Abstract
Metastatic recurrence is still a major challenge in breast cancer treatment, but the underlying mechanisms remain unclear. Here, we report that a small adaptor protein, SH3BGRL, is upregulated in the majority of breast cancer patients, especially elevated in those with metastatic relapse, indicating it as a marker for the poor prognosis of breast cancer. Physiologically, SH3BGRL can multifunctionally promote breast cancer cell tumorigenicity, migration, invasiveness, and efficient lung colonization in nude mice. Mechanistically, SH3BGRL downregulates the acting-binding protein profilin 1 (PFN1) by accelerating the translation of the PFN1 E3 ligase, STUB1 via SH3BGRL interaction with ribosomal proteins, or/and enhancing the interaction of PFN1 with STUB1 to accelerate PFN1 degradation. Loss of PFN1 consequently contributes to downstream multiple activations of AKT, NF-kB, and WNT signaling pathways. In contrast, the forced expression of compensatory PFN1 in SH3BGRL-high cells efficiently neutralizes SH3BGRL-induced metastasis and tumorigenesis with PTEN upregulation and PI3K-AKT signaling inactivation. Clinical analysis validates that SH3BGRL expression is negatively correlated with PFN1 and PTEN levels, but positively to the activations of AKT, NF-kB, and WNT signaling pathways in breast patient tissues. Our results thus suggest that SH3BGRL is a valuable prognostic factor and a potential therapeutic target for preventing breast cancer progression and metastasis.
Similar content being viewed by others
Introduction
Breast cancer is the most prevalent malignancy in women and the second leading cause of cancer-related deaths worldwide [1]. Despite the improvements in early detection as well as the effective adjuvant and targeted therapies, metastasis and recurrence still largely remain as the major cause of breast cancer death [2, 3]. As known, multiple genes and signaling pathways have been unveiled to be responsible for the progression and metastasis of breast cancer, Including HER2, AKT, NF-kB, and Wnt signaling pathways that are constitutively and abnormally activated in most types of cancers. Hyperactivation of Wnt signaling generally augments tumor cell motility and invasiveness by transcriptional upregulation of multiple Wnt downstream genes related to the epithelial-mesenchymal transition [4, 5]. Likewise, hyperactivation of NF-κB signaling also elicits tumor cell migration, invasion, and proliferation through transcriptional upregulation of multiple NF-kB downstream genes, such as matrix metalloproteinase (MMPs) and c-Myc [6,7,8]. However, the underlying mechanisms on the multiple activations of AKT, NF-kB, and WNT pathways remain largely unclear.
During tumor progression and metastasis, tumor cells acquire the invasive capability to enable their dissemination and distant spreading. Disruption of the cell cytoskeleton is another hallmark of the transformed cells, which contributes to the aberrant function or dysfunction of signaling pathways that govern cytoskeleton homeostasis. For instance, insufficient filamentous actin (F-actin) density is associated with the worse breast cancer grade, indicating that cytoskeletal disruption leads to tumor cell aggressiveness in breast cancer [9]. Profilin1 (PFN1) is a small evolutionarily conserved ubiquitous actin-binding protein composed of 139 amino acids and two motifs, an actin-binding domain and a poly-L-proline (PLP)-binding domain [10, 11]. PFN1 plays multiple roles in protein–protein interaction, cell–cell adhesion, gene splicing/transcription, nuclear export of actin and membrane trafficking [12,13,14], as well as cell morphology and migration [14,15,16] through actin polymerization and dynamics. It has been shown that PFN1 depletion enhances cell migration of normal human mammary epithelial cells and breast cancer cells [17, 18]. Meanwhile, PFN1 also functions as a tumor suppressor and is downregulated in various cancers, including breast, bladder, pancreatic and hepatic cancers, and low PFN1 expression is correlated with poor prognosis of pancreatic and bladder cancer patients [19,20,21,22]. Mechanistically, PFN1 overexpression can coordinately upregulate PTEN and inhibit AKT activation for breast cancer repression [23, 24]. Conversely, PFN1 knockdown enhances motility, migration, and metastasis of breast cancer cells [25]. Although the E3 ligase STUB1 (also known as CHIP) is reported to interact with PFN1 to ubiquitinate it for proteasome-mediated degradation [26], the intricate mechanism of PFN1 downregulation by STUB1 expression alteration and the consequent physiological role in metastasis of breast cancer remains unknown.
SH3 domain binding glutamate-rich protein-like (SH3BGRL) was first identified in down syndrome [27], which encodes a protein of 114 amino acids with a conserved proline-rich PLPPQIF sequence for the Homer SH3-binding and EVH1-binding motifs [28]. As a scaffold protein, SH3BGRL plays a crucial role in various physiological events of a cell through protein–protein interaction [29]. For instance, SH3BGRL involves in the formation of the central nervous system and intestine of zebrafish and Parkinson’s disease [30, 31]. However, the function of SH3BGRL in human cancer is largely unclear. Previously, we indicated that wild-type human SH3BGRL could repress metastasis of triple-negative breast cancer cells [32] which is in line with SH3BGRL as an inhibitor of v-Rel-induced transformation in chicken cells [33] and a suppressor in leukemogenesis [34]. In contrast, clinical data indicate that SH3BGRL is robustly upregulated in BRCA1 mutation-positive and lymphocytic infiltration-positive breast tumors [35] and in ER-positive and triple-negative breast cancers based on proteomics analysis [36, 37]. Recently, we disclosed that SH3BGRL confers innate drug resistance to HER2-targeted therapy of breast cancer by stabilizing and activating HER2 on cell membranes [38]. Thus, the function of SH3BGRL in breast cancer remains controversial and elusive.
Here, we disclosed that SH3BGRL can promote breast cancer cell metastasis by enhancing proteasome degradation of the actin-binding protein PFN1 and subsequently activate multiple downstream signals. This novel SH3BGRL-STUB1-PFN1 axis along with the downstream aberrant PTEN/AKT, NF-kB, and WNT pathways would be potential targets for clinical breast cancer therapy.
Results
High SH3BGRL expression correlates to metastasis and poor prognosis of breast cancer
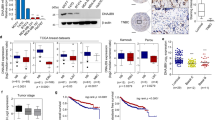
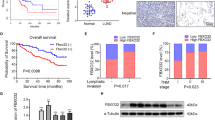
To thoroughly investigate the correlationship between SH3BGRL expression and the metastatic relapse of breast cancer, we first analyzed the public datasets from TCGA and GEO databases (Table S1) and observed that SH3BGRL mRNA level was increased in the majority of breast cancer tissues compared to their adjacent normal tissues (Fig. 1A; Fig. S1A–D). Moreover, SH3BGRL was significantly upregulated in breast cancer patient’s tissues with distant metastasis and relapse compared with those without metastasis or relapse in datasets GSE6532 and GSE12276 (Fig.1B). Importantly, patients with high SH3BGRL mRNA expression had shorter 5-year metastasis-free survival, 5-year relapse-free survival, and overall survival by analysis of other patient cohorts (Fig.1C; Fig. S1E–J). To validate these results, we examined the expression of SH3BGRL protein in 14 fresh breast patient tumors with paired adjacent tissues, in which 10 patients showed no metastasis and the other 4 had distant metastasis. Results revealed that SH3BGRL protein expression was not only remarkably elevated in all tumor tissues, but further increased in the metastatic patients’ tissues (Fig. 1D). Besides, we checked SH3BGRL protein in several breast cancer cell lines and another more 32 pairs of patient samples and found that SH3BGRL protein is almost upregulated in the assayed breast cancer cell lines and patient tissues (Fig.1E, F). To investigate the relevance of SH3BGRL on breast cancer patient prognosis, we conducted the Kaplan–Meier analysis and manifested that patients with higher SH3BGRL protein expression suffered worse overall survival, relapse-free survival and metastasis-free survival by retro-analyzing 100 breast cancer cases (Fig. 1G). Together, results here indicated that SH3BGRL upregulation is positively correlated with the poor prognosis as well as the distant metastatic relapse of breast cancer patients.
