Abstract
The dynamics of mitochondrial biogenesis regulation is critical in maintaining cellular homeostasis for immune regulation and tumor prevention. Here, we report that mitochondrial biogenesis disruption through TFAM reduction significantly impairs mitochondrial function, induces autophagy, and promotes esophageal squamous cell carcinoma (ESCC) growth. We found that TFAM protein reduction promotes mitochondrial DNA (mtDNA) release into the cytosol, induces cytosolic mtDNA stress, subsequently activates the cGAS-STING signaling pathway, thereby stimulating autophagy and ESCC growth. STING depletion or mtDNA degradation by DNase I abrogates mtDNA stress response, attenuates autophagy, and decreases the growth of TFAM depleted cells. In addition, autophagy inhibitor also ameliorates mitochondrial dysfunction-induced activation of the cGAS-STING signaling pathway and ESCC growth. In conclusion, our results indicate that mtDNA stress induced by mitochondria biogenesis perturbation activates the cGAS-STING pathway and autophagy to promote ESCC growth, revealing an underappreciated therapeutic strategy for ESCC.
Similar content being viewed by others
Introduction
Mitochondria are the major intracellular organelles for energy generation and cellular metabolism. Mitochondria are important for cellular homeostasis through regulating cellular calcium concentrations, controlling ROS production, and regulating apoptosis and immune responses [1]. It has been demonstrated that the dysfunction of mitochondria plays multiple roles in cancer initiation and progression [2, 3]. Mitochondrial transcription factor A (TFAM) is a pivotal factor for regulating mitochondrial DNA (mtDNA) replication, transcription and biogenesis [4, 5]. The aberrant expression of TFAM was reported in many tumor tissues and multiple cell lines [6, 7]. In addition, mtDNA copy number was significantly lower in resected cancerous tissues, and low mtDNA number is correlated with tumor size, invasion, pathological stage and poor prognosis in esophageal squamous cell carcinoma (ESCC) [8, 9]. However, the role of TFAM in ESCC tumorigenesis and the underlying mechanisms remain largely unknown.
Mitochondrial damage-associated molecular patterns (DAMPs), which are released from mitochondria under stress, are involved in cancer initiation and progression [3, 10]. Mitochondrial DAMPs, including mtDNA, extracellular ATP, TFAM, ROS, cardiolipin and N-formyl peptide, are recognized by membrane-bound or cytoplasmic-pattern recognition receptors (PRRs) [10]. Previous studies had shown that TFAM deficiency markedly induces mtDNA instability and cytosolic mtDNA stress. Cytosolic mtDNA is recognized as DAMP, and subsequently triggers cGAS-STING-IRF3-dependent signaling to facilitate the production of type I interferons and inflammatory cytokines [11,12,13]. Hypoxia induces cytosolic mtDNA accumulation, which in turn associates with HMGB1 released from the nucleus. The mtDNA-HMGB1 complex activates TLR9 signaling pathways to promote HCC cells proliferation [14]. Further, our recently study found that increased mitochondrial fission through upregulated Drp1 expression induces cytosolic mtDNA stress in HCC cells and cytokine CCL2 secretion, which in turn promotes tumor associated macrophage infiltration in HCC [15]. Furthermore, STING activation by cytosolic DNA is also involved in innate immune recognition of immunogenic tumors [16]. Recent studies found that the activation of the cGAS-STING pathway is involved in anticancer immune responses as well as cancer immune escape [17,18,19]. However, whether abnormal expression of TFAM induces mitochondrial dysfunction and mtDNA stress in ESCC is not well understood. Furthermore, the role of the cGAS-STING pathway in ESCC has not been investigated.
In this study, we investigated the role of TFAM in ESCC in vitro and in vivo, including the effect of TFAM-mediated mitochondrial dysfunction on the cytosolic mtDNA stress, the cGAS-STING signaling pathway, the autophagy process and their involvement in cancerous growth. Our results unveil a new molecular mechanism underlying mtDNA stress and autophagy induction in promoting ESCC growth.
Results
TFAM downregulation accelerates ESCC cancer growth in cell culture and xenograft tumors
To investigate whether TFAM-mediated mitochondrial biogenesis is involved in ESCC, we analyzed the expression of TFAM in a cohort of 57 ESCC patients. We found that TFAM expression was remarkably downregulated in ESCC tissues and ESCC patients with low TFAM expression had a significantly poorer prognosis than those with high TFAM expression (Fig. 1A–C, Fig. S1A, B). In addition, we found a low TFAM expression in poorly differentiated KYSE-70 and moderately KYSE-140 ESCC cells (Fig. S1C, D). In contrast, TFAM expression is high in well differentiated KYSE-30 ESCC cell line and immortalized normal esophageal NE-3 cells (Fig. S1C, D). These results suggest that TFAM expression likely contributes to malignancy in ESCC. To explore the potential roles of TFAM in ESCC survival, ESCC cells with TFAM overexpression or knockdown were established (Fig. S1E, F, S2A, B, S3A, B). EdU incorporation assay showed that ESCC cells with TFAM deficiency had significantly more EdU incorporation than control cells (Fig. 1D, S2C). In contrast, the EdU incorporation was much reduced in ESCC cells with high TFAM expression (Fig. S3C). Furthermore, cell viability and proliferation were remarkably increased in ESCC cells with TFAM deficiency in MTS and colony formation assays (Fig. 1E, F, S2D, E), while TFAM overexpression had the opposite effects (Fig. S3D, E). The effects of TFAM levels on tumor growth were further tested in vivo in subcutaneous nude mice xenograft model. As shown in Fig. 1G, H, the growth rate of xenograft tumors derived from ESCC cells with TFAM deficiency was significantly increased in comparison with control tumors. Whereas xenograft tumors derived from ESCC cells with TFAM overexpression exhibited an opposite effect (Fig. S3F, G). In accordance with xenograft tumor weights and growth rates, the Ki-67 immunostaining showed that the levels of TFAM inversely correlate with Ki-67 staining signals (Fig. 1I, S3H). TFAM is a pivotal factor for mitochondrial biogenesis and mass by regulating mtDNA replication and transcription. Therefore, we investigated the mitochondrial mass change by translocase of outer mitochondrial membrane 20 (TOMM20) expression. As shown in Fig. S4A–C, the expression of TOMM20 was remarkably downregulated in ESCC tissues than peritumor tissues. The correlations between the expression of TFAM and TOMM20 in ESCC from the Cancer Genome Atlas (TCGA) database was analyzed using Assistant for Clinical Bioinformatics (www.aclbi.com). We found that the expression of TFAM has significantly positive correlation with that of TOMM20 in ESCC (Fig. S4D).
