Abstract
Tumor angiogenesis plays vital roles in tumorigenesis and development; regulatory mechanism of angiogenesis is still not been fully elucidated. NSD2, a histone methyltransferase catalyzing di-methylation of histone H3 at lysine 36, has been proved a critical molecule in proliferation, metastasis, and tumorigenesis. But its role in tumor angiogenesis remains unknown. Here we demonstrated that NSD2 promoted tumor angiogenesis in vitro and in vivo. Furthermore, we confirmed that the angiogenic function of NSD2 was mediated by STAT3. Momentously, we found that NSD2 promoted the methylation and activation of STAT3. In addition, mass spectrometry and site-directed mutagenesis assays revealed that NSD2 methylated STAT3 at lysine 163 (K163). Meanwhile, K to R mutant at K163 of STAT3 attenuated the activation and angiogenic function of STAT3. Taken together, we conclude that methylation of STAT3 catalyzed by NSD2 promotes the activation of STAT3 pathway and enhances the ability of tumor angiogenesis. Our findings investigate a NSD2-dependent methylation–phosphorylation regulation pattern of STAT3 and reveal that NSD2/STAT3/VEGFA axis might be a potential target for tumor therapy.
Similar content being viewed by others
Introduction
Angiogenesis is required in both tumor growth and metastasis and is considered as one of the ten hallmarks of cancer [1]. Because of the rapid proliferation signature of cancer, newborn blood vessels arise from pre-existing microvasculature to meet the demand of tumor progression [2]. Normally, angiogenesis is strictly under the control of angiogenic and anti-angiogenic factors. However, in some conditions such as inflammation, wound healing, and tumorigenesis, the balance is disrupted and blood vessel formation is promoted [3]. Among the most common factors influencing angiogenesis including VEGF, EGF, HGF, b-FGF, etc., VEGF seems to be the most important molecule [4,5,6]. VEGF antagonists, such as bevacizumab, significantly attenuate angiogenesis and have been used in the treatment of multiple carcinomas [7, 8]. However, similar to most targeted drugs, VEGF antagonists also encounter both primary and secondary resistance, which is associated with the failure of VEGF antagonists [9]. Therefore, an in-depth exploration of the mechanism of tumor angiogenesis as well as its regulation is crucial and may be of great help in guiding the clinical targeted therapy.
Signal transducers and activators of transcription (STATs) are a vital protein family of transcription factors widely studied in cancer research. STAT3 was identified at first as an acute phase response factor activated by the cytokine interleukin-6 (IL-6) [10, 11]. STAT3 plays a key role in several biological processes in carcinoma, including angiogenesis, proliferation, resisting apoptosis, evading the immune response, and so on, and nearly all the hallmarks of cancer proposed by Weinberg are under the regulation of STAT3 [12, 13]. In recent years, non-classical post-translational modifications, such as methylation, acetylation, ubiquitination, and SUMOylation, have gained considerable attention in recent years [14]. STAT3 is reported to be methylated by SET9 and EZH2 at different residue sites resulting in different transcription activating status of STAT3 [15,16,17,18]. Acetylation of STAT3 catalyzed by acetyltransferase p300 is reported to be essential in the formation of STAT3 homodimer, which is thought to play an important transcription factor function [19].
NSD2 (also known as WHSC1 and MMSET) is a histone methyltransferase catalyzing di-methylation at lysine 36 of histone 3 (H3K36me2) and is associated with active transcription of a series of genes [20]. NSD2 plays a significant role in cell development, and NSD2 haploinsufficiency is associated with Wolf–Hirschhorn syndrome, which is a multiple malformation syndrome [21]. Meanwhile, NSD2 knocked-out mice exhibited embryonic development disorder [22]. In recent years, the function of NSD2 in all cancer types has been gradually revealed. Multiple myeloma (MM), one of the most fatal hematologic malignancies, is often characterized by chromosomal translocation [23]. Among them, the t (4;14) translocation, which is one of the major types of chromosomal translocation in MM, is associated with the overexpression of NSD2 and leads to a poor prognosis [24,25,26]. NSD2 is overexpressed in invasive prostate cancer, especially in metastases and proven to be related to the unfavorable prognosis in prostate malignant patients [26]. NSD2 can also directly methylate PTEN and enhance the DNA damage repair ability in colorectal cancer, and as a result, enhance the resistance of cancer cells to chemotherapy [27]. Similar results have been reported in other solid carcinomas such as esophagus carcinoma, stomach carcinoma, hepatocellular carcinoma, lung cancer, corpus uteri malignancy, etc. [28]. However, the function of NSD2 in angiogenesis remains to be explored.
Here, we demonstrated that NSD2 interacted with STAT3 enhancing its methylation and phosphorylation and resulting in transcription activation of VEGFA and promotion of angiogenesis. Moreover, this study identified Lysine 163 (K163) of STAT3 as the NSD2-dependent methylation site, and this residue could be of significance in the activation process of STAT3 signaling pathway. The findings indicate that NSD2 plays a crucial part in tumor angiogenesis and could be a potential therapeutic targeted against tumor angiogenesis.