A Analysis of SH3BGRL expression in breast tumor tissues and normal breast tissues or the paired breast tissues from datasets of GSE15852, GSE45827, GSE26304, and TCGA dataset. B Analysis of SH3BGRL in breast cancer patients with metastasis or relapse, compared to those without metastasis or relapse based on the GSE6532 and GSE12276 datasets. C Kaplan–Meier plots for distant metastasis-free survival, and relapse-free survival of breast cancer patients with low versus high SH3BGRL expression in GSE2990. D, E Immunoblots of SH3BGRL in 10 paired non-metastatic breast cancer samples (T) and 4 paired metastatic ones (TM), compared with adjacent normal counterparts (N) (D), and in MCF-10A and 7 cultured breast cancer cell lines (E). β-actin served as the loading control. F IHC staining of SH3BGRL expression in 32 paired human breast cancer tissues, compared with their adjacent normal ones. Statistical SH3BGRL expression is shown in the lower panel. Scale bars are shown; P < 0.0001. G Kaplan–Meier curves for overall survival, distant metastasis-free survival and relapse-free survival of breast cancer patients with low versus high SH3BGRL protein level based on the Kaplan–Meier Plotter Liu_2014 dataset (n = 100; P = 0.046).
SH3BGRL upregulation enhances invasiveness and migration of cancer cells
To explore the physiological role of SH3BGRL in breast cancer, we established an SH3BGRL-overexpressing stable cell line derived from MCF-7 cells that have a low level of endogenous SH3BGRL, and SH3BGRL-knockdown cell lines from MDA-MB-453 cells with high SH3BGRL expression (Fig. S2A). SH3BGRL overexpression renders MCF-7 cells to present a spindle-like shape (Fig. 2A), indicating its invasion-promoting role. We then carried out a gene set enrichment analysis (GSEA) and found that high SH3BGRL expression is positively correlated to gene signatures of metastasis and invasion in breast cancer patients (Fig. 2B). As expected, SH3BGRL upregulation experimentally enhanced breast cancer cell motility and invasiveness, by contrast, SH3BGRL depletion attenuated these features by wound-healing and transwell assays (Fig. 2C, D). SH3BGRL also promoted tumorigenesis of breast cancer cells for colony formation by using the normal and anchorage-independent growth culture systems, respectively (Fig. 2E, F). These results are consistent with the previously reported in both MCF-7 and MDA-MB-453 cells [38]. We additionally determined the role of SH3BGRL in breast cancer metastasis for lung colonization and observed that SH3BGRL overexpression significantly enhanced lung metastasis of MCF-7 cells (Fig. 2G; Fig. S2B), indicating the metastatic role of SH3BGRL in breast cancers.
A Representative morphologies of the indicated MCF-Vector and MCF-SH3BGRL cells. B GSEA plot of SH3BGRL expression in breast cancer patients, which is positively correlated with metastatic EMT gene signatures based on the published dataset GSE15852 (n = 41). C Representative wound-healing of the indicated cells. Experiments were triply repeated independently. Wound closure states were photographed at the indicated time. ****P < 0.0001. D Representative views of invaded or migrated cells assayed by Matrigel-coated or non-coated transwell assays. The invaded or migrated cells were quantified from three independent experiments.****P < 0.0001. E Representative images of colonies generated by indicated cells for 14–20 days. The right panel showed the quantification of colonies from three individual experiments. ****P < 0.0001. F Soft agar colony formation of the indicated cells. Colonies larger than 0.1 mm diameter was quantified after 16 days and analyzed. Experiments were triply repeated independently. ***P < 0.001; ****P < 0.0001. G Representative bright-field images of the lungs and visible surface metastatic lesions of nude mice injected intravenously via the caudal vein with indicated cells. n = 6; ****P < 0.0001.
SH3BGRL downregulates PFN1 through direct interaction with PFN1 in breast cancer cells
To explore the underlying mechanism of SH3BGRL on promoting breast tumor metastasis, we performed immunoprecipitation with anti-SH3BGRL antibodies and identified the SH3BGRL-specific binding proteins with mass spectrometry (MS) (Fig. S3A, B). MS identification disclosed about 165 specific SH3BGRL-interacting proteins in one tumor tissue, but not in the adjacent tissue (Fig. S3C), among which many proteins are ribosomal subunits and some involved in the translation machinery (Fig. S3D). Moreover, we figured out SH3BGRL-bound proteins in another two tumor tissues and observed 27 commonly shared SH3BGRL-bound proteins unique in these tumor tissues (Fig. 3A). Based on the well-known function of these identified proteins, only the actin-binding profilin1 (PFN1) is directly associated with cell morphology and cancer metastasis (Table S2). Thus, we first sought to characterize the interaction between SH3BGRL and PFN1 by reciprocal co-immunoprecipitation. Results demonstrated that SH3BGRL interacted with PFN1 in various breast cancers and 293 T cells (Fig. 3B). Meanwhile, SH3BGRL overexpression dramatically decreased PFN1 protein expression, whereas SH3BGRL silence with its specific shRNAs increased it (Fig. 3C; Fig. S3E). We then noted that PFN1 protein expression alteration was not affected either on the transcriptional or translational level by SH3BGRL (Fig. S4A–D).
A Schematic of SH3BGRL-bound proteins in three breast tumor (T1, T5, and T6) tissues with high SH3BGRL protein expression and the adjacent tissue of T6 (N6) by immunoprecipitation and MS peptide sequencing. PFN1 is one of the 27 shared proteins specifically identified in all tumor tissues. B Mutual co-immunoprecipitation of SH3BGRL with PFN1 in HEK293T(293T), MDA-MB-231, MCF-7, and MDA-MB-453 cells. C Western blots of PFN1 and SH3BGRL expression in the indicated cells. shRNA1+2 represents a stable cell pool mixed by two individual stable SH3BGRL-knockdown lines by two different SH3BGRL-specific shRNAs 1 and 2 as shown in Fig. S2A. β-actin was used as a loading control. Statistical PFN1 expression is shown from four independent immunoblots. Error bars represent mean±s.d.*P < 0.05, **P < 0.01. D, E Interaction of PFN1 with SH3BGRL verified by the indicated SH3BGRL mutations in 293T (D) and MCF-7 (E) cells transfected with SH3BGRL or its truncated mutants Δα1, Δα2, Δα3, Δβ3, Δα4, Δα5, and Δα6. The SH3BGRL protein structure is schematically shown under blots in (E). F Immunoblot of PFN1 expression affected by the indicated SH3BGRL mutations as above. MCF-7 cells were transiently transfected with HA-SH3BGRL and truncated mutants, β-actin was used as a loading control. The protein band intensity was scanned and measured by Image J software. Relative PFN1 level against β-actin is further normalized against HA-GFP as shown.
To verify the specific interaction of SH3BGRL to PFN1, we individually deleted all 6 α helixes and the β3 sheet (containing SH3B domain) in SH3BGRL protein to construct 7 HA-tagged SH3BGRL truncated mutants. The co-immunoprecipitation in both 293T and MCF-7 cells revealed that deletion of the β3 sheet domain of SH3BGRL notably abolished its binding ability with PFN1, and deletion of the α3 helix just attenuated the interaction (Fig. 3D, E). Immunoblots accordingly confirmed that the breakdown of SH3BGRL β3 sheet or α3 helix efficiently deterred PFN1 downregulation, compare to wild-type SH3BGRL and other mutants (Fig. 3F). Thus, the downregulation of PFN1 seems to be attributed to the interaction of SH3BGRL with PFN1 through the α3 helix and β3 sheet.