A Representative immunohistochemical (IHC) staining images of TFAM in 57 paired tumor and peritumor tissues of ESCC patients. Scale bar: 100 μm. B TFAM expression in 57 paired tumor and peritumor tissues of ESCC patients was determined by western blot analyses. T, tumor; P, peritumor. C Kaplan–Meier curve analysis of overall survival in ESCC patients by the expression of TFAM in tumor tissues. Patients were divided into two groups by the IHC score median value of TFAM for further analysis. D–F Cell proliferation potential was evaluated by EdU incorporation, colony formation and MTS cell viability assay in ESCC cells with TFAM deficiency. G, H Tumor growth curves and weight of subcutaneous xenograft tumor model established from ESCC cells with TFAM deficiency (n = 6). I The expression of Ki67 in xenograft tumors established from ESCC cells with TFAM deficiency was detected by IHC staining. Scale bar: 100 μm. shTFAM, shRNA expression vector against TFAM; shCtrl, control shRNA. *P < 0.05; **P < 0.01; ***P < 0.001.
TFAM deficiency induces mitochondrial dysfunction of ESCC cells
The involvement of TFAM in the regulation of ESCC growth prompted us to analyze the effects of TFAM level changes on mitochondrial function. First, using JC-1 fluorescent molecule staining, we found that shRNA depletion of TFAM decreased mitochondrial membrane potential in KYSE-30 cells (Fig. 2A), while forced TFAM expression increased mitochondrial membrane potential in KYSE-70 cells (Fig. 2B). Second, shRNA depletion of TFAM decreased ATP levels in KYSE-30 cells (Fig. 2C, left panel), while forced TFAM expression increased ATP levels in KYSE-70 cells (Fig. 2C, right panel). Third, shRNA depletion of TFAM increased cellular ROS levels in KYSE-30 cells (Fig. 2D, left panel), while forced TFAM expression decreased ROS levels in KYSE-70 cells (Fig. 2D, right panel). Above results suggest that TFAM levels are critical of the cellular homeostasis of mitochondrial functions including membrane potential, ATP production, and ROS generation.
A, B Mitochondrial membrane potential in ESCC cells with TFAM deficiency or overexpression was analyzed by JC-1 staining and observed by fluorescence microscope. Scale bars: 100 μm. C ROS levels were investigated by DCFH-DA fluorescence with flow cytometry in ESCC cells with TFAM deficiency or overexpression. D ATP production of ESCC cells with TFAM deficiency or overexpression were detected. shTFAM, shRNA expression vector against TFAM; and shCtrl, control shRNA; TFAM, expression vector encoding TFAM; EV, empty vector. *P < 0.05; **P < 0.01; ***P < 0.001.
TFAM depletion promotes autophagy in ESCC cells
To analyze the cell biology functions of TFAM, we performed TEM analyses of KYSE-30 cells without or with TFAM depletion. We found that TFAM depletion induced the formation of double-membrane structures with features of autophagic vesicles in KYSE-30 cells (Fig. 3A). This observation prompted us to analyze several key factors involved in autophagy by western blot. We found that shTFAM depletion increased the level of LC3II with a concomitant decrease in p62/SQSTM (Fig. 3B), indicating that TFAM depletion stimulated the autophagy mediated protein degradation. Using a GFP-LC3B autophagy reporter gene, we found that the number of autophagic vesicle puncta significantly increased after TFAM depletion (Fig. 3C). The autophagy inhibitor BafA significantly blocked degradation of p62 and increased the number of autophagic vesicle puncta (Fig. S5A, B). Moreover, we found a significant increase in LC3B immunostaining in xenograft tumors derived from TFAM depletion cells (Fig. 3D). Further, we found a significant increase in LC3B staining in ESCC tumor tissues than peritumor tissues (Fig. 3E). When chloroquine (CQ) was used to block autophagy, we found that CQ treatment decreased the colony formation ability as well as EdU incorporation in KYSE30 cells after TFAM depletion (Fig. 3F, G). Above results support a notion that autophagy plays a stimulatory role in the growth of ESCC tumors.
A Double-membrane autophagosomes in ESCC cells with TFAM deficiency was detected by transmission electron microscopy. Scale bar: 0.5 μm. B Western blot analyses were performed to detect the expression of autophagy-related proteins in ESCC cells with TFAM deficiency. C Representative images of fluorescent LC3B puncta (green) in ESCC cells with TFAM deficiency. Scale bar: 10 μm. D Representative immunohistochemical (IHC) staining images of LC3B in xenograft tumors developed from ESCC cells with TFAM deficiency. Scale bar: 100 μm. E Representative immunohistochemical (IHC) staining images of LC3B in 57 paired tumor and peritumor tissues of ESCC patients. Scale bar: 100 μm. F, G Cell proliferation potential was evaluated by colony formation and EdU incorporation assay in ESCC cells treatment as indicated. shTFAM, shRNA expression vector against TFAM; and shCtrl, control shRNA; CQ, chloroquine. *P < 0.05; **P < 0.01; ***P < 0.001.
