and apoptosis rates, as measured by Annexin V FACS (Fig. 5E, F), more significantly than observed for either treatment alone. Similarly, upregulation of pro-apoptotic markers, cleaved PARP, and cleaved caspase 3, in LBH KD cells was enhanced by AD6738 treatment, whereas anti-apoptotic proteins, Bcl-2 and Bcl-XL, were further decreased (Fig. 5G, H).
MTS assays in MDA-MB-231 (A) and HCC1806 (B) cells showing normalized cell viability in LBH KD compared to NT control cells treated with DMSO vehicle or 0.5 µM of AZD6738 for 72 h. C, D Bar graphs showing active caspase 3/7 activity upon treatment with DMSO or 0.5 µM of AZD6738. Representative Annexin-V FACS plots (top), with quantification of Annexin-V+ apoptotic cells (bottom), in LBH KD MDA-MB-231 (E) and HCC1806 (F) cells with the indicated treatments. G, H Western blot showing expression of pro- and anti-apoptotic markers −/+ AZD6738 treatment in LBH KD compared to NT control cells. I Synergy maps of MDA-MB-231 (left) and HCC1806 (right) treated with increasing concentrations of LBH siRNA and AZD6738. The 3D synergy matrix was generated with SynergyFinder Plus (n = 3 biological replicates). The color code spans from statistically significant synergism (dark red, CI < 1, P < 0.001) to statistically significant antagonism (dark green, CI > 1, P < 0.001). Error bars represent the mean ± s.e.m. (n = 3 biological replicates). ***P < 0.001; ns not significant (one-way ANOVA in A, B; two-tailed Student’s t-test in C–E).
To unequivocally test whether LBH KD and ATRi act synergistically in promoting TNBC cell death, we established the individual dose-response matrixes for each TNBC line testing pairwise combinations of increasing concentrations of LBH siRNA and six doses of AD6738 (Fig. 5I; Fig. S3A–F). Calculation of the synergy distribution for each combination using SynergyFinder confirmed that LBH KD and ATRi had synergistic effects in both MDA-MD-231 and HCC1806, although the concentrations of LBH siRNA and AZD6738, when synergy was achieved, varied (Fig. 5I; Fig. S3C–F). These results reinforce the notion that LBH downmodulation in combination with ATRi may represent a viable new therapeutic strategy to effectively induce TNBC cell killing.
ATR inhibition exacerbates genome instability induced by LBH loss
ATR activation normally facilitates DNA repair, and ATR inhibition results in the accumulation of DNA damage [45]. Thus, we also characterized the effects of LBH KD and ATR inhibition on genome stability. Comet assays revealed that AZD6738 treatment significantly increased DNA damage in LBH KD TNBC cells compared to LBH KD or AZD6738 treatment alone (Fig. 6A, B). Western Blot analysis confirmed that AZD6738 successfully blocked activation of the ATR-CHK1 ssDNA damage response in LBH KD cells (Fig. 6C, D). We further investigated the accumulation of replication stress-induced DNA damage upon ATRi by examining the expression of ɣH2AX and phosphorylated RPA32. Both markers were significantly increased upon AZD6738 treatment compared to LBH KD without ATRi (Fig. 6E, F). Confocal analysis, moreover, showed profound increases in ɣH2AX- and p-53BP1-positive DNA damage foci in AZD6738 treated LBH KD cells (Fig. 6G, H). These results demonstrate that the ssDNA damage induced by LBH KD is exacerbated by ATR inhibition.
Alkaline comet assays showing synergistic effects of LBH KD and AZD6738 treatment (0.5 µM for 72 h) on ssDNA breaks in MDA-MB-231 (A) and HCC1806 (B) cells. Representative comet images are displayed on the left; scatter dot plots with quantification of the olive tail movement on the right (N = 50 random nuclei from n = 3 samples/group were scored). C, D Western blot analysis of phosphorylated vs. total ATR, CHK1 protein expression in LBH KD compared to NT control cells −/+ 0.5 µM AZD6738 treatment. E, F Western blot showing ɣH2AX and RPA32 expression in LBH KD and NT control cells −/+ 0.5 µM AZD6738 treatment. Red arrows highlight the active, phospho-RPA32 protein band. G, H Representative immunofluorescence images of ɣH2AX and p-53BP1 nuclear foci formation with single nuclei close-ups (insets) in LBH KD compared to NT control cells treated with DMSO or 0.5 µM AZD6738 for 72 h. DAPI was used as nuclear counter stain. Scale bars, 20 µm. Error bars represent the mean ± s.e.m.. *P < 0.05; **P < 0.01; ***P < 0.001; and ns not significant (two-tailed Student’s t-test). All data are representative of experiments repeated three times.
