Abstract
Thyroid hormone receptor α1 (TRα1) mediates the genomic actions of thyroid hormone (T3). The biology of TRα1 in growth and development has been well studied, but the functional role of TRα1 in cancers remains to be elucidated. Analysis of the human thyroid cancer database of The Cancer Genome Atlas (TCGA) showed that THRA gene expression is lost in highly dedifferentiated anaplastic thyroid cancer (ATC). We, therefore, explored the effects of TRα1 on the progression of ATC. We stably expressed TRα1 in two human ATC cell lines, THJ-11T (11T-TRα1 #2, #7, and #8) and THJ-16T (16T-TRα1 #3, #4, and #8) cells. We found that the expressed TRα1 inhibited ATC cell proliferation and induced apoptosis. TCGA data showed that THRA gene expression was best correlated with the paired box gene 8 (PAX8). Consistently, we found that the PAX8 expression was barely detectable in parental 11T and 16T cells. However, PAX8 gene expression was elevated in 11T- and 16T-TRα1-expressing cells at the mRNA and protein levels. Using various molecular analyses, we found that TRα1 directly regulated the expression of the PAX8 gene. Single-cell transcriptomic analyses (scRNA-seq) demonstrated that TRα1 functions as a transcription factor through multiple signaling pathways to suppress tumor growth. Importantly, scRNA-seq analysis showed that TRα1-induced PAX8, via its transcription program, shifts the cell landscape of ATC toward a differentiated state. The present studies suggest that TRα1 is a newly identified regulator of thyroid differentiation and could be considered as a potential therapeutic target to improve the outcome of ATC patients.
Similar content being viewed by others
Introduction
Thyroid hormone receptor α1 (TRα1) is a ligand-dependent transcription factor that mediates the genomic actions of thyroid hormone (T3) in growth, development, and differentiation. TRα1 cDNA was first cloned from the chicken embryonic cDNA library [1]. Subsequently, cloning of the mammalian TRα1 cDNAs was shortly followed afterward [2,3,4,5]. Since then, extensive studies have demonstrated that TRα1 is critical in maintaining normal physiological T3-mediated homeostasis, cell proliferation, and organ development of many mammalian species [6]. The important role of TRα1 in differentiation was demonstrated in promoting intestinal stem cells into epithelia cells [7,8,9] and in the differentiation of embryonic stem cells into cardiomyocytes [10]. TRα1 is known to differentiate multipotent neural stem cells and to mature oligodendrocytes [11,12,13,14]. These findings indicate that TRα1 is a transcription factor that could initiate and drive cellular programs to promote differentiation in different cell types.
However, it is unknown whether TRα1 could act as a differentiation factor in highly dedifferentiated human cancer cells. We chose to study this question by using anaplastic thyroid cancer (ATC). ATC is extremely fast-growing, and at the time of diagnosis, the disease is too far advanced to be successfully treated through surgery or chemotherapy. ATC is highly dedifferentiated, as indicated by the loss of thyroid differentiation transcription factors, such as the paired box gene 8 (PAX8). Due to the loss of thyroid differentiation transcription factors in ATC, suppressed expression of the sodium iodide symporter (NIS) gene makes radioiodine therapy ineffective.
These dedifferentiated phenotypic characteristics of ATC provided us with a viable model to explore the role of TRα1 in the re-differentiation of highly dedifferentiated thyroid cancer. We showed that TRα1, when exogenously introduced into two ATC cell lines established from human ATC tumors (THJ-11T and THJ-16T cells; hereafter, simply referred to as 11T and 16T cells), inhibited ATC cell proliferation and induced apoptosis in vitro and in vivo in xenograft tumors. Analysis of The Cancer Genome Atlas (TCGA) database showed that THRA gene expression was lost in poorly differentiated thyroid cancer, most notably in the highly dedifferentiated ATC. Notably, analysis of thyroid differentiation scores demonstrated that THRA gene expression was significantly positively correlated with the expression of PAX8. Using single-cell transcriptomic analyses (scRNA-seq), we found that TRα1 induced the expression of the PAX8 gene, which can shift the cell landscape of ATC towards a more differentiated state through its transcription program. The present studies identified one mechanism by which TRα1 acted to re-differentiate ATC. Thus, TRα1 is a newly identified regulator of thyroid differentiation. Our findings could have important clinical implications in that the re-differentiation of ATC by TRα1 could make the disease more amendable to treatment through radioiodine therapies. In this context, TRα1 could be considered as a potential therapeutic target.
Results
TRα1 inhibits human ATC cell proliferation in vitro and growth within in vivo xenograft tumors
We made ATC lines stably expressing TRα1 to explore how TRα1 affects cell growth. In the parental 11T and 16T cells, TRα1 protein signals were barely visible (Fig. 1A-I-i, lanes 4 and 8, respectively). We used the three cloned cell lines expressing TRα1 similarly in 11T (11T-TRα1 clones #2, #7, and #8, shown in Fig. 1A-I-ii-a) and in 16T (16T-TRα1 clones #3, #4, and #8, Fig. 1A-I-ii-b) for characterization. The expression of TRα1 led to the suppression of cell growth in 11T (Fig. 1B-I-i) and 16T cells (Fig. 1B-I-ii). We further used immunohistochemical analysis (IHC) on the proliferation marker Ki-67 to demonstrate that the expression of TRα1 inhibited cell proliferation in 11T cells. Cells positively stained by Ki-67 were markedly decreased in 11T-TRα1 #2, #7, and #8 (Fig. 1C-I-i, panels e, h, and k) versus parental 11T (panel b). Quantitative analysis showed that TRα1 decreased Ki-67 positively-stained cells in 11T cells by 63–76% (Fig. 1C-I-ii-a). TRα1 also decreased the Ki-67 positively-stained cells in 16T (Fig. 1D-I-i, panels e, h, and k) versus parental 16T (panel b). Quantitative analysis showed that TRα1 decreased Ki-67 positively-stained cells in 16T cells by 63–74% (Fig. 1D-I-ii-a). Further, we have used CellTiter-Glo luciferase assay to further confirm that TRα1 inhibited the proliferation of 11T and 16T cells (Supplementary Fig. 1).
A Representative western blot analysis showing expression of TRα1 protein in ATC cells (n = 3). The cells were seeded in 6-well plates at a density of 500,000 cells per well and culutured for 2-3 days before the cells were lysated for analysis. (B), Cell proliferation curves for 11T and 11T-TRα1 #2, #7, and #8, as well as 16T and 16T-TRα1 #3, #4, and #8 cells, respectively. The cells were seeded in 6-well plates at a density of 50,000 cells per well and counted daily for 5 days. C, D The cells were expanded in 3–7 days before the cells were pelleted for IHC analysis. Representative IHC images for Ki-67 (b, e, h, k) and cleaved caspase-3 (c, f, i, l) (i) and quantitative analysis of IHC results (ii, n = 3). TRα1 inhibits cell migration of ATC cells (E-I-i, E-I-ii, F-I-i and F-I-ii). E, F The migration capacity was assayed for 11T and 11T-TRα1 #2, #7, and #8, as well as for 16T and 16T-TRα1 #3, #4, and #8 cells, respectively. Significant differences are indicated by asterisks (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 . Data represent the mean±SD (n = 3).