TFAM depletion induced cytosolic release of mtDNA through the mitochondrial permeability transition pore (mPTP)
It had been demonstrated that TFAM deficiency induces mtDNA instability and cytosolic release [12, 15]. However, the role of TFAM in mediating mtDNA dynamics remained unexplored in ESCC cells. As shown in Fig. 4A and S6A, B, mitochondrial nucleoids in TFAM depletion ESCC cells exhibited an abnormal distribution, indicating that the packaging and organization of mtDNA were altered. To test the cytosolic mtDNA release, the cytosolic fraction lacking mitochondria was isolated and quantified for the cytosolic mtDNA using droplet digital PCR. We found that TFAM depletion markedly increased mtDNA content in the cytosolic compartment than control cells (Fig. 4B). DNase I but not heat-inactivated DNase I treatment dramatically reduced cytosolic mtDNA content in ESCC cells after TFAM depletion (Fig. 4A, B). The molecular mechanisms underlying the escape of mtDNA into cytosol under stress have been reported, including BAX/BAK macropores, mitochondrial permeability transition pores (mPTPs) and voltage-dependent anion channel 1 (VDAC1) oligomerization [11, 13, 20, 21]. The formation of BAX/BAK macropores frequently happens during apoptosis to facilitate mitochondrial contents including mtDNA and cytochrome C to release into the cytosol [21]. Our results indicated that the levels of apoptosis were reduced in ESCC cells with TFAM depletion by western blot and flow cytometry analyses (Fig. 4C, D), suggesting that apoptosis is not the major cause of the cytosolic release of mtDNA. In contrast, to prevent mPTP formation by pharmacological (cyclosporin A, CsA) or siRNA targeting the mPTP component PPID significantly decreased cytosolic mtDNA concentrations in KYSE30 cells after TFAM depletion (Fig. 4E, F, S6C). Moreover, to inhibit VDAC1 oligomerization by the VBIT-4 inhibitor or siRNA targeting VDAC1 decreased TFAM deficiency-mediated cytosolic mtDNA release (Fig. 4E, F, S6C). Our results indicate that mPTP pathways are involved in cytosolic release of mtDNA after TFAM depletion in ESCC cells.
A Representative confocal microscopy images of ESCC cells stained with Picogreen (green, DNA) and MitoTracker (red, mitochondria) treatment as indicated. Scale bar: 20 μm. B Droplet digital PCR was performed to measure the cytosolic mtDNA copy number in ESCC cells treatment as indicated. C Western blot analyses were performed to detect the expression of apoptosis-related proteins in ESCC cells with TFAM deficiency. D The apoptosis of ESCC cells treatment as indicated was detected by flow cytometry analysis using Annexin V and PI staining. The cells were treated with CCCP (150 mM) for 4 h before apoptosis analysis. E, F Droplet digital PCR and confocal microscopy was performed to measure the cytosolic mtDNA copy number in ESCC cells treatment as indicated. shTFAM, shRNA expression vector against TFAM; shCtrl, control shRNA; NC, negative control; siPpid, siRNAs against Ppid; siVDAC1, siRNAs against VDAC1; CsA, cyclosporin A; VBIT, VDAC1 oligomerization inhibitor VBIT-4; HI DNase I, heat inactivated DNase I. *P < 0.05; **P < 0.01; ***P < 0.001.
Cytosolic mtDNA is involved in autophagy induction and cell growth
Next, we investigated the role of cytosolic mtDNA in autophagy and cell growth. Firstly, we found that cytosolic mtDNA degradation by DNase I but not heat-inactivated DNase I treatment decreased LC3B-II and increased p62/SQSTM1 accumulation in TFAM shRNA treated cells in western blot assays (Fig. 5A). Secondly, DNase I but not heat-inactivated DNase I treatment dramatically decreased autophagic vesicles in TFAM depleted cells detected by TEM (Fig. 5B) or the GFP-LC3 reporter gene (Fig. 5C). In addition, DNase I but not heat-inactivated DNase I treatment dramatically decreased the colony formation ability (Fig. 5D) and EdU incorporation (Fig. 5E) in TFAM depleted cells. Taken together, above data strongly support that cytosolic mtDNA at least partially contributed to autophagy induction and EdU incorporation after TFAM depletion.
A Western blot analyses were performed to detect the expression of autophagy-related proteins in ESCC cells treatment as indicated. B Double-membrane autophagosomes in ESCC cells treatment as indicated was detected by transmission electron microscopy. Scale bar: 0.5 μm. C Representative images of fluorescent LC3B puncta (green) in ESCC cells treatment as indicated. Scale bar: 10 μm. D, E Cell proliferation potential was evaluated by colony formation and EdU incorporation assay in ESCC cells treatment as indicated. shTFAM, shRNA expression vector against TFAM; shCtrl, control shRNA; NC, negative control; HI DNase I, heat inactivated DNase I. *P < 0.05; **P < 0.01; ***P < 0.001.
Cytosolic mtDNA stress promotes autophagy by cGAS-STING signaling pathway
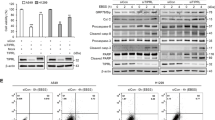
To explore the molecular mechanisms underlying TFAM depletion-mediated autophagy induction, RNA sequencing analyses were performed. Transcriptome profiling identified 909 upregulated and 1591 downregulated genes after TFAM depletion (Table S5). PI3K-Akt, MAPK, autophagy and mTOR signaling pathway, cytosolic DNA-sensing pathway were enriched in the KEGG pathway analysis of differentially expressed genes (Fig. 6A). It had been demonstrated that cGAS-STING can be activated by cytosolic mtDNA [12, 15], while STING activation is implicated in cancer progression and antitumor immune response [17]. Interestingly, RNA sequencing analysis found the expression of TMEM173, which encodes STING, is significantly downregulated in ESCC cells after TFAM depletion (Fig. 6B). Although the STING protein was decreased, TFAM depletion markedly increased the phosphorylation of STING and TBK1, suggesting a kinase cascade is involved to activate STING and autophagy (Fig. 6C). We also found that inhibition of autophagy by chloroquine increased the levels of STING, p62/SQSTM1 and LC3B-II, suggesting these proteins are degraded by autophagy (Fig. 6D). Moreover, in ESCC cells after TFAM depletion, siRNA knockdown of cGAS, the upstream cytosolic DNA sensor in the STING pathway, abolished LC3B-II accumulation and blocked STING and p62/SQSTM1 degradation, while siRNA knockdown of STING blocked p62/SQSTM1 degradation and LC3B-II increase (Fig. 6E). In addition, STING knockdown significantly decreased autophagic vesicle formation in the TEM and GFP-LC3B reporter assays (Fig. 6F, G). Consistent with a role of cytosolic mtDNA in the activation of the cGAS-STING pathway, we found DNase I but not the heat inactivated DNase I treatment attenuated STING and TBK1 phosphorylation and blocked STING degradation after TFAM depletion (Fig. 6H). Together, these results indicate that cytosolic mtDNA promotes autophagy in ESCC cells through the cGAS-STING signaling pathway.