LBH depletion synergizes with ATR inhibition to suppress in vivo TNBC tumor growth
A major aim of this study was to identify new treatment strategies to more effectively control aggressive TNBC cancers, for which therapy options are limited. Hence, we next examined the potential synergistic effects between LBH KD and ATRi on in vivo TNBC tumor growth. For these studies we generated orthotopic TNBC tumor models by inoculating MDA-MB-231 and HCC1806 cells that were stably transduced with validated LBH-targeted shRNA (shKD) or a non-target shRNA (shCtrl) (Fig. S4A, B) into NOD-SCID IL2Rgamma−/− (NSG) mice, as indicated in the experimental outline (Fig. 7A).
TNBC lines, MDA-MB-231 and HCC1806, were stably transduced with validated LBH targeted shRNA (shKD) or a non-targeted control shRNA (shCtrl) (see Fig. S2). 1 ×106 cells were injected orthotopically into NSG mice. A Schematic representation of timeline of experiment indicating cell implantation, start point and concentration of AZD6738 drug treatment, and the experiment endpoint. B, C Normalized tumor volumes in MDA-MB-231 (A) and HCC1806 (B) Xenografts over the time of drug treatment (n = 6 tumors/group). D, E Tumor weights at the treatment end point (n = 6 tumors/group). F, G Representative images of tumors (top) and Hematoxylin-Eosin (H&E) stained tumor sections (bottom). Scale bars, 100 µm. H, I Representative IHC images of cleaved Caspase-3-stained tumor sections (H), with quantification of cleaved caspase-3-positive (CC3+) cells (n = 4 tumors/group) (I). Error bars represent the mean ± s.e.m. *P < 0.05; **P < 0.01; ***P < 0.001; ns not significant (one-way ANOVA in B, C; two-tailed Student’s t-test in D, E, I).
While AZD6738 treatment alone had no or only a modest effect in reducing in vivo tumor growth of shCtrl-transduced MDA-MB-231 and HCC1806 control cells (Fig. 7B–G), LBH KD, and more profoundly, LBH KD in combination with ATRi significantly reduced tumor volumes over time (Fig. 7B, C), tumor weights (Fig. 7D, E) and size (Fig. 7F, G - top panels). Notably, hematoxylin-eosin (H-E) staining of tumor sections showed increased necrosis in LBH KD, and LBH KD plus AZD6738 treated Xenografts (Fig. 7F, G - bottom panels), indicating that the regression of tumors under these conditions was due to increased cell death. Moreover, immunohistochemical (IHC) analysis revealed a significant increase in cleaved caspase-3 positive apoptotic cells in LBH KD tumors, which was further elevated by ATRi (Fig. 7H, I). Thus, LBH KD and ATRi synergize to induce TNBC cell lethality in vivo, blocking tumor growth.
LBH downmodulation enhances the efficacy of ATRi in other cancer types
Lastly, we asked if the effect of LBH downmodulation in stimulating the efficacy of ATRi is TNBC-specific. To test this, we performed LBH knockdown studies in an ER-negative, HER2+ luminal breast cancer cell line, SK-BR-3, and in normal-derived MCF10A, expressing low to medium levels of LBH, respectively (Fig. 8A, B; Fig. S5A, B) [12]. We also depleted LBH in cell line models for other cancer types, i.e., pancreatic (MiaPaca-2), colon (Caco-2), and lung (H358, A549) (Fig. 8C, F; Fig. S5A, B). While LBH KD and AZD6738 treatment alone or in combination did not impair viability of normal MCF10A breast epithelial cells, synergistic effects between LBH KD and ATRi in reducing cell viability were observed in SK-BR-3, MiaPaca-2, and Caco-2 (Fig. 8A–D). In contrast, H358 and A549 lung adenoma lines, in which LBH has been shown to have tumor suppressive activity [24], were resistant to ATRi, despite significant LBH downmodulation (Fig. 8E, F; Fig. S5B). Thus, LBH loss enhances the efficacy of ATRi also in non-TNBC cancers, depending on the tissue type.
MTS assays in: A normal-derived MCF10A breast epithelial cells; B SK-BR-3, a luminal HER2+ER- breast cancer cell line; and in cell line models for: C pancreatic (MiaPaca-2); D colorectal (CaCo-2); and E, F lung (H358, A549) cancer. Cells after transient transfection with NT or LBH KD siRNA were treated with DMSO vehicle or 0.5 µM of AZD6738 for 72 h. Error bars represent the mean ± s.e.m. (n = 4 biological replicates). *P < 0.05; **P < 0.01; and ***P < 0.001; ns not significant (two-tailed Student’s t-test).
Collectively, our results identify LBH as an effective molecular target to induce TNBC cell lethality and to increase the efficacy of clinical ATR inhibitors, providing a novel rationale for inhibiting LBH in combination with ATR to improve anti-cancer therapy.