SH3BGRL accelerates PFN1 degradation by enhancing E3 ligase STUB1 translation and interaction with PFN1
To further determine the mechanism of SH3BGRL on PFN1 downregulation, we first examined PFN1 expression change induced by SH3BGRL and found no apparent PFN1 mRNA level alteration. Cycloheximide (CHX) chase assay showed that when blocking the novel protein synthesis, SH3BGRL overexpression efficiently reduced PFN1 protein level, while SH3BGRL silence attenuated the extent of PFN1 decrease (Fig. 4A, B; Fig. S5A, B). To check whether SH3BGRL promotes PFN1 degradation through the autophagic lysosome degradation or the ubiquitin-proteasome process, we treated the cells with the known lysosome inhibitor CQ and proteasome inhibitor MG132, respectively. Results showed that PFN1 protein level change was abrogated in either SH3BGRL-overexpressing or its silence cells upon MG132 treatment, but CQ treatment showed no such effect (Fig. 4C–E), indicating the existence of the proteasome-dependent PFN1 degradation by SH3BGRL. Ubiquitination analysis demonstrated that the ubiquitin-conjugated PFN1 level was increased in SH3BGRL-overexpressing cells, while decreased in SH3BGRL knockdown cells (Fig. 4F). Given that PFN1 is ubiquitinated and degraded by the co-chaperone E3 ligase STUB1 (CHIP) [26], we investigated whether PFN1 downregulation results from STUB1 expression change through SH3BGRL overexpression in breast cancer cells. Immunoblots revealed that STUB1 protein level was reduced upon SH3BGRL depletion in MDA-MB-453 (Fig. 4G), suggestive of the degradation of PFN1 mediated by STUB1 expression level. However, STUB1 protein level was not clearly impaired by SH3BGRL in both MCF-7 and MDA-MB-231 cells (Fig. 4H), hinting the STUB1 expression-independent PFN1 degradation by SH3BGRL in some types of cells.
A, B Immunoblots of PFN1 expression in the indicated cells for 0, 4, 8, 12 h (A). Statistical PFN1 protein stability curve was obtained from three independent experiments (B), ****P < 0.0001. 50 μg/ml cycloheximide (CHX) was used to treat cells. C–E Immunoblots of PFN1 expression in the indicated cells treated with or without MG132 or CQ for 12 h (C, D). Statistical PFN1 expression by quantification of PFN1 intensity is presented as histograms (E). Three independent experiments were performed for each type of cells, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. F Immunoblots of the ubiquitinated PFN1 in the indicated cells treated with or without MG132 for 12 h. Cell lysates were immunoprecipitated with PFN1 antibody and immunoblotted against ubiquitin antibody. G, H Immunoblots of SH3BGRL, STUB1, and PFN1 in the indicated cells. β-actin was used as a loading control. I Polyribosome profiling of the STUB1 in indicated cells. β-actin was used as an internal negative loading control. The marked numbers are equivalent to the harvested fractions after gradient centrifuge. Fraction 1 represents the top (10%) of the gradient, fraction 10 represents the bottom (50%) of the gradient. J Immunoprecipitation of PFN1 with STUB1 along with SH3BGRL expression alteration in the indicated cells. Interaction between PFN1 and STUB1 was evaluated by the binding ratio of STUB1 to PFN1. The protein band intensity was scanned and measured by Image J software, Ratio of STUB1 to PFN1 is further normalized against that of the vector as shown.
We first investigated the underlying mechanism of STUB1 downregulation by SH3BGRL in MDA-MB-453 cells. As we have identified abundant proteins involved in protein translation machinery, we thus asked whether SH3BGRL interacts with ribosomes to modulate the universal protein translation, including STUB1. We then carried out the GST-pull down combined with mass spectrometry (MS) analysis and confirmed the existence of various ribosome-building proteins in the precipitated SH3BGRL-interacting proteins, including ribosomal subunits, translation initiation, and extension factors (Fig. S5C), indicating that SH3BGRL might be assembled in ribosomes to modulate protein translation. Besides, co-immunoprecipitation and immunofluorescence demonstrated the co-localization of SH3BGRL with another ribosomal subunit, RPL7a (Fig. S5D, E), verifying the interaction of SH3BGRL with ribosomes. Therefore, we proposed that SH3BGRL involves in the general translation machinery to regulate protein production, including STUB1 translation. To clarify this hypothesis, we performed a whole-cell polyribosome profiling with mRNA sequencing analysis in MDA-MB-453 cells with SH3BGRL knockdown and observed a universal protein translational alteration featured by the polyribosome fraction shift (Fig. S6A; Table S3). Polyribosome fraction profiling analysis indicated that STUB1 expression was obviously enhanced by SH3BGRL on the translational level, with a rightward shift of about 3 fractions (Fig. S6B–G; Table S4). Semi-quantitative RT-PCR confirmed that STUB1 mRNA was mainly enriched in the heavier polyribosome fractions in the parental cells with SH3BGRL expression, but SH3BGRL knockdown resulted in the STUB1 mRNA shift to lighter/fewer polyribosome fractions in the same cells (Fig. 4I), indicating the translational enhancement of STUB1 expression by SH3BGRL in MDA-MB-453 cells.
Meanwhile, we explored the possible mechanism on how SH3BGRL downregulates PFN1 in both MCF-7 and MDA-MB-231 cells, since STUB1 protein level was not increased by SH3BGRL in those cells as shown above. Interestingly, we noted that SH3BGRL overexpression evidently enhanced the binding affinity of STUB1 to PFN1 in both MCF-7 and MDA-MB-231 cells, whereas SH3BGRL knockdown reduced the bound amounts of PFN1 to STUB1 in MDA-MB-453 cells by immunoprecipitation (Fig. 4J). Taken together, we demonstrated that SH3BGRL could promote PFN1 degradation through either STUB1 translational upregulation or/and the stronger binding of STUB1 to PFN1, leading to the accelerated PFN1 ubiquitin-proteasome degradation in breast cancer cells.
SH3BGRL activates downstream AKT, NF-kB and WNT pathways via PFN1 depletion
To further dissect the downstream molecular events of the SH3BGRL-STUB1-PFN1 axis in breast tumor cell invasion and tumorigenesis, we retrieved the public datasets again and found that SH3BGRL upregulation was positively correlated with activations of the PI3K/AKT, NF-kB, and WNT signaling pathways (Fig. 5A), which is in line with the result that PFN1 increases PTEN protein to inactivate PI3K/AKT signaling pathway in breast cancer [23]. To verify this, we conducted immunoblotting and noted that SH3BGRL overexpression markedly repressed PTEN expression along with PFN1 downregulation, while SH3BGRL knockdown increased PTEN expression with elevated PFN1 level and the inhibition of the subsequent PI3K-AKT signaling (Fig. 5B; Fig. S7A, B). SH3BGRL also increased β-catenin expression and enhanced the nuclear translocation of β-catenin (N) and p65(N) (Fig. 5B, C), indicating the activations of the NF-kB and WNT signaling pathways. We further examined the targeted genes of NF-kB and WNT signaling and demonstrated that the NF-kB genes, including MMP2, MMP9, and c-Myc and WNT target genes, including Slug, Vimentin (VIM) and N-Cadherin were all upregulated (Fig. 5D; Fig. S7C–E). To screen the signaling pathway alteration induced by SH3BGRL in breast cancer cells, we performed a transcriptome RNA sequencing and conducted a signaling pathway enrichment analysis in MDA-MB-453 cells with SH3BGRL knockdown. Results revealed that SH3BGRL is possibly involved in cell cycle, ribosome biogenesis and some oncogenic signaling pathways (Fig. S8A, B). Further analysis suggested that SH3BGRL could lead to a proto-oncogene, c-Myc expression increase, which is a known target gene of NF-kB (Fig. S8C).