We also found that TRα1 acted to increase apoptosis, as shown by an increase in cells stained with cleaved caspase-3 antibodies in 11T (Fig C-I-i; compare panels f, i, and l, respectively, for clones #2, #7, and #8, with the parental cells in panel c) and in 16T (Fig. 1D-I-i; compare panels f, i, and l, respectively, for clones #3, #4 and #8, with the parental cells in panel c). Quantitative analysis shows that TRα1 increased cleaved caspase-3 positively-stained cells in 11T cells by 45–82% (Fig. 1C-I-ii-b) and in 16T cells by 75–65% (Fig. 1D-I-ii-b).
To understand whether TRα1 could affect the migration capacity of 11T and 16T cells, we carried out wound healing assays. As shown in Fig. 1E-I-i, the migration capacity of all three clones of 11T-TRα1-expression cells (#2, #7 and #8) was all retarded as compared with the parental 11T cells (Fig. 1E-I-ii). Similarly, we also found that the migration capacity of all three clones of 16T-TRα1-expression cells (#3, #4 and #8) was all retarded as compared with the parental 11T cells (Fig. 1F-I-i and -ii). These results indicated that TRα1 could delay the migration of ATC cells.
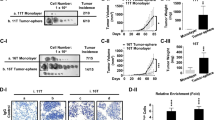
We next evaluated how the expression of TRα1 affected in vivo tumor growth in xenograft models. Compared to the parental 11T cells, expression of TRα1 in the three clones (#2, #7, and #8) resulted in the suppression of tumor growth, as shown in the growth curves (Fig. 2A-I). At the experimental endpoint (28 days), only skin bumps were observed (Fig. 2A-II), indicating that TRα1 could act to block tumor growth in 11T cells (Fig. 2A-III). In 16T cells, expression of TRα1 in the three clones (#3, #4, and #8) also led to the suppression of tumor development (Fig. 2 B-I). We noted that there were some minor variabilities in the inhibitory effects among the three 16T-TRα1 expression cells. However, at the endpoint of 48 days, the suppression effects of TRα1 were clearly shown, as the tumor weights of the three clones of 16T-TRα1 were significantly lower than those of tumors induced by the parental 16T cells (Figs. 2B-II and 2B-III). It would be of interest to point out that tumors induced by TRα1-expressing cells were much less vascularized than their respective parental tumors (Figs. 2A-II and 2 B-II), indicating TRα1 could inhibit angiogenesis in ATC.
A 11T- and 11T-TRα1 #2, #7, and #8-induced xenograft tumors. B 16T- and 16T-TRα1 #3, #4, and #8-induced xenograft tumors. Growth curves (I), gross images (II), and weight (III) of induced xenograft tumors. Significant differences are indicated by asterisks (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Data represent the mean±SD.
The inhibitory effect of TRα1 in tumor growth was also demonstrated by IHC analysis using proliferation marker Ki-67. TRα1 expression in 11T cells (clones #2, #7, and #8) had decreased immuno-stained Ki-67 intensities (Fig. 3A-I, compare panels e, h, and k with that of tumor induced by parental 11T cells). Similar decreased Ki-67 intensities were also detected in the 16T-TRα1 cells (#3, #4, and #8; compare panels e, h, and k with that of tumor induced by parental 16T cells). Quantitative analysis showed that tumor cells positively stained with Ki-67 were decreased by 44–49% in 11T-TRα1 cells and 18–45% in 16T-TRα1 cells (Figs. 3A-II-a and 3B-II-a, respectively).
A The cells were expanded in 3–7 days before the cells were pelleted for IHC analysis. Representative IHC images for Ki-67 (b, e, h, k) and cleaved caspase-3 (c, f, i, l) (i) and quantitative analysis of IHC results (ii, n = 3) for 11T- and 11T-TRα1 #2, #7, and #8-induced xenograft tumors and B IHC analysis for Ki-67 and cleaved caspase-3 in 16T- and 16T-TRα1 #3, #4, and #8-induced xenograft tumors. Significant differences are indicated by asterisks (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Data represent the mean ± SD.
Consistent with the observations in cultured ATC cells (Figs. 1C, D), we found that TRα1 induced apoptosis of tumor cells, as evidenced by an increased number of cells positively stained with cleaved caspase-3 in 11T-TRα1 cells (Fig. 3A-I, compare panels in the third column with those on the first left column) and in 16T-TRα1 cells (Fig. 3B-I, compare panels in the third column with those on the first left column). Quantitative analysis showed that tumor cells positively stained with cleaved caspase-3 were increased by 31–57% in 11T-TRα1 cells and 22–68% in 16T-TRα1 cells (Figs. 3A-II-b and 3B-II-b, respectively). Taken together, these in vitro and in vivo data demonstrated that TRα1 acted to inhibit ATC tumor growth by suppressing cell proliferation and inducing apoptosis.
The loss of THRA gene expression is associated with dedifferentiation of thyroid cancer
The finding that the expression of TRα1 acts to suppress ATC tumor growth prompted us to ask whether its loss is related to thyroid cancer progression. To this end, we analyzed THRA gene expression using gene expression data in TCGA. When compared with normal thyroid tissues (n = 105), the THRA gene expression in 505 papillary thyroid cancer (PTC) patients was significantly lower (Fig. 4A-I). More detailed analysis shows that THRA gene expression in the well-differentiated PTC was not significantly different from the normal thyroid (Fig. 4A-II, compare data set in the middle versus data set on the left). However, THRA gene expression was significantly lower in poorly differentiated PTC (Fig. 4A-II, compare the third data set on the right versus the first data set on the left), indicating that decreased THRA gene expression is associated with dedifferentiation in thyroid cancer. This notion was further supported by the analysis comparing the normal thyroid and PTC, where the THRA gene expression was the lowest in ATC using the GSE33630 dataset (Fig. 4A-III).
A-I Downregulation of THRA mRNA expression in PTC, compared to normal. A-II Loss of THRA expression in poorly differentiated PTC, compared to normal and well-differentiated PTC. The PTC samples from TCGA-THCA were classified into well- and poorly-differentiated PTC according to thyroid differentiation score (TDS) [17], a single metric that summarized the expression levels of 16 thyroid metabolism and function genes. A-III Gradual loss of THRA mRNA expression over the progression of thyroid cancer from normal to ATC. B Association of the expression of BRAF mutations (B-I), TERT mutations (B-II), RAS mutations (B-III) and combined BRAF and TERT mutations (B-IV) with the expression of the THRA gene expression analyzed by TCGA database. C Positive correlation between THRA and PAX8 mRNA expression in human tissues (I and II) analyzed by Pearson’s correlation coefficient, and loss of both THRA and PAX8 mRNA expression in ATC (II). D-I Downregulation of PAX8 mRNA expression in PTC, compared to normal. D-II Loss of PAX8 mRNA expression in poorly differentiated PTC, compared to normal and well-differentiated PTC. D-III Gradual loss of PAX8 mRNA expression over the progression of thyroid cancer from normal to ATC. Data were obtained from TCGA-THCA (n = 505) [17] and GSE33630 [18]. Significant differences are indicated by asterisks (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
We further analyzed the relationship of the expression of the THRA gene with common driver mutations of thyroid cancer: BRAF, RAS, and TERT. Interestingly, the THRA expression was significantly lower in tumors with either BRAF mutation (Fig. 4B-I) or TERT mutation (Fig. 4B-II) while showing no significant association with RAS mutation (Fig. 4B-III). Surprisingly, tumors harboring both mutations in BRAF and TERT exhibited the lowest expression of THRA (Fig. 4B-IV), compared to tumors harboring no mutation or a single mutation (BRAF or TERT). These results suggest that the loss of THRA expression is closely related to BRAF and TERT mutations for thyroid cancer progression given these mutations have been known to increase mortality in patients with thyroid cancer [15].