Discussion
In this study, we identified that LBH is an essential pro-survival factor in TNBC that protects tumor cells from genome instability and cell death. Depletion of LBH causes replication stress and ssDNA damage, triggering an ATR DNA damage response and inducing apoptosis. Importantly, LBH downmodulation synergizes with ATR inhibition, which inhibits DNA repair, to boost TNBC cell killing both in vitro and in vivo.
Investigating novel specific treatment strategies to overcome treatment resistance in TNBC is essential to improve the life expectancy of patients. Targeting genomic instability, e.g., with PARP inhibitors, has proven to be a successful approach in TNBC patients with BRCA1/2 DNA repair deficiencies and is widely used in the clinic for a variety of cancers [48]. However, only ca. 19% of TNBC exhibit genetic or somatic mutations in BRCA genes [7, 9]. Recent genomic and transcriptomic analyses suggest that TNBC develop complex survival strategies through transcriptional/epigenetic changes and expression of cell survival factors to cope with the genotoxic stress associated with genomic instability [49, 50]. Targeting these survival factors may represent an alternative strategy.
Our study identifies LBH as one such survival factor. LBH is overexpressed in nearly 50% of basal-like TNBC [12], which have the worst prognosis and show the highest genome complexity and chemoresistance among the different TNBC subgroups [6, 9, 51]. We show that depletion of LBH, without any other treatment, was sufficient to induce DNA damage and increase TNBC cell death. Increased DNA damage in LBH-deficient TNBC cells was evident by phosphorylation and recruitment of ɣH2AX and 53BP1 to nuclear DNA damage foci and occurred predominantly in S-phase, linking these events to defective DNA replication. Alkaline comet, in combination with DNA fiber assays, further revealed that the DNA damage caused by LBH downmodulation was due to ssDNA breaks resulting from DNA replication fork stalling and replicative stress, which was confirmed by hyper-phosphorylation and increased genome occupancy of RPA32.
RPA32 is a major sensor of replication stress [38, 39], and ssDNA-bound RPA activates the ATR DDR by recruiting ATR via its binding partner ATRIP to the sites of ssDNA damage [42,43,44,45]. Indeed, we found that LBH depletion in TNBC cells caused increased genome occupancy and phosphorylation of ATR and its target CHK1, but not of ATM and CHK2. Thus, loss of LBH specifically triggers the ATR/CHK1 ssDNA damage response, which is known to halt cell cycle progression in S/G2 phase to allow DNA repair or induce apoptosis [45].
Previous studies in rheumatoid arthritis (RA) models have also suggested a role of LBH in maintaining genome stability [20]. Like our observations in TNBC cells, LBH depletion in primary RA fibroblast-like synovitis (FLS) cells caused S-phase cell cycle arrest due to delays in DNA replication and increased DNA damage, activating phosphorylation of CHK1 [20]. Increased CHK1 phosphorylation was also observed in Lbh−/− knockout mice in vivo after arthritis inducing K/BxN serum transfer [20]. Moreover, DNA damage gene signatures were enriched in transcriptomes of Lbh−/− null inner ear cells, in which LBH has an essential pro-survival function and protects from progressive hearing loss [22]. Thus, LBH expression protects not only cancer cells from DNA damage, but also normal, non-transformed cells under certain pathologic conditions.
At present it is unclear how LBH, as transcription co-regulator, contributes to genome stability and influences DNA fork progression. LBH KD studies in RA FLS cells have shown that LBH deficiency decreases the expression of the catalytic subunit of DNA polymerase alpha (POLA1), which initiates de novo DNA synthesis at replication forks and discontinuously replicates DNA during lagging strand DNA synthesis [20]. Our recent pathway analysis in cancers in which LBH is under-expressed compared to normal tissues (i.e., lung, melanoma) suggests that LBH deficiency may affect de novo nucleotide synthesis [26], which can also lead to stalled replication forks [45]. Alternatively, LBH may affect the expression of DNA repair genes [22]. A major future effort will be to identify how LBH prevents replicative stress and DNA damage.
Importantly, since we found LBH downmodulation specifically triggers the ATR damage response, we explored, for the first time, possible strategies for inhibiting ATR activation in LBH-deficient cancer cells. We selected AZD6738 (ceralasertib), an ATR inhibitor in phase II clinical trials that is orally administered with low toxicity and side effects [46, 47]. Moreover, AZD6738 can be used in combination with other therapies to increase effectiveness [45, 47]. Our results, in aggregate, demonstrate that by inducing DNA damage, depletion of LBH sensitizes TNBC cells to AZD6738 drug treatment. In fact, AZD6738 treatment, which inhibits DNA repair, potently synergized with LBH downmodulation to increase the accumulation of DNA damage, enhancing programmed cell death and TNBC lethality both in vitro and in vivo.