Results
NSD2 is overexpressed in altered carcinomas
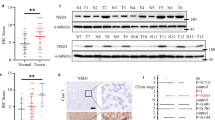
To clarify the role of NSD2 in tumorigenesis and progression, we have analyzed the expression level of NSD2 in adjacent and carcinoma tissues in TCGA database using UALCAN analyses tool [29]. As shown in Fig. 1a–d, the expression of NSD2 was upregulated in various forms of cancer. Next, we have collected 12 pairs of colon cancer tissues and adjacent tissues for further proof. A real-time qPCR assay was performed to detect the mRNA level, and the results showed that the mRNA expression level of NSD2 in carcinoma tissues was higher than that in para-carcinoma specimens (Fig. 1e). To investigate further, a western blot assay was carried out with a consistent result obtained (Fig. 1f, g). Immunohistochemistry (IHC) staining results also showed that NSD2 expression was elevated in tumor tissues (Fig. 1h, i).
a–d NSD2 expression level in TCGA database in various forms of cancer using UALCAN analyses tool (http://ualcan.path.uab.edu/). e Expression level of NSD2 mRNA in carcinoma and para-carcinoma tissues. f, g Expression level of NSD2 protein in tumor and adjacent specimens. h, i NSD2 staining of paired clinical specimens and the statistic quantification results. Results are presented as mean ± SD, **p < 0.01.
Inhibition of NSD2 moderates tumor-induced angiogenesis in vivo and in vitro
Now that, we have convinced that NSD2 was upregulated in tumor tissues. However, until now, little has been revealed about the role of NSD2 in tumor angiogenesis. In this study, we performed a GSEA analysis using public datasets and found that the upregulation of NSD2 was related to angiogenesis (Supplementary Fig. S1a). And then, we constructed stable NSD2 shRNA-NSD2(shNSD2)-expressing SW48 and SW480 cell lines using lentiviruses containing two altered shRNA sequences against NSD2 (Supplementary Fig. S1b,c). A mouse xenograft model was set up using indicated cell lines. A tumor growth curve was drawn, which indicated that tumors in the shNSD2 group grew more slowly (Fig. 2a). After 28 days, mice were sacrificed and the xenografts were gathered and weighed. Xenografts formed by shNSD2 cells were smaller and pallid in appearance meanwhile lighter in weight (Fig. 2b, c). Next, we detected the mRNA expression level of several angiogenic factors such as VEGFA, PDGFB, etc. with the intervention of NSD2. Surprisingly, loss of NSD2 was found to inhibit the expression of VEGFA (Supplementary Fig. S1d). Consistent with the above results, IHC staining was performed and revealed that knockdown of NSD2 inhibited the expression of VEGFA (Fig. 2d). IHC staining results of CD31 revealed that tumor vessel density decreased with NSD2 intervention (Fig. 2e). Besides, the xenografts’ protein detection results showed that the loss of NSD2 attenuated the expression of VEGFA (Fig. 2f). Next, we performed some experiments to explore the angiogenic function of NSD2 in vitro. The western blot results showed that VEGFA protein expression decreased with the intervention of NSD2 (Supplementary Fig. S1e). As only secretory VEGFA could promote angiogenesis in the tumor microenvironment, the enzyme-linked immunosorbent assay (ELISA) revealed that NSD2 knockdown decreased the levels of secretory VEGFA (Fig. 2g, h). A tube-formation assay was performed using HUVECs incubated with indicated condition mediums (CMs). Less tube-formation was observed in the shNSD2 groups (Fig. 2i, j and Supplementary Fig. S1f). Besides, migration and proliferation ability of HUVECs incubated with altered CMs was evaluated by Cell Counting Kit-8 (CCK8) and transwell assays respectively (Supplementary Fig. S1g–k). VEGFA mRNA expression level was also attenuated upon the inhibition of NSD2 (Supplementary Fig. S2a). To furthermore investigate the function of NSD2 upon angiogenesis in vivo, we have also performed in vivo Matrigel plug assays. Some researchers performed this assay with the addition of cancer cells while others not. We thought that both methods had advantages and disadvantages. The former might avoid the effect of intra-tumor VEGFA, while the latter could better simulate the tumor microenvironment. Through deliberation, we decided to have a try using the two methods to complement each other. We have performed in vivo Matrigel plug assay in two methods. FITC-dextran was injected through the caudal vein before the mice were sacrificed. After the plugs were obtained, autofluorescence were observed through the frozen section (Supplementary Fig. S2b, c), Hemoglobin of these plugs was also measured to evaluate the angiogenesis (Supplementary Fig. S2d). Those plugs with cancer cells were fixed and stained with CD31 antibody to observe the angiogenesis (Supplementary Fig. S2e–g). Taken together, inhibition of NSD2 moderates tumor-induced angiogenesis in vivo and in vitro.
a The growth curve of mice xenografts formed by shNC or shNSD2 SW480 cells (n = 6). b Mice xenografts formed by SW480 with or without NSD2 intervention. c Tumor weights after sacrificing the mice (n = 6). d IHC staining of xenografts using antibodies against NSD2, VEGFA, and CD31. e Microvascular densities of the tumors in the three groups as quantified by CD31 (n = 6).f Western blot results of xenografts homogenates, g, h Detection of VEGFA concentration of CMs from indicated cells by ELISA (n = 3). i, j HUVECs tube-formation assay incubated with CMs from indicated cells (n = 3). Results are presented as mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
NSD2 influences the activation of the STAT3 signaling pathway
INCB054329, a bromodomain and extra-terminal domain inhibitor, could be regarded as a potent inhibitor of NSD2 and was reported to suppress the JAK-STAT pathway [30]. Meanwhile, GSEA analysis was performed in two different GEO datasets and results showed that the upregulation of NSD2 was significantly associated with the IL-6/STAT3 pathway (Fig. 3a, b). Therefore, we put forward the hypothesis that NSD2 activates the STAT3 signaling pathway to upregulate the expression of VEGFA. To test this hypothesis, we have performed western blot assays, and the results revealed that inhibition of NSD2 could impair the phosphorylation of STAT3, while overexpression of NSD2 led to an increase in STAT3 phosphorylation (Fig. 3c–f). The nuclear translocation extent of STAT3 can reflect its functional status. Through a nuclear and cytosol protein separation assay, we could conclude that NSD2 promoted the nuclear translocation of STAT3 (Fig. 3g–j). Consistently, an immunofluorescence assay showed that the aggregation of STAT3 in the nucleus decreased with NSD2 intervention (Fig. 3k). Taken together, NSD2 influenced the activation of the STAT3 signaling pathway.