A GSEA plots of SH3BGRL expression positively correlated with PI3K/AKT signaling and Wnt pathway-driven gene signatures in breast cancer patient cohorts (GSE15852, n = 41), as well as the positive correlation with NF-kB pathway-driven gene signatures (GSE45827, n = 130). B Immunoblots of PFN1, PTEN and AKT, NFkB, and WNT key signaling genes in the indicated breast cancer cell lines. C Immunofluorescence staining of localizations of P65 and β-catenin in indicated cells. The scale bar is 10 μm. D Immunoblots of the expression of indicated EMT signaling-related proteins and c-Myc in the indicated cells. E Immunoblots of the expression of PFN1, PTEN, and the indicated AKT, NF-kB, and WNT signaling-related proteins, EMT signaling proteins, and c-Myc in the indicated cells.
Moreover, to investigate the mediator function of PFN1 in SH3BGRL’s oncogenic effect, we additionally knocked down PFN1 expression in the SH3BGRL-silenced MDA-MB-453 or compensatorily overexpressed PFN1 in the SH3BGRL-overexpressing MCF-7 cells. Results demonstrated that supplemental PFN1 overexpression blocked the activating effect of SH3BGRL on AKT, NF-kB, and WNT signalings in the SH3BGRL-overexpressing MCF-7 cells (Fig. 5E; Fig. S9A–C). Conversely, the forced PFN1 depletion in MDA-MB-453-SH3BGRL-KD cells mimicked the PTEN suppression from SH3BGRL overexpression, while activated PI3K/AKT, NF-kB, and WNT signalings (Fig. 5E; Fig. S9D–F). Results here revealed that SH3BGRL exerts its tumorigenic and metastatic functions through PFN1 as a mediator in breast cancer cells.
PFN1 is an essential bridge for SH3BGRL-mediated metastasis and tumorigenesis
To validate whether PFN1 is an indispensable mediator for SH3BGRL-promoted metastasis and tumorigenesis, we then determined the roles of the SH3BGRL-PFN1 axis in the motility and invasion of breast cancer cells for metastasis and tumorigenesis. The compensatory PFN1 overexpression in MCF-7 cells with SH3BGRL overexpression counteracted the promoting effects of SH3BGRL on cell capabilities in wound healing (Fig. 6A; Fig. S10A), cell migration (Fig. 6B), colony formation (Fig. 6C) and the anchorage-independent growth (Fig. 6D), compared to their parental cells. Likewise, the deliberately PFN1 knockdown in SH3BGRL-silenced MDA-MB-453 cell almost restored the all above processes (Fig. 6A–D; Fig. S10B). The dependence of SH3BGRL on PFN1 in tumorigenesis was also validated by in vitro soft agar assay (Fig. 6E). Furthermore, the tail vein injection of cells for lung metastasis demonstrated that the metastatic role of SH3BGRL in cells could be only achieved along with PFN1 downregulation, as the additionally artificial PFN1 depletion could effectively reverse the attenuated metastatic feature of cells with SH3BGRL depletion, compared with the parental MDA-MB-453 cells (Fig. 6F; Fig. S10C). Kaplan–Meier analysis demonstrated that knockdown of SH3BGRL alone dramatically prolonged the survival time of the xenografted nude mice, compared to the control mice, while the additional depletion of PFN1 in these SH3BGRL-silenced cells neutralized this survival time prolongation (Fig. 6G). Immunoblots of lysates of those metastatic mice lung tumor nodules revealed that SH3BGRL silence repressed the demonstrated downstream AKT, NF-kB, WNT, and EMT pathways, whereas, the additional knockdown of PFN1 in SH3BGRRL-silenced tumors restored them (Fig. 6H; Fig. S10D, E). Thus, our results experimentally verified that PFN1 is a mediator of SH3BGRL to promote tumorigenesis and metastasis of breast cancer cells both in vitro and in vivo.
A Wound healing analysis of the indicated cells. The wound closure was measured with the gap width. The experiment was repeated three times independently, ****P < 0.0001. B Transwell assays of the indicated cells for cell invasiveness or migration ability. Representative views of invaded or migrated are presented from either matrigel-coated or non-coated transwell assays, respectively, and quantified. Experiments were repeated three independent times. ****P < 0.0001. C Representative colonies (up panel) and the quantification (down panel) by the indicated breast cancer cells 14 days after the colony formation assay. D, E Soft agar colony formation of indicated cells 16 days post-inoculation (D) and quantifications of them presented as histograms. Experiments were repeated three independent times. **P < 0.01; ***P < 0.001; ****P < 0.0001. F, G Lung metastasis of the indicated cells in nude mice. Cells were injected via the tail vein. Representative bright-field images of the lung (E) and the visible surface metastatic lesions of tumors were counted, n = 6, **P < 0.01; ****P < 0.0001. F Kaplan–Meier survival curves of nude mice with the above lung metastatic tumor (G). H Immunoblots of expression of SH3BGRL, PFN1, STUB1, PTEN, and indicated AKT, NF-kB, WNT, and EMT signaling-related proteins and c-Myc in lysates of metastatic lesions of mice lung tissues. β-actin was used as a loading control.
SH3BGRL is negatively correlated with PFN1 expression, while positive with AKT, NF-kB, and WNT signaling activations in breast cancers
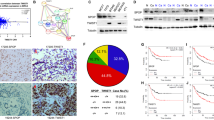
To validate the relevance of SH3BGRL with PFN1 and the downstream AKT, NF-kB, WNT, and EMT events in breast cancer patients, we performed IHC staining of SH3BGRL and PFN1 in 32 pairs of the paraffin-embedded breast cancer tissues (Fig. 7A). The statistical results of IHC analysis revealed that SH3BGRL was highly expressed with the low level of PFN1 in breast cancer tissues, which was opposite to that in the matched adjacent normal tissues (Fig. 7B, C; Fig. S11A). Statistical analysis showed that SH3BGRL protein level was negatively correlated with that of PFN1 (Fig. 7D). This result was further validated in 14 breast cancer samples by immunoblots, in which SH3BGRL protein level was observed to be negatively correlated with both PFN1 and PTEN, whereas positively to activations of AKT, NF-kB, WNT, and EMT signaling pathways (Fig. 7E, F; Fig. S11B). Moreover, analysis of another 74 breast cancer cases from Breast Invasive Carcinoma datasets manifested that SH3BGRL protein level was also positively correlated with N-cadherin and MMP2, but inversely correlated with E-cadherin expression (Fig. S11C–E).
A–C Representative IHC staining of SH3BGRL and PFN1 expressions in 32 primary breast cancer specimens along with the matched adjacent normal tissues, scale bar is 50 mm (A). PFN1 is downregulated (B), while SH3BGRL is upregulated (C) by statistical analysis. n = 32, P < 0.0001. D The negative correlationship of SH3BGRL protein expression with that of PFN1 in (A). E Immunoblots of expression of the indicated proteins in 14 fresh breast cancer tissues. β-actin was used as a loading control. Protein band intensity was quantified and analyzed with densitometry and Image J software and the heatmap of all protein expression is shown on the right panel. F Correlationships of SH3BGRL with PFN1, PTEN, p-AKT, p-GSK3β, p-IKKα, β-catenin(N), P65(N), E-cadherin, N-cadherin, SLUG, VIM, MMP9, and c-Myc protein expressions are presented. G Schematic mechanism of SH3BGRL-PFN1 axis in breast tumor progression and metastasis.