Collectively, our results yield vital new mechanistic insights into the pro-survival mechanisms of poor prognosis TNBC by identifying LBH as a key survival factor and protector of cancer genomes. Importantly, they provide a novel rationale for inhibiting LBH in combination with ATR inhibitors to improve outcomes in TNBC, and other aggressive cancers with LBH overexpression [26]. A future goal will be to identify LBH inhibiting drug molecules that can be used with ATR inhibitors to develop more effective treatment options for cancer patients.
Materials and methods
Cell lines
Cell lines, if not indicated otherwise, were purchased from ATCC (Manassas, VA, USA). MDA-MB-231 (# HTB-26), HCC1806 (# CRL-2335), MIAPaCa-2 (# CRL-1420), and CaCo-2 (#HTB-37) were grown in DMEM (Corning, NY, USA #10-013-CV) with 10% FBS (Thermo Scientific, Waltham, MA, USA #16000044), Non-Essential Amino Acids (Thermo Scientific #11140050) and Pen-Strep (Thermo Scientific #15140122). HCC1395 (# SC-CRL-2324) was grown in RPMI (Thermo Scientific #A10491-01) with 10% FBS, Pen-Strep; SK-BR-3 (# HTB-30) in McCoy’s 5A (Thermo Scientific #16600082) plus 10% FBS, Pen-Strep; and MCF10A (# CRL-10317) in HuMEC ready mix (Thermo Scientific #12752010). A549 (# CCL-185) and H358 (# CRL-5807) were grown in RPMI 1640 (Corning, NY, USA #10-040-CV) with 10% FBS, Pen-Strep. Cell lines were tested routinely for mycoplasma infection. All cells were cultured in a 5% CO2 incubator at 37 °C.
Small interfering RNA (siRNA) and short hairpin RNA (shRNA)-mediate gene knockdown
For RNAi studies, triplicate samples of cells were transiently transfected with 2 nM of either scrambled non-targeting siRNA pool (Horizon Discovery, Waterbeach, UK #D-001810-10) or two individual LBH-specific siRNAs (Horizon Discovery #J-014248-10/LBH KD1 and #J-014248-11/LBH KD2), using Dharmafect #1 transfection reagent (Horizon Discovery #T-2001-01) according to the manufacturer’s protocol. Cells were incubated with siRNA containing media for 72 h prior to splitting for other studies. For stable LBH knockdown, MDA-MB-231-luciferase [52] and HCC1806 were transduced with ready-made Mission shRNA lentiviral particles (Sigma) expressing two different LBH-specific shRNAs (#TRCN0000107525-shLBH-1 and custom made shLBH-2: 5′-AGAGAGTGAGCCGCAATTGTT-3′) or a non-targeting control shRNA (Sigma-Aldrich, Burlington, MA, USA #SHC002V) at MOI = 5 in the presence of 8 µg/ml polybrene. Individual MDA-MB-231 and HCC1806 polyclonal cultures stably expressing LBH shRNA or control shRNA were obtained by selection with 1–2 µg/ml puromycin.
Quantitative real-time PCR (qPCR)
Total RNA was extracted using Trizol reagent (Thermo Scientific #15596018), and 1 µg of total RNA was reverse transcribed with M-MLV Reverse Transcriptase (Promega, Madison, MI, USA #M1708). qPCR was performed using SsoFast Evagreen PCR master Mix (Bio-Rad Laboratories, Hercules, CA, USA #1725201) and a Bio-Rad CFX96 Thermal Cycler. mRNA expression was normalized to the expression of GAPDH using the 2^-ΔΔCT method. The sequences of qPCR primers are: LBH-forward 5′-TCACTGCCCCGACTATCTG-3′, LBH-reverse 5′-GGTTCCACCACTATGGAGG-3′; GAPDH-forward 5′-GGTTCCACCACTATGGAGG-3′, and GAPDH-reverse 5′-GACAAGCTTCCCGTTCTCAG-3′.
Western blot analysis
Cells were harvested in RIPA lysis buffer (Thermo Scientific #89901, MA, USA) supplemented with protease (EMD-Millipore, Burlington, MA, USA #5391341) and phosphatase (Thermo Scientific, #1862495) inhibitor cocktails. Cell lysates were passed 5-8 times through a 26-gauge needle before centrifugation at high-speed. Cleared lysates were snap frozen until further use. Total protein was quantified using BCA reagent (Thermo Scientific #23227). 20–50 μg of total protein lysates were separated under reducing conditions (2.5% β-mercaptoethanol) by SDS-Polyacrylamide Gel Electrophoresis and transferred to nitrocellulose membrane (Biorad) using a BioRad turbo blotter. Membranes were blocked in TBS + 0.1% Tween 20 (TBST) + 5% milk for 1 h and incubated with primary antibodies (see Supplementary Table 1) in blocking solution overnight at 4 °C. Blots were washed with TBST buffer and incubated with HRP-coupled secondary antibodies (Santa Cruz, Dallas, TX, USA 1:10,000). Protein bands were detected using West Femto Super Signal Kit (Thermo Scientific #34095) on X-ray film and quantified by densitometry using ImageJ analysis.