We next sought to understand which thyroid differentiation genes are most affected by the reduced THRA gene expression. Because the paired domain transcription factor PAX8 is known to play a fundamental role in the maintenance of functional differentiation in thyroid cells [16], we explored whether there was an association of expression of the THRA gene with the PAX8 gene in TCGA database. Notably, we found that the expression of the THRA gene was significantly positively correlated with the expression of the PAX8 gene in PTC tumors (r = 0.4909, p < 0.0001; Fig. 4C-I). The positive correlation of these two genes was also demonstrated in ATC using the dataset in GSE33630 (r = 0.4909, p < 0.0001; Fig. 4C-II). Using the TCGA Thyroid Cancer Collection (THCA) database [17], we found that the PAX8 gene was significantly lower in PTC (Fig. 4D-I) and was lowest in poorly differentiated PTC (Fig. 4D-II). Using GSE33630 [18], we found that, when compared with normal thyroid and PTC, the expression of the PAX8 gene was lowest in ATC (Fig. 4D-III). The decreased expression of the THRA gene was associated the dedifferentiation of thyroid cancer, and the positive correlation of the PAX8 gene with the THRA gene suggested to us that TRα1 could regulate the expression of the PAX8 gene.
TRα1 directly regulates the expression of the PAX8 gene to modulate the differentiation of thyroid cancer cells
To test our hypothesis that TRα1 could regulate the expression of the PAX8 gene, we analyzed PAX8 protein in the three cloned 11T and 16T TRa1 expression cell lines. Concurrent with elevated TRα1 protein levels (Fig. 5A-I-a, group i, A-II-a), the PAX8 protein was also elevated in 11T-TRα1 expression cells (Fig. 5A-I-b, group i, A-II-b). Similarly, when compared to the expression of TRα1 proteins (Fig. 5A-I-a, group ii, A-II-a), the PAX8 protein levels were also elevated in 16T-TRα1 expression cells (Fig. 5A-I-b, group ii, A-II-b). We further analyzed the PAX8 protein levels through IHC analysis of fixed-cell pellets. Consistent with western blot analysis, cells strongly stained with anti-PAX8 antibody were detected in all three clones of 11T-TRα1 expression cells (Fig. 5B-I-i) and 16T-TRα1 expression cells (Fig. 5B-I-ii). The positively-stained cells were counted, and quantitative data indicated that PAX8-expressing cells were 43–68% higher in the TRα1-expressing cells than the control parental 11T cells (Fig. 5B-II-a) and 50–65% higher in the TRα1-expressing cells than the parental 16T cells (Fig. 5B-II-b). These cell-based findings were further confirmed in tumors induced by 11T-TRα1 cells or 16T-TRα cells. IHC showed that cells expressing TRα1 had elevated PAX8 in tumors induced by the three clones of 11T-TRα1 cells and 16T-TRα1 cells, respectively (Fig. 5C-I-i and -ii). The quantitative analysis showed that PAX8 protein levels, as detected by IHC analysis, were increased by 49–70% in 11T-TRα1-expressing cells and 61–82% in 16T-TRα1-expressing cells (Fig. 5C-II-a and -b). Using anti-TRα1 antibody, we showed that compared with the xenograft tumors induced by 16T parental cells (Figs. 5D-I-b and 5D-II), TRα1 proteins were clearly detected in xenograft tumors induced by TRα1-expressing cells (clone #3, #4, and #8 in panels c, d, and e, Figs. 5D-I and 5D-II). Taken together, these findings indicate that increased TRα1 protein level is associated with elevated level of PAX8 proteins.
A The cells were expanded in 3–7 days before the cells were pelleted for IHC analysis. Representative western blot analysis showing high expression of TRα1 and PAX8 proteins in ATC cells (n = 3). B, C Representative IHC images and quantitative analysis of IHC results for PAX8 in 11T, 11T-TRα1 #2, #7, and #8 and 16T, 16T-TRα1 #3, #4 cultured cells (B-I and B-II); and xenograft tumors (C-I and C-II) (n = 3). D Analysis of the TRα1 protein levels by IHC in tumors induced by 16 T cells (D-I-i and D-II) and by 16T-TRα1-expressing cells (D-I-i and D-II). Significant differences are indicated by asterisks (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Data represent the mean±SD.
The expression of the PAX8 gene is directly regulated by TRα1
The findings showing that PAX8 protein levels were elevated in TRα1-expressing cells prompted us to ask whether the PAX8 gene is directly regulated by TRα1. TRα1 regulates target gene expression through binding with thyroid hormone response elements (TREs) on the promoter of target genes [19, 20]. Toward this end, we searched for potential TREs in the promoter region of the PAX8 gene. As shown in Fig. 6A-I, we identified a putative TRE with the sequence of AGGCCAGAGGTCA (a DR1 TRE) located in the −778 to −790 region on the PAX8 promoter. To validate that this TRE was the TRα1 binding site, we carried out ChIP assays by using monoclonal antibody C4, which recognizes the C-terminal sequences of TRα1. We first carried out ChIP assays in the parental 11T cells, which did not express TRα1 proteins, no significant binding was detected (Fig. 6A-II-a). However, a significant binding of TRα1 to the TRE was detected in the 11T-TRα1 #2 (Fig. 6A-II-b), 11T TRα1 #7 (Fig. 6A-II-c), 11T TRα1 #8 (Fig. 6A-II-d). Similarly, no apparent binding was detected in the parental 16T cells (Fig. 6A-II-e), but a significant binding of TRα1 to the TRE was detected in the 16T TRα1 #3 cells (Fig. 6A-II-f).
A-I Schematic representation of an upstream promoter of the PAX8 gene locus. The promoter region encompassed −1779 bp upstream and +549 bp downstream from the transcription starting site. Location of the putative PAX8 TRE, transcription start site (TS), or ATG site is indicated. A-II ChIP assays show the binding of TRα1 to the PAX8 promoter in the 11T parental, 11T-TRα1 #2, #7, #8, 16T parental and 16T-TRα1 #3 cells. The antibody IgG was a control, and monoclonal anti-TRα1 antibody, C4, which recognizes TRα1, was used in the ChIP assays. Bar 1: input; bar 2: IgG negative controls; bar 3: C4. The significant differences are indicated by P values (n = 3). B Transcriptional activity mediated by the PAX8 promoter (B-I-i) or a PAX8 TRE (B-I-ii) was measured by luciferase reporter assay in the CV1 (B-II-a), FTC133 (B-II-b), and FTC236 (B-II-c) cells. The luciferase activity was measured after CV1, FTC133 or FTC236 cells were transfected with a luciferase reporter and treated with vehicle or T3 for 24 h. Control is the pGL3 luciferase reporter vector. PAX8 TRE luciferase reporter contains the TRE sequence AGGCCAGAGGTCA of the PAX8 promoter (pGL3-TRE reporter). pGL3-PAX8 luciferase reporter contains the 2328 bp DNA fragment from −1779 to +549 of the PAX8 promoter (pGL3-PAX8 reporter). Reporter assays were carried out with vehicle or in the presence of T3 (10 nM).