To evaluate the PFN1 and SH3BGRL expressions, their mRNA and protein levels were determined in 10 of the former 32-paired breast cancer samples. Results demonstrated that both SH3BGRL mRNA and protein levels were positively correlated to activations of the analyzed signaling pathways, but there was a non-strict relationship between PFN1 mRNA and its protein level (Fig. S12A–C). The inconsistent expression levels of PFN1 mRNA and its protein were also examined by analyses of other different public breast cancer datasets (Fig. S13A–H; Table S5), verifying the non-transcriptional regulation of PFN1 in breast cancers. By analyzing another 421 (TCGA, Cell 2015) and 168 (SMC 2018) breast cancer cohorts, SH3BGRL mRNA expression was also shown to be positively correlated with the downstream target gene expression of NF-kB and WNT, including slug, VIM, N-Cadherin and MMP2, and negatively with E-Cadherin, respectively (Fig. S14A–E).
Finally, Kaplan–Meier Plotter showed that patients with high SH3BGRL protein expression, low PFN1 and E-cad protein levels suffered poorer 5-year overall survival, relapse-free survival, and metastasis-free survival, based on the follow-up of 126 cases of breast cancer patients (Fig. S15A, B). The downstream MMP2, MMP9, and VIM protein levels were also shown to be positive with the poor prognosis in another 65 cases (Fig. S15C–E). Taken together, these results strongly indicated that SH3BGRL-PFN1 and the downstream AKT, NF-kB, and WNT signaling axis play crucial roles in breast cancer metastasis, relapse, and prognosis (Fig. 7G).
Discussions
Breast cancer is the most frequent and leading cause of cancer-related death among women, mainly attributed to its metastasis and relapse [39]. Unfortunately, the current advances on the clinical and histopathologic characters or biomarkers are still unsatisfactory for the early diagnosis and late prognosis of breast cancer patients who eventually develop metastasis. It is imperative to identify effective novel molecular biomarkers and therapeutic targets for the efficient early diagnosis and efficient therapy for those patients. Here we uncover that SH3BGRL would be such a promising biomarker. Given that depletion of either SH3BGRL or PFN1 can effectively inhibit tumorigenesis and metastasis of breast cancer cells, we expect that SH3BGRL itself or its downstream AKT, NF-kB, or WNT signaling would have potent therapy value for the SH3BGRL-overexpressing breast cancer. We recently demonstrated that SH3BGRL confers HER2-targeted therapy resistance and the silence of SH3BGRL, in turn, benefits the therapy efficacy [38]. Accumulating evidence also shows that SH3BGRL is overexpressed in various types of cancer, including ER-positive breast cancer, BRCA1 mutation breast tumors, lymphocytic infiltrate-positive breast cancer, and squamous oral carcinoma [35,36,37]. Taken results here together, we at least partially discover the exact reason why breast cancer patients with SH3BGRL overexpression usually experience frequent relapse and poor prognosis.
As a scaffold protein, it is reasonable to predict that SH3BGRL involves in protein–protein interaction and signal transduction crosstalk to modulate cell cytoskeletal rearrangement, membrane trafficking, and other important cellular processes [29]. Recently, we biochemically disclosed the interaction between SH3BGRL with HER2 to stabilize HER2 on cell membranes for the prolonged HER2 activation [38]. Consistently, here we similarly observed that SH3BGRL enhances the interaction of PFN1 with STUB1 to promote PFN1 ubiquitination-mediated proteasome degradation. Besides, SH3BGRL can simultaneously enhance PFN1 degradation via the translation efficiency of the E3 ligase, STUB1 for PFN1 ubiquitination. In this process, the intricate interaction of SH3BGRL with various ribosomal subunit proteins is unveiled, which accounts for the translation enhancement of STUB1, but the underlying mechanism of this interaction on gene translation efficiency or gene specificity is unclear, and need to be further dissected. Tentatively, we propose that binding of SH3BGRL to ribosome subunits would modulate the assembly or the allosteric conformation of ribosomes, leading to the biased recognition or binding of ribosomes to mRNA molecules for varied translation efficiency, as complex RNA secondary structures in mRNA molecules can modulate the binding mode of RNA with protein. Nevertheless, this hypothesis should be clarified by structural analysis.
Given that PFN1 is a small evolutionarily conserved ubiquitous actin-binding protein in humans [10, 11], it is reasonable to understand that PFN1 works for actin polymerization on cell morphology and migration[14, 18]. PFN1 also involves in tumorigenesis as a tumor suppressor and is downregulated in various cancer types, including breast, bladder, pancreatic and hepatic cancer [19, 22], but the mechanism on PFN1 downregulation is still obscure. Our results here at least partially explain this post-translational downregulation of PFN1 in breast cancers, as SH3BGRL alteration doesn’t impair PFN1 mRNA level, but on its protein level via protein degradation manners. In light of PFN1 knockdown enhances motility, migration, and metastasis of the breast cancer cell [18, 23, 24], we additionally confirm that PFN1 overexpression can upregulate PTEN while inhibiting AKT activation to suppress tumorigenic and metastatic capability of breast cancer cells, which is in line with the previous result that PFN1 can post-transcriptionally upregulate PTEN [23]. Moreover, we manifest that PFN1 can repress other signaling pathways related to tumorigenesis and metastasis, including the well-known WNT and NF-kB signaling cascades. Downregulation of PFN1 leads to the activations of either β-catenin target genes, N-cadherin, slug, Vim, and E-cadherin downregulation for tumor cell epithelial-mesenchymal transition, or the NF-kB target genes MMP2, MMP9, and c-MYC for the tumor cell-extracellular matrix degradation and metastasis. All these results indicate that this SH3BGRL-induced PFN1 downregulation plays pivotal and multiple roles in breast cancer cell metastasis, and SH3BGRL would be a novel upstream suppressor of PFN1.
Controversially, some previous reports indicate that SH3BGRL may also act as a tumor suppressor to inhibit v-Rel-induced transformation in chicken cells [33] and leukemogenesis [34]. We also have shown that SH3BGRL physiologically suppresses the metastasis of the triple-negative breast cancer cells, MDA-MB-231 in mice models through inhibition of Src activation [32]. Unexpectedly, here we still note that SH3BGRL downregulates PFN1 in the same cells, indicating the intricate and flexible function of SH3BGRL as an adaptor protein that usually works on the cell type- or cellular context-dependent manners to render the overall effect. Therefore, the exact and eventual role of SH3BGRL in a particular type of cancer should be intensively investigated.
We previously revealed that SH3BGRL is a marker for poor prognosis of HER2-positive breast cancers based on its mRNA level by analyzing the information of TCGA database [38]. However, when analysis of the breast cancer data from KM plotter or some GEO datasets, we note that SH3BGRL mRNA level in turn is a marker for good prognosis (Fig. S16). When only focusing on SH3BGRL protein level, we consistently observe that SH3BGRL protein is robustly overexpressed in all types of breast cancers, endorsing it as a marker for poor prognosis, which is consistent with our protein analysis of SH3BGRL level with different methods. Currently, the high-throughput RNA-sequencing is widely approached to measure a gene’s expression [40], but the inconsistency of its mRNA abundance with protein level should not be negligible [41]. Thus the eventual protein amount of a functional protein would be the key factor to its physiological functions, and the combination of RNA-sequencing with the translatome would be a whole-view strategy for a specific gene expression.
Nevertheless, our results here thoroughly unveil this novel SH3BGRL-mediated STUB1-PFN1 axis in breast cancers along with the downstream events, which interprets the underlying mechanism of SH3BGRL upregulation on breast cancer progression and metastasis. Therefore, SH3BGRL would be a novel diagnostic and prognostic factor for breast cancers. Accordingly, targeting either SH3BGRL or the downstream activated signaling would have potent clinical value in the prevention of breast cancer progression and metastasis.