A KEGG pathway enrichment analyses of differentially expressed genes in RNA sequencing data. B Heatmap analysis of gene expression by RNA sequencing assay in ESCC cells with TFAM deficiency. C–E Western blot analyses were performed to detect the proteins expression of cGAS-STING signaling pathway in ESCC cells treatment as indicated. F Double-membrane autophagosomes in ESCC cells treatment as indicated was detected by transmission electron microscopy. Scale bar: 0.5 μm. G Representative images of fluorescent LC3B puncta (green) in ESCC cells treatment as indicated. Scale bar: 10 μm. H Western blot analyses were performed to detect the proteins expression of cGAS-STING signaling pathway in ESCC cells treatment as indicated. shTFAM, shRNA expression vector against TFAM; shCtrl, control shRNA; NC, negative control; CQ, chloroquine; siSTING, siRNAs against STING; sicGAS, siRNAs against cGAS; HI DNase I, heat inactivated DNase I. *P < 0.05; **P < 0.01.
The mtDNA-cGAS-STING pathway and autophagy accelerate cancer growth
After siRNA depletion of cGAS and STING in TFAM depleted cells, we found that EdU incorporation and colony formation were decreased (Fig. 7A, B), suggesting that the mtDNA-cGAS-STING pathway is important for cancerous growth. Upon activation, STING elicits two downstream functions including the activation of type I interferon expression and autophagy induction. RNA sequencing analyses indicated that the classical interferon stimulated genes were significantly downregulated in the TFAM depleted cells (Fig. S7A), supporting that STING degradation attenuated the expression of type I IFN genes. After TFAM depleted cells were stimulated with cGAMP to trigger STING activation, we found that the classical interferon stimulated genes were remarkable increased after cGAMP treatment but decreased upon TFAM depletion (Fig. S7B). Next, we applied STING covalent inhibitor H-151 to treat KYSE30 cells after TFAM depletion. H-151 is an inhibitor for STING by suppressing STING palmitoylation and TBK1 phosphorylation, which in turn reduces IFN-β expression [22]. Consistent with this previous report, we also found that H-151 suppressed the phosphorylation of TBK-1 (Fig. S8A) and IFN-β expression (Fig. S8B). However, STING and SQSTM1/p62 degradation, and LC3B-II increase were not inhibited by H-151 treatment (Fig. S8A). Further, after H-151 treatment of TFAM depleted cells, more autophagic vesicles were detected in TEM assays and the GFP-LC3B reporter assays (Fig. S8C, D), suggesting that H-151 inhibited STING-mediated interferon expression but elevated autophagy. After H-151 treatment of TFAM depleted cells, EdU incorporation and colony formation were significantly increased (Fig. 7A, B). Consistent with the in vitro results, H-151 treatment of TFAM depleted cells further elevated the growth rate of xenograft tumors and the weight of dissected tumors (Fig. 7C, D), suggesting that autophagy increase after H-151 treatment favors cancerous growth. To further test the role of autophagy in tumor growth, we treated TFAM depleted tumors with both H-151 and chloroquine, and found that the tumor promotion effect of H-151 was significantly nullified by chloroquine treatment (Fig. 7C, D). We also found a significant increase in LC3B immunostaining in xenograft tumors derived from TFAM depletion cells treated with H-151 and chloroquine (Fig. S8E). Moreover, Ki67 staining of the dissected tumor sections found that Ki67+ cells were slightly increased after H-151 treatment in TFAM depleted cells, while were markedly reduced in tumors after a combined treatment with H-151 and chloroquine (Fig. 7E). We also investigated the effect of cGAS selective inhibitor G140 on TFAM depletion-mediated ESCC cells proliferation. Our results indicated that STING degradation and autophagy were significantly blocked by G140 (Fig. S9A). The EdU incorporation and colony formation were remarkable decreased in TFAM depleted ESCC cells after G140 treatment (Fig. S9B, C). Above results indicated that autophagy plays an important role in cancerous growth, especially in the cells under mtDNA stress in the context of the cGAS-STING pathway activation.
A, B Cell proliferation potential was evaluated by colony formation and EdU incorporation assay in ESCC cells treatment as indicated. C, D Tumor growth curves and weight of subcutaneous xenograft tumor model developed from ESCC cells treatment as indication (n = 5). E The expression of Ki67 in xenograft tumors established from ESCC cells treatment as indication was detected by IHC staining. Scale bar: 100 μm. shTFAM, shRNA expression vector against TFAM; and shCtrl, control shRNA; siSTING, siRNAs against STING; sicGAS, siRNAs against cGAS; siCtrl, control siRNA; NC, negative control; CQ, chloroquine. *P < 0.05; **P < 0.01; ***P < 0.001.
Discussion
Accumulating evidence indicates that mitochondrial biogenesis is involved in tumorigenesis and cancer progression [2, 3]. In this study, we demonstrated that TFAM expression is significantly downregulated in ESCC tissue and low expression of TFAM is correlated with poor overall survival of ESCC patients. Furthermore, TFAM depletion significantly induces mitochondrial dysfunction and cytosolic mtDNA stress in ESCC cells. Mechanistically, we found that cytosolic mtDNA stress promoted autophagy of ESCC cells by triggering cGAS-STING signaling pathway and consequently promoted ESCC progression (see also the Graphical Abstract).