To further evaluate if the PAX8 promoter mediates the TRα1-regulated PAX8 expression, we performed luciferase assays using two different luciferase reporters. For one reporter, designated as pGL3-PAX8 (see Fig. 6B-I-i), the expression of the luciferase was driven by the large segment of the PAX8 upstream promoter encompassing −1779 to +549 bp, including DR1 TRE (located in −790 to −778 bp). For the other reporter, designated as pGL3-TRE, the expression of the luciferase activity was driven by the DR1 TRE located at −790 to – 778 bp (see Fig. 6B-I-ii). We found that the luciferase activity mediated by GL3-TRE and GL3-PAX8 was increased 1.6- and 1.4-fold by T3, respectively, in the CV1 cells (Bar 4 vs bar 3, bar 6 vs bar 5, Fig. 6B-II-a). In human FTC133 cells, T3 activated the luciferase activity mediated by GL3-TRE and GL3-PAX8 by 2- and 1.4-fold, respectively (Bar 4 vs bar 3, bar 6 vs bar 5, Fig. 6B-II-b) and by 2.5- and 1.4-fold, respectively, in human FTC236 cells (Bar 4 vs bar 3, bar 6 vs bar 5, Fig. 6B-II-c). These results indicate that TRα1 directly regulates the PAX expression through the interaction with the PAX8 promoter. These findings demonstrate that the PAX8 gene is a newly identified T3 target gene.
Single-cell transcriptomic analysis reveals reversal of dedifferentiation state of ATC by TRα1-activated expression of PAX8
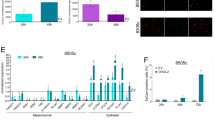
To understand the functional consequences of the TRα1-activated expression of the PAX8 gene in vivo, we performed scRNA-seq analysis using the single cells dissociated from 16T parental and 16T-TRα1 tumors (Fig. 7A). We obtained single-cell transcriptome data for 10,961 and 10,584 cells from each of two 16T parental tumors and for 9064 and 7556 cells from each of two 16T-TRα1 tumors. Cell types for the clusters were annotated against a built-in reference within the SingleR Bioconductor package [21], the Human Primary Cell Atlas [22]. Notably, in 16T parental tumors (Fig. 7B-I), we identified tumor cells (73.8%), mesenchymal stem cells (MSCs, 20.6%), and undifferentiated stem cells (5.5%). By contrast, in 16T-TRα1 tumors (Fig. 7C-I), we found reduced number of tumor cells (56.1%), epithelial cells (35.5%), and fibroblasts (8.5%). The contrasting cell clusters of MSCs and undifferentiated stem cells identified in 16T-induced tumors versus cell clusters of the fibroblasts, epithelial cells, and reduced number of tumor cells in 16T-TRα1-indunced cells suggest the remodeling of the cell landscape toward a tumor-free milieu and re-differentiated state by the expression of TRα1. Figures 7B-II and 7C-II show heatmaps displaying the expression of top gene markers used to characterize each cluster of 16T parental cell-induced tumors and 16T-TRα1 cell-induced tumors, respectively. These data clearly showed that TRα1 could act to change the differentiation state in ATC cells.
A 16T parental or 16T-TRα1 cells were injected onto the flank of nude mice, each in duplicates. After 49 days, tumors were dissected and dissociated into single cells for single-cell transcriptomic analysis. (B-I and C-I), TSNE plots of single-cell RNA sequencing of 16T and 16T-TRα1 xenograft tumors, respectively, and a pie chart showing the different cell types identified for the clusters in 16T (B-I) and 16T-TRα1 (C-I) xenograft tumors. (B-II and C-II) show heap maps displaying the top gene markers used to characterize each clusters of 16T parental cell-induced tumors and 16T-TRα1 cell-induced tumors, respectively.
TRα1 increases the NIS protein levels in ATC-cultured cells and xenograft tumors
To provide additional evidence that the expression of TRα1 protein could lead to re-differentiation of ATC cells, we carried out experiments to demonstrate that the sodium iodide symporter (NIS) encoded by the SLC5A5 gene, could be re-expressed in TRα1-expressing cells and the induced tumors. NIS is a plasma membrane glycoprotein localized at the basolateral membrane of thyroid follicular cells mediating the active transport of iodide into the thyroid gland as an important prerequisite for the biosynthesis of thyroid hormones. Figure 8 shows that the expression of the SLC5A5 gene at the protein levels by western blot analysis and IHC in 11T and 16T cells in vitro and in vivo using xenograft tumors. As shown by the western blot analysis, NIS protein levels in 11T and 16T cells were increased in the TRα1-expressing cells (Fig. 8A-I-i and -ii, respectively and Fig. 8A-II-b). The elevated NIS levels were further demonstrated by IHC in 11T and 16T cell pellets (Figs. 8B-I-i and 8B-II-a for 11T cells; Figs. 8B-I-ii and 8B-II-b for 16T cells). Further, using the tumors induced by parental 11T and 16T and their TRα1-expressing cells, we also found that the NIS proteins were elevated in tumor-induced by TRα1 (Figs. 8C-I-i and 8C-II-a for 11T cells and Figs. 8C-I-ii and 8C-II-b for 16T cells). These findings indicate that re-differentiation by TRα1 in ATC cells could lead to re-expression of the SLC5A5 gene, which could have important implications for clinical application for treatment of ATC patients.
A The cells were expanded in 3–7 days before the cells were pelleted for IHC analysis. Representative western blot analysis showing high expression of TRα1 and NIS proteins in ATC cells (n = 3). B, C Representative IHC images and quantitative analysis of IHC results for NIS proteins in 11T, 11T-TRα1 #2, #7, and #8,16T and 16T-TRα1 #3, #4, and #8 cultured cells (B-I and B-II) and for xenograft tumors (C-I and C-II) (n = 3). Significant differences are indicated by asterisks (*P < 0.05], **P < 0.01, ***P < 0.001 and ****P < 0.0001. Data represent the mean±SD.
Discussion
Much is already known about how TRα1 could function as a transcription factor to mediate differentiation in target organs, such as the intestine, the heart, and the brain [7,8,9,10,11,12,13,14]. However, how TRα1 could function to change the differentiation state in cancer cells has been less studied. We sought to answer this question by using ATC cells as a model in our studies. ATC is a highly dedifferentiated thyroid cancer, reflected by the lost expression of thyroid transcription factors, such as PAX8. Indeed, the expression of the THRA gene was markedly lower in ATC as compared to normal thyroid and differentiated PTC (see Fig. 4). Exogenous expression of the THRA gene in the two human ATC cells resulted in the inhibition of proliferation and induction of apoptosis in vitro and in vivo in xenograft tumors. Notably, scRNA-seq analysis revealed that tumors induced by 16T parental cells could be subclassed into three major types of cells: 73.8% tumor cells, 20.6% MSC cells, and 5.5% undifferentiated stem cells. In contrast, tumors induced by TRα1-expressing cells, in addition to reduction of tumor cells (56.1%), also showed two clusters of well-differentiated epithelial cells (35.5%) and fibroblasts (8.4%). These findings show that TRα1 can mediate re-differentiation of ATC tumor cells. We further demonstrated that one mechanism by which TRα1 mediated the re-differentiation was at least in part via the activated expression of the PAX8 gene (see Fig. 5 and 7). Additional evidence to further support that TRα1 could activate the expression of PAX8 gene in the tumors induced by the parental 16T cells and 16T-TRα1-expressing cells is shown Supplementary Fig. 2A. More cells expressed the THRA gene in 16-TRα1 tumors (6.14%) than in 16T tumors (3.5%), representing a 1.8-fold cells expressing the THRA gene (Supplementary Fig. 2A, bar d versus bar c). Further, more cells expressed the PAX8 gene in 16-TRα1 tumors (32.1%) than the parental 16T tumor (21.6%) (Supplementary Fig. 2A, bar b versus bar a), representing a 1.5-fold increase of cells expressing the PAX8 gene in the TRα1-expressing tumors. These findings are consistent with the IHC data in that the elevated expression of the THRA gene at the protein level in 16T-TRα1 tumors is associated with a higher PAX8 protein level than in 16T-parental tumors (Figs. 5C-II-b and 5D-II). Further, we showed that TRα1 directly regulated PAX8 via interaction of DR1 TRE (Fig. 6). Thus, the present studies identified TRα1 as a novel regulator of thyroid differentiation.