Materials and methods
Cell culture and reagents
Human breast cancer cell lines MCF-7, MDA-MB-453, MDA-MB-231, MDA-MB-468, ZR-75-1, SK-BR-3, BT-549, BT-474, and human embryonic kidney 293 cells (HEK293T) were purchased from the Shanghai Cell Bank of Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin with 5% CO2 at 37 °C in a humidified incubator. MCF-10A, a non-tumorigenic epithelial cell line, was purchased from the American Type Culture Collection (ATCC, Manassas, VA) and cultured in keratinocyte serum-free medium (KSFM) supplemented with0.1 ng/ml human recombinant epidermal growth factor and 20 μg/ml bovine pituitary extract (Invitrogen, Carlsbad, CA), All cell lines were verified by short-tandem-repeat typing and regularly examined for mycoplasma contamination by using the MycoAlert Mycoplasma Detection Kit (Lonza, Basel, Switzerland). For cell treatment, chloroquine diphosphate salts (CQ, Sigma-Aldrich) were dissolved in DMSO at a concentration of 50 μM and directly added to the cell culture medium, and an equal volume of DMSO alone was used as a negative control. Likewise, Cycloheximide (CHX, 50 μg/ml, Selleck, China) or MG132 (20 μM, Sigma) was also used to treat cells at the indicated time points.
Tissue specimens and Immunohistochemical (IHC) staining
Thirty-two pairs of paraffin-embedded and 10 pairs of freshly frozen breast tumor tissues, as well as 14 individual breast cancer tissues, were collected, according to Sun Yat-Sen University Health Regulation with informed consent. This study was further approved by the Research Ethics Committee. The fresh tissues were formalin-fixed, paraffin-embedded and all samples were sliced into 5 μm-thin sections with Leica BM 2135 microtome, and subsequently stained with anti-SH3BGRL (1:200 dilution, Sigma), anti-PFN1 (1:200 dilution, CST) antibodies. Slices were developed with Polink-1 HRP DAB Detection System (ZSGB-BIO) and Images were captured and confirmed by two independent professional pathologists under a microscope (Nikon, Tokyo, Japan). The color intensity of slides was divided into four grades (points) to score SH3BGRL and PFN1 expression levels for statistical analysis.
Plasmids and stable cell line construction
The MCF-7-SH3BGRL, MDA-MB-453-shSH3BGRL stable cell lines and HA-tagged SH3BGRL mutants were established as previously described (Table S6) [38]. To silence PFN1, PFN1 shRNA plasmids containing a pool of 4 target-specific shRNAs were purchased (Santa Cruz Biotechnologies, CA, USA) and transfected with Lipofectamine 3000 (Invitrogen, Waltham, USA) following the manufacturer’s instructions. PFN1 overexpression in MCF-7-SH3BGRL stable cell was established with pEnter-PFN1 plasmid transfection. Similarly, PFN1 knockdown in MDA-MB-453-shSH3BGRL cells was done with pAV-U6-GFP-shRNAs plasmids. After transfection, cells were selected with 500 μg/ml G418 (Sigma, St. Louis, USA) or 1 μg/ml Puromycin (Sigma, St. Louis, USA) for 4–5 weeks, and the single stable cell clone was picked under a fluorescence microscope (Nikon, Tokyo, Japan) to make the stable cell pools.
Wound healing assay
The indicated cells were sub-cultured on 6-well plates with DMEM containing 10% FBS. When cells reached about 85% confluence, the cell layer was scratched to form a gap on the bottom of a plate with a pipette tip. The cells were further incubated for 24 to 48 h to observe the gap healing. Phase-contrast images of the wound healing were obtained digitally using an inverted Olympus microscope with a ×10 objective lens at 0, 24, and 48 h after scratching. The length of the healed gap was measured and compared with that in the initial ones. All experiments were carried out in triplicates.
Transwell migration assay and Transwell matrix penetration assay
For the Transwell assay or Transwell matrix penetration assay, the indicated cells (1 × 104) were plated on the upper side of a polycarbonate Transwell filter with or without Matrigel in the upper chamber of the BioCoat™ invasion chambers (BD, Bedford, MA). After 48 h of incubation at 37 °C, the cells in the upper chamber were removed with cotton swabs, and the migrated and invaded cells on the lower membrane surface were fixed in 4% paraformaldehyde and stained with 0.5% crystal violet. The cells were counted (five random 100× fields per well) and expressed as the mean number of cells per field of view. All the experiments were repeated three times.
Anchorage-independent growth ability assay
One thousand cells were trypsinized and suspended in a 1.5 ml complete medium plus agar (0.35% w/v, Sigma-Aldrich). The agar cell mixture was plated on top of a bottom layer with 0.6% agar complete medium mixture. For 16 to 24 days, viable colonies that were larger than 0.1 mm were counted. The experiment was carried out for each cell line in triplicates.
Colony formation assay
Colony formation assay was initiated by seeding two thousand cells in 6-well plates. Cells were continued to grow in a fresh medium for 14 to 20 days. Colonies were fixed with 4% formaldehyde for 15 min, stained with 0.5% crystal violet for another 10 min, and counted under an inverted microscope. All experiments were repeated at least three times.
Experimental metastasis assays
BALB/c-nude mice (female, 4–5 weeks of age, 18–20 g) were purchased from Guangdong Medical Laboratory Animal Center. All mouse experiments were conducted under the Institutional Animal Care and Use Committee (IACUC) approved protocols by Sun Yat-sen University. Six nude mice in each group were injected intravenously via the tail vein with 6 × 106 MCF-7-Vector, MCF-7-SH3BGRL cells and 5 × 105 MDA-MB-453-shRNA-V, MDA-MB-453-shRNA1+2, MDA-MB-453-shRNA1+2-shPFN1 cells to examine the metastatic of the experimental cells in vivo. After 12 weeks, mice were executed and Lungs were photographed for gross morphology and scored for the number of metastatic nodules using the formula N/3, where N is the diameter of each visible nodule in millimeter.
Western blotting (WB) analysis
Equal quantities of cellular proteins, prepared in sample lysis buffer and heated for 10 min at 100 °C, were electrophoresed through a 10% SDS/polyacrylamide gel and transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA), followed by incubation with the indicated antibodies and visualized by chemiluminescent substrates (Millipore) and exposed on radiography films. Protein band intensity was quantified and analyzed with densitometry and Image J software. The membranes were incubated with Anti-SH3BGRL (1:1000; #HPA051248; Sigma-Aldrich), anti-β-actin (1:1000; #4967; CST), anti-PFN1 (1:1000;#137235;SantaCruz), anti-STUB1 (1:1000; #2080; CST), anti-HA (1:1000; #3724; CST), anti-PTEN (1:1000; #9559; CST), anti-phospho-Akt(Ser473) (1:1000; #9271; CST), anti-Akt (1:1000; #9272; CST), anti-phospho-GSK-3β (Ser9) (1:1000;#9323;CST), anti-GSK-3β (1:1000; #9315; CST), anti-β-catenin (1:1000; #9582; CST), anti-phospho-IKKα/β(Ser176/180) (1:1000; #2697; CST), anti-IKKα (1:1000; #2682; CST), anti-IKBα (1:1000; #4812; CST), anti-P65(1:1000; #8242; CST), anti-Histone H3 (1:1000; #4499; CST), anti-E-cadherin(1:1000; #3195; CST), anti-N-cadherin (1:1000; #4061; CST), anti-Slug (1:1000; #AP2053a; Abgent), anti-Vimentin (1:1000; #ab3978 Abcam), anti-MMP1(1:1000; #54376; CST), anti-MMP2(1:1000; #40994; CST), anti-MMP9 (1:1000; #13667; CST), anti-ubiquitin antibody (1:1000; #3933 S; CST), anti-RPL7a (1:1000; #2415; CST) and anti-c-Myc (1:1000; #9402; CST) for Western blots analyses, cells were lysed with lysing buffer (10 mM Tris-HCl, pH 7.4; 150 mM NaCl; 0.5 %NP-40; 1 mM EDTA; 0.2 mM PMSF and protease inhibitor cocktail).