Mitochondrial biogenesis homeostasis is under coordinated regulation of nuclear and mitochondrial genes to adapt to different physiological and pathological conditions. Accumulating evidence suggests that mitochondrial biogenesis is associated with tumor initiation, progression and metastasis [23, 24]. TFAM is a vital mtDNA binding protein and is essential for mitochondrial biogenesis by regulating packaging, stability, and replication of mitochondrial genome [4]. Some research reported that TFAM is highly expressed in tumor tissue and associated with poor progression of patients in microsatellite stable colorectal cancer, hepatocellular carcinoma and non-small cell lung cancer [25, 26]. However, other study found that elevation TFAM expression inhibited tumor growth in microsatellite-unstable colorectal cancer and breast cancer [27, 28]. In the present study, our data found that TFAM expression was remarkably downregulated in ESCC tumors and patients with lower TFAM expression had significantly shorter overall survival. Furthermore, TFAM deficiency significantly increased ESCC cell growth rate, colony formation, and EdU incorporation in vitro and increased tumor growth rate, tumor weight and percentage of Ki67+ cells in subcutaneous xenograft tumor. These results suggested that TFAM plays pivotal roles in promoting ESCC progression.
Mitochondrial dysfunction contributes to tumorigenesis and tumor cell growth by modulating cell cycle, gene expression and coordinately regulating apoptosis and autophagy [2, 3, 29]. Previous studies found that autophagy inhibitors are potential experimental therapeutics to overcome treatment resistance in ESCC [30]. Here, we observed that TFAM deficiency significantly reduced ATP production, decreased mitochondrial membrane potential and elevated ROS production. Consistent with previous findings, we found that mitochondrial dysfunction induced by TFAM deficiency significantly promoted autophagy in ESCC cells, and the autophagy inhibitors markedly alleviated TFAM deficiency-induced survival of ESCC cells.
TFAM is essential for mtDNA stability and knockdown expression of TFAM markedly elicits mtDNA instability and promotes escape of mtDNA into cytoplasm, which is recognized as cytosolic mtDNA stress [12, 31]. However, whether TFAM deficiency can induce mtDNA stress in cancer cells is largely unknown. In this study, we found that TFAM deficiency may influence architecture of mitochondrial nucleoids and increase the mtDNA content in cytosol of ESCC cells, which was commonly recognized as cytosolic mtDNA stress. In addition, we found that cytosolic mtDNA stress played a vital role for TFAM deficiency-induced ESCC survival, while depleting cytosolic mtDNA after DNase I treatment significantly reversed TFAM deficiency-induced EdU incorporation and proliferation. Although our current and previous studies demonstrate that mitochondrial dysfunction is implicated in cytosolic mtDNA stress, we note that the underling mechanism for mtDNA release in cancer remains unknown. Under different cell stress, mtDNA can escape to cytosol through BAX/BAK macropores, mitochondrial outer membrane permeabilization (MOMP), or VDAC1 oligomerization [11, 13, 20, 21]. Our results found that TFAM may regulate mtDNA release through decreasing mitochondrial membrane potential and promoting VDAC1 oligomerization in ESCC cells. Pharmacological inactivation of the mPTP using CsA or VDAC1 oligomerization inhibitor VBIT-4 significantly inhibited TFAM deficiency-mediated cytosolic mtDNA accumulation. In addition, TFAM is a mitochondrial DNA binding protein therefore can directly contribute to mitochondrial nucleoid structure maintenance and mtDNA distribution [32, 33]. Therefore, further studies are need to reveal other molecular mechanisms underlying this process.
Our recently study found that increased mitochondrial fission significantly induced cytosolic mtDNA stress in HCC cells and promoted the tumor associated macrophages infiltration and hepatocellular carcinoma progression by activating TLR9-mediated NF-κB signaling pathway [15]. However, the role of mtDNA stress in tumor progression is largely unknown. In present study, our data indicated that cytosolic mtDNA stress-induced by TFAM deficiency is involved in ESCC progression through triggering autophagy. DNase I treatment significantly reversed TFAM deficiency-induced EdU incorporation, proliferative potential and autophagy. Cytosolic mtDNA mediates the activation of TLR9, AIM2, NLRP3 and STING signaling pathway [34]. Using RNA sequencing analysis, we found that STING is downregulated, autophagy and cytosolic DNA-sensing pathway is activated in ESCC cells with TFAM deficiency. Recently, some research has demonstrated that activation of STING signaling was also promotes autophagy [35, 36]. Meanwhile, our results demonstrated that TFAM deficiency in ESCC cells markedly increased phosphorylation of STING and TBK1 and induced STING degradation, which indicated STING mediated autophagy were activated. These finding promotes us to further explore whether STING mediated autophagy is involved in TFAM deficiency induce ESCC survival. We found that knockdown of cGAS or STING significantly alleviated p62 degradation and LC3B conversion, autophagosomes formation and GFP-LC3B fluorescence puncta accumulation, which indicated autophagy abrogation. The decrease in autophagy is accompanied by attenuating EdU incorporation and proliferative potential in cGAS or STING knockdown ESCC cells. Furthermore, we investigated the effect of H-151 on TFAM deficiency-induced ESCC survival. Consistent with previous research, our results discovered that H-151 reduced IFN-β expression by inhibiting TBK1 phosphorylation, which indicated the suppression of STING-mediated immune responses. However, the activation of autophagy by TFAM deficiency was not suppressed by H-151. Collectively, our results demonstrated that autophagy facilitated by STING pathway under mtDNA stress conditions promotes ESCC cell and cancer growth, suggesting that blocking STING-mediated autophagy and simultaneous awakening STING-mediated immune response is a better therapeutic strategy for cancer.
Materials and methods
Animals
Xenograft model was established using 4-week-old female BALB/c nude mice by subcutaneous injection ESCC cells (5 × 106 cells per mice) into the back right flanks of nude mice, all mice were randomly divided and at least 5 each indicated group. Tumor volume was measured weekly and calculated according to (Width2 × Length) / 2. The mice were sacrificed 6 weeks after injection, and the tumors nodules were harvested, photographed and weighted. No animals were not excluded in this study and any abnormality in weight or apparent disease symptoms were not observe before performing experiments in animals. Animal study was approved by the Institutional Animal Care and Use Committee of Henan University.