Cell viability-apoptosis assays
For MTS assays, 2 × 103 cells were plated in triplicates on 96-well plates. Viable cells were quantified daily over 5 days using the CellTiter 96 AQueous One Cell Proliferation Assay (Promega #G3580) according to the manufacturer’s instructions. For apoptosis assays, 2 × 103 cells were seeded on 96-well plates in triplicates and grown for 3 days in normal growth medium. Caspase 3/7 activity was measured at day 0 and day 3 using the Caspase Glo 3/7 Assay kit (Promega #G811C). All values were normalized to the readings obtained on day 0. For Annexin-V FACS analysis, 1.5 × 105 cells were seeded on 6-well plate in triplicates and grown for 5 days in normal growth medium. Cells were harvested by trypsinization, washed twice in ice-cold PBS, and co-stained with Annexin V-FITC-conjugated antibodies and Propidium Iodide using the Alexa Fluor 488 Annexin V/Dead cell apoptosis kit (Thermo Scientific #V13241). Immunostained cells were quantified on a BD LSRII FACS analyzer with BD FACS Diva version 6.3.1 and analyzed with FCS Express 7 software.
EdU incorporation cell cycle assay
Sub-confluent cells were pulse labeled with 10 µM EdU for 2 h. Incorporated EdU was detected using Click-iT EdU Alexa Fluor 488 flow cytometry assay kit (Thermo Scientific #C10632). FxCycle Violet dye (Thermo Scientific #F10347) was used as DNA stain. For experiments involving ɣH2AX, EdU pulse-labeled cells were incubated with ɣH2AX antibody (CST, Danvers, MA, USA 1:200) for 20–30 min on ice prior to Click-iT EdU detection. Ten thousand labeled cells were captured using BD LSRII Flow Cytometer with BD FACS Diva version 6.3.1 and analyzed with FCS Express 7 software.
Immunofluorescence
Cells were seeded on coverslips and, after growth for 3 days in normal growth media, fixed with 4% paraformaldehyde for 20 min followed by washes in PBS and permeabilization with 0.1% Triton X-100 in 1% BSA/PBS. Coverslips were washed and blocked in 1% BSA/PBS for 1 h followed by incubation with primary antibodies (see Supplementary Table 1) at 4 °C overnight. Anti-rabbit Alexa Fluor 488 or anti-mouse Alexa Fluor 546 conjugated secondary antibodies were incubated in the dark for 1 h at room temperature. Coverslips were washed, mounted onto a glass slide in Fluoroshield Mounting Media with DAPI (Sigma), and imaged using an Olympus confocal microscope. Images were processed with Olympus FluoView software FV10-ASW 3.1.
DNA fiber assay
Six days after transient transfection of MDA-MB-231 and HCC1806 with 2 nM of LBH-targeted (KD) or non-targeted (NT) control siRNA, subconfluent cells were pulse labeled with 25 µM CldU (Sigma #C6891) for 20 min, washed twice briefly with growth media, followed by pulse labeling with 25 µM IdU (Sigma #I7125) for 20 min. Cells were trypsinized to single cell suspension and, after centrifugation, re-suspended in PBS to 4 × 105 cells/ml. Two µl of cell suspension was dropped onto the top side of a saline-coated glass slide (Sigma #S4651-72) and allowed to partially dry for 5 min. Ten µl of lysis buffer (200 mM Tris-HCL, pH 7.4, 50 mM EDTA and 0.5% SDS) was added, mixed by gentle swirling with a pipette tip, and incubated at room temperature for 10 min. The slides were tilted at a 15° angle to allow the drop to slowly roll down the slide length. When the drop reached the bottom edge, the slides were put horizontally and allowed to dry. Slides were then fixed in 3:1 methanol/acetic acid and air-dried overnight. The next day, slides were treated with 2.5 M HCl for 1 h, washed three times with PBST (PBS with 0.1% Tween 20), and blocked with PBST + 2% BSA, followed by incubation with anti-BrdU antibodies (BD Bioscience: 347580 1:500; and Abcam: ab6326 1:1000) for 1 h at room temperature. After three washes in PBST, slides were incubated with 488 and 555 Alexa Fluor secondary antibodies (Thermo Scientific) for 1 h at RT. Slides were washed three times with PBST, allowed to drain, and mounted in fluoroshield mounting media (EMD Millipore #JA1750). DNA fibers were visualized using an Olympus confocal microscope under 60X oil objective and images were analyzed with FluoView software. DNA fiber lengths were measured using ImageJ.
Comet assay
Alkaline comet assays were performed using Trevigen single cell electrophoresis kit (R&D Systems, Minneapolis, MN, USA #4250-050-K) according to the manufacturer’s protocol. DNA was stained with SYBR Gold (Invitrogen), and comet olive tail movement was determined by counting 50 cells per sample using TriTek CometScore software 2.0.0.32.