It is of interest to note that the expression of the PAX8 gene was found in the MSC and undifferentiated stem cells in 16T parental tumors (Supplementary Fig. 6B-a). In contrast to previous reports of a more restricted expression pattern for the PAX8 gene, recent studies have shown that the PAX8 gene is expressed in all the tissues investigated using the Common Fund’s Genotype-Tissue Expression (GTEx) Program (https://gtexportal.org) [23]. PAX8 expression has been shown to associate with tumors of kidney, prostate, breast, cervix, lung, esophagus, pancreas, skin, thyroid, and ovary [24,25,26,27]. Consistent with our findings, the PAX8 gene was reported to express in embryonic stem cells [28] and adipose-derived MSC cells [29]. As found by TSNE analysis showing the expression of the PAX8 gene in the epithelial cells of the 16T-TRα1 tumors (Supplementary Fig. 2B-b), the PAX8 gene was also reported to express in non-neoplastic tissue such as epithelial cells of thyroid, endocervix, endometrium, fallopian, seminal vesicle, epididymis, pancreatic islet cells, lymphoid cells, and glomerular parietal cells [30, 31]. PAX8 was found to act as a transcription factor to convert human and mouse fibroblasts into renal tubular epithelial cells, indicating the PAX8 can function in the fibroblasts [32]. Together with these recent reports, our studies have further expanded the scope in the PAX8 gene expression patterns.
Early association studies have shown that the THRA locus undergoes frequent loss of heterozygosity (LOH) in sporadic breast cancer, and rearrangement of the THRA gene has also been reported in leukemia, breast, and stomach cancer [33,34,35]. These associated findings suggest that the loss of the THRA gene could benefit cancer development. This notion is supported by our observations in the present studies that the expression of the THRA gene is lost during thyroid carcinogenesis, as shown from analysis of TCGA and GSE33630 data (see Fig. 4). Moreover, when TRα1 was exogenously expressed, ATC proliferation and tumor growth were blocked (Figs. 1 and 2). Single-cell RNA-seq data showed that there were 25% fewer tumor cells in 16T-TRα1 cell-induced tumors than in 16T parental cell-induced tumors (See Fig. 7B, C). While there were 667 common marker genes in the two tumor cell clusters, there were also unique genes between the two (Supplementary Fig. 3). Analysis of the significant pathways mediated by these unique genes exhibited distinct pathways (Supplementary Table 1A, B) between the two tumor cell clusters. These data indicated that TRα1 could mediate direct and/or indirect signaling to reduce tumor growth via multiple cellular pathways. In this context, TRα1 could be considered as a tumor suppressor.
In the past decades, differentiation therapy has emerged as a potential therapeutic avenue to treat cancers. Instead of aiming to kill tumor cells by traditional chemotherapy and radiation, differentiation therapy alters the tumor cells to be more like normal cells. Differentiation therapy reactivates endogenous differentiation programs in cancer cells to eliminate cancer phenotypes. A notable example is the use of all-trans-retinoic acid to induce rapid clearance of acute promyelocytic leukemia (APL) [36,37,38]. Recently, drugs that trigger differentiation in a variety of primary tumors have been identified [39]. In the case of thyroid cancer, re-differentiation therapy, followed by I-131 treatment, is a potential option to treat radioiodine refractory differentiated thyroid cancer patients. Various agents have been tested, such as retinoic acids, histone deacetylase inhibitors, peroxisome proliferative-activated receptor γ agonists, mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinase (MEK/ERK) inhibitors and phosphatidylinositol 3-kinase (PI3K)/AKT inhibitors [40]. Selumetinib, MAPK kinase (MEK)1/2 inhibitor, and Dabrafinib, a B-RAF (BRAF) inhibitor, have been used in clinical trials. Both inhibitors stimulate radioiodine uptake in patients [41, 42], which supports differentiation therapy as a potentially viable option to improve the treatment outcome of poorly differentiated thyroid cancer. However, no complete response has been observed by using the re-differentiation agents identified so far. Furthermore, the potential adverse toxicity of the inhibitors used in differentiation therapy would also need to be considered if long-term used is warranted. In view of these considerations, there is a need to search for and develop other novel re-differentiation therapeutic targets. The present studies show that TRα1, via its transcription activity to initiate multiple signaling pathways, not only suppresses tumor growth, but it also converts the highly dedifferentiated tumor cells to a more differentiated cellular landscape via one of its reactivated thyroid differentiation transcription factors, PAX8. These findings suggested that TRα1 could be further tested as a novel differentiation target for poorly differentiated thyroid cancer.
Using various animal models of single Thra or Thrb gene knockout, as well as double knockouts, investigators have demonstrated the TRα1 and TRβ have isoform-specific and redundant functions in target tissues [43,44,45,46,47]. It is of interest to note that the TR isoform also has common functions in cancer biology. We and others have shown that TRβ acts as a tumor suppressor in many cancers, such as breast cancer and thyroid cancer [48,49,50]. The present studies showed that TRα1 suppresses proliferation and induces apoptosis to inhibit ATC tumor growth. Recently, we showed that TRβ suppresses cancer stem cell activity in ATC in vitro and in vivo and that the expression of the THRB gene is negatively correlated with stemness, as shown by TCGA THCA database [51]. Interestingly we found that there was no significant correlation of THRA gene expression with the stemness of thyroid cancer (Supplementary Fig. 4). Our present findings indicated that TRα1 acted to re-differentiate ATC (Fig. 7C), suggesting that these two TR sub-type function in an isoform-dependent way to regulate thyroid cancer progression. Analysis of the TCGA THCA database showed low lymph node metastasis incidence was associated with high expression of both THR genes. Conversely, incidence of high lymph node metastasis was associated with low expression of both THR genes (Supplementary Fig. 3). Low THR gene expression was detrimental to patient survival (Supplementary Fig. 5). It is of interest to note that there was no significant association with clinical outcome of lymph metastasis nor with disease-free survival, if only the expression of a single TR gene was used in the analysis. These findings suggested that each TR isoform would need to contribute its subtype-specific functions, i.e., TRβ acts to suppress cancer stem cells and TRα1 functions to induce re-differentiation. These distinct anti-tumor activities would need to act concordantly and collaboratively, to lead to positive clinical outcomes. Taken together, our findings have uncovered that in cancer cells, TR also acts via specific and redundant functions. Further, we have broadened our understanding of TRs in the biology of thyroid cancer.