Human tissue samples
Total 57 paired ESCC tissue specimens were obtained from Nanyang central Hospital of Henan University. The demographic and clinical information was collected and distributed in Supplementary Table S1. All patients were clinically and pathologically diagnosed to have ESCC. The latest follow-up date was August 2017. This study was approved by Ethics Committee of Henan University, and the informed consents were signed by all participants.
Cell culture and reagents
Reagents and antibodies used in this study were provided in Supplementary Table S3 and S4. Human immortalized esophageal epithelial cells NE-3 and ESCC cell lines with different degrees of malignancy KYSE-30, KYSE-70, KYSE-140 were routinely cultured in RPMI-1640 (Corning, 10040-CVR) supplemented with 10% Fetal Bovine Serum (BI, 04-001-1Acs). The cells were verified based on cell morphology and authenticated using short tandem repeat (STR) DNA testing. KYSE-30 with TFAM knockdown and KYSE-70 with TFAM overexpression were established by stable transfection using the Lipofectamine 2000 reagent (Invitrogen, 11668019) and selection with G418 (Invitrogen, 10131027). Two clones for KYSE-30 with different shTFAM were used in this study. The siRNA sequences used in this study were synthesized by GenePharma (Shanghai, China) and provided in Supplementary Table S2.
Cytosolic mtDNA content detection
Mitochondria isolation kit (Beyotime, C3601) was used to isolate the cytosolic fraction of ESCC cells following the manufacturer’s instructions as previously described [15]. Cytosolic mtDNA content was measured using ND1 gene by droplet digital PCR (ddPCR) and normalized with same cell number. Briefly, the 20 μl ddPCR reaction mixture consisted of 2 × ddPCR Super Mix for probe, 900 nM primers and 500 nM probe for ND1. The 70 μl of droplet generation oil and ddPCR reaction mixture were subjected into a DG8 cartridge (Bio-Rad Laboratories, QX200™) to generated water-in-oil droplet by droplet generator (Bio-Rad Laboratories, QX200™). The water-in-oil droplet was transferred to 96-well PCR plate (Bio-Rad Laboratories, QX200™). After heat-sealed using PX1TM PCR plate sealer (Bio-Rad Laboratories, QX200™), the PCR reaction was performed in C1000 Touch Thermal Cycler (Bio-Rad Laboratories, QX200™) with a condition as follows: 95 °C for 10 min; 40 cycles of 95 °C for 30 s, 55 °C for 1 min; and a final step at 72 °C for 10 min. The mtDNA content was read on the QX200 droplet reader (Bio-Rad Laboratories, QX200™) and analyzed by QX200 droplet reader software, version 1.6.6.0320 (Bio-Rad Laboratories, QX200™).
RNA extraction and qPCR analysis
Total RNA of ESCC cells or human tissue samples were extracted using TRIzol Reagent and complementary DNA was synthesized using PrimeScript RT Reagent kit. qPCR analysis was performed as previously described [15]. Primer sequences of qPCR were provided in Supplementary Table S2.
RNA sequencing analysis
Total RNA of ESCC cells were extracted as previously described. The RNA integrity was firstly examined using Bioanalyzer 2100 (Agilent, Santa Clara, CA) before sequence analysis, and the samples with RNA integrity number more than 7 were subjected to construction of sequencing sample. Finally, the library was sequenced on Illumina novaseq by Genewiz Suzhou, In. We estimated the differentially expressed of mRNAs, GO analysis and KEGG pathway annotation as previous reported [37].
Western blot analysis
Proteins from ESCC tissues and cell lines were separated by electrophoresis and transferred to nitrocellulose membranes by electroblotting. The nitrocellulose membranes were blocked with 5% skim milk for 1 h at room temperature (RT). Subsequently, membranes incubation with primary antibody diluted in 5% skim milk overnight at 4 °C. After washing with PBS 3 times, membranes were incubated with HRP-conjugated secondary antibody for 1 h at RT. Membranes were visualized using ECL (Fisher Scientific) and viewed using a FluorChem Multifluor system (ProteinSimple).
Immunohistochemistry and H&E staining
Immunohistochemistry (IHC) and H&E staining were processed on tumor tissue from patients and mice model [15]. IHC and H&E staining images were reviewed and photographed under Olympus IX53 microscope (Olympus Corporation). The expression of indicated protein was evaluated independently by two pathologists who were blinded of to the specimens and clinicopathological features of patients.
Cell viability and proliferation assays
ESCC cells were seeded into 96-well culture plate at a density of 2 × 103 cells/well to determine the cell viability by MTS assay. (Promega Corporation, G3580). After 1, 2, 3, 4, 5 days of inoculated, 20 μl MTS solution was added according to the manufacturer’s instructions and incubation for another 3 h. The absorbance of optical density (OD) at wavelength of 490 nm was determined using PerkinElmer EnSpire microplate reader.
Colony formation and ethynyl deoxyuridine (EdU) incorporation assay were performed to evaluate proliferation potential of ESCC cells. For colony formation assay, ESCC cells were seeded into 6-well plate at a density of 1 × 103 cells/well and inculcated for 15 days. The colonies were fixed with 4% paraformaldehyde for 15 min and stained with 0.5% crystal violet solution for another 5 min. The number of colonies were calculated and statistics assays from three independent experiments. After cultured with 24 h, the cells inculcated with EdU according to the manufacturer’s instructions (Ribbio, C10310-1) and fixed with 4% paraformaldehyde. Subsequently, the cells were inculcated with glycine and permeabilized with 0.5% Triton X-100. Finally, the cells were incubated with Apollo® 567 staining solution and the DNA of cells was stained with Hoechst 33342. The stained cells were visualized with a fluorescence microscope (Olympus Corporation, U-LH100HG).