Immunofluorescence microscopy
Cells seeded on coverslips were fixed with 4% paraformaldehyde (PFA) in 1× PBS for 25 min and permeabilized with 0.2% Triton X-100 (BIOSHARP) for another 10 min at room temperature. Cells were then blocked with 5% BSA in PBS for 1 h at room temperature and incubated with primary antibodies overnight at 4 °C. After washing 4 times with 1× PBS, cells were stained with Alexa-488, Alexa-594, and Alexa-647 conjugated secondary antibodies (Abcam, Cambridge, MA) at room temperature for 1 h. Cells were then washed with 1× PBS and incubated with DAPI for 5 min and mounted with anti-fade reagent and observed under LSM710 confocal microscope (Carl Zeiss AG).
Co-immunoprecipitation assay and MS analysis
Cells grown in 100-mm culture dishes and Breast tumor tissues were lysed using 800 mL of lysis buffer [25 mM HEPES (pH 7.4), 150 mM NaCl,1% NP-40, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF)]. After being maintained on ice for 10 min, the lysates were clarified by microcentrifugation at 10,000 rpm for 10 min. And then the supernatants were incubated with 25 μl of antibody or IgG-cross-linked protein G beads overnight at 4 °C.The beads were then washed four times with wash buffer [25 mM HEPES (pH 7.4), 150 mM NaCl, 0.5% NP-40,1 mM EDTA, 1 mM PMSF]. After removing all the liquid, the pelleted beads were resuspended in 30 μl of 1 M glycine (pH 3.0) and denatured for SDS-PAGE separation for either immunoblot or Coomassie blue or silver staining. The specific bands were cut out and subjected to MS analysis.
Ubiquitination and protein half-life analyses
To detect the ubiquitination levels of endogenous PFN1, MCF-7-GFP, MCF-7-SH3BGRL, MDA-MB-453-shRNA-V, and MDA-MB-453-shRNA1+2 cells were treated with 20 μM MG132 (Sigma) for 12 h to block proteasomal degradation. The cells were lysed and immunoprecipitated overnight at 4 °C with Protein G Magnetic Beads (Thermo) loaded or bound with anti-PFN1 (CST) antibodies according to the immunoprecipitation assay described above. The immunoprecipitated proteins were subjected to immunoblotting analysis with antibodies against Ubiquitin (CST). For protein half-life analysis, 50 μg/ml Cycloheximide (Sigma-Aldrich) was used to treat cells for 0, 4, 8, and 12 h, respectively, and protein levels were measured based on the protein band intensity from immunoblots.
Nuclear protein extraction assay
The nuclear protein extraction assay was performed using a Nuclear Extraction Kit (Thermo Fisher Scientific, MA). Briefly, cells (5 × 106) were collected, gently resuspended with 500 μl of 1× Hypotonic Buffer, and incubated on ice for 15 min. Then, 25 μl detergent buffer (10% NP40) was added and the homogenate was vortexed for 10 s. The homogenate was centrifuged for 10–15 min at 3000 rpm, 4 °C. The pellet was the nuclear fraction. The pellet was resuspended in 50 μl of complete Cell Extraction Buffer for 30 min on ice with vortexing at 10 min intervals. The sample was centrifuged for 30 min at 14,000 × g, 4 °C. The supernatant (nuclear fraction) was transferred to a clean microcentrifuge tube and store at −80 °C until use.
RNA extraction, cDNA synthesis, and real-time quantitative PCR
Total RNA was extracted from cells and fresh-frozen tumor specimens using the TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. Equal amounts of mRNA were used to generate cDNAs with a Revert Aid First Strand cDNA Synthesis Kit (Thermo), and the generated cDNAs were used for semi-quantitative analysis (RT-PCR) and real-time quantitative-PCR (qPCR). qPCR was performed using a 480 SYBR Green I Master kit (Roche) and a LightCycler 480 Detection System (Roche). The primer sequences used for real-time PCR are listed in Table S7. The data were normalized to β-actin and were expressed as relative mRNA levels. DNA band intensity was quantified and analyzed with densitometry and Image J software.
Ribosome mRNA profiling and sequencing
For polyribosome isolation [42], cells were incubated with 90 mg/ml cycloheximide (Sigma) for 10 min followed by trypsinization and harvest. Twenty million cells were resuspended in RSB (20 mM Tris-HCl [pH 7.4], 20 mM NaCl, 30 mM MgCl2, RNasin, 100 mg/ml Heparin, and 5 mg/ml cycloheximide). An equal volume of lysis buffer (1.2% Triton X-100, 1.2% deoxycholate) was added and then incubated on ice for 5 min. The nuclei and cell debris were removed by centrifugation for 3 min at 12,000 rpm. The supernatant was then diluted with an equal volume of dilution buffer (25 mM Tris-HCl [pH 7.4], 25 mM NaCl, 25 mM MgCl2, 0.05% Triton X-100, and 500 mg/ml heparin) and 0.4 ml of the extract was loaded onto 11.5 ml linear 10 to 50% sucrose gradients and centrifuged at 36,000 rpm for 2 h in a SW41 rotor (Beckman). Ten (1.2 ml each) fractions were collected with the BioComp piston gradient fractionator linked to an EM-1 UV Monitor (BioRad). The fractions were incubated in 1% SDS and proteinase K at 42 °C for 30 min. RNA was purified by Phenol Chloroform extraction followed by ethanol precipitation, and PFN1, STUB1, and β-actin mRNA were detected by semi-quantitative analysis. DNA band intensity was quantified and analyzed with densitometry and Image J software. Meanwhile, mRNA from each fraction was subjected to RNA-sequencing analysis. Gene fraction shift represents the value of the marked fraction number with maximum target RNA minus that of the control group from sucrose density-gradient centrifugation. The marked numbers are equivalent to the collected fractions after gradient centrifuge, for instance, fraction 1 represents the top (10%) of the gradient, fraction 8 represents the bottom (50%) of the gradient. β-actin, GAPDH, α-tubulin, and G6PD were used as negative controls for polyribosome profiling analysis.
Gene expression profiling and GSEA analysis
Analyses were performed with human breast cancer data from The Cancer Genome Atlas (TCGA) data https://portal.gdc.cancer.gov/projects/TCGA-BRCA), UALCAN (http://ualcan.path.uab.edu/analysis.html), cBioPortal (https://www.cbioportal.org/datasets), Kaplan–MeierPlotter (https://kmplot.com/analysis/) and Gene Expression Omnibus (GEO) datasets, including GSE26304, GSE45827, GSE15852, GSE28784, GSE5764, GSE21422, GSE12276, GSE2990, GSE26971, and GSE16293 in the NCBI database (https://www.ncbi.nlm.nih.gov/geo/). SH3BGRL expression was treated as a numeric variable. Then, a continuous-type CLS file of the SH3BGRL profile to phenotype labels in GSEA was applied. The metric for ranking genes in GSEA was set as “Pearson”, and the other parameters were set to their default values. GSEA was performed using GSEA 2.0.9 software (http://www.broadinstitute.org/gsea/).
Statistical analysis
Statistical analyses were performed using GraphPad Prism 7. For comparison of two groups, P-values were calculated with a Student’s t-test. For comparison of more than two groups, P-values were calculated using the ANOVA test. The relationships between SH3BGRL expression and the clinicopathological characteristics were tested using the v2 test. Survival curves were plotted with the Kaplan–Meier method and compared by the log-rank test. Survival data were evaluated using univariate and multivariate Cox regression analyses. Bivariate correlations between variables were calculated using Spearman’s rank correlation coefficients. P < 0.05 was considered statistically significant in all cases.
Change history
20 December 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41388-021-02129-1
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: Cancer J Clin. 2015;65:5–29.
Loibl S, Poortmans P, Morrow M, Denkert C, Curigliano G. Breast cancer. Lancet. 2021;S0140-6736:32381–3.
Waks AG, Winer EP. Breast cancer treatment: a review. JAMA. 2019;321:288–300.