Materials and methods
Cell cultures
ATC cells (11T and 16T) were derived from human anaplastic thyroid tumors. 11T cells carry KRAS mutation, whereas THJ-16T cells have PI3KCA, RET, and TP53 mutations. The TRα1-expressing 11T or 16T cell lines were established as described. Briefly, the cells were seeded in the wells of a 6-well plate at the density of 6 × 105 one day before transfection with a TRα1-expressing vector pcDNA3.1-TRα1 using Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instruction. Twenty-four hours after transfection, the cells were selected with 200 μg/ml G418 for 14 days. The selected single colonies were picked and expanded for the analysis. The cells were cultured in RPMI-1640 media supplemented with 10% FBS, 1% non-essential amino acid, 1% sodium pyruvate, and 1% antibiotic-antimycotic solution (Thermo Fisher Scientific, Waltham, MA, USA). 11T-TRα1 #2, #7, and #8 and 16T-TRα1 #3, #4, and #8 cells were cultured in the same media as for ATC cells. African green monkey kidney fibroblast Cells (CV-1) were plated 24 h before transfection in DMEM media containing10% (vol/vol) hormone-depleted fetal calf serum, 100 U/ml penicillin,100 μg/ml streptomycin in six-well plates at a density of 0.5 × 106 cells/well. FTC-133 and FTC-236 cells, kindly provided by Dr. Orlo H. Clark (UCSF Mount Zion Medical Center, San Francisco, CA), were derived from a primary human follicular thyroid carcinoma and neck lymph node metastases, respectively [48]. FTC133 cells carry NF1, PTEN and TP53 mutations while FTC236 cells carry RB1, PTEN and TP53 mutations. These cells were plated 24 h before transfection in DMEM/Ham’s F12 (1:1) medium supplemented with 10% hormone-depleted fetal bovine serum, 2 mM Glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen), or DMEM/Ham’s F12 (1:1) medium (Invitrogen) supplemented with 10% hormone-depleted fetal bovine serum, 10 μg/ml bovine insulin (Sigma-Aldrich), 1 mIU/ml bovine TSH (Sigma-Aldrich), 2 mM Glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) in six-well plates at a density of 0.5 × 106 cells/well. All cells were cultured in a 37 °C, 5% CO2 incubator.
Cell migration assay
11 T, 11T-TRα1 #2, #7, and #8, and 16 T, 16T-TRα1 #3, #4, and #8, cells were plated in 6-well plates at a density of 500,000 cells per well. After 48 h, a pipette tip was used to make a scratch. Cells were washed with phosphate-buffered saline. Cell migration was visualized under a microscope at ×100 magnification at 0 and 24 h.
In vivo mouse xenograft study
All animal experiments were performed under protocols approved by the National Cancer Institute Animal Care and Use Committee. For xenograft studies, 6- to 8-week-old female athymic nude mice were used in the studies. Parental 11T, and 16T cells and their TRα1 expressing cells (5 × 106 cells) were each added in 200 μl 50% Matrigel basement membrane matrix and subcutaneously inoculated into the right flank of mice. We then monitored the frequency of tumor initiation and tumor growth for about 8 weeks and then euthanized the mice to dissect the tumors for further analyses. When the longest diameter of the biggest tumor exceeded 20 mm, we simultaneously euthanized all the mice to dissect the tumors for further analyses.
Western blot analysis
Cell Extraction Buffer (Thermo Fisher Scientific, Waltham, MA, USA) and phenylmethylsulfonyl fluoride (PMSF) were added to the cells, followed by centrifugation at 13,000 rpm for 10 min to collect the supernatant. After protein quantification, the lysates were heated at 100 °C for 5 min. Proteins were loaded on Tris-glycine gels (Invitrogen, Thermo Fisher Scientific) and transferred to 0.45 μm nitrocellulose membranes (Millipore, Burlington, MA). They were then incubated with the primary antibody overnight at 4 °C. PAX8 (Proteintech, Rosemont, IL) antibodies were used. Membranes were washed with TBS-T and incubated with secondary antibodies for 1 h at room temperature. After washing with TBS-T, proteins were detected using the ECL system (Enzo Life Sciences, New York, USA). Band intensities were quantified by the ImageJ software (ImageJ 1.48v; Wayne Rasband, NIH).
Cell proliferation assays
11T, 11T-TRα1 #2, #7, and #8, and 16T, 16T-TRα1 #3, #4, and #8, cells were plated in 6-well plates at a density of 50,000 cells per well. Cells were counted daily for 5 days using a cell counter (Countess II, Thermo Fisher Scientific, Waltham, MA, USA). Cell proliferation was also analyzed by CellTiter-Glo 2.0 luciferase assay. ATC 11T-3.1, 16T-3.1, 11T-TRα1#2, #7, #8, 16T-TRα1#3, #4, and #8 cells were seeded in 96-well plates at a density of 1000 cells per well. On the day of assay, each well was replaced with 25 μl of fresh medium and then added with 25 μl CellTiter-Glo 2.0 luciferase reagent to quantify ATP for the evaluation of the number of viable cells. CellTiter-Glo 2.0 luciferase activity (Promega, Madison, Wisconsin) was measured daily for 5 days. In addition, the proliferation was also assayed by Ki-67 immunostaining using slides from fixed cell pellet with anti-Ki-67 antibodies (cell signaling technology, Massachusetts, MA).
Immunohistochemistry
Cells or tissue from xenografted tumors were fixed in 10% neutral buffered formalin and then embedded in paraffin. Sections 5 µm thick were prepared and stained with hematoxylin and eosin. Immunohistochemistry (IHC) was performed as previously [48]. Anti-Ki-67 (cell signaling technology, Massachusetts, MA), cleaved caspase-3 (cell signaling technology, Massachusetts, MA), and PAX8 (Proteintech, Rosemont, IL) antibodies were used. Tissue slides were visualized under a microscope at ×200 magnification, and the relative percentage of positive cells were determined with NIH ImageJ software.
RNA extraction and real-time RT-PCR analysis
Total RNA extraction was performed using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. cDNA was generated using oligo dT primers and GoScript™ Reverse Transcriptase (Promega, Wisconsin-Madison, WI). PCR was determined using QuantStudio 3 (A28567, Applied Biosystems).
Analysis of TCGA data
A transcriptome and its matched clinical patient data from The Cancer Genome Atlas (TCGA-THCA, normal thyroid, n = 59; thyroid cancer, n = 505) and GSE33630 (normal thyroid, n = 45; papillary thyroid cancer (PTC), n = 49; anaplastic thyroid cancer (ATC), n = 11) were analyzed to investigate clinical implication of THRA gene in human thyroid cancer. We also used a thyroid differentiated score (TDS), a single measure that summarized the expression levels of 16 thyroid metabolism and function genes, to divide PTC into well- and poorly differentiated PTC in the TCGA-THCA.