Mitochondrial DNA staining
The distribution of mitochondrial DNA was investigated to evaluate mitochondrial DNA stress. After washing with HBSS for 3 times, the mitochondria of ESCC cells were labeled with MitoTracker Red FM (Invivogen, M7512) for 0.5 h at 37 °C, followed by washing 3 times with HBSS and the mitochondrial DNA were stained with Picogreen (UE, P2023) for another 1.5 h at 37 °C. For TOMM20 staining, ESCC cells first fixed with 4% paraformaldehyde in PBS at room temperature for 15 min and permeabilized with 0.2% Triton X-100 for 15 min, and then incubated with Picogreen (UE, P2023) and TOMM20 antibody and Alexa Fluor 647-labeled secondary antibody. Then, the distribution of mitochondrial DNA and mitochondrial morphology was immediately monitored using Nikon N-SIM Structured Illumination microscope (Nikon, Nikon/A1 + N-SIM).
Autophagy formation analysis
The autophagic vesicles formation was analyzed by transmission electron microscopy. Briefly, ESCC cells were first treated with 4% glutaraldehyde and then fixed with 1% OsO4 in 0.1 M cacodylate buffer containing 0.1% CaCl2. The samples were then stained with 1% uranyl acetate, dehydrated in increasing concentrations of ethanol and embedded in araldite. Thin sections were stained with uranyl acetate and lead citrate and analyzed with an electron microscope (FEI, Tecnai G2), at 11,500 magnifications.
To evaluate LC3B fluorescent puncta accumulation, ESCC cells were first transiently transfected with pcDNA3.1-GFP-LC3B (GenePharma, B5500) for 24 h. After staining with Hoechst 33,342 (UE, H4047), ESCC cells were viewed by Nikon N-SIM Structured Illumination microscope (Nikon, Nikon/A1 + N-SIM) and the number of LC3B fluorescent puncta was calculated.
Mitochondrial function measurement
We analyzed the change of mitochondrial function after TFAM knockdown through measuring ROS levels, ATP production and mitochondrial membrane potential. ROS levels of ESCC cells were measured after incubation with 10 μM DCFH-DA (Beyotime Biotechnology, S0033S) by flow cytometry according to the manufacturer’s protocols. Cell Titer-Glo Luminescent assay (Promega, G7570) was used to determine the total ATP levels following the manufacturer’s instructions, and the results were normalized to cell number. To determine effects of TFAM knockdown on mitochondrial membrane potential, we utilized JC-1 dye (Beyotime Biotechnology, C2006). Briefly, ESCC cells were first stained with 5 mg/L JC-1 dye for 20 min and were viewed under fluorescence microscope (Olympus Corporation, U-LH100HG) and the ratio of JC-1 monomers to JC-1 aggregates (ratio of green to orange fluorescence emission).
DNase I treatment
To digest the cytosolic mtDNA, PULSin™ protein delivery reagent (Polyplus Transfection, 501-01) was used to deliver DNase I according the manufacturer’s instructions. ESCC cells were firstly washed with serum-free RPMI-1640 and incubated with DNase I/PULSin mixture containing 3 μg DNase I or heat inactive DNase I for 4 h at 37 °C. Then, removing the mixture and the cells were cultured with complete medium for another 24 h for further studies.
Statistical analysis
Unpaired two-sided Student’s t tests were used for comparisons two groups where appropriate. All data were expressed as the standard error of mean. The IHC score of TFAM were dichotomized to perform prognosis analysis. The overall survival of subgroup patients was distinguished by Kaplan–Meier survival curve and log-rank test. All experiments were independently performed and repeated at least three times for statistical analyses. The investigator was blinded to patients’ clinical data and no blinding was done in animal studies. SPSS 20.0 software (IBM, Chicago, IL) was used to perform statistical analyses and p < 0.05 was considered to indicate a statistically significant.
Data availability
All data are available in the main text or the Supplementary Materials. Sequencing data have been deposited in GEO under accession No. GSE182710.
References
West AP, Shadel GS, Ghosh S. Mitochondria in innate immune responses. Nat Rev Immunol. 2011;11:389–402.
Vyas S, Zaganjor E, Haigis MC. Mitochondria and Cancer. Cell. 2016;166:555–66.
Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–98.
Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, et al. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–44.
Hillen HS, Morozov YI, Sarfallah A, Temiakov D, Cramer P. Structural basis of mitochondrial transcription initiation. Cell. 2017;171:1072–81.
Huang Q, Li J, Xing J, Li W, Li H, Ke X, et al. CD147 promotes reprogramming of glucose metabolism and cell proliferation in HCC cells by inhibiting the p53-dependent signaling pathway. J Hepatol. 2014;61:859–66.
Wen YA, Xiong X, Scott T, Li AT, Wang C, Weiss HL, et al. The mitochondrial retrograde signaling regulates Wnt signaling to promote tumorigenesis in colon cancer. Cell Death Differ. 2019;26:1955–69.
Masuike Y, Tanaka K, Makino T, Yamasaki M, Miyazaki Y, Takahashi T, et al. Esophageal squamous cell carcinoma with low mitochondrial copy number has mesenchymal and stem-like characteristics, and contributes to poor prognosis. PLoS ONE. 2018;13:e0193159.
Tan DJ, Chang J, Liu LL, Bai RK, Wang YF, Yeh KT, et al. Significance of somatic mutations and content alteration of mitochondrial DNA in esophageal cancer. BMC Cancer. 2006;6:93.
Krysko DV, Agostinis P, Krysko O, Garg AD, Bachert C, Lambrecht BN, et al. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends Immunol. 2011;32:157–64.
Rongvaux A, Jackson R, Harman CC, Li T, West AP, de Zoete MR, et al. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 2014;159:1563–77.
West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520:553–7.
White MJ, McArthur K, Metcalf D, Lane RM, Cambier JC, Herold MJ, et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 2014;159:1549–62.
Liu Y, Yan W, Tohme S, Chen M, Fu Y, Tian D, et al. Hypoxia induced HMGB1 and mitochondrial DNA interactions mediate tumor growth in hepatocellular carcinoma through Toll-like receptor 9. J Hepatol. 2015;63:114–21.