Giles RH, Van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653;1–24. https://doi.org/10.1016/s0304-419x(03)00005-2.
Nguyen DX, Chiang AC, Zhang XH, Kim JY, Kris MG, Ladanyi M, et al. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009;138:51–62.
Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–6.
Pikarsky E, Porat RM, Stein I, et al. NF-kappaB functions as a tumor promoter in inflammation-associated cancer. Nature. 2004;431:461–6.
Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–62.
Mouneimne G, Hansen SD, Selfors LM, Petrak L, Hickey MM, Gallegos LL, et al. Differential remodeling of actin cytoskeleton architecture by profilin isoforms leads to distinct effects on cell migration and invasion. Cancer Cell. 2012;22:615–30.
Carlsson L, Nyström LE, Sundkvist I, Markey F, Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J Mol Biol. 1977;115:465–83.
Birbach A. Profilin, a multi-modal regulator of neuronal plasticity. BioEssays. 2008;30:994–1002.
Jockusch BM, Murk K, Rothkegel M. The profile of profilins. Rev Physiol Biochem Pharmacol. 2007;159:131–49.
Carlsson L, Nyström LE, Sundkvist I, Markey F, Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J Mol Biol. 1977;115:465–83.
Witke W. The role of profilin complexes in cell motility and other cellular processes. Trends cell Biol. 2004;14:461–9.
Yarmola EG, Bubb MR. Profilin: emerging concepts and lingering misconceptions. Trends biochemical Sci. 2006;31:197–205.
Hohenester E, Yurchenco PD. Laminins in basement membrane assembly. Cell Adhes Migr. 2013;7:56–63.
Bae YH, Ding Z, Das T, Wells A, Gertler F, Roy P. Profilin1 regulates PI(3,4)P2 and lamellipodin accumulation at the leading edge thus influencing motility of MDA-MB-231 cells. Proc Natl Acad Sci USA. 2010;107:21547–52.
Bae YH, Ding Z, Zou L, Wells A, Gertler F, Roy P. Loss of profilin-1 expression enhances breast cancer cell motility by Ena/VASP proteins. J Cell Physiol. 2009;219:354–64.
Gronborg M, Kristiansen TZ, Iwahori A, Chang R, Reddy R, Sato N, et al. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteom. 2006;5:157–71.
Janke J, Schluter K, Jandrig B, Theile M, Kolble K, Arnold W, et al. Suppression of tumorigeniciy in breast cancer cells by the microfilament protein profilin 1. J ExpMed. 2000;191:1675–85.
Zoidakis J, Makridakis M, Zerefos PG, Bitsika V, Esteban S, Frantzi M, et al. Profilin 1 is a potential biomarker for bladder cancer aggressiveness. Mol Cell Proteom. 2012;11:M111. 009449.
Wu N, Zhang W, Yang Y, Liang YL, Wang LY, Jin JW, et al. Profilin 1 obtained by proteomic analysis in all-trans retinoic acid-treated hepatocarcinoma cell lines is involved in inhibition of cell proliferation and migration. Proteomics. 2006;6:6095–106.
Das T, Bae YH, Wells A, Roy P. Profilin-1 overexpression upregulates PTEN and suppresses AKT activation in breast cancer cells. J Cell Physiol. 2009;218:436–43.
Zou L, Ding Z, Roy P. Profilin-1 overexpression inhibits proliferation of MDA-MB-231 breast cancer cells partly through p27kip1 upregulation. J Cell Physiol. 2010;223:623–9.
Ding Z, Joy M, Bhargava R, et al. Profilin-1 downregulation has contrasting effects on early vs late steps of breast cancer metastasis. Oncogene. 2014;33:2065–74.
Choi YN, Lee SK, Seo TW, Lee JS, Yoo SJ. C-Terminus of Hsc70-interacting protein regulates profilin1 and breast cancer cell migration. Biochem. Biophys Res Commun. 2014;446:1060–6.
Egeo A, Mazzocco M, Arrigo P, Vidal-Taboada JM, Oliva R, Pirola B, et al. Identification and characterization of a new human gene encoding a small protein with high homology to the proline-rich region of the SH3BGR gene. Biochem Biophys Res Commun. 1998;247:302.
Yin L, Xiang Y, Zhu DY, et al. Crystal structure of human SH3BGRL protein: the first structure of the human SH3BGR family representing a novel class of thioredoxin fold proteins. Proteins. 2005;61:213–6.
Cesareni G, Panni S, Nardelli G, et al. Can we infer peptide recognition specificity mediated by SH3 domains? Febs Lett. 2002;513:38–44.
Tong F, Zhang M, Guo X, et al. Expression patterns of SH3BGR family members in zebrafish development. Dev genes evolution. 2016;226:287–95.
Werner CJ, Heyny-von Haussen R, Mall G, Wolf S. Proteome analysis of human substantia nigra in Parkinson’s disease. Proteome Sci. 2008;6:8.
Wang H, Liu B, Al-Aidaroos AQ, Shi H, Li L, Guo K, et al. Dual-faced SH3BGRL: oncogenic in mice, tumor suppressive in humans. Oncogene. 2016;35:3303–13.
Majid SM, Liss AS, You M, Bose HR. The suppression of SH3BGRL is important for v-Rel-mediated transformation. Oncogene. 2005;25:756–68.
Xu L, Zhang M, Li H, Guan W, Liu B, Liu F, et al. SH3BGRL as a novel prognostic biomarker is down-regulated in acute myeloid leukemia. Leuk Lymphoma. 2018;59:918–30.
van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6.
Abba MC, Hu Y, Sun H, et al. Gene expression signature of estrogen receptor alpha status in breast cancer. BMC Genomics. 2005;6:37.
Muniz Lino MA, Palacios-Rodriguez Y, Rodriguez-Cuevas S, et al. Comparative proteomic profiling of triple-negative breast cancer reveals that up-regulation of RhoGDI-2 is associated to the inhibition of caspase 3 and caspase 9. J Proteom. 2014;111:198–211.
Li H, Zhang M, Wei Y, Haider F, Lin Y, Guan W, et al. SH3BGRL confers innate drug resistance in breast cancer by stabilizing HER2 activation on cell membrane. J Exp Clin cancer Res. 2020;39:81.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, et al. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011;7:548.
Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–73.
Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63.
Funding
This work was supported by National Science Foundation of China (No. 81672704), the Guangdong Provincial Key Laboratory of Precision Medicine and Clinical Translation Research of Hakka Population (2018B030322003KF02), and Natural Science Foundation of Guangdong Province (No. 2021A1515010999) to WH.
Author information
Authors and Affiliations
Contributions
S Zhang, S Yang, and H Wang conceived the research and designed the experiments, S Zhang performed the most experiments and data analyses; X Guo collect samples and performed IHC staining with analysis, X Liu performed colony formation assay, XZ Zhong performed Immunoblots of genes in breast cancer tissues; S Zhang, H Wang and S Yang wrote the draft. All authors approved the final manuscript submission.
Corresponding authors
Ethics declarations
Competing interests
Drs. Haihe Wang and Shulan Yang, researchers on the study team, are co-inventors on Patent 201710512823.6 (Sun Yat-sen University) which deals with the application of SH3BGRL as a diagnostic and therapy target. No other authors have competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, S., Guo, X., Liu, X. et al. Adaptor SH3BGRL promotes breast cancer metastasis through PFN1 degradation by translational STUB1 upregulation. Oncogene 40, 5677–5690 (2021). https://doi.org/10.1038/s41388-021-01970-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01970-8
- Springer Nature Limited
This article is cited by
-
Biomarker discovery with quantum neural networks: a case-study in CTLA4-activation pathways
BMC Bioinformatics (2024)
-
Quantitative proteomics reveals cellular responses to individual mAb expression and tunicamycin in CHO cells
Applied Microbiology and Biotechnology (2024)