Abstract
Connective tissue growth factor (CTGF), also known as CCN2, is a member of the CCN protein family of secreted proteins with roles in diverse biological processes. CTGF regulates biological functions such as cell proliferation, migration, adhesion, wound healing, and angiogenesis. In this study, we demonstrate a mechanistic link between CTGF and enhanced aerobic glycolysis in triple-negative breast cancer (TNBC). We found that CTGF is overexpressed in TNBC and high CTGF expression is correlated with a poor prognosis. Also, CTGF was required for in vivo tumorigenesis and in vitro proliferation, migration, invasion, and adhesion of TNBC cells. Our results indicate that extracellular CTGF binds directly to integrin αvβ3, activating the FAK/Src/NF-κB p65 signaling axis, which results in transcriptional upregulation of Glut3. Neutralization of CTGF decreased cell proliferation, migration, and invasion through downregulation of Glut3-mediated glycolytic phenotypes. Overall, our work suggests a novel function for CTGF as a modulator of cancer metabolism, indicating that CTGF is a potential therapeutic target in TNBC.
Similar content being viewed by others
Introduction
Connective tissue growth factor (CTGF, also known as CCN2), first identified as a mitogen secreted by human vascular endothelial cells, is a member of the CCN protein family [1]. As a secreted matricellular protein in the extracellular matrix (ECM), CTGF functions primarily by interacting with cell surface receptors, growth factors, and cytokines [2]. CTGF is involved in cell adhesion, proliferation, migration, and differentiation, and its pathological role in fibrosis has been studied extensively [3, 4]. CTGF is also known to promote tumor growth in pancreas, prostate, brain, and breast cancer, and to be involved in angiogenesis, metastasis, and drug resistance [5, 6].
Breast cancer is the most common cancer diagnosed in women, and the leading cause of death in women worldwide [7]. Triple-negative breast cancer (TNBC), defined by the absence of the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2), is an aggressive subtype of breast cancer with high rates of recurrence and distant metastases [8]. Compared with breast cancer patients with other subtypes, TNBC patients have a poorer prognosis, in part because there are no effective targeted therapies for TNBC [9]. Therefore, there is an unmet need to identify therapeutic targets in TNBC.
Cancer cells are known to have an altered metabolism; particularly, the rate of glycolysis is enhanced, even in the presence of oxygen. This form of aerobic glycolysis, termed the Warburg effect, is known to promote the uncontrolled proliferation of cancer cells by allowing rapid ATP synthesis and biosynthesis [10]. TNBC cells achieve enhanced glycolysis through increased glucose uptake and upregulation of glycolysis-related enzymes such as hexokinase 2, pyruvate kinase 2, and lactate dehydrogenase A.
Various studies have investigated the roles of CTGF in breast cancer. A quantitative real-time PCR-based analysis of 44 primary breast tumors and seven paired normal breast tissues revealed that CTGF was overexpressed in advanced stages of breast cancer and lymph node-positive breast tumors [11]. CTGF-mediated signaling through integrin αvβ3 induced expression of S100A4, leading to increased migration and invasion of TNBC cells [12]. Also, CTGF expression was inversely correlated with drug sensitivity in breast cancer patients and conferred chemoresistance in TNBC patients [13]. These reports collectively indicate that elevated levels of CTGF are associated with the aggressive phenotype of TNBCs. However, it remains unclear whether CTGF is associated with the highly glycolytic phenotype of TNBCs.
In this study, we demonstrate that CTGF induces cancer progression in TNBC by upregulating glycolysis. We found that CTGF was overexpressed in TNBCs and that CTGF transcriptionally upregulated Glut3 through the integrin αvβ3/FAK/Src/NF-κB p65 signaling axis. We also found that the CTGF-induced increase in Glut3 expression functionally supported the enhanced glycolytic phenotype and aggressive phenotype of TNBC. Neutralization of extracellular CTGF using CTGF-specific antibodies resulted in decreased Glut3 expression, followed by decreased cell proliferation, migration, and invasion. Our results indicate a novel mechanism by which CTGF contributes to the aggressive phenotype of TNBC through upregulation of aerobic glycolysis.