a, b GSEA analysis was performed in two different GEO datasets to reveal the association between the expression of NSD2 and the activation of IL-6/JAK/STAT3 pathway. c, d Western blot to detect the phosphorylation status changes of STAT3 upon the intervention of NSD2 in SW480 and SW48 cell lines. e, f Western blot to detect the STAT3 phosphorylation changes with upregulation of NSD2 in SW480 and LoVo cell lines. g–j Nuclear and plasma proteins were separated and detected by western blot with intervention or overexpression of NSD2 in SW480 and SW48 cell lines. k Immunofluorescence assay was performed to reflect the nuclear-location status changes of STAT3 with the intervention of NSD2.
The angiogenic function of NSD2 is mediated by STAT3 signaling pathway
STAT3 signaling pathway plays a vital role in the regulation of VEGFA secretion and angiogenesis. In this study, we proved that NSD2 promoted angiogenesis, VEGFA expression, and STAT3 activation. We then determined if the function of NSD2 was dependent on the STAT3 signaling. A western blot assay was performed. and increased STAT3 phosphorylation and upregulation of VEGFA were reversed by STAT3 knockdown (Fig. 4a, b). ELISA results showed that the concentration of VEGFA in CM was elevated in NSD2-overexpressing cells but was reversed by downregulating STAT3 expression (Fig. 4c, d). Furthermore, the angiogenic effect of CMs from cells expressing NSD2 was enhanced, while the angiogenic function of NSD2 was reversed by attenuating STAT3 expression (Fig. 4e–g). The migration ability of HUVECs incubated with indicated CMs was measured by stretch and transwell assay, respectively. Consistently, HUVECs incubated with CMs from NSD2-overexpressing cells exhibited better migratory ability and this effect was inhibited by STAT3 intervention (Supplementary Fig. S3a–g). The CCK8 results showed that HUVECs incubated with CMs from NSD2-overexpressing cells grew faster, but these results were reversed by STAT3 intervention (Supplementary Fig. S3h, i). Therefore, we conclude that the angiogenic function of NSD2 is mediated by STAT3 signaling pathway.
a, b Western blot evaluating whether STAT3 was dependent in the NSD2-mediated VEGFA expression promotion by overexpressing NSD2 and silencing STAT3 at the same time. c, d VEGFA concentration of CMs from indicated cells detected by ELISA (n = 3). e–g HUVECs tube-formation assay incubated with CMs from indicated cells (n = 3). Results are presented as mean ± SD, **p < 0.01, ***p < 0.001, ****p < 0.0001.
STAT3 inhibitor STATTIC abolishes the angiogenic function of NSD2 in vivo
The transcription factor STAT3 plays an important role in tumorigenesis and tumor development; hence, targeted drugs against the STAT3 signaling pathway have been widely studied. Here, STATTIC, which is considered an effective STAT3 inhibitor, was selected to observe the therapeutic effect in NSD2-overexpressing carcinomas. Surprisingly, NSD2 was found to upregulate the phosphorylation of STAT3 and the expression of VEGFA, and these effects were reversed by STATTIC (Fig. 5a). Meanwhile, as STAT3 is a critical transcriptional factor, it is vital to make certain the influence of NSD2 upon the mRNA expression level of VEGFA. The quantitative real-time PCR (RT-qPCR) results showed that NSD2 could upregulate the mRNA expression level of VEGFA while blockade STAT3 signaling pathway using siRNA against STAT3 or STAT3 inhibitor, the VEGFA upregulation effect was reversed (Supplementary Fig. S4a, b). Next, a xenograft model was set up using LoVo cell line. A growth curve was drawn to determine the growth status of the xenograft, which indicated that upregulation of NSD2 promoted tumor proliferation but this promotion effect was blocked by the application of STATTIC (Fig. 5b). After the mice were sacrificed, the xenografts were collected and the tumor volume and weight determined. Xenografts formed by NSD2-overexpressing cells were bigger and heavier, and the effects were reversed by STATTIC (Fig. 5c, d). Furthermore, IHC staining was performed, and the results showed that phosphorylation of STAT3, expression of VEGFA, and CD31-positive cell counts were elevated when NSD2 was overexpressed; however, these were reversed by STATTIC (Fig. 5e). According to CD31 staining status, microvessel density was elevated with NSD2 overexpression and was blocked with the addition of STATTIC (Fig. 5f). Besides, the xenograft homogenate was obtained and a western blot assay was performed (Fig. 5g). To better investigate the NSD2/STAT3 signaling axis in vivo, We have performed Matrigel plug assays. Autofluorescence through frozen section and hemoglobin measurement indicated an increased blood perfusion with NSD2 overexpression, while STAT3 blockade could obviously reverse this effect (Supplementary Fig. S4c–e). For those plugs with cancer cells, the vessels on the surfaces of the plugs visible to the naked eyes, the CD31 staining condition also revealed that NSD2 promoted angiogenesis in a STAT3-dependent manner (Supplementary Fig. S4f–h). Taken together, we conclude that NSD2 promotes tumor proliferation and angiogenesis in vivo, while these effects are blocked by STAT3 inhibitor STATTIC.
a STAT3 inhibitor STATTIC (10 μM, 48 h) was added into LoVo cells with or without NSD2 overexpression, indicated protein expression levels were detected by western blot. b Xenograft model was set up using vesicle or STATTIC (3.75 mg/kg, every 2 days) by intratumoral injection after xenografts were touchable, Tumor volumes were measured after every 4 days and a growth curve drawn (n = 6). c The images of tumors from LoVo xenograft mice with altered treatments (n = 6). d Indicated tumor weight was measured (n = 6). e, f IHC staining of xenografts using antibodies against NSD2, p-STAT3, VEGFA, and CD31, according to the staining status of CD31, MVD was statistically analyzed (n = 6). g Tissue homogenates were obtained from these xenografts and a western blot assay was performed. Results are presented as mean ± SD, **p < 0.01, ***p < 0.001, ****p < 0.0001.
NSD2 directly interacts with and methylates STAT3 to activate STAT3 signaling pathway
To further investigate the mechanism by which NSD2 regulates the phosphorylation of STAT3, we performed an immunoprecipitation (IP) assay and explored the physical interaction between NSD2 and STAT3 endogenously (Fig. 6a). Furthermore, we constructed the overexpressing plasmid HA-NSD2 and Flag-STAT3, and both were co-transfected in the HEK293T cell line. An IP assay was performed to verify the exogenous interaction, and a consistent result was obtained (Fig. 6b). To further support this result, immunofluorescence was performed to observe the co-localization status of NSD2 and STAT3 (Fig. 6c). NSD2 is a histone methyltransferase, hence we put forward our hypothesis that NSD2 could methylate STAT3 protein. To confirm this hypothesis, through a quantified immunoprecipitation (qIP) assay, we concluded that downregulation of NSD2 resulted in the loss of methylation of STAT3 protein, while overexpression of NSD2 was associated with the hyper-methylation of STAT3 protein (Fig. 6d, e). We further tried to explain the relationship between STAT3 methylation and phosphorylation. DZNep, an inhibitor reported to be global histone methylation rather than an EZH2 inhibitor, was used to confirm the influence of STAT3 methylation upon phosphorylation. Surprisingly, we found that NSD2 upregulated both the phosphorylation and methylation of STAT3, while DZNep successfully blocked the methylation of STAT3 protein and resulted in a decrease in STAT3 phosphorylation (Fig. 6f). To find out the specific binding domain between NSD2 and STAT3 and the NSD2-dependent methylation residue of STAT3, we constructed three fragment plasmids of STAT3 based on the natural structural domain (Fig. 6g). Next, through overexpressing these plasmids in HEK293T, the IP results showed that Fragment1 (AA1-320) of STAT3 interacted with NSD2 (Fig. 6h). These finds indicate that NSD2 interacts with and methylates STAT3 to activate STAT3 signaling pathway.
a In SW480 cell line, IP assays using antibodies against NSD2 and STAT3 respectively were performed. b In HEK293T cell line, after co-transfection of HA-NSD2 and Flag-STAT3, IP assays were performed to enrich HA-NSD2 and Flag-STAT3. c Immunofluorescence assay was performed to observe the co-localization conditions of STAT3 and NSD2 in SW480 cell line, plot profile qualitative analysis was used to describe the co-localization status. d, e qIP assays and western blot to detect the methylation levels of STAT3 with NSD2 upregulation or attenuation. f Using global methylation inhibitor DZNep, a qIP assay was performed to elucidate the methylation and phosphorylation status of STAT3 with NSD2 overexpression. g The full-length STAT3 was divided into three parts and three fragment plasmids were constructed. h Using three Flag-tagged fragment plasmids of STAT3, an IP assay was performed.
NSD2 methylates STAT3 at K163
In this study, we just proved that NSD2 directly interacted with and methylated STAT3. To find out the NSD2-dependent STAT3 methylation site, IP was used to enrich the STAT3 protein for mass spectrometry (Fig. 7a). We identified 14 lysine residues in our spectrometry result (Supplementary Table S2). Among them, some residues such as K49 and K140 that were already reported to be methylated residues were also observed in our mass spectrometry results. Therefore, by analyzing the mass spectrometry result and searching for the lysing residues located in the first fragment of the STAT3 protein and excluding the lysine residues that were already reported to be catalyzed by other enzymes, we have found two possible lysine residues, K163 and K244 as NSD2-dependent methylation sites (Fig. 7b). A secondary mass spectrometry result is shown to prove the K163 methylation status of STAT3 (Fig. 7c). Next, we investigated the target site of the NSD2-dependent STAT3 methylation residue between the two possible sites. We constructed K to R mutant plasmid at K163 and K244, respectively, by overexpressing or downregulating NSD2, and IP used to detect the methylation status of STAT3. The results showed that at first both site mutations attenuated the methylation level of STAT3. The changes in NSD2 expressing level influenced the methylation status of K244R mutant rather than K163R mutant (Fig. 7d, e). These results implied that NSD2 influenced the methylation of STAT3 at lysine 163. We also analyzed the conservative property between different species and concluded that 163 of STAT3 is conservative in most species (Fig. 7f). In conclusion, we prove that NSD2 directly interacts with STAT3 and methylates the latter at K163 residue.
a Using IP assay for the enrichment of STAT3 protein, staining with Coomassie bright blue and verification by mass spectrometry. b Screening strategy for predicting the possible methylation residues of STAT3 that is dependent on NSD2 function. c Secondary mass spectrometry result of one possible methylation residue. d, e qIP assays were performed by detecting the methylation changes of wild-type STAT3, K163R, and K244R mutants, respectively, with NSD2 overexpression or intervention. f The STAT3 K163 site amino acid in different species. g Schematic diagram of our hypothesis about this project.
Inhibition of methylation at K163 of STAT3 partially abolishes the angiogenic function of STAT3 in vitro and in vivo
Now that, we have proven that NSD2 methylated STAT3 at K163 and this residue might take part in the activation of STAT3 signaling pathway. Next, we need to identify function of methylation at K163 of STAT3. At first, we used lentivirus containing shRNA against STAT3 3’UTR to set up a STAT3 endogenously knockdown cell line and rescued them with wild-type STAT3 and STAT3 with K to R mutant at K163 (K163R). Here, K to R mutant was often thought to be a methylation defective mutant. Western blot results showed that K163R attenuated the STAT3 activation and VEGFA expression partially compared with wild-type STAT3 (Fig. 8a, b). Meanwhile, a nuclear protein separation assay was performed and the results indicated that the methylation defective status at K163 resulted in the less translocation to the nuclear (Supplementary Fig. S5a). CMs from indicated cells were gathered, and VEGFA concentration was measured by ELISA, and the results showed that compared with wild-type STAT3, STAT3 K163R decreased the secretory VEGFA in CMs (Fig. 8c, d). The HUVECs tube-formation assay and CCK8 assay results indicated that STAT3 K163 partially attenuated angiogenic function of STAT3 (Fig. 8e–g and Supplementary Fig. S5b, c). Next, we set up mice xenograft model using indicated SW480 cells. The tumor volume was measured in vivo, and a growth curve was drawn. The results showed that K to R mutation at K163 inhibited tumor proliferation (Fig. 8h). After the mice were sacrificed, xenografts were gathered and weighed. The results indicated that STAT3 K163R mutant inhibited tumor growth compared with wild-type STAT3 (Fig. 8i, j). IHC staining results also showed that defective methylation status at K163 attenuated both phosphorylation of STAT3 and expression of VEGFA in vivo (Supplementary Fig. S5d). Microvessel density quantified by CD31 staining results also indicated an inhibition effect on angiogenesis of STAT3 K163R (Supplementary Fig. S5e). Besides, the xenograft homogenate was obtained and a western blot assay was performed, and similar results with IHC were conducted (Supplementary Fig. S5f). Taken together, we conclude that methylation defective mutant K163R of STAT3 partially inhibits angiogenic function of STAT3 in vitro and in vivo.
a, b Using lentivirus containing shRNA against STAT3 3’UTR to knockdown endogenous STAT3 expression level and then to rescue with lentivirus containing wild-type STAT3 or methylation defective mutant K163R, western blot assays were performed. c, d VEGFA concentration of CMs from indicated cells detected by ELISA (n = 3). e–g HUVECs tube-formation assay incubated with CMs from indicated cells (n = 3). h–j SW480 cells were used lentivirus containing shRNA against STAT3 3’UTR to knockdown endogenous STAT3 expression level and rescued with indicated lentivirus, and then they were subcutaneous injected to set up a mice xenograft model. h A growth curve was drawn here (n = 6). i Xenografts were gathered and the image was shown here (n = 6). j These xenografts were weighed (n = 6). Results are presented as mean ± SD, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Discussion
In this study, we confirmed that NSD2 is associated with STAT3 to methylate and change its phosphorylation level. Moreover, we identified K163 as an NSD2-dependent methylation site of STAT3, which is also responsible forSTAT3 phosphorylation. Both EZH2 and SET9 are reported to have the ability to methylate STAT3 at different amino residues upon IL-6 stimulation [15, 16]. In this study, NSD2 was found to also take part in this process. Therefore, this kind of methylation–phosphorylation crosstalk may be a common phenomenon in the regulation of altered types of signaling pathways, which guarantees the precise regulation of various biological behaviors.
Post-translational modification is of great significance in regulating protein activities. As one of post-translational modifications, methylation has been greatly researched in recent years. The methylation of non-histone protein may be involved in the regulation of the signaling pathway by influencing protein stability, affecting the activities of transcription factors, influencing other post-translational modifications, altering the protein–protein interaction, and so on [31,32,33]. For example, AKT was reported to be methylated by SETDB1 at K64 and K140/142, respectively, promoting the activity of PI3K/AKT signaling pathway [34, 35]. Even more important, mithramycin, an anti-neoplastic antibiotic, is reported to effectively inhibit SETDB1 expression and function, and overcome the resistance of KRAS mutant CRC cells to cetuximab both in vitro and in vivo [36]. NSD2 is a histone methyltransferase responsible for di-methylation of histone 3 at lysine 36 (H3K36me2). NSD2 possesses the capacity to methylate histones, hence could have the ability to methylate non-histone proteins as well. A previous study elucidated that NSD2 methylates PTEN at K349, promoting its phosphorylation by ATM under the DNA double-strand breaks condition and promotes DNA damage repair [27]. In another study, NSD2 was found to methylate Aurora kinase A (AURKA) at K14 and K117 with changes in its kinase activity mediating cell proliferation via the p53 signal pathway [37]. In our study, we identified STAT3 as an original non-histone protein substrate of NSD2 and elucidated the function of a specific methylation amino residue. These findings provided theoretical basis for the usage of potential NSD2 inhibitor and the exploitation of inhibitory peptide to block the interaction between NSD2 and STAT3 and the methylation of specific residue.
Mounting evidence suggests that NSD2 plays a key role in tumorigenesis and tumor progression [38,39,40,41,42,43,44]. However, the role of NSD2 in tumor angiogenesis is in distinct. In this study, loss of NSD2 was found to impair cancer angiogenesis both in vitro and in vivo. Angiogenesis is a process controlled by angiogenic and anti-angiogenic factors [3]. Here, we revealed that VEGFA rather than EGF, FGF, HGF, and other factors took part in the NSD2-mediated tumor angiogenesis.
In conclusion, this study has elucidated the function of NSD2 in regulating VEGFA-mediated tumor angiogenesis. Mechanistically, we have identified an original methylation amino residue of STAT3 that is dependent on NSD2. Besides, the non-histone protein methylation function of NSD2 in regulating STAT3 signal pathway transduction is reported. These findings provide evidence that NSD2 antagonists or NSD2–STAT3 interfering peptides can be used as therapeutic targets in targeting angiogenesis.
Methods
Clinical samples collecting and immunohistochemistry (IHC) assay
Paired carcinoma and adjacent specimens were gathered from patients diagnosed with colon cancer. Four-millimeter thick section slides were stained with antibodies against NSD2, STAT3, p-STAT3, and CD31. Immunoreactive score was used to quantify IHC staining results. The percentage of positively stained was scored as follows, 1 (<10%), 2 (10–50%), 3 (50–75%), and 4 (>75%). The intensity was defined as 0–3: 0, no staining; 1, weak staining; 2, moderate staining; 3, strong staining. The staining index was obtained by multiplying these two scores. For analyses, scores between 8 and 12 were considered the high expression while scores between 0 and 6 were regarded as low expression.
Cell Counting Kit-8 (CCK8)
The cell line HUVECs (2.5 × 103/100 μl) were seeded in 96-well plates and treated with CMs from altered disposed cell lines. At indicated times, 10 μl of CCK8 solution was mixed with the medium. After incubation for 1.5 h, the cell proliferative rate was detected by measuring the absorbance at 450 nm.
Transwell assay
The cell line HUVECs (5 × 104/200 μl) were resuspended using serum-free medium. The cells were then seeded in the upper chamber, and 650 μl of CM was put into the lower chamber. After incubation for 16 h and staining with crystal violet solution, cells attached to the back side of upper chamber were observed using light microscope.
Tube-formation assay
Matrigel with reduced growth factor was added into 96-microwell plates 50 μl per well and placed at 37 °C for 30 min for solidification. Then, 4 × 104 HUVECs were resuspended with CMs from different cells and added in the microwells. They were incubated for 8 h and then observed using an inverted microscope. Meanwhile number of branch points per field was counted.
Immunofluorescence assay
Cells were seeded on coverslips. The cells were fixed with neutral tissue fixator. The fixed cells were permeabilized with 0.1% Triton X-100 for 2 min, blocked with 1% bovine serum albumin for 1 h, and treated with the indicated primary antibodies at 4 °C overnight. The cells were then treated with secondary antibodies and incubated for 1 h and DAPI for 15 min at room temperature. Visualization of the cells was done using a confocal laser scanning microscope (Olympus FLUOVIEW FV1000).
Enzyme-linked immunosorbent assay (ELISA)
Secretory VEGFA levels were measured using ELISA (#RK00023, Abclonal Technology, Wuhan, China) kits according to the protocols.
Western blot
Protein samples were electrophoretically separated and transferred to PVDF membranes. Molecular weight-specific bands were incubated with specific primary antibodies at 4 °C overnight and were then incubated with HRP-conjugated secondary antibodies for 2 h at room temperature. The bands were visualized with ECL reagents (Thermo Fisher, MA, USA).
Immunoprecipitation (IP) and quantified immunoprecipitation (qIP) assay
Cells were lysed in NP40 solution on the ice for 30 min. After centrifuged, supernatants were mixed with 20 μl of blank magnetic beads (#HY-K0202, MCE, NJ, USA) for 2 h. Then, 3–5 μl antibodies were added into 40 μl magnetic beads that had been resuspended in 500 μl NP40 lysis buffer. The mixture was incubated at 4 °C overnight. The magnetic beads were isolated from the mixture and incubated with protein supernatants at 4 °C. After 4 h, the beads were then boiled with an SDS-loading buffer. Samples were collected for subsequent experiments.
Quantitative real-time PCR (RT-qPCR)
Total RNA was extracted using RNAiso (Takara, Japan) according to the manufacturer’s instructions. Synthesis of cDNA was done using ABScript II RT Master Mix (RK20403, Abclonal Technology, Wuhan, China). Quantification of the mRNA for the indicated genes was done using an ABI 7300 QuantStudio3 PCR (RT-PCR) System using Genious 2X SYBR Green Fast qPCR Mix (RK21206, Abclonal Technology, Wuhan, China). Gene-specific primer sequences were designed from the PrimerBank database (Supplementary Table S1) [45].
Mouse tumor xenograft model
Four-week-old male nude mice were obtained from Huafukang Bio-Technology (Beijing, China). These mice were randomly separated in different groups. The mice were subcutaneously injected with 5 × 105 cancer cells. In some experiments, vehicle or STATTIC was intra-tumor injected every 2 days. After 24 or 28 days, mice were sacrificed and xenografts were collected for subsequent tests.
Modification residue identification by mass spectrometry detection
The STAT3 protein sample enriched by the IP assay was sent to the National Protein Science Facility, School of Life Science, Tsinghua University. The gel band was digested, dried, and redisolved in 0.1% trifluoroacetic acid. Peptides were analyzed by an Orbitrap Fusion mass spectrometer.
The MS data were searched against the target protein database from UniProt using an in-house Proteome Discoverer (Version PD1.4, Thermo Fisher Scientific, USA). The search criteria were: full trypsin specificity was required; carbamidomethylation (C) were set as the fixed modifications; the oxidation (M) and methyl (K/R) were set as the variable modification.
Statistical analysis
All statistical analyses were implemented with GraphPad Prism 7.0 (LaJolla, CA, USA). Student’s t-tests and ANOVA analysis were used. Statistical significance was set at p < 0.05.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) with the dataset identifier PXD021336.
References
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Adams RH, Alitalo K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol cell Biol. 2007;8:464–78.
De Palma M, Biziato D, Petrova TV. Microenvironmental regulation of tumour angiogenesis. Nat Rev Cancer. 2017;17:457–74.
Canavese M, Ngo DT, Maddern GJ, Hardingham JE, Price TJ, Hauben E. Biology and therapeutic implications of VEGF-A splice isoforms and single-nucleotide polymorphisms in colorectal cancer. Int J Cancer. 2017;140:2183–91.
Stratman AN, Schwindt AE, Malotte KM, Davis GE. Endothelial-derived PDGF-BB and HB-EGF coordinately regulate pericyte recruitment during vasculogenic tube assembly and stabilization. Blood. 2010;116:4720–30.
Stratman AN, Davis MJ, Davis GE. VEGF and FGF prime vascular tube morphogenesis and sprouting directed by hematopoietic stem cell cytokines. Blood. 2011;117:3709–19.
Saltz LB. Bevacizumab in colorectal cancer: it should have worked. Lancet Oncol. 2016;17:1469–70.
Smeets D, Miller IS, O’Connor DP, Das S, Moran B, Boeckx B, et al. Copy number load predicts outcome of metastatic colorectal cancer patients receiving bevacizumab combination therapy. Nat Commun. 2018;9:4112.
Haibe Y, Kreidieh M, El Hajj H, Khalifeh I, Mukherji D, Temraz S, et al. Resistance mechanisms to anti-angiogenic therapies in cancer. Front Oncol. 2020;10:221.
Darnell JE Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–21.
Minami M, Inoue M, Wei S, Takeda K, Matsumoto M, Kishimoto T, et al. STAT3 activation is a critical step in gp130-mediated terminal differentiation and growth arrest of a myeloid cell line. Proc Natl Acad Sci USA. 1996;93:3963–6.
Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–46.
Yu H, Jove R. The STATs of cancer-new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105.
Bharadwaj U, Kasembeli MM, Robinson P, Tweardy DJ. Targeting Janus kinases and signal transducer and activator of transcription 3 to treat inflammation, fibrosis, and cancer: rationale, progress, and caution. Pharmacol Rev. 2020;72:486–526.
Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci USA. 2010;107:21499–504.
Dasgupta M, Dermawan JK, Willard B, Stark GR. STAT3-driven transcription depends upon the dimethylation of K49 by EZH2. Proc Natl Acad Sci USA. 2015;112:3985–90.
Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–52.
Luo J, Wang K, Yeh S, Sun Y, Liang L, Xiao Y, et al. LncRNA-p21 alters the antiandrogen enzalutamide-induced prostate cancer neuroendocrine differentiation via modulating the EZH2/STAT3 signaling. Nat Commun. 2019;10:2571.
Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–73.
Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J, et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell. 2011;44:609–20.
Stec I, Wright TJ, van Ommen GJ, de Boer PA, van Haeringen A, Moorman AF, et al. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet. 1998;7:1071–82.
Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, et al. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–91.
Hideshima T, Mitsiades C, Tonon G, Richardson PG, Anderson KC. Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer. 2007;7:585–98.
Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92:3025–34.
Keats JJ, Maxwell CA, Taylor BJ, Hendzel MJ, Chesi M, Bergsagel PL, et al. Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients. Blood. 2005;105:4060–9.
Foltz SM, Gao Q, Yoon CJ, Sun H, Yao L, Li Y, et al. Evolution and structure of clinically relevant gene fusions in multiple myeloma. Nat Commun. 2020;11:2666.
Zhang J, Lee YR, Dang F, Gan W, Menon AV, Katon JM, et al. PTEN methylation by NSD2 controls cellular sensitivity to DNA damage. Cancer Discov. 2019;9:1306–23.
Hudlebusch HR, Santoni-Rugiu E, Simon R, Ralfkiær E, Rossing HH, Johansen JV, et al. The histone methyltransferase and putative oncoprotein MMSET is overexpressed in a large variety of human tumors. Clin Cancer Res. 2011;17:2919–33.
Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–58.
Stubbs M, Burn T, Sparks R, Maduskuie T, Diamond S, Rupar M. et al. The novel bromodomain and extraterminal domain inhibitor INCB054329 induces vulnerabilities in myeloma cells that inform rational combination strategies. Clin Cancer Res. 2019;25:300–11.
Biggar KK, Li SS. Non-histone protein methylation as a regulator of cellular signalling and function. Nat Rev Mol Cell Biol. 2015;16:5–17.
Zhang X, Huang Y, Shi X. Emerging roles of lysine methylation on non-histone proteins. Cell Mol Life Sci. 2015;72:4257–72.
Hamamoto R, Saloura V, Nakamura Y. Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat Rev Cancer. 2015;15:110–24.
Wang G, Long J, Gao Y, Zhang W, Han F, Xu C, et al. SETDB1-mediated methylation of Akt promotes its K63-linked ubiquitination and activation leading to tumorigenesis. Nat Cell Biol. 2019;21:214–25.
Guo J, Dai X, Laurent B, Zheng N, Gan W, Zhang J, et al. AKT methylation by SETDB1 promotes AKT kinase activity and oncogenic functions. Nat cell Biol. 2019;21:226–37.
Hou Z, Sun L, Xu F, Hu F, Lan J, Song D, et al. Blocking histone methyltransferase SETDB1 inhibits tumorigenesis and enhances cetuximab sensitivity in colorectal cancer. Cancer Lett. 2020;487:63–73.
Park JW, Chae YC, Kim JY, Oh H, Seo SB. Methylation of Aurora kinase A by MMSET reduces p53 stability and regulates cell proliferation and apoptosis. Oncogene. 2018;37:6212–24.
Lhoumaud P, Badri S, Rodriguez-Hernaez J, Sakellaropoulos T, Sethia G, Kloetgen A, et al. NSD2 overexpression drives clustered chromatin and transcriptional changes in a subset of insulated domains. Nat Commun. 2019;10:4843.
Cheong CM, Mrozik KM, Hewett DR, Bell E, Panagopoulos V, Noll JE, et al. Twist-1 is upregulated by NSD2 and contributes to tumour dissemination and an epithelial-mesenchymal transition-like gene expression signature in t(4;14)-positive multiple myeloma. Cancer Lett. 2020;475:99–108.
He C, Liu C, Wang L, Sun Y, Jiang Y, Hao Y. Histone methyltransferase NSD2 regulates apoptosis and chemosensitivity in osteosarcoma. Cell Death Dis. 2019;10:65.
Wang JJ, Zou JX, Wang H, Duan ZJ, Wang HB, Chen P, et al. Histone methyltransferase NSD2 mediates the survival and invasion of triple-negative breast cancer cells via stimulating ADAM9-EGFR-AKT signaling. Acta Pharmacologica Sin. 2019;40:1067–75.
Aytes A, Giacobbe A, Mitrofanova A, Ruggero K, Cyrta J, Arriaga J, et al. NSD2 is a conserved driver of metastatic prostate cancer progression. Nat Commun. 2018;9:5201.
Xie Z, Chooi JY, Toh SHM, Yang D, Basri NB, Ho YS, et al. MMSET I acts as an oncoprotein and regulates GLO1 expression in t(4;14) multiple myeloma cells. Leukemia. 2019;33:739–48.
Swaroop A, Oyer JA, Will CM, Huang X, Yu W, Troche C, et al. An activating mutation of the NSD2 histone methyltransferase drives oncogenic reprogramming in acute lymphocytic leukemia. Oncogene. 2019;38:671–86.
Wang X, Spandidos A, Wang H, Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012;40:D1144–9. Database issue.
Acknowledgements
We are grateful to the members in Guihua Wang’s lab and Junbo Hu’s lab for the critical inputs and suggestions. This work is supported by NSFC (No. 81773113 GW, No. 81922053 GW, No.81702264 XC, No. 81974432 GW, and No. 81874186 JH).
Author information
Authors and Affiliations
Contributions
GW conceived the project. GW and JH acquired funding and designed the majority of experiments. ZC and XL supervised the project and gave some advice. DS wrote the manuscript and performed most of the molecular biological experiments. JL analyzed the results. YC, AL, and QW performed most of the phenotype experiments. CZ did the mass spectrometry detection and analysis. YF and JW made their efforts in the bioinformatics analysis. XC made contributions and provided support in the process of revision.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethics approval and consent to participate
All patient specimens mentioned in this study were approved by the Ethics Committee of Tongji Hospital following the Declaration of Helsinki and informed consents were signed before the operation. Animal experiments were performed strictly following the Animal Study Guideline of Huazhong University of Science and Technology.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Song, D., Lan, J., Chen, Y. et al. NSD2 promotes tumor angiogenesis through methylating and activating STAT3 protein. Oncogene 40, 2952–2967 (2021). https://doi.org/10.1038/s41388-021-01747-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01747-z
- Springer Nature Limited
This article is cited by
-
Discovery of NSD2 non-histone substrates and design of a super-substrate
Communications Biology (2024)
-
ETHE1 dampens colorectal cancer angiogenesis by promoting TC45 Dephosphorylation of STAT3 to inhibit VEGF-A expression
Cell Death & Disease (2024)
-
METTL3 and STAT3 form a positive feedback loop to promote cell metastasis in hepatocellular carcinoma
Cell Communication and Signaling (2023)
-
PRMT5 methylating SMAD4 activates TGF-β signaling and promotes colorectal cancer metastasis
Oncogene (2023)
-
Comprehensive pan-cancer analysis of STAT3 as a prognostic and immunological biomarker
Scientific Reports (2023)