Single-cell dissociation from xenografts and single-cell transcriptomic analysis
We dissociated the xenograft tumors into single cells using a tumor dissociation kit. The tumors dissected from 16T parental or 16T- TRα1 cell-induced xenograft were cut into small pieces of 2–4 mm, and these tissues were transferred into the gentleMACS C Tube containing an enzyme mix (2.35 mL of RPMI-1640, 100 µL of Enzyme D, 50 µL of Enzyme R, and 12.5 µL of Enzyme A), followed by the tumor dissociation using a gentle MACS Dissociator. The dissociated cells were then centrifuged, resuspended, and applied to a cell strainer (70 µm) placed on a 15 mL tube, followed by washing the cell strainer with 10 mL of RPMI-1640. The cells were then centrifuged at 300 x g for 7 min, resuspended with the AKC lysing buffer to remove erythrocytes, washed with phosphate buffer saline. These single cells were analyzed by single-cell RNA sequencing (scRNA-seq). Further detailed methods and bioinformatics analyses are included in the Supplementary Materials and Methods section.
Construction of luciferase reporters
The PAX8 promoter luciferase reporter in the pGL3 vector (pGL3-PAX8) was purchased from Addgene (Plasmid #11994). For the construction of a PAX8 thyroid hormone response (TRE) containing Luc reporter, the AGGCCAGAGGTCA sequence of the PAX8 promoter was cloned into the pGL3 luciferase reporter. The constructed PAX8 TRE luciferase reporter was confirmed by Sanger DNA sequencing analysis.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed as described previously [52]. To evaluate the binding of TRα1 to the PAX8 promoter, real-time quantitative PCR was conducted using precipitated chromatin DNA with a pair of primers: PAX8TREF, caccagtggaccccttctta and PAX8-TRE-R, aactcctcgcctctctcctc.
Statistical analysis
Continuous variables were compared using a two-tailed Student’s t-test, or Mann–Whitney U test when comparing less than 5 biological replicates, and categorical data were compared using a two-tailed χ2 test or Fisher’s exact test. Two-way analysis of variance with Bonferroni’s post-hoc test was used to compare tumor growth in the xenograft model according to the time and groups. Disease-free was compared between high and low THRA groups. Statistical analysis was performed using SPSS version 23.0 or GraphPad Prism. Data are presented as the mean+SD. All P values were two-sided throughout, and those less than 0.05 were considered statistically significant.
Further detailed methods are included in Supplementary Materials and Methods section.
Data availability
scRNA-seq data have been submitted to NCBI GEO. The GEO accession number is GSE235216 for accessing the data.
References
Sap J, Munoz A, Damm K, Goldberg Y, Ghysdael J, Leutz A, et al. The c-erb-A protein is a high-affinity receptor for thyroid hormone. Nature. 1986;324:635–40.
Benbrook D, Pfahl M. A novel thyroid hormone receptor encoded by a cDNA clone from a human testis library. Science. 1987;238:788–91.
Thompson CC, Weinberger C, Lebo R, Evans RM. Identification of a novel thyroid hormone receptor expressed in the mammalian central nervous system. Science. 1987;237:1610–4.
Hodin RA, Lazar MA, Wintman BI, Darling DS, Koenig RJ, Larsen PR, et al. Identification of a thyroid hormone receptor that is pituitary-specific. Science. 1989;244:76–9.
Murray MB, Zilz ND, McCreary NL, MacDonald MJ, Towle HC. Isolation and characterization of rat cDNA clones for two distinct thyroid hormone receptors. J Biol Chem. 1988;263:12770–7.
Pascual A, Aranda A. Thyroid hormone receptors, cell growth and differentiation. Biochim Biophys Acta. 2013;1830:3908–16.
Sun G, Roediger J, Shi YB. Thyroid hormone regulation of adult intestinal stem cells: Implications on intestinal development and homeostasis. Rev Endocr Metab Disord. 2016;17:559–69.
Giolito MV, Plateroti M. Thyroid hormone signaling in the intestinal stem cells and their niche. Cell Mol Life Sci. 2022;79:476.
Shi YB, Hasebe T, Fu L, Fujimoto K, Ishizuya-Oka A. The development of the adult intestinal stem cells: Insights from studies on thyroid hormone-dependent amphibian metamorphosis. Cell Biosci. 2011;1:30.
Lee YK, Ng KM, Chan YC, Lai WH, Au KW, Ho CY, et al. Triiodothyronine promotes cardiac differentiation and maturation of embryonic stem cells via the classical genomic pathway. Mol Endocrinol. 2010;24:1728–36.
Chen C, Zhou Z, Zhong M, Zhang Y, Li M, Zhang L, et al. Thyroid hormone promotes neuronal differentiation of embryonic neural stem cells by inhibiting STAT3 signaling through TRalpha1. Stem Cells Dev. 2012;21:2667–81.
Deng C, Zhang Z, Xu F, Xu J, Ren Z, Godoy-Parejo C, et al. Thyroid hormone enhances stem cell maintenance and promotes lineage-specific differentiation in human embryonic stem cells. Stem Cell Res Ther. 2022;13:120.
Fernandez M, Pirondi S, Manservigi M, Giardino L, Calza L. Thyroid hormone participates in the regulation of neural stem cells and oligodendrocyte precursor cells in the central nervous system of adult rat. Eur J Neurosci. 2004;20:2059–70.
Gothie JD, Sebillot A, Luongo C, Legendre M, Nguyen Van C, Le Blay K, et al. Adult neural stem cell fate is determined by thyroid hormone activation of mitochondrial metabolism. Mol Metab. 2017;6:1551–61.
Liu R, Bishop J, Zhu G, Zhang T, Ladenson PW, Xing M. Mortality Risk Stratification by Combining BRAF V600E and TERT Promoter Mutations in Papillary Thyroid Cancer: Genetic Duet of BRAF and TERT Promoter Mutations in Thyroid Cancer Mortality. JAMA Oncol. 2017;3:202–8.
Pasca di Magliano M, Di Lauro R, Zannini M. Pax8 has a key role in thyroid cell differentiation. Proc Natl Acad Sci USA. 2000;97:13144–9.
Cancer Genome Atlas Research N. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–90.
Tomás G, Tarabichi M, Gacquer D, Hebrant A, Dom G, Dumont JE, et al. A general method to derive robust organ-specific gene expression-based differentiation indices: application to thyroid cancer diagnostic. Oncogene 2012;31:4490–8.
Cheng SY, Leonard JL, Davis PJ. Molecular aspects of thyroid hormone actions. Endocr Rev. 2010;31:139–70.
Brent GA. Mechanisms of thyroid hormone action. J Clin Invest. 2012;122:3035–43.
Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–72.
Mabbott NA, Baillie JK, Brown H, Freeman TC, Hume DA. An expression atlas of human primary cells: inference of gene function from coexpression networks. BMC Genomics. 2013;14:632.
Thompson B, Davidson EA, Liu W, Nebert DW, Bruford EA, Zhao H, et al. Overview of PAX gene family: analysis of human tissue-specific variant expression and involvement in human disease. Hum Genet. 2021;140:381–400.
Dupain C, Ali HM, Mouhoub TA, Urbinati G, Massaad-Massade L. Induction of TTF-1 or PAX-8 expression on proliferation and tumorigenicity in thyroid carcinomas. Int J Oncol. 2016;49:1248–58.
Laury AR, Perets R, Piao H, Krane JF, Barletta JA, French C, et al. A comprehensive analysis of PAX8 expression in human epithelial tumors. Am J Surg Pathol. 2011;35:816–26.
