Abstract
The principle of synthetic lethality, which refers to the loss of viability resulting from the disruption of two genes, which, individually, do not cause lethality, has become an attractive target approach due to the development and clinical success of Poly (ADP-ribose) polymerase (PARP) inhibitors (PARPi). In this review, we present the most recent findings on the use of PARPi in the clinic, which are currently approved for second-line therapy for advanced ovarian and breast cancer associated with mutations of BRCA1 or BRCA2 (BRCA1/2) genes. PARPi efficacy, however, appears to be limited by acquired and inherent resistance, highlighting the need for alternative and synergistic targets to eliminate these tumors. Here, we explore other identified synthetic lethal interactors of BRCA1/2, including DNA polymerase theta (POLQ), Fanconi anemia complementation group D2 (FANDC2), radiation sensitive 52 (RAD52), Flap structure-specific endonuclease 1 (FEN1), and apurinic/apyrimidinic endodeoxyribonuclease 2 (APE2), as well as other protein and nonprotein targets, for BRCA1/2-mutated cancers and their implications for future therapies. A wealth of information now exists for phenotypic and functional characterization of these novel synthetic lethal interactors of BRCA1/2, and leveraging these findings can pave the way for the development of new targeted therapies for patients suffering from these cancers.
Similar content being viewed by others
Introduction
Breast cancer susceptibility gene 1 (BRCA1) and breast cancer susceptibility gene 2 (BRCA2) are two tumor suppressor genes, first linked to breast and ovarian cancer over two decades ago. Elimination of patents on BRCA1 or BRCA2 (BRCA1/2) for diagnostic testing in 2013 has significantly reduced the cost of genetic evaluation for these genes and led to their inclusion onto many multigene panels. BRCA1/2 proteins play a crucial role in DNA double-strand break (DSB) repair mediated by the error-free homologous recombination (HR) pathway [1]. Additionally, they have important roles in the protection of stalled replication forks, transcription regulation, chromatin modulation, cell cycle regulation, checkpoint enforcement, telomere maintenance, and transcription-coupled repair [1,2,3]. Given their indispensable contribution in maintaining genomic stability, it is unsurprising that mutations in BRCA1/2 confer high-penetrance susceptibility to breast and ovarian cancers [4]. These mutations increase the risk of developing breast cancer by 49–57% and ovarian cancer by 18–40% [4]. Notably, BRCA1 mutations frequently give rise to the aggressive, higher-grade, triple-negative breast cancer subtype which has poorer prognosis compared to other breast cancer subtypes [5]. Breast tumors that arise from BRCA2 mutations are predominantly high-grade invasive ductal carcinoma presenting as a luminal subtype of breast tumor harboring HR-deficiencies [6]. BRCA1/2 mutations account for ~15% of all ovarian cancer cases [7]. Histologically, these tumors stratify under the high-grade serous ovarian cancer subtype. Globally, breast and ovarian cancers are a leading cause of cancer-related deaths in women. Annually, approximately over 3 million women are diagnosed with breast or ovarian cancer. While ~5% of breast and 15% of ovarian cancer cases are thought to arise due to mutations in BRCA1/2, a recently proposed weighted model, HRDetect, which predicts BRCA1/2-deficiency with a 98.7% sensitivity, has estimated that up to 22% of breast cancer tumors may harbor mutations in these genes [8] and identified HR-deficiency in 69% of triple-negative breast tumors [9]. It is also noteworthy that therapeutic strategies that benefit cancer patients with BRCA1/2 mutations have also been shown to help patients with mutations in other genes necessary for the HR pathway. These include mutations in Ataxia Telangiectasia Mutated, Fanconi anemia (FA) genes, and checkpoint kinase 2 [10]. Also, while BRCA1/2 mutations are commonly associated with breast and ovarian cancers, inherited and sporadic mutations in these genes are also found in other cancer types, including melanoma and cancer of the prostate and pancreas [11]. These mutations can be advantageous for targeted therapy and synthetic lethal strategies have proven to be a promising approach to targeting these tumors [12].
Synthetic lethality refers to a genetic interaction in which the loss of two genes results in loss of viability, whereas the perturbation of either one of the genes does not. In recent years, through genetic screening and other methods, synthetic lethal partners have been identified for several major tumor suppressor genes, including BRCA1, BRCA2, and PTEN. This review will focus specifically on synthetic lethal interactions of BRCA1/2, including PARP, POLQ, RAD52, FANCD2, FEN1, and APEX2. We will highlight the physiological functions of these proteins, their status in cancer and explore the essentiality of these genes in BRCA1/2-deficient models. We will also briefly discuss non-protein synthetic lethal interactions that have been identified thus far and discuss strategies that have been, and continue to be, exploited to target BRCA1/2- and HR-deficient tumors.
Poly (ADP-ribose) polymerase (PARP)
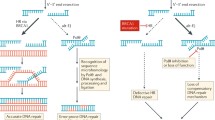
PARP inhibition was the first identified therapeutic synthetic lethal interaction with BRCA1/2-deficiency. PARP1 and PARP2 are DNA damage sensing and transducing enzymes that bind DNA via two zinc finger motifs and transfer poly ADP-ribosyl (PAR) moieties onto DNA repair effector proteins—a posttranslational modification process known as PARylation. At DNA damage sites, including single-strand break (SSB) sites, allosteric changes in PARP1 activates its catalytic function, contributing to chromatin remodeling and the recruitment of proteins involved in HR, nucleotide excision repair, and base excision repair (BER) [12,13,14]. Ultimately, PARP1 autoPARylation results in its release from repaired DNA [12]. Targeting PARP as a novel therapeutic strategy for eliminating BRCA1/2-mutated tumors first entered the spotlight after two studies in 2005 reported a synthetic lethal interaction between PARP inhibition and BRCA1/2 mutations [15, 16]. PARP inhibition using very potent small-molecule inhibitors leads to replication fork collapse, chromosomal instability, cell cycle arrest in G2, and subsequent apoptosis in BRCA1/2-deficient cells. This finding, coupled with previous studies that reported viable and fertile Parp1−/− mice [17], despite known PARP1 function in response to genomic assaults, garnered significant interest in targeting PARP for the elimination of BRCA1/2-deficient tumors. The initial model of the mechanism underlying this synthetic lethal interaction was reported to be caused by the accumulation of DSBs due to replication fork collapse when the fork encounters persistent SSBs caused by PARP inhibition [15] (Fig. 1). However, several studies have since reported that some PARP inhibitors (PARPi; olaparib, rucaparib, niraparib, and talazoparib) lead to the trapping of PARP molecules at sites of DNA damage by preventing autoPARylation [18,19,20].
PARP1/2 are DNA damage sensing and transducing enzymes. PARP depletion kills HR-deficient cells by compromising base excision repair (BER), a compensatory pathway in the absence of homologous recombination. Chemical inhibition of PARP leads to PARP trapping, resulting in replication stress and genomic instability, resulting in loss of viability in HR-deficient cells.
Currently, several FDA-approved PARPi are available for treatment or maintenance of tumors originating at various sites including breast, primary peritoneum, fallopian tubes, ovaries, and pancreas [21, 22]. PARPi differ in their PARP-trapping potency. In order from lowest to highest PARP-trapping ability are veliparib, rucaparib, olaparib, niraparib, and talazoparib. Veliparib is currently being investigated for the treatment of solid triple-negative breast cancer and high-grade serous ovarian tumors with documented mutations in BRCA1/2, PALB2, and other HR genes (NCT03123211). It has been and is also currently under investigation in other cancers including gastric (NCT01123876), rectal (NCT01589419), SCLC (NCT03227016), and other cancer types in combination with chemotherapeutics or radiotherapy. As of writing this review, veliparib has not yet gained FDA approval. Details discussing clinical development of PARPi, their mechanism of action, and adverse effects have been reviewed elsewhere [23,24,25]. FDA-approved PARPi (rucaparib, olaparib, niraparib, and talazoparib) and their combinations in conjunction with other anticancer agents are summarized in Table 1 according to PARP-trapping potency, cancer site, genomic variant, eligibility, and treatment purpose.
It is also noteworthy that given the compelling evidence surrounding the interplay between the host immune system, DNA damage, and inflammation in tumors [26], several clinical trials are currently examining the efficacy of combining PARPi with immunotherapy. Active combinatorial trials in gynecologic cancers, as well as breast, urothelial, lung, and prostate cancer, are discussed elsewhere [27]. However, PARPi are limited due to intrinsic and acquired resistance to these drugs. Briefly, PARPi resistance mechanisms in HR-deficient settings include reversion mutations in mutant BRCA1/2 genes, restoration of HR via enhanced end resection, DNA replication fork protection, restoration of PARylation, as well as loss of PARP1. These mechanisms are reviewed in detail elsewhere [28, 29]. This highlights the need of identifying new synthetic lethal interactions of BRCA1/2 and other HR genes, which may serve as novel targets or perhaps exhibit synergy when targeted in combination with PARPi or current chemo-, radio-, or immunotherapy regimens.
Radiation sensitive 52 (RAD52)
RAD52 is a DNA-binding protein that was first identified in Saccharomyces cerevisiae through a genetic screen for mutants sensitive to ionizing radiation (IR) [30]. Given the essentiality of Rad52 for HR and DNA repair in S. cerevisiae, surprisingly, RAD52−/− mice are viable, fertile, exhibit only moderate HR deficiency, no DNA damage sensitivity, and no cancer predisposition [30]. Functionally, RAD52 can bind ssDNA, playing a significant role in single-strand annealing (SSA) and HR repair of DSBs [30]. Studies have demonstrated that in mammalian cells, HR operates with at least two alternative sub-pathways including the dominant BRCA1/2-dependent canonical pathway and RAD52-dependent repair pathway [31, 32]. In the RAD52-dependent HR pathway, RAD52 facilitates the loading of RAD51 onto ssDNA coated with replication protein A (RPA), leading to homologous pairing, and strand invasion [33, 34]. Canonically, BRCA2 mediates assembly of RAD51 on ssDNA through a complex consisting of BRCA1/BRCA2/PALB2; however, in a BRCA1/2-deficient background, RAD52 can compensate for this essential component of HR [31, 32, 35]. Moreover, RAD52 interacts with RPA-coated overhangs, aligns the complementary regions, and trims 3′ overhangs in association with the ERCC1/XPF endonuclease complex, a crucial sequence of events that promote the error-prone SSA repair of DSBs [36, 37]. RAD52 also has been implicated in transcription-coupled HR [38]. In the G0/G1 phase of the cell cycle, Cockayne syndrome B protein detects DSB at sites of active transcription and recruits RAD51, RAD51C, and RAD52 to carry out HR-mediated repair using the newly synthesized RNA as a template [38]. Additionally, RAD52 also limits excessive remodeling of stalled replication forks [39].
As previously mentioned, RAD52 might not be an essential protein for healthy tissues; however, there is evidence that suggests RAD52 is critical for maintaining tumor genome integrity. Using The Cancer Genome Atlas (TCGA) database, a study found a significant association between amplification of the genomic region containing the RAD52 gene and the development of lung squamous cell carcinoma [40]. Additionally, two single nucleotide polymorphisms were found to associate with higher RAD52 expression, platinum resistance, and poor clinical outcome in cervical squamous cell carcinoma patients [41,42,43]. Furthermore, due to its compensatory roles in HR and SSA, RAD52 has been reported to be essential for BRCA1/2-deficient cancers [32, 33] (Fig. 2). Targeting RAD52 in BRCA1/2- and HR-deficient cancer cells sensitizes tumors to excess DSBs, while healthy cells and tissues with intact BRCA1/2-dependent HR remain unharmed [31, 32]. Mechanistically, it has been reported that the synthetic lethal relationship between RAD52 and BRCA1/2 is dependent on the endonuclease/exonuclease/phosphatase family domain-containing protein 1 (EEPD1) [44]. EEPD1 creates toxic intermediates post-cleavage of stressed replication forks, which require either BRCA1/2- or RAD52-dependent HR [44]. However, the absence of RAD52 and BRCA1/2 results in DSB accumulation and subsequent cell death. This synthetic lethal relationship has encouraged several groups to develop small-molecule inhibitors of RAD52 (RAD52i). Several RAD52i, including D-I03, 6-hydroxy-DL-dopa, epigallocatechin, and F779-0434, have been developed and have demonstrated selective in vitro killing of BRCA1/2-deficient cells [45,46,47]. Several of these inhibitors, including D-I03 and F779-0434, have been shown to cause synergistic accumulation of DSBs and eradication of BRCA-deficient tumor cells in immunodeficient mice in combination with PARPi with minimal toxicity to normal cells and tissues [48]. These findings indicate that inhibition of RAD52 could potentially improve the therapeutic outcome of BRCA-deficient malignancies treated with PARPi and limit the emergence drug-induced clones and harm to normal tissues.
RAD52 facilitates the loading of RAD51 onto ssDNA and also promotes the error-prone SSA to repair DNA double-strand breaks in the context of BRCA1/2-deficiency. Loss of RAD52 leads to loss of a compensatory DNA repair pathway resulting in genomic instability and cell death in BRCA1/2-deficient cells.
DNA polymerase theta (POL θ)
POL θ is encoded by the POLQ gene and is broadly expressed in normal tissues. POL θ belongs to the A family of DNA polymerases and is a crucial component of the alternative end-joining (Alt-EJ) repair pathway of DSBs [49, 50]. Alt-EJ occurs independently of the Ku protein complexes, and it serves as a backup pathway for DNA repair when HR, NHEJ, or BER become compromised [51]. Furthermore, the Alt-EJ pathway joins unprotected telomeres, which are generally joined by NHEJ in a POL θ-dependent manner [52]. POL θ contains a C-terminal polymerase domain and an N-terminal helicase-like domain. POL θ promotes Alt-EJ by limiting RAD51 nucleation onto ssDNA and stabilizing the annealing of two long 3′ ssDNA overhangs (post-resection dependent on MRN and CtBP-interacting protein (CtIP)) with as little as two bp of homology and using one 3′ ssDNA overhang as a synthesis primer [50, 53]. The helicase-like domain of POL θ displaces RPA from ssDNA tails and its polymerase domain promotes microhomology and subsequent gap filling [54]. Thus, the occurrence of microhomologies at sites of break is considered a defining signature of Alt-EJ [49]. Interestingly, POL θ has also been shown to add nucleotides to provide microhomology due to its terminal transferase activity [55]. Ultimately, processing by POL θ creates a stable, annealed intermediate that is sealed by DNA ligase I or III. POL θ has also been implicated in DNA replication [56]. It has been shown to function during the earliest steps of DNA replication and influences the timing of replication initiation [56]. This function is modulated through its interaction with the Orc2 and Orc4, components of the origin recognition complex. Based on previously outlined functions of POL θ, it is not surprising that Polq−/− mice and cells display elevated levels of spontaneous DNA damage, increased formation of micronuclei, and sensitivity to DNA damaging agents such as IR and mitomycin C [57, 58].
Overexpression of POL θ has been reported in several cancers, including stomach, lung, colon, breast, and ovarian cancers [49]. Furthermore, patients with higher expression of POL θ had shorter relapse-free survival compared to patients with relatively lower POL θ expression [49]. These studies suggest that low POL θ expression may potentially confer a selective disadvantage to tumors. POL θ may drive tumorigenesis and lead to aggressive tumors through its functions in Alt-EJ. Alt-EJ is intrinsically mutagenic, typically generating deletions at the repair junction, and potentially being a major driving force of genomic instability in human cancers [59]. Specifically, in HR-deficient cells, Alt-EJ acts as a compensatory form of DNA repair [60]. Deficiency in one DNA repair pathway can result in cellular hyper-dependence on a second compensatory DNA repair pathway. This dependency is evident in HR-deficient tumors, which present a genomic profile in which there are substantial deletions with microhomology due to overreliance on Alt-EJ [60] (Fig. 3). Consistent with this notion, the abrogation of Alt-EJ through POL θ-depletion preferentially kills HR-defective cells in vitro and in vivo. Furthermore, POL θ-depletion reduced the survival of HR-deficient cells exposed to PARPi, cisplatin, or MMC resulting in increased interest for POL θ as a potential therapeutic target in breast and ovarian cancers with BRCA1/2-deficiency and other HR deficiencies [60]. Several companies are currently in the process of developing POL θ inhibitors. Furthermore, using CRISPR-based genetic screens, a study identified 140 synthetic lethal genes with POL θ, many of which are involved in DNA damage repair pathways [61]. Interestingly, this study identified 275 out of 926 (29.7%) breast cancers in TCGA cohort having a deficiency in one or more of the 140 genes that are synthetically lethal with POL θ, highlighting the impact and benefit an effective POL θ inhibitor might have on patient outcomes [61].
Fanconi anemia complementation group D2 (FANCD2)
The FANCD2 gene encodes FANCD2 protein. The FA complementation group comprised of 22 proteins, including FANCD2, do not share sequence similarity but rather are related by their assembly into protein complexes that function in similar signal transduction pathways. This FA pathway is activated either during DNA replication or upon DNA damage and serves to maintain genomic stability through the repair of interstrand crosslinks and stabilization of stalled replication forks [62, 63]. Unsurprisingly, FA is an autosomal recessive disorder associated with mutations in genes from the FA complementation group [63]. This rare disorder is characterized by chromosomal abnormalities, increased chromosomal breakage, defective DNA repair, and hypersensitivity to DNA crosslinking agents [63]. Of the FA proteins, FANCD2 is the most evolutionarily conserved gene [64]. Functionally, upon FA pathway activation, FANCD2 is monoubiquitinated at lysine 561 and with its partner FANCI, coordinate several repair proteins and pathways such as HR (through interaction with BRCA1/2, RAD51, etc.), translesion DNA synthesis (through interaction with pol η), and nucleotide excision repair [64,65,66,67]. Importantly, monoubiquitinated FANCD2 is regarded as a functional representation of activated FA signaling. FANCD2 also localizes to stalled replication forks to protect nascent strands from excessive nucleolytic degradation and to modulate the replication origin firing rate [68,69,70], indicating that it also functions independently of the FA pathway.
Patients with FANCD2 mutations have increased risk of developing myelogenous malignancies, squamous cell carcinomas, congenital abnormalities, and reduced fertility [63]. These findings are also recapitulated in Fancd2−/− mice [71]. Even though FANCD2 is classically identified as a tumor suppressor, a study found that FANCD2 is significantly overexpressed in subgroups of ovarian, breast, and uterine cancers associated with HR-deficiency and high genomic instability [72]. Mechanistically, in BRCA1/2-deficient cells, FANCD2 localizes to stalled replication forks and recruits POL θ and CtIP, highlighting its requirement for Alt-EJ [72]. In the absence of BRCA1/2, replication stress is observed and the stability of the replication fork is compromised (Fig. 4). Reduced FANCD2 expression in BRCA1/2-deficient cells exacerbates replicative stress and impairs Alt-EJ, leading to genomic instability and substantially reduced cell viability [72]. Additionally, FANCD2 overexpression was found to confer resistance to PARPi through replication fork stabilization, independently of HR restoration. Thus, targeting FANCD2 in specific contexts, such as in HRD tumors and PARPi resistance tumors, could be an important therapeutic strategy. Currently, no FANCD2 inhibitors have been identified; however, high-throughput assays are being utilized to identify small-molecule inhibitors for this protein [73].
cFANCD2 plays a role in the alternative end-joining repair of DNA double-strand breaks by recruiting POL θ and PARP1, critical mediators of Alt-EJ. Additionally, FANCD2 also localizes to stalled replication forks to protect nascent strands from excessive degradation. Loss of FANCD2 leads to loss of a compensatory DNA repair pathway and destabilization of the replication forks, resulting in genomic instability and cell death in BRCA1/2-deficient cells. Ub ubiquitin, Star site of genome instability.
Flap structure-specific endonuclease 1 (FEN1)
FEN1 is a highly conserved structure-specific endonuclease that catalyzes the cleavage of bifurcated RNA or DNA structures called 5′ flaps [74]. Such structures are generated during long-patch BER or the synthesis of the lagging strand. Removal of these 5′ flaps by FEN1 creates a single nicked product that can be sealed by a DNA ligase. Additionally, FEN1 also plays a crucial role in telomere maintenance and processing of stalled replication forks [75]. Due to its function in replication and maintenance of genomic integrity, FEN1 is vital in highly proliferative tissues and its deletion in mice leads to early embryonic lethality [76]. FEN1 is overexpressed in many cancers [77], including cancer of the testis, lung, and brain [78]. In an evolutionarily conserved synthetic lethal interaction network, FEN1 was identified as a broad-spectrum target for anticancer therapy [79]. Genomic and protein expression analyses have identified FEN1 as a critical biomarker in the breast, ovarian, lung, liver, and other cancers [77, 80, 81]. FEN1 overexpression has been associated with poor prognosis in estrogen-receptor-positive and negative breast tumors and high grade, high stage ovarian cancer. In lung cancer, it was shown that C20, a small-molecule inhibitor of FEN1, sensitizes lung cancer cells in vitro and mouse models to cisplatin and other DNA damage-inducing agents [82]. Similar observations have been reported in breast cancer using another small-molecule inhibitor (SC13) of FEN1.
Recently, using DNA repair-focused shRNA and CRISPR screening in BRCA2-isogenic cell lines, FEN1 was identified as a synthetic lethal target for BRCA1/2 [83]. Mechanistically, it was shown that FEN1 plays a role in microhomology-mediated end joining (Fig. 5). However, given the multifaceted functions of FEN1 in maintaining genome integrity through its role in the processing of Okazaki fragments, BER, among others [75], the phenotype exhibited by loss of FEN1 in BRCA1/2-deficient cells is likely due to impairment of several of its functions. Another study has also since proposed targeting FEN1 in human cancers with defects in HR [84]. It is worth mentioning that in a BRCA1/2-proficient setting, concomitant inhibition of FEN1 with SC13 and PARP leads to DSB accumulation [85]. Since the toxic PARP-SSB intermediates that form due to PARP trapping are resolved primarily through BER, and the association of FEN1 loss with impaired BER, targeting this enzyme should sensitize BRCA1/2-deficient tumors to PARPi. Several small-molecule inhibitors of FEN1 exist and drugs targeting FEN1 are currently in production by several companies. Thus, targeting FEN1 may be an important therapeutic strategy that is efficacious alone, and may be synergistic in combination with current treatment modalities including cisplatin and PARPi.
FEN1 play a role in 5′ flap processing following microhomology-mediated end joining of DNA double-strand breaks through the compensatory DSB repair pathway. It also plays a role in DNA repair through the long-patch SSB repair pathway. Loss of FEN1 or its inhibition leads to replication stress as a result of compromised 5′ flap processing and base excision repair in BRCA1/2-deficient cells, leading to genomic instability and cell death.
Apurinic/apyrimidinic endodeoxyribonuclease 2 (APE2)
Apurinic/apyrimidinic (AP) sites are pre-mutagenic lesions primarily repaired through BER [86]. APEX2 encodes apurinic/apyrimidinic endodeoxyribonuclease 2 (APE2), an AP endonuclease crucial for this repair process. APE2 has weaker endonuclease activity compared to APE1, the major AP endonuclease; however, APE2 has strong 3′ phosphodiesterase and 3′−5′ exonuclease activity [87]. In addition to BER, APE1 and APE2 have also been implicated in HR [88]. While both proteins were shown to regulate RAD51 expression, APE2 was found to interact with various HR regulating proteins, including BRCA1, BRCA2, BARD1, RAD52, and TP53. Short-hairpin mediated loss and chemical inhibition of these AP nucleases have been shown to inhibit HR in multiple myeloma cells [88]. In normal fibroblasts, overexpression of these nucleases was shown to increase HR. APE2 also has a role in class-switch recombination, cell cycle progression during proliferation of lymphocytes, and the ATR-Chk1 DNA damage response pathway [89, 90]. Additionally, APE2 is important in proliferating cell nuclear antigen (PCNA)-dependent repair of H2O2-induced oxidative DNA damage [91]. A recent study has identified that genomic alterations in APE2 occur at a ~17% frequency using data available from TCGA with upregulation of APE2 expression in kidney, uterine, liver, lung, as well as breast cancer [92].
The aforementioned genetic screen conducted using DNA repair-focused shRNA and CRISPR libraries also identified APEX2 as a BRCA1/2 synthetic lethal target [83]. Validation studies found increased γH2AX foci formation, elevated CHK1 and RPA32 phosphorylation, and growth defects following APE2 depletion in BRCA1/2-mutant cells. Experiments to rescue cell proliferation suggested that the nuclease activity of APE2 was critical for the survival of BRCA2-mutant cells. Reconstitution of these APE2-depleted BRCA2-mutant cells with wild-type APE1 or its nuclease-deficient point mutant was unable to rescue cell growth. It should be noted that, unlike APE1, APE2 has been reported to be strongly associated with replication forks using stable isotope labeling with amino acids in cell culture and isolation of protein on nascent DNA followed by mass spectrometry(iPOND-SILAC-MS) [93]. According to the model recently proposed in this study, APE2–PCNA interaction enables AP site processing at replication forks, which would otherwise lead to fork stalling, potentially contributing to DNA damage and genomic instability in the absence of BRCA1/2 functions [83] (Fig. 6). Interestingly, despite its high sequence similarity with APE2, and its role as the major BER AP endonuclease, APE1 was not identified to be synthetically lethal with BRCA1/2 loss of function and its overexpression was unable to rescue loss of viability in cells lacking APE2 and BRCA2. A second study has also recently identified APEX2 as a synthetic lethal interactor of BRCA1/2 using a genome-wide CRISPR screen [94]. The mechanistic model proposed in this study suggests that the primary function of APE2 is to reverse blocked 3′ DNA ends, thereby preventing endogenously arising 3′ blocking lesions, which are a significant vulnerability in BRCA1/2-deficient cells [94]. While the dual loss of APE1 and APE2 sensitized multiple myeloma cells to the PARPi PJ34, studies evaluating the synergy between loss of APE2 function and PARP inhibition have not yet been reported. However, given that PARP-SSB intermediates are removed through BER, and APE2 function is essential for this repair pathway, it is plausible that loss of APE2 will lead to sensitization to PARPi.
Other protein and nonprotein targets
With advances in genomic editing and genetic screenings, more synthetic lethal interactors of BRCA1/2 have been identified. A study utilized an epigenetic compounds library to identify the histone deacetylases (HDACs) inhibition as synthetically lethal for BRCA1-mutated breast cancers [95]. Class I HDAC inhibition leads to global transcriptional changes [95]. More specifically, it increases expression of the thioredoxin-interacting protein, an inhibitor of the critical antioxidant, thioredoxin. This leads to reactive oxygen species-mediated DNA damage accumulation and exacerbation of genomic instability in BRCA1-deficient cells, thus resulting in cell death [95]. Another study using kinase inhibitor screen has identified polo-like kinase 1 (PLK1) as a potential synthetic lethal partner of BRCA1. Since BRCA1 plays a role in centrosome duplication and cytokinesis [96], loss of PLK1 activity in BRCA1-deficient cells led to giant multinucleated cells with large aggregates of centrosomes, causing mitotic catastrophes and cytokinesis failure, thereby, leading to cell death. Additionally, volasertib, a PLK1 inhibitor currently in late-stage clinical trials for adult acute myeloid leukemia patients, was used to demonstrate the in vivo killing of BRCA1-deficient cancer cells. Although volasertib has shown to have cardiovascular side-effects [97], it still has the potential to be used for the therapy of BRCA1-deficient tumors. It should be noted that PLK1 inhibition triggered mild synthetic lethal phenotypes in BRCA2-deficient cells compared to a more robust phenotype observed in BRCA1-deficient cells [98].
More recently, it has been demonstrated that targeting DNA damage orchestrator E3 ubiquitin ligase ring finger protein 168 (RNF168) can induce synthetic lethality in BRCA1-deficient tumors [99,100,101]. Mechanistically, RNF168 was shown to act redundantly with BRCA1 to load PALB2-BRCA2-RAD51 onto damaged DNA [99]. Therefore, inhibition of RNF168-mediated chromatin ubiquitylation pathway in the absence of BRCA1 further compromised residual HR and resulted in elevated genomic instability and cell death. Interestingly, overexpression of RNF168 also led to cell death in BRCA1-null cells [100]; this was due to robust recruitment of 53BP1 resulting in inhibition of end resection, a key process required for HR. In another recent study, RNF168 was shown to maintain genomic stability in BRCA-deficient cells through its role in R-loop resolution [101]. Mechanistically, it was shown that RNF168 ubiquitylates helicase DHX9 to recruit it to R-loops in BRCA-deficient cells [101]. Thus, RNF168 deficiency resulted in R-loop exacerbation, which then led to DSBs, senescence, and consequently cell death in BRCA-mutant cells.
Depletion of oncogenic helicase amplified in liver cancer 1 (ALC1) was also recently shown in several studies to confer synthetic lethality/sickness to HR-deficient cells and sensitize cells to PARPi [102,103,104,105]. One study showed that ALC1 catalytic activity is required for the release of trapped PARP2, but not PARP1, from damaged chromatin [102]. Thus, depletion of ALC1 resulted in PARP2 trapping and impaired SSB repair response, subsequently leading to PARPi hypersensitivity and synthetic lethality with BRCA1/2. Similarly, another study demonstrated that ALC1 is a key determinant of PARPi toxicity and its deficiency causes synthetic sickness in BRCA-mutant cells [103]. In this study, ALC1 was shown to modulate chromatin accessibility to allow processing of damaged bases. Thus, ALC1 loss decreased chromatin accessibility and association of base damage factors, resulting in the accumulation of replication-associated damage, increased trapping of PARP1/2, and dependence on HR [103]. ALC1 loss significantly reduced the survival of BRCA-mutant cells and enhanced PARPi sensitivity by up to 250-fold [103]. An additional study also recently reported that ALC1 underlies PARPi resistance and showed that ALC1 deficiency enhances trapping of inhibited PARP1, which results in impaired binding of both HR and NHEJ repair proteins at DNA lesions [104]. The synthetic lethal interaction between defective ALC1 nucleosome remodeling and HRD was also documented in another study [105]. This study reported that ALC1 loss resulted in incomplete processing of BER intermediates, which consequently resulted in DNA gaps after replication and a critical dependence on repair via HR [105]. Taken together, these studies provide plentiful evidence for targeting ALC1 alone or as a PARPi sensitizer to augment current therapeutic strategies employed to treat HR-deficient cancers.
Synthetic lethality in the context of BRCA1 has also been expanded to nonprotein targets. The silencing of microRNA 223-3p (miR223-3p) enhanced Alt-EJ repair of stressed replication forks. Thus, reconstituting expression of miR223-3p in BRCA1-deficient cancers caused synthetic lethality due to the Alt-EJ dependency, as previously highlighted in the context of POL θ and FANCD2 [106].
While the bona fide synthetic lethal targets of BRCA1/2 have some function in maintaining genomic stability through several different mechanisms, there remains a wealth of unexplored pathways that could be essential in a BRCA1/2-deficient background. For instance, the aforementioned study in which DNA repair-focused genetic screens identified FEN1 and APE1 as synthetic lethal partners with BRCA1/2, also highlighted mRNA splicing as an essential pathway in BRCA2-mutant cells [83]. Specifically, splicing factor 3B subunit 2, a component of the U2 small nuclear ribonucleoprotein particle necessary for forming the spliceosome [107], was found to be essential in BRCA2-mutant cells in vitro. Furthermore, two spliceosome inhibitors, spliceostatin-A and sudemycin D6, also resulted in loss of viability of BRCA2-mutant cells. This highlights the potential synthetic lethal relationship between the loss of U2 spliceosome and BRCA2; however, this finding was not investigated in the context of BRCA1-deficiency, and the mechanism of synthetic lethality has not yet been explored.
Due to the functions of BRCA1/2 in the prevention of genomic instability through the protection of stalled replication forks from unscheduled nucleases, a vulnerability that could be exploited has emerged in BRCA1/2-deficient cells [69, 108]. Several groups have identified synthetic lethal partners of BRCA1/2 that are involved in replication fork protection such as CtIP, exonuclease 3′−5′ domain-containing 2 (EXD2) and ubiquitin-specific peptidase 1 (USP1) [109,110,111]. CtIP has been shown to protect nascent DNA strands following fork reversal by disrupting DNA2 nuclease activity [109]. This allows BRCA1/2-mediated assembly of RAD51 nucleofilaments to prevent meiotic recombination 11 (MRE11) fork degradation, ensuring fork restart, thus maintaining genomic integrity. In the absence of CtIP and BRCA1/2 proteins, both nascent strands at reversed forks are vulnerable to over-resection by DNA2 and MRE11, thus resulting in genomic instability [109]. However, it should also be noted that a recent study that identified CtIP as a crucial regulator of fork integrity suggested that germline CtIP mutations may predispose to early onset breast cancer due to deleterious fork degradation events rather than changes in DSB resection efficiency [112]. Another factor, EXD2, was also shown to counteract fork reversal, preventing uncontrolled degradation of nascent DNA, thereby allowing for efficient fork restart [110]. Combined deficiency of EXD2 and BRCA1/2 resulted in the loss of fork protection and lead to collapsed forks adversely impacting genome stability and cell viability [110]. It should also be noted that the loss of EXD2 hampered Alt-EJ; thus, the mechanism for the observed synthetic lethality is multifactorial. Another group demonstrated that loss of USP1, a factor that stabilizes replication forks by deubiquitinating PCNA and replacing the translesion synthesis polymerase, leads to cell death in BRCA1-deficient cells due to excessive replication fork degradation [111].
While on the topic of replication stress, it should be noted that four-stranded secondary DNA structures known as G-quadruplexes (G4) also contribute to replication stress [113]. These structures have regulatory functions in different regions of the genome during transcription, translation, etc., and their stabilization has deleterious effects on genomic stability as they can act as a potent barrier to replication machinery. Several stabilizers of G4 structures, such as pyridostatin, have been shown to reduce the proliferation of HR-defective cells by exacerbating defects associated with HR-deficiency, including accumulation of DSBs, checkpoint activation, and deregulating G2/M progression [114]. It is also important to note that pyridostatin sensitivity has also been observed in HR-deficient cells that have acquired resistance to the PARPi olaparib due to loss of REV7 or 53BP1. These results highlight the therapeutic potential of G4-stabilizing drugs for the elimination of HR-deficient tumors, including tumors that exhibit PARPi resistance. Two additional small-molecule drugs, CX-5461 and CX-3543, have been identified as G4 stabilizers that selectively kill BRCA1/2-deficient tumors [115]; providing further evidence supporting stabilization of G4 structures for the elimination of BRCA1/2-deficient tumors. CX-5461 is currently in an advanced phase I clinical trial (NCT02719977) to assess tolerability in patients with recurrent, metastatic, locally advanced, or unresectable breast cancer with known BRCA1/2 or other HR germline aberrations.
Concluding remarks
Despite recent success in clinical trials, the benefit of PARPi is limited by inherent and acquired resistance, highlighting the need for the identification of alternative and perhaps synergistic targets. In this review, we have presented an integrative perspective on newly identified protein and nonprotein synthetic lethal interactions of BRCA1/2. We present evidence that suggest that these findings could also target other DNA repair deficiencies that present as HR-deficiency. Novel targets are being identified at an unprecedented rate using high-throughput approaches and efforts are underway to map cancer dependencies systematically (DepMap Portal; https://depmap.org/portal/); however, translation to the clinic remains limited. Additionally, inherent assay limitations (2D vs. 3D cell culture) should also be contemplated as they may obscure novel potent synthetic lethal targets. Furthermore, while current assays focus on identifying synthetic lethal interactions at the gene level, limited research exists at the domain or amino acid level for BRCA1/2. Future efforts should consider synthetic lethal interactions specifically to the domain or hotspot mutations to improve precision cancer therapy and limit toxicities. As of writing this review, given the hyper-dependence of cancer cells on replication and continuous proliferative signaling, in addition to the possibility of selective elimination of BRCA1/2-deficient cancer cells, therapeutic strategies targeting the replication stress response have emerged at the forefront of the next wave of synthetic lethal targets.
References
Chen C-C, Feng W, Lim PX, Kass EM, Jasin M. Homology-directed repair and the role of BRCA1, BRCA2, and related proteins in genome integrity and cancer. Annu Rev Cancer Biol. 2018;2:313–36.
Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2012;12:68–78.
Wei L, Lan L, Yasui A, Tanaka K, Saijo M, Matsuzawa A, et al. BRCA1 contributes to transcription-coupled repair of DNA damage through polyubiquitination and degradation of Cockayne syndrome B protein. Cancer Sci. 2011;102:1840–7.
Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007;25:1329–33.
Chen H, Wu J, Zhang Z, Tang Y, Li X, Liu S, et al. Association between BRCA status and triple-negative breast cancer: a meta-analysis. Front Pharmacol. 2018;9:909–909.
Bane AL, Beck JC, Bleiweiss I, Buys SS, Catalano E, Daly MB, et al. BRCA2 mutation-associated breast cancers exhibit a distinguishing phenotype based on morphology and molecular profiles from tissue microarrays. Am J Surg Pathol. 2007;31. https://journals.lww.com/ajsp/Fulltext/2007/01000/BRCA2_Mutation_associated_Breast_Cancers_Exhibit_a.15.aspx.
Neff RT, Senter L, Salani R. BRCA mutation in ovarian cancer: testing, implications and treatment considerations. Ther Adv Med Oncol. 2017;9:519–31.
Davies H, Glodzik D, Morganella S, Yates LR, Staaf J, Zou X, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. 2017;23:517–25.
Chopra N, Tovey H, Pearson A, Cutts R, Toms C, Proszek P, et al. Homologous recombination DNA repair deficiency and PARP inhibition activity in primary triple negative breast cancer. Nat Commun. 2020;11:2662.
Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373:1697–708.
Mersch J, Jackson MA, Park M, Nebgen D, Peterson SK, Singletary C, et al. Cancers associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer. 2015;121:269–75.
Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–8.
Pines A, Vrouwe MG, Marteijn JA, Typas D, Luijsterburg MS, Cansoy M, et al. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J Cell Biol. 2012;199:235–49.
Horton JK, Stefanick DF, Prasad R, Gassman NR, Kedar PS, Wilson SH. Base excision repair defects invoke hypersensitivity to PARP inhibition. Mol Cancer Res. 2014;12:1128.
Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21.
Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7.
Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, et al. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–20.
Pommier Y, O’Connor MJ, de Bono J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci Transl Med. 2016;8:362ps17.
Murai J, Huang S-YN, Renaud A, Zhang Y, Ji J, Takeda S, et al. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther. 2014;13:433–43.
Murai J, Huang SN, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012;72:5588–99.
Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med. 2019;381:317–27.
Yi M, Dong B, Qin S, Chu Q, Wu K, Luo S. Advances and perspectives of PARP inhibitors. Exp Hematol Oncol. 2019;8:29.
Morales J, Li L, Fattah FJ, Dong Y, Bey EA, Patel M, et al. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit Rev Eukaryot Gene Expr. 2014;24:15–28.
Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–8.
Mateo J, Lord CJ, Serra V, Tutt A, Balmaña J, Castroviejo-Bermejo M, et al. A decade of clinical development of PARP inhibitors in perspective. Ann Oncol. 2019;30:1437–47.
Mouw KW, Goldberg MS, Konstantinopoulos PA, D’Andrea AD. DNA damage and repair biomarkers of immunotherapy response. Cancer Discov. 2017;7:675.
Vikas P, Borcherding N, Chennamadhavuni A, Garje R. Therapeutic potential of combining PARP inhibitor and immunotherapy in solid tumors. Front Oncol. 2020;10:570.
Noordermeer SM, van Attikum H. PARP inhibitor resistance: a tug-of-war in BRCA-mutated cells. Trends Cell Biol. 2019;29:820–34.
Li H, Liu Z-Y, Wu N, Chen Y-C, Cheng Q, Wang J. PARP inhibitor resistance: the underlying mechanisms and clinical implications. Mol Cancer. 2020;19:107.
Hanamshet K, Mazina OM, Mazin AV. Reappearance from obscurity: mammalian Rad52 in homologous recombination. Genes. 2016;7:63.
Lok BH, Carley AC, Tchang B, Powell SN. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene. 2013;32:3552–8.
Feng Z, Scott SP, Bussen W, Sharma GG, Guo G, Pandita TK, et al. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc Natl Acad Sci USA. 2011;108:686–91.
Ma CJ, Kwon Y, Sung P, Greene EC. Human RAD52 interactions with replication protein A and the RAD51 presynaptic complex. J Biol Chem. 2017;292:11702–13.
Gibb B, Ye LF, Kwon Y, Niu H, Sung P, Greene EC. Protein dynamics during presynaptic-complex assembly on individual single-stranded DNA molecules. Nat Struct Mol Biol. 2014;21:893–900.
Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–83.
Bhargava R, Onyango DO, Stark JM. Regulation of single-strand annealing and its role in genome maintenance. Trends Genet. 2016;32:566–75.
Motycka TA, Bessho T, Post SM, Sung P, Tomkinson AE. Physical and functional interaction between the XPF/ERCC1 endonuclease and hRad52. J Biol Chem. 2004;279:13634–9.
Wei L, Nakajima S, Böhm S, Bernstein KA, Shen Z, Tsang M, et al. DNA damage during the G0/G1 phase triggers RNA-templated, Cockayne syndrome B-dependent homologous recombination. Proc Natl Acad Sci USA. 2015;112:E3495–504.
Malacaria E, Pugliese GM, Honda M, Marabitti V, Aiello FA, Spies M, et al. Rad52 prevents excessive replication fork reversal and protects from nascent strand degradation. Nat Commun. 2019;10:1412.
Lieberman R, Xiong D, James M, Han Y, Amos CI, Wang L, et al. Functional characterization of RAD52 as a lung cancer susceptibility gene in the 12p13.33 locus. Mol Carcinog. 2016;55:953–63.
Gossage L, Madhusudan S. Cancer pharmacogenomics: role of DNA repair genetic polymorphisms in individualizing cancer therapy. Mol Diagn Ther. 2007;11:361–80.
Zhang L, Ma W, Li Y, Wu J, Shi GY. Pharmacogenetics of DNA repair gene polymorphisms in non-small-cell lung carcinoma patients on platinum-based chemotherapy. Genet Mol Res. 2014;13:228–36.
Shi T-Y, Yang G, Tu X-Y, Yang J-M, Qian J, Wu X-H, et al. RAD52 variants predict platinum resistance and prognosis of cervical cancer. PLoS ONE. 2012;7:e50461.
Hromas R, Kim H-S, Sidhu G, Williamson E, Jaiswal A, Totterdale TA, et al. The endonuclease EEPD1 mediates synthetic lethality in RAD52-depleted BRCA1 mutant breast cancer cells. Breast Cancer Res. 2017;19:122.
Huang F, Goyal N, Sullivan K, Hanamshet K, Patel M, Mazina OM, et al. Targeting BRCA1- and BRCA2-deficient cells with RAD52 small molecule inhibitors. Nucleic Acids Res. 2016;44:4189–99.
Hengel SR, Malacaria E, Folly da Silva Constantino L, Bain FE, Diaz A, Koch BG, et al. Small-molecule inhibitors identify the RAD52-ssDNA interaction as critical for recovery from replication stress and for survival of BRCA2 deficient cells. eLife. 2016;19:5.
Cramer-Morales K, Nieborowska-Skorska M, Scheibner K, Padget M, Irvine DA, Sliwinski T, et al. Personalized synthetic lethality induced by targeting RAD52 in leukemias identified by gene mutation and expression profile. Blood. 2013;122:1293–304.
Sullivan-Reed K, Bolton-Gillespie E, Dasgupta Y, Langer S, Siciliano M, Nieborowska-Skorska M, et al. Simultaneous targeting of PARP1 and RAD52 triggers dual synthetic lethality in BRCA-deficient tumor cells. Cell Rep. 2018;23:3127–36.
Wood RD, Doublié S. DNA polymerase θ (POLQ), double-strand break repair, and cancer. DNA Repair. 2016;44:22–32.
Yousefzadeh MJ, Wyatt DW, Takata K-I, Mu Y, Hensley SC, Tomida J, et al. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 2014;10:e1004654.
Thompson LH. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: the molecular choreography. Mutat Res. 2012;751:158–246.
Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature. 2015;518:254–7.
Kent T, Chandramouly G, McDevitt SM, Ozdemir AY, Pomerantz RT. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase θ. Nat Struct Mol Biol. 2015;22:230–7.
Mateos-Gomez PA, Kent T, Deng SK, McDevitt S, Kashkina E, Hoang TM, et al. The helicase domain of Polθ counteracts RPA to promote alt-NHEJ. Nat Struct Mol Biol. 2017;24:1116–23.
Kent T, Mateos-Gomez PA, Sfeir A, Pomerantz RT. Polymerase θ is a robust terminal transferase that oscillates between three different mechanisms during end-joining. eLife. 2016;17:5.
Fernandez-Vidal A, Guitton-Sert L, Cadoret J-C, Drac M, Schwob E, Baldacci G, et al. A role for DNA polymerase θ in the timing of DNA replication. Nat Commun. 2014;5:4285.
Shima N, Munroe RJ, Schimenti JC. The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol Cell Biol. 2004;24:10381–9.
Goff JP, Shields DS, Seki M, Choi S, Epperly MW, Dixon T, et al. Lack of DNA polymerase theta (POLQ) radiosensitizes bone marrow stromal cells in vitro and increases reticulocyte micronuclei after total-body irradiation. Radiat Res. 2009;172:165–74.
Boboila C, Jankovic M, Yan CT, Wang JH, Wesemann DR, Zhang T, et al. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc Natl Acad Sci USA. 2010;107:3034–9.
Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MIR, et al. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature. 2015;518:258–62.
Feng W, Simpson DA, Carvajal-Garcia J, Price BA, Kumar RJ, Mose LE, et al. Genetic determinants of cellular addiction to DNA polymerase theta. Nat Commun. 2019;10:4286.
Nakanishi K, Cavallo F, Perrouault L, Giovannangeli C, Moynahan ME, Barchi M, et al. Homology-directed Fanconi anemia pathway cross-link repair is dependent on DNA replication. Nat Struct Mol Biol. 2011;18:500–3.
Nalepa G, Clapp DW. Fanconi anaemia and cancer: an intricate relationship. Nat Rev Cancer. 2018;18:168–85.
Nepal M, Che R, Ma C, Zhang J, Fei P. FANCD2 and DNA damage. Int J Mol Sci. 2017;18:1804
Joo W, Xu G, Persky NS, Smogorzewska A, Rudge DG, Buzovetsky O, et al. Structure of the FANCI-FANCD2 complex: insights into the Fanconi anemia DNA repair pathway. Science. 2011;333:312–6.
Knipscheer P, Räschle M, Smogorzewska A, Enoiu M, Ho TV, Schärer OD, et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–701.
Renaud E, Rosselli F. FANC pathway promotes UV-induced stalled replication forks recovery by acting both upstream and downstream Polη and Rev1. PLoS ONE. 2013;8:e53693.
Lossaint G, Larroque M, Ribeyre C, Bec N, Larroque C, Décaillet C, et al. FANCD2 binds MCM proteins and controls replisome function upon activation of s phase checkpoint signaling. Mol Cell. 2013;51:678–90.
Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–16.
Schwab RA, Nieminuszczy J, Shah F, Langton J, Lopez Martinez D, Liang C-C, et al. The Fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Mol Cell. 2015;60:351–61.
Yang Q, Xie H, Zhong Y, Li D, Ke X, Ying H, et al. Severe Fanconi anemia phenotypes in Fancd2 depletion mice. Biochem Biophys Res Commun. 2019;514:713–9.
Kais Z, Rondinelli B, Holmes A, O’Leary C, Kozono D, D’Andrea AD, et al. FANCD2 maintains fork stability in BRCA1/2-deficient tumors and promotes alternative end-joining DNA repair. Cell Rep. 2016;15:2488–99.
Sharp MF, Murphy VJ, Twest SV, Tan W, Lui J, Simpson KJ, et al. Methodology for the identification of small molecule inhibitors of the Fanconi anaemia ubiquitin E3 ligase complex. Sci Rep. 2020;10:7959.
Grasby JA, Finger LD, Tsutakawa SE, Atack JM, Tainer JA. Unpairing and gating: sequence-independent substrate recognition by FEN superfamily nucleases. Trends Biochem Sci. 2012;37:74–84.
Saharia A, Teasley DC, Duxin JP, Dao B, Chiappinelli KB, Stewart SA. FEN1 ensures telomere stability by facilitating replication fork re-initiation. J Biol Chem. 2010;285:27057–66
Larsen E, Gran C, Saether BE, Seeberg E, Klungland A. Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol Cell Biol. 2003;23:5346–53.
Singh P, Yang M, Dai H, Yu D, Huang Q, Tan W, et al. Overexpression and hypomethylation of flap endonuclease 1 gene in breast and other cancers. Mol Cancer Res. 2008;6:1710–7.
Nikolova T, Christmann M, Kaina B. FEN1 is overexpressed in testis, lung and brain tumors. Anticancer Res. 2009;29:2453–9.
van Pel DM, Barrett IJ, Shimizu Y, Sajesh BV, Guppy BJ, Pfeifer T, et al. An evolutionarily conserved synthetic lethal interaction network identifies FEN1 as a broad-spectrum target for anticancer therapeutic development. PLoS Genet. 2013;9:e1003254.
Abdel-Fatah TMA, Russell R, Albarakati N, Maloney DJ, Dorjsuren D, Rueda OM, et al. Genomic and protein expression analysis reveals flap endonuclease 1 (FEN1) as a key biomarker in breast and ovarian cancer. Mol Oncol. 2014;8:1326–38.
Zhang Y, Liu X, Liu L, Chen J, Hu Q, Shen S, et al. Upregulation of FEN1 is associated with the tumor progression and prognosis of hepatocellular carcinoma. Dis Markers. 2020;2020:2514090.
He L, Luo L, Zhu H, Yang H, Zhang Y, Wu H, et al. FEN1 promotes tumor progression and confers cisplatin resistance in non-small-cell lung cancer. Mol Oncol. 2017;11:640–54.
Mengwasser KE, Adeyemi RO, Leng Y, Choi MY, Clairmont C, D’Andrea AD, et al. Genetic screens reveal FEN1 and APEX2 as BRCA2 synthetic lethal targets. Mol Cell. 2019;73:e6.
Guo E, Ishii Y, Mueller J, Srivatsan A, Gahman T, Putnam CD, et al. FEN1 endonuclease as a therapeutic target for human cancers with defects in homologous recombination. Proc Natl Acad Sci. 2020;117:19415.
He L, Zhang Y, Sun H, Jiang F, Yang H, Wu H, et al. Targeting DNA flap endonuclease 1 to impede breast cancer progression. EBioMedicine. 2016;14:32–43.
Krokan HE, Bjørås M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012583.
Burkovics P, Szukacsov V, Unk I, Haracska L. Human Ape2 protein has a 3′−5′ exonuclease activity that acts preferentially on mismatched base pairs. Nucleic Acids Res. 2006;34:2508–15.
Kumar S, Talluri S, Pal J, Yuan X, Lu R, Nanjappa P, et al. Role of apurinic/apyrimidinic nucleases in the regulation of homologous recombination in myeloma: mechanisms and translational significance. Blood Cancer J. 2018;8:92.
Guikema JEJ, Linehan EK, Tsuchimoto D, Nakabeppu Y, Strauss PR, Stavnezer J, et al. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J Exp Med. 2007;204:3017–26.
Willis J, Patel Y, Lentz BL, Yan S. APE2 is required for ATR-Chk1 checkpoint activation in response to oxidative stress. Proc Natl Acad Sci. 2013;110:10592.
Burkovics P, Hajdú I, Szukacsov V, Unk I, Haracska L. Role of PCNA-dependent stimulation of 3′-phosphodiesterase and 3′–5′ exonuclease activities of human Ape2 in repair of oxidative DNA damage. Nucleic Acids Res. 2009;37:4247–55.
Jensen KA, Shi X, Yan S. Genomic alterations and abnormal expression of APE2 in multiple cancers. Sci Rep. 2020;10:3758.
Dungrawala H, Rose KL, Bhat KP, Mohni KN, Glick GG, Couch FB, et al. The replication checkpoint prevents two types of fork collapse without regulating replisome stability. Mol Cell. 2015;59:998–1010.
Alvarez-Quil¢n A, Wojtaszek JL, Mathieu M-C, Patel T, Appel CD, Hustedt N, et al. Endogeous DNA 3' blocks are vulnerabilities for BRCA1 and BRCA2 deficiency and are reversed by the APE2 nuclease. Mol Cell. 2020;78:1152–1165.
Zhang B, Lyu J, Yang EJ, Liu Y, Wu C, Pardeshi L, et al. Class I histone deacetylase inhibition is synthetic lethal with BRCA1 deficiency in breast cancer cells. Acta Pharm Sin B. 2020;10:615–27.
Kais Z, Parvin JD. Regulation of centrosomes by the BRCA1-dependent ubiquitin ligase. Cancer Biol Ther. 2008;7:1540–3.
Raab M, Kappel S, Krämer A, Sanhaji M, Matthess Y, Kurunci-Csacsko E, et al. Toxicity modelling of Plk1-targeted therapies in genetically engineered mice and cultured primary mammalian cells. Nat Commun. 2011;2:395.
Carbajosa S, Pansa MF, Paviolo NS, Castellaro AM, Andino DL, Nigra AD, et al. Polo-like kinase 1 inhibition as a therapeutic approach to selectively target BRCA1-deficient cancer cells by synthetic lethality induction. Clin Cancer Res. 2019;25:4049–62.
Zong D, Adam S, Wang Y, Sasanuma H, Callén E, Murga M, et al. BRCA1 haploinsufficiency is masked by RNF168-mediated chromatin ubiquitylation. Mol Cell. 2019;73:1267–81. e7.
Krais JJ, Wang Y, Bernhardy AJ, Clausen E, Miller JA, Cai KQ, et al. RNF168-mediated ubiquitin signaling inhibits the viability of BRCA1-null cancers. Cancer Res. 2020;80:2848–60.
Patel PS, Abraham KJ, Guturi KKN, Halaby M-J, Khan Z, Palomero L, et al. RNF168 regulates R-loop resolution and genomic stability in BRCA1/2-deficient tumors. J Clin Investig. 2021;131:e140105.
Blessing C, Mandemaker IK, Gonzalez-Leal C, Preisser J, Schomburg A, Ladurner AG. The oncogenic helicase ALC1 regulates PARP inhibitor potency by trapping PARP2 at DNA breaks. Mol Cell. 2020;80:862–75.e6.
Verma P, Zhou Y, Cao Z, Deraska PV, Deb M, Arai E, et al. ALC1 links chromatin accessibility to PARP inhibitor response in homologous recombination-deficient cells. Nat Cell Biol. 2021. https://doi.org/10.1038/s41556-020-00624-3.
Juhász S, Smith R, Schauer T, Spekhardt D, Mamar H, Zentout S, et al. The chromatin remodeler ALC1 underlies resistance to PARP inhibitor treatment. Sci Adv. 2020;6:eabb8626.
Hewitt G, Borel V, Segura-Bayona S, Takaki T, Ruis P, Bellelli R, et al. Defective ALC1 nucleosome remodeling confers PARPi sensitization and synthetic lethality with HRD. Mol Cell. 2021(81):767–83.
Srinivasan G, Williamson EA, Kong K, Jaiswal AS, Huang G, Kim H-S, et al. MiR223-3p promotes synthetic lethality in BRCA1-deficient cancers. Proc Natl Acad Sci. 2019;116:17438.
Sun C. The SF3b complex: splicing and beyond. Cell Mol Life Sci. 2020. https://doi.org/10.1007/s00018-020-03493-z.
Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–42.
Przetocka S, Porro A, Bolck HA, Walker C, Lezaja A, Trenner A, et al. CtIP-mediated fork protection synergizes with BRCA1 to suppress genomic instability upon DNA replication stress. Mol Cell. 2018;72:568–82. e6.
Nieminuszczy J, Broderick R, Bellani MA, Smethurst E, Schwab RA, Cherdyntseva V, et al. EXD2 protects stressed replication forks and is required for cell viability in the absence of BRCA1/2. Mol Cell. 2019;75:605–19. e6.
Lim KS, Li H, Roberts EA, Gaudiano EF, Clairmont C, Sambel LA, et al. USP1 is required for replication fork protection in BRCA1-deficient tumors. Mol Cell. 2018;72:925–41. e4.
Zarrizi R, Higgs MR, Voßgröne K, Rossing M, Bertelsen B, Bose M, et al. Germline RBBP8 variants associated with early-onset breast cancer compromise replication fork stability. J Clin Investig. 2020. https://doi.org/10.1172/JCI127521.
Bryan TM. Mechanisms of DNA replication and repair: insights from the study of G-quadruplexes. Molecules. 2019;24:3439.
Zimmer J, Tacconi EMC, Folio C, Badie S, Porru M, Klare K, et al. Targeting BRCA1 and BRCA2 deficiencies with G-quadruplex-interacting compounds. Mol Cell. 2016;61:449–60.
Xu H, Di Antonio M, McKinney S, Mathew V, Ho B, O’Neil NJ, et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat Commun. 2017;8:14432.
Acknowledgements
We thank M.A. Pujana for critical reading of the paper. We would also like to thank all the members of the Hakem laboratory for helpful discussions. We apologize to investigators we were unable to reference due to length limitations. RH holds the Lee K. and Margaret Lau Chair in Breast Cancer Research, which is a joint project with the University of Toronto and The Princess Margaret hospital. RH is supported by the Canadian Institutes of Health Research (FDN 143214), and the Canadian Cancer Society (705367 and 706439). PSP is supported by STARS21, Terry Fox Foundation, Princess Margaret Cancer Foundation, Department of Medical Biophysics, University of Toronto, and the Ontario Graduate Scholarship, Government of Ontario. AA is supported by Canada Graduate Scholarships for Master’s program, Ontario Graduate Scholarship, and the Department of Laboratory Medicine and Pathobiology, University of Toronto.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Patel, P.S., Algouneh, A. & Hakem, R. Exploiting synthetic lethality to target BRCA1/2-deficient tumors: where we stand. Oncogene 40, 3001–3014 (2021). https://doi.org/10.1038/s41388-021-01744-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01744-2
- Springer Nature Limited
This article is cited by
-
Polθ: emerging synthetic lethal partner in homologous recombination-deficient tumors
Cancer Gene Therapy (2024)
-
Combination of PARP Inhibitors and Androgen Receptor Pathway Inhibitors in Metastatic Castration-Resistant Prostate Cancer
Drugs (2024)
-
2-Hydroxy-3-methylanthraquinone inhibits homologous recombination repair in osteosarcoma through the MYC-CHK1-RAD51 axis
Molecular Medicine (2023)
-
Genomic and molecular landscape of homologous recombination deficiency across multiple cancer types
Scientific Reports (2023)
-
PBRM1, SETD2 and BAP1 — the trinity of 3p in clear cell renal cell carcinoma
Nature Reviews Urology (2023)