Results
CTGF is overexpressed in triple-negative breast cancer
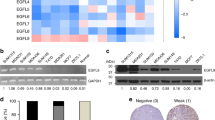
In a publicly available gene expression database, the Cancer Cell Line Encyclopedia portal, we observed that CTGF was highly expressed in breast cancer cells with the TNBC subtype (Fig. 1A, Supplementary Fig. S1A). Furthermore, RNA sequencing data of breast cancer cell lines revealed that CTGF is overexpressed in TNBC subtype cell lines compared to luminal subtype cell lines (GSE100878) (Supplementary Fig. S1B). To confirm that CTGF is overexpressed in TNBC, we evaluated the expression of CTGF in breast cancer cells and observed that CTGF was overexpressed in TNBC subtype cell lines compared to normal (MCF10A) or other subtype cell lines (Fig. 1B). Furthermore, immunohistochemical analysis of a human breast cancer tissue array revealed that CTGF was overexpressed in TNBC tissues compared to normal tissues adjacent to the tumor, benign tissues, and tissues of other breast cancer subtypes (Fig. 1C left and middle panel). CTGF expression was also increased in advanced stages of breast cancer (Fig. 1C right panel). Kaplan–Meier survival analysis using a public clinical database revealed that TNBC patients with high CTGF expression had significantly lower recurrence-free survival rates (RFS) than patients with low CTGF expression (Fig. 1D). These results suggest that CTGF may be associated with cancer progression in TNBC. To further dissect the role of CTGF in TNBC, we generated stable CTGF knockdown cell lines by introducing shRNAs into MDA-MB-231 and Hs578T, two TNBC cell lines with high CTGF expression. To exclude possible effects of clonal selection, two clones were selected from each shRNA-transfected cell line (MDA-MB-231; clones 2 and 3 were derived from shRNA1, clones 10 and 21 were derived from shRNA2; and Hs578T; clones 1 and 3 were derived from shRNA1, clones 7 and 13 were derived from shRNA2). CTGF knockdown was confirmed at both the protein and mRNA levels (Fig. 1E, F). When we subcutaneously injected equal numbers of MDA-MB-231 control shRNA (Cont sh) cells and CTGF shRNA cells into the right flanks of immunodeficient mice, we found that CTGF knocked-down cells were unable to form solid tumors in contrast to control cells, indicating that CTGF is required for in vivo tumorigenicity (Fig. 1G, Supplementary Fig. S1C).
A CTGF mRNA expression values (log2 RPKM) of breast cancer cell lines were clustered according to breast cancer subtype (luminal A, n = 12; luminal B, n = 5; HER2 + , n = 9; TNBC, n = 23). B Upper, western blot analysis of total cell lysates with the indicated antibodies. MCF10A was loaded twice to show equal exposure of the separate blots. β-tubulin was used as a loading control. Middle, protein band densities were relatively quantified using ImageJ software (n = 3, triplicates were repeated for three independent experiments). Values were normalized to MCF10A. Statistical significance was compared to MCF10A. Lower, quantitative real-time PCR showing transcript levels of CTGF (n = 3, triplicates were repeated for three independent experiments). Values were normalized to GAPDH. Statistical significance was compared to MCF10A. C Immunohistochemical analysis of a breast cancer patient tissue array. Expression of CTGF was quantified using ImageJ, (NAT: normal tissue adjacent to the tumor) (n = 75). Histology grading was based on WHO standard. Evaluation was performed with the three aspects: Degree of glandular duct formation, Nuclear pleomorphism, and Nuclear fission counting. D Survival analysis of TNBC patients based on CTGF expression levels using a public patient database (https://kmplot.com/analysis/) (high, n = 65; low, n = 190). E Western blot analysis of total cell lysates and culture media with the indicated antibodies. β-tubulin was used as a loading control for cell lysates and Ponceau S staining of bovine serum albumin was used as a loading control for secreted proteins. F Quantitative real-time PCR showing transcript levels of CTGF (n = 3, triplicates were repeated for three independent experiments). Values were normalized to GAPDH. G Growth of MDA-MB-231 control and CTGF knockdown cell (clone 2) xenografts was monitored over 43 days (n = 3). *P < 0.05; **P < 0.005; ***P < 0.0005. P values in A and C were calculated using one-way ANOVA, post hoc Tukey’s multiple comparisons test; B and F, one-way ANOVA, post hoc Dunnett’s multiple comparisons test; D, log-rank test; G, two-tailed t test.
CTGF regulates cell proliferation, migration, and adhesion
Since we confirmed that CTGF is overexpressed in TNBC and plays a role in in vivo tumor formation, we next sought to further investigate the role of CTGF in TNBC using CTGF knockdown cell lines. We found that CTGF knockdown reduced cell proliferation and colony formation ability in both MDA-MB-231 and Hs578T cells (Fig. 2A, B). To further substantiate these results, we performed flow cytometric cell cycle analyses and found that the decreased cell growth of CTGF knockdown cells was due to cell cycle arrest in the G1 phase (Supplementary Fig. S2). Also, silencing of CTGF in both MDA-MB-231 and Hs578T cells resulted in decreased cell migration and invasion (Fig. 2C). In addition, CTGF knockdown decreased cell adhesion (Fig. 2D). Taken together, these results suggest that CTGF contributes to cancer progression in TNBC by enhancing oncogenic properties of TNBC cells such as cell proliferation, migration, invasion, and adhesion.