Sangoi AR, Cassarino DS. PAX-8 expression in primary and metastatic Merkel cell carcinoma: an immunohistochemical analysis. Am J Dermatopathol. 2013;35:448–51.
Tacha D, Zhou D, Cheng L. Expression of PAX8 in normal and neoplastic tissues: a comprehensive immunohistochemical study. Appl Immunohistochem Mol Morphol. 2011;19:293–9.
Ma R, Latif R, Davies TF. Human embryonic stem cells form functional thyroid follicles. Thyroid. 2015;25:455–61.
Chu Y, Zhu C, Wang Q, Liu M, Wan W, Zhou J, et al. Adipose-derived mesenchymal stem cells induced PAX8 promotes ovarian cancer cell growth by stabilizing TAZ protein. J Cell Mol Med. 2021;25:4434–43.
Ozcan A, Shen SS, Hamilton C, Anjana K, Coffey D, Krishnan B, et al. PAX 8 expression in non-neoplastic tissues, primary tumors, and metastatic tumors: a comprehensive immunohistochemical study. Mod Pathol. 2011;24:751–64.
Fu DJ, De Micheli AJ, Bidarimath M, Ellenson LH, Cosgrove BD, Flesken-Nikitin A, et al. Cells expressing PAX8 are the main source of homeostatic regeneration of adult mouse endometrial epithelium and give rise to serous endometrial carcinoma. Dis Model Mech. 2020;13.
Kaminski MM, Tosic J, Kresbach C, Engel H, Klockenbusch J, Muller AL, et al. Direct reprogramming of fibroblasts into renal tubular epithelial cells by defined transcription factors. Nat Cell Biol. 2016;18:1269–80.
Futreal PA, Soderkvist P, Marks JR, Iglehart JD, Cochran C, Barrett JC, et al. Detection of frequent allelic loss on proximal chromosome 17q in sporadic breast carcinoma using microsatellite length polymorphisms. Cancer Res. 1992;52:2624–7.
Yokota J, Yamamoto T, Miyajima N, Toyoshima K, Nomura N, Sakamoto H, et al. Genetic alterations of the c-erbB-2 oncogene occur frequently in tubular adenocarcinoma of the stomach and are often accompanied by amplification of the v-erbA homologue. Oncogene. 1988;2:283–7.
Dayton AI, Selden JR, Laws G, Dorney DJ, Finan J, Tripputi P, et al. A human c-erbA oncogene homologue is closely proximal to the chromosome 17 breakpoint in acute promyelocytic leukemia. Proc Natl Acad Sci USA. 1984;81:4495–9.
Huang ME, Ye YC, Chen SR, Chai JR, Lu JX, Zhoa L, et al. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood. 1988;72:567–72.
Degos L, Dombret H, Chomienne C, Daniel MT, Miclea JM, Chastang C, et al. All-trans-retinoic acid as a differentiating agent in the treatment of acute promyelocytic leukemia. Blood. 1995;85:2643–53.
Warrell RP Jr., de The H, Wang ZY, Degos L. Acute promyelocytic leukemia. N Engl J Med. 1993;329:177–89.
de The H. Differentiation therapy revisited. Nat Rev Cancer. 2018;18:117–27.
Hong CM, Ahn BC. Redifferentiation of Radioiodine Refractory Differentiated Thyroid Cancer for Reapplication of I-131 Therapy. Front Endocrinol (Lausanne). 2017;8:260.
Ho AL, Grewal RK, Leboeuf R, Sherman EJ, Pfister DG, Deandreis D, et al. Selumetinib-enhanced radioiodine uptake in advanced thyroid cancer. N Engl J Med. 2013;368:623–32.
Rothenberg SM, McFadden DG, Palmer EL, Daniels GH, Wirth LJ. Redifferentiation of iodine-refractory BRAF V600E-mutant metastatic papillary thyroid cancer with dabrafenib. Clin Cancer Res. 2015;21:1028–35.
Forrest D, Hallbook F, Persson H, Vennstrom B. Distinct functions for thyroid hormone receptors alpha and beta in brain development indicated by differential expression of receptor genes. EMBO J. 1991;10:269–75.
Gothe S, Wang Z, Ng L, Kindblom JM, Barros AC, Ohlsson C, et al. Mice devoid of all known thyroid hormone receptors are viable but exhibit disorders of the pituitary-thyroid axis, growth, and bone maturation. Genes Dev. 1999;13:1329–41.
Dellovade TL, Chan J, Vennstrom B, Forrest D, Pfaff DW. The two thyroid hormone receptor genes have opposite effects on estrogen-stimulated sex behaviors. Nat Neurosci. 2000;3:472–5.
Mendoza A, Hollenberg AN. New insights into thyroid hormone action. Pharmacol Ther. 2017;173:135–45.
Forrest D, Vennstrom B. Functions of thyroid hormone receptors in mice. Thyroid. 2000;10:41–52.
Kim WG, Zhu X, Kim DW, Zhang L, Kebebew E, Cheng SY. Reactivation of the silenced thyroid hormone receptor beta gene expression delays thyroid tumor progression. Endocrinology. 2013;154:25–35.
Martinez-Iglesias O, Garcia-Silva S, Tenbaum SP, Regadera J, Larcher F, Paramio JM, et al. Thyroid hormone receptor beta1 acts as a potent suppressor of tumor invasiveness and metastasis. Cancer Res. 2009;69:501–9.
Davidson CD, Gillis NE, Carr FE.Thyroid Hormone Receptor Beta as Tumor Suppressor: Untapped Potential in Treatment and Diagnostics in Solid Tumors. Cancers (Basel). 2021;13:4254.
Lee WK, Zhu X, Park S, Zhu YJ, Zhao L, Meltzer P, et al. Regulation of cancer stem cell activity by thyroid hormone receptor beta. Oncogene. 2022;41:2315–25.
Park S, Han CR, Park JW, Zhao L, Zhu X, Willingham M, et al. Defective erythropoiesis caused by mutations of the thyroid hormone receptor alpha gene. PLoS Genet. 2017;13:e1006991.
Acknowledgements
This research was supported by the Intramural Research Programs of the Center for Cancer Research of the National Cancer Institute, National Institutes of Health. We thank Dr. Michael Kelly and his team for carrying out single-cell RNA sequencing at the Single Cell Analysis Facility, Cancer Research Technology Program, Frederick National Laboratory (Leidos Biomed), NCI. We also thank Joelle Mornini, NIH Library, for manuscript editing assistance.
Author information
Authors and Affiliations
Contributions
Conception and designs were performed by S-YC, EH, WKLD, YJZ and XZ. Development of methodology and acquisition of data were carried out by EH, WKLD, YJZ, XZ, LZ, and YY. Analysis and interpretation of data were performed by EH, WKLD, YJZ, XZ, YY and S-YC. Administrative, technical, or material support were performed by S-YC and LZ.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Hwang, E., Doolittle, W.K.L., Zhu, Y.J. et al. Thyroid hormone receptor α1: a novel regulator of thyroid cancer cell differentiation. Oncogene 42, 3075–3086 (2023). https://doi.org/10.1038/s41388-023-02815-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-023-02815-2
- Springer Nature Limited
We’re sorry, something doesn't seem to be working properly.
Please try refreshing the page. If that doesn't work, please contact support so we can address the problem.