Bao D, Zhao J, Zhou X, Yang Q, Chen Y, Zhu J, et al. Mitochondrial fission-induced mtDNA stress promotes tumor-associated macrophage infiltration and HCC progression. Oncogene. 2019;38:5007–20.
Woo SR, Fuertes MB, Corrales L, Spranger S, Furdyna MJ, Leung MY, et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–42.
Barber GN. STING: infection, inflammation and cancer. Nat Rev Immunol. 2015;15:760–70.
Khoo LT, Chen LY. Role of the cGAS-STING pathway in cancer development and oncotherapeutic approaches. EMBO Rep. 2018;19:e46935.
Su T, Zhang Y, Valerie K, Wang XY, Lin S, Zhu G. STING activation in cancer immunotherapy. Theranostics. 2019;9:7759–71.
Kim J, Gupta R, Blanco LP, Yang S, Shteinfer-Kuzmine A, Wang K, et al. VDAC oligomers form mitochondrial pores to release mtDNA fragments and promote lupus-like disease. Science. 2019;366:1531–6.
McArthur K, Whitehead LW, Heddleston JM, Li L, Padman BS, Oorschot V, et al. BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflflux during apoptosis. Science. 2018;359:eaao6047.
Haag SM, Gulen MF, Reymond L, Gibelin A, Abrami L, Decout A, et al. Targeting STING with covalent small-molecule inhibitors. Nature. 2018;559:269–73.
LeBleu VS, O’Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, et al. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16:992–1003.
Luo C, Widlund HR, Puigserver P. PGC-1 coactivators: shepherding the mitochondrial biogenesis of tumors. Trends Cancer. 2016;2:619–31.
Sun X, Zhan L, Chen Y, Wang G, He L, Wang Q, et al. Increased mtDNA copy number promotes cancer progression by enhancing mitochondrial oxidative phosphorylation in microsatellite-stable colorectal cancer. Signal Transduct Target Ther. 2018;3:8.
Xie D, Wu X, Lan L, Shangguan F, Lin X, Chen F, et al. Downregulation of TFAM inhibits the tumorigenesis of non-small cell lung cancer by activating ROS-mediated JNK/p38MAPK signaling and reducing cellular bioenergetics. Oncotarget. 2016;7:11609–24.
Guo J, Zheng L, Liu W, Wang X, Wang Z, Wang Z, et al. Frequent truncating mutation of TFAM induces mitochondrial DNA depletion and apoptotic resistance in microsatellite-unstable colorectal cancer. Cancer Res. 2011;71:2978–87.
Ni H, Guo M, Zhang X, Jiang L, Tan S, Yuan J, et al. VEGFR2 inhibition hampers breast cancer cell proliferation via enhanced mitochondrial biogenesis. Cancer Biol Med. 2021;18:139–54.
Boland ML, Chourasia AH, Macleod KF. Mitochondrial dysfunction in cancer. Front Oncol. 2013;3:292.
Khan T, Relitti N, Brindisi M, Magnano S, Zisterer D, Gemma S, et al. Autophagy modulators for the treatment of oral and esophageal squamous cell carcinomas. Med Res Rev. 2020;40:1002–60.
Wu Z, Oeck S, West AP, Mangalhara KC, Sainz AG, Newman LE, et al. Mitochondrial DNA stress signalling protects the nuclear genome. Nat Metab. 2019;1:1209–18.
Chan DC. Mitochondrial fusion and fission in mammals. Annu Rev Cell Dev Biol. 2006;22:79–99.
Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, et al. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol Biol Cell. 2007;18:3225–36.
Fang C, Wei X, Wei Y. Mitochondrial DNA in the regulation of innate immune responses. Protein Cell. 2016;7:11–6.
Gui X, Yang H, Li T, Tan X, Shi P, Li M, et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019;567:262–6.
Prabakaran T, Bodda C, Krapp C, Zhang BC, Christensen MH, Sun C, et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 2018;37:e97858.
Gao W, Guo H, Niu M, Zheng X, Zhang Y, Xue X, et al. circPARD3 drives malignant progression and chemoresistance of laryngeal squamous cell carcinoma by inhibiting autophagy through the PRKCI-Akt-mTOR pathway. Mol Cancer. 2020;19:166.
Acknowledgements
We thank Dr. Jinliang Xing of State Key Laboratory of Cancer Biology and Department of Physiology and Pathophysiology, Fourth Military Medical University for providing the plasmid vectors with TFAM.
Funding
Project funded by China Postdoctoral Science Foundation (NO. 2020M682286, 2021T140180) to DB, the Young Core Instructor of Henan (NO. 2021GGJS027) to DB. Thanks for startup fund from Henan University to YW and DB.
Author information
Authors and Affiliations
Contributions
This study was conceived and led by DB. YL, QY and HC performed the cell culture experiments. QY, HC, JH, XY, and XY performed the animal studies. YW, XW, JS, HY, HL, HY, and LW helped with the data analysis. LW, QY and YL performed the histological analysis and pathological diagnosis. DB, YW and HY wrote the manuscript and organized figures. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Li, Y., Yang, Q., Chen, H. et al. TFAM downregulation promotes autophagy and ESCC survival through mtDNA stress-mediated STING pathway. Oncogene 41, 3735–3746 (2022). https://doi.org/10.1038/s41388-022-02365-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02365-z
- Springer Nature Limited
This article is cited by
-
Focusing on mitochondria in the brain: from biology to therapeutics
Translational Neurodegeneration (2024)
-
Blockade of the mitochondrial DNA release ameliorates hepatic ischemia-reperfusion injury through avoiding the activation of cGAS-Sting pathway
Journal of Translational Medicine (2024)
-
FGF21 attenuates neuroinflammation following subarachnoid hemorrhage through promoting mitophagy and inhibiting the cGAS-STING pathway
Journal of Translational Medicine (2024)
-
Mitochondria transcription and cancer
Cell Death Discovery (2024)
-
Aerobic Exercise Ameliorates Cognitive Disorder and Declined Oxidative Stress via Modulating the Nrf2 Signaling Pathway in d-galactose Induced Aging Mouse Model
Neurochemical Research (2024)