Drug inhibition studies
2.5 × 103 MDA-MB-231 cells transiently transfected with 2 nM of NT or LBH KD siRNAs were seeded onto 96-well plates in triplicates and grown in complete growth medium. After 24 h, media was replaced with media containing inhibitors to ATR (AZD6738, Selleck Chemicals, Houston, TX, USA #S7693) or ATM (KU55933, Selleck Chemicals #S1092) at different concentrations (100, 33.3, 11.1, 3.7, 1.2, or 0.41 µM). Cells were grown for an additional 72 h, after which absorbance (495 nm) was quantified using the CellTiter 96 AQueous One Cell Proliferation Assay (Promega). IC50 concentrations were calculated by the standard dose response curve method using GraphPad Prism software 7.05. In subsequent ATR inhibition (ATRi) studies, cells were treated with 0.5 µM AZD6738 for 72 h. For synergy analysis, MDA-MB-231 and HCC1806 cells were transfected with increasing concentrations of siLBH#2 siRNA (0; 0.1; 0.39; 1.5; 6.26 or 25 nM) using Dharmafect#1 transfection reagent. Three days post transfections, cells were seeded onto 96 well plates and each siLBH KD sample set was treated with six different doses of AZD6738 (0; 62.5; 125; 250; 500 and 1000 nM) for 72 h. Cell viability was assessed by MTS assay and the results analyzed with different algorithms to determine the synergism between LBH loss and ATRi using SynergyFinder Plus (http://www.synergyfinderplus.org/).
Mouse studies
MDA-MB-231-Luciferase or HCC1806 cells (1 × 105) stably transduced with validated shLBH-2 (shKD) or control nontargeted shRNA (shCtrl) were suspended in 100 µl PBS containing 50% Matrigel (BD) and injected bilaterally into the inguinal mammary fat pads of 6–7-week-old NOD-SCID IL2Rgammanull (NSG) female mice [Stock No. 005557; Jackson Laboratories (Bar Harbor, ME, USA)] (n = 10 NSG mice/group). Primary tumor growth was quantified weekly by caliper measurements. Once the tumor size reached 200–300 mm3, mice in each study group were randomly divided into two subgroups: vehicle treated, or AZD6738 treated. AZD6738 suspended in 5% Carboxy Methyl Cellulose (CMC) was orally administered daily for 21 days at 50 mg/kg body weight. Mice receiving 5% CMC vehicle only over the same period served as controls. Tumors were harvested at treatment endpoint and analyzed as stated in Results. All experiments and procedures involving mice were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Miami in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Immunohistochemistry
Tumor paraffin sections (4 µm) were immunostained with antibodies to Cleaved Caspase 3 (see Supplementary Table 1), as in ref. [27], using an automated Leica Bond RXM slide stainer. Quantification of marker positive cells was performed for four tumors per study group and n > 5 different areas per tumor using ImageJ software.
Statistical analyses
Statistical analyses were performed using GraphPad Prism V7 and Microsoft Excel software. Comparisons of two groups were made using two-tailed unpaired Student’s t-test. Analysis of variance (ANOVA) was used for more than two groups. P < 0.05 was considered statistically significant. Data are expressed as mean ± standard error of the mean (s.e.m.).
Data availability
All relevant data are available from the authors upon request.
References
Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–48.
Isakoff SJ. Triple-negative breast cancer: role of specific chemotherapy agents. Cancer J. 2010;16:53–61.
Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–34.
Nofech-Mozes S, Trudeau M, Kahn HK, Dent R, Rawlinson E, Sun P, et al. Patterns of recurrence in the basal and non-basal subtypes of triple-negative breast cancers. Breast Cancer Res Treat. 2009;118:131–7.
Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52.
Staaf J, Glodzik D, Bosch A, Vallon-Christersson J, Reutersward C, Hakkinen J, et al. Whole-genome sequencing of triple-negative breast cancers in a population-based clinical study. Nat Med. 2019;25:1526–33.
Duijf PHG, Nanayakkara D, Nones K, Srihari S, Kalimutho M, Khanna KK. Mechanisms of genomic instability in breast cancer. Trends Mol Med. 2019;25:595–611.
Derakhshan F, Reis-Filho JS. Pathogenesis of triple-negative breast cancer. Annu Rev Pathol. 2022;17:181–204.
Briegel KJ, Joyner AL. Identification and characterization of Lbh, a novel conserved nuclear protein expressed during early limb and heart development. Dev Biol. 2001;233:291–304.
Al-Ali H, Rieger ME, Seldeen KL, Harris TK, Farooq A, Briegel KJ. Biophysical characterization reveals structural disorder in the developmental transcriptional regulator LBH. Biochem Biophys Res Commun. 2010;391:1104–9.
Rieger ME, Sims AH, Coats ER, Clarke RB, Briegel KJ. The embryonic transcription cofactor LBH is a direct target of the Wnt signaling pathway in epithelial development and in aggressive basal subtype breast cancers. Mol Cell Biol. 2010;30:4267–79.
Lindley LE, Curtis KM, Sanchez-Mejias A, Rieger ME, Robbins DJ, Briegel KJ. The WNT-controlled transcriptional regulator LBH is required for mammary stem cell expansion and maintenance of the basal lineage. Development. 2015;142:893–904.
Briegel KJ, Baldwin HS, Epstein JA, Joyner AL. Congenital heart disease reminiscent of partial trisomy 2p syndrome in mice transgenic for the transcription factor Lbh. Development. 2005;132:3305–16.
Conen KL, Nishimori S, Provot S, Kronenberg HM. The transcriptional cofactor Lbh regulates angiogenesis and endochondral bone formation during fetal bone development. Dev Biol. 2009;333:348–58.
Powder KE, Cousin H, McLinden GP, Craig Albertson R. A nonsynonymous mutation in the transcriptional regulator lbh is associated with cichlid craniofacial adaptation and neural crest cell development. Mol Biol Evol. 2014;31:3113–24.
Weir E, McLinden G, Alfandari D, Cousin H. Trim-Away mediated knock down uncovers a new function for Lbh during gastrulation of Xenopus laevis. Dev Biol. 2021;470:74–83.
Liu Q, Guan X, Lv J, Li X, Wang Y, Li L. Limb-bud and heart (LBH) functions as a tumor suppressor of nasopharyngeal carcinoma by inducing G1/S cell cycle arrest. Sci Rep. 2015;5:7626.
Ekwall AK, Whitaker JW, Hammaker D, Bugbee WD, Wang W, Firestein GS. The rheumatoid arthritis risk gene LBH regulates growth in fibroblast-like synoviocytes. Arthritis Rheumatol. 2015;67:1193–202.
Matsuda S, Hammaker D, Topolewski K, Briegel KJ, Boyle DL, Dowdy S, et al. Regulation of the cell cycle and inflammatory arthritis by the transcription cofactor LBH gene. J Immunol. 2017;199:2316–22.
Jiang Y, Zhou J, Zou D, Hou D, Zhang H, Zhao J, et al. Overexpression of Limb-Bud and Heart (LBH) promotes angiogenesis in human glioma via VEGFA-mediated ERK signalling under hypoxia. EBioMedicine. 2019;48:36–48.
Liu H, Giffen KP, Grati M, Morrill SW, Li Y, Liu X, et al. Transcription co-factor LBH is necessary for the survival of cochlear hair cells. J Cell Sci. 2021;134:jcs254458.
Chen J, Huang C, Chen K, Li S, Zhang X, Cheng J, et al. Overexpression of LBH is associated with poor prognosis in human hepatocellular carcinoma. Onco Targets Ther. 2018;11:441–8.
Deng M, Yu R, Wang S, Zhang Y, Li Z, Song H, et al. Limb-bud and heart attenuates growth and invasion of human lung adenocarcinoma cells and predicts survival outcome. Cell Physiol Biochem. 2018;47:223–34.
Yu R, Li Z, Zhang C, Song H, Deng M, Sun L, et al. Elevated limb-bud and heart development (LBH) expression indicates poor prognosis and promotes gastric cancer cell proliferation and invasion via upregulating Integrin/FAK/Akt pathway. PeerJ. 2019;7:e6885.
Young IC, Brabletz T, Lindley LE, Abreu M, Nagathihalli N, Zaika A, et al. Multi-cancer analysis reveals universal association of oncogenic LBH expression with DNA hypomethylation and WNT-Integrin signaling pathways. Cancer Gene Ther. 2023;30:1234–48.
Ashad-Bishop K, Garikapati K, Lindley LE, Jorda M, Briegel KJ. Loss of Limb-Bud-and-Heart (LBH) attenuates mammary hyperplasia and tumor development in MMTV-Wnt1 transgenic mice. Biochem Biophys Res Commun. 2019;508:536–42.
Liu L, Luo Q, Xu Q, Xiong Y, Deng H. Limb-bud and heart development (LBH) contributes to glioma progression in vitro and in vivo. FEBS Open Bio. 2022;12:211–20.
Lamb R, Ablett MP, Spence K, Landberg G, Sims AH, Clarke RB. Wnt pathway activity in breast cancer sub-types and stem-like cells. PLoS One. 2013;8:e67811.
Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–26.
Honeth G, Bendahl PO, Ringner M, Saal LH, Gruvberger-Saal SK, Lovgren K, et al. The CD44+/CD24- phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;10:R53.
Khramtsov AI, Khramtsova GF, Tretiakova M, Huo D, Olopade OI, Goss KH. Wnt/{beta}-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am J Pathol. 2010;176:2911–20.
Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–9.
Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009;106:13820–5.
Frontini M, Kukalev A, Leo E, Ng YM, Cervantes M, Cheng CW, et al. The CDK subunit CKS2 counteracts CKS1 to control cyclin A/CDK2 activity in maintaining replicative fidelity and neurodevelopment. Dev Cell. 2012;23:356–70.
Gu Y, Rosenblatt J, Morgan DO. Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. EMBO J. 1992;11:3995–4005.
Mass G, Nethanel T, Kaufmann G. The middle subunit of replication protein A contacts growing RNA-DNA primers in replicating simian virus 40 chromosomes. Mol Cell Biol. 1998;18:6399–407.
Liaw H, Lee D, Myung K. DNA-PK-dependent RPA2 hyperphosphorylation facilitates DNA repair and suppresses sister chromatid exchange. PLoS One. 2011;6:e21424.
Ashley AK, Shrivastav M, Nie J, Amerin C, Troksa K, Glanzer JG, et al. DNA-PK phosphorylation of RPA32 Ser4/Ser8 regulates replication stress checkpoint activation, fork restart, homologous recombination and mitotic catastrophe. DNA Repair (Amst). 2014;21:131–9.
Toledo L, Neelsen KJ, Lukas J. Replication catastrophe: when a checkpoint fails because of exhaustion. Mol Cell. 2017;66:735–49.
Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9.
Marechal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol. 2013;5:a012716.
Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res. 2010;108:73–112.
Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–8.
Lecona E, Fernandez-Capetillo O. Targeting ATR in cancer. Nat Rev Cancer. 2018;18:586–95.
Kwon M, Kim G, Kim R, Kim KT, Kim ST, Smith S, et al. Phase II study of ceralasertib (AZD6738) in combination with durvalumab in patients with advanced gastric cancer. J Immunother Cancer. 2022;10:e005041.
McMullen M, Karakasis K, Loembe B, Dean E, Parr G, Oza AM. DUETTE: a phase II randomized, multicenter study to investigate the efficacy and tolerability of a second maintenance treatment in patients with platinum-sensitive relapsed epithelial ovarian cancer, who have previously received poly(ADP-ribose) polymerase (PARP) inhibitor maintenance treatment. Int J Gynecol Cancer. 2020;30:1824–8.
Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–8.
Garrido-Castro AC, Lin NU, Polyak K. Insights into molecular classifications of triple-negative breast cancer: improving patient selection for treatment. Cancer Discov. 2019;9:176–98.
Marine JC, Dawson SJ, Dawson MA. Non-genetic mechanisms of therapeutic resistance in cancer. Nat Rev Cancer. 2020;20:743–56.
Lehmann BD, Jovanovic B, Chen X, Estrada MV, Johnson KN, Shyr Y, et al. Refinement of triple-negative breast cancer molecular subtypes: implications for neoadjuvant chemotherapy selection. PLoS One. 2016;11:e0157368.
Minn AJ, Kang Y, Serganova I, Gupta GP, Giri DD, Doubrovin M, et al. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J Clin Invest. 2005;115:44–55.
Acknowledgements
We thank the Analytic Imaging Core of the Miami Project to Cure Paralysis for confocal imaging. This work was supported by NIH/NIGMS Grant R01GM113256 (K.J.B.), the Department of Defense (DoD)/Breast Cancer Research Program (BCRP) Breakthrough Award W81XWH-19-1-0255 (K.J.B.); a Tumor Biology Program Trainee Award (K.G.), and research funds from the Sylvester Comprehensive Cancer Center (K.J.B.). Research reported in this publication was performed in part at the Flow Cytometry Shared Resource (FCSR; RRID: SCR_022501) and the Cancer Modeling Shared Resource (CMSR; RRID: SCR_022889) of the Sylvester Comprehensive Cancer Center at the University of Miami Miller School of Medicine, which is supported by the National Cancer Institute Cancer Center Support Grant (CCSG) P30-CA240139.
Author information
Authors and Affiliations
Contributions
K.G. and K.J.B. conceived the study and designed the experiments. K.G. performed most experiments, analyzed the data, prepared the figures, and wrote the first draft of the manuscript. I.-C.Y. and S.H. generated stable LBH knockdown cell lines. I.-C.Y. performed in vivo Xenograft experiments and data analysis. P.R. provided essential reagents and feedback on the study. C.J. reviewed and edited the manuscript. K.J.B. directed the study, reviewed the data, wrote the manuscript, and secured funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Garikapati, K., Young, IC., Hong, S. et al. Blocking LBH expression causes replication stress and sensitizes triple-negative breast cancer cells to ATR inhibitor treatment. Oncogene 43, 851–865 (2024). https://doi.org/10.1038/s41388-024-02951-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-024-02951-3
- Springer Nature Limited