A Cell proliferation assays were performed with MDA-MB-231 and Hs578T control and CTGF knockdown cells. B Colony formation assays were performed with MDA-MB-231 and Hs578T control and CTGF knockdown cells. C Transwell migration and invasion assays were performed with MDA-MB-231 and Hs578T control and CTGF knockdown cells. D Adhesion assays were performed with MDA-MB-231 and Hs578T control and CTGF knockdown cells. Data are presented as means ± SDs (n = 3, triplicates were repeated for three independent experiments). ***P < 0.0005. P values in A were calculated using two-way ANOVA, post hoc Dunnett’s multiple comparisons test; B, C, and D, one-way ANOVA, post hoc Dunnett’s multiple comparisons test.
CTGF promotes aerobic glycolysis
TNBCs exhibit altered glucose metabolic phenotypes that support their enhanced rates of proliferation, migration, and invasion [14]. Since our analyses above indicated that CTGF promotes cell proliferation, migration, and invasion, we next investigated the effect of CTGF on the metabolic phenotypes of TNBCs. First, we measured the viability of cells after glucose starvation or treatment with rotenone (20 µM), an oxidative phosphorylation (OXPHOS) inhibitor, for 24 h. CTGF knockdown cells showed decreased sensitivity to glucose starvation (Fig. 3A). Conversely, CTGF knockdown cells had increased sensitivity to rotenone treatment (Fig. 3A). These results indicated that CTGF knockdown cells had decreased dependency on glycolysis and increased dependency on OXPHOS. We also observed that CTGF knockdown cells had decreased glycolytic phenotypes such as glucose uptake, ATP production, and lactate production (Fig. 3B). To directly confirm that the metabolic shift was caused by CTGF knockdown, we examined mitochondrial respiratory capacity and glycolytic flux in CTGF knockdown cells. Consistent with our previous results, basal respiration, ATP-linked respiration, and maximal respiratory capacity of CTGF knockdown cells were higher than those of control cells, while the rate of glycolysis and glycolytic capacity of CTGF knockdown cells were lower than those of control cells (Fig. 3C). Native in-gel lactate dehydrogenase (LDH) isoenzyme assays revealed that while control cells mainly expressed LDH isoforms favoring conversion of pyruvate to lactate (LDH5 and LDH4), CTGF knockdown cells mainly expressed a LDH isoform that favors the reverse reaction (LDH2) (Fig. 3D). These data collectively suggest that CTGF promotes aerobic glycolysis in TNBC cells, resulting in glycolytic phenotypes.
A Cell viability assays were performed with the indicated cell lines after 24 h of glucose starvation or rotenone treatment (20 µM). B Upper, glucose uptake assays; middle, ATP assays; lower, lactate assays were performed with the indicated cell lines. C Upper left, oxygen consumption rate (OCR) of MDA-MB-231 control and CTGF knockdown cells (clone 2) was measured using a Seahorse XFp analyzer. Basal respiration, ATP production, and maximal respiration were determined by the sequential addition of 1 μM oligomycin, 0.5 μM FCCP, and 0.5 μM rotenone-antimycin A. Lower left, extracellular acidification rate (ECAR) of MDA-MB-231 control and CTGF knockdown cells (clone 2) was measured using a Seahorse XFp analyzer. Glycolysis, glycolytic capacity, and glycolytic reserve were determined by the sequential addition of 1 mM glucose, 2 mM oligomycin, and 1 mM 2-deoxy-D-glucose. Upper right, basal respiration, ATP production, and maximal respiration were compared between MDA-MB-231 control and CTGF knockdown cells. Lower right, glycolysis, glycolytic capacity, and glycolytic reserve were compared between MDA-MB-231 control and CTGF knockdown cells. D Native in-gel lactate dehydrogenase (LDH) isoenzyme assays were performed with the indicated cell lines. Data are presented as means ± SDs (n = 3, triplicates were repeated for three independent experiments). *P < 0.05; **P < 0.005; ***P < 0.0005. P values in A and B were calculated using one-way ANOVA, post hoc Dunnett’s multiple comparisons test; C, two-tailed t test.
CTGF is required for focal adhesion and stress fiber formation
Focal adhesions are structures that anchor the actin cytoskeleton of cells to the ECM [15]. Focal adhesions, composed of multiple proteins including integrins, adaptor proteins, nonreceptor tyrosine kinases, small GTPases, and cytoskeletal proteins, serve as signaling scaffolds that modulate numerous signals related to cancer progression [16]. Based on our finding that CTGF knockdown resulted in decreased cell adhesion, we monitored the focal adhesion of cells through F-actin staining with Alexa Fluor 488 conjugated phalloidin (Alexa Fluor 488-phalloidin). We found that CTGF knockdown cells had fewer focal adhesions and stress fibers than control cells (Supplementary Fig. S3A). To quantitatively assess the amount of F-actin contents, we analyzed Alexa Fluor 488-phalloidin stained cells by flow cytometry. We observed that F-actin content was significantly diminished in CTGF knockdown cells compared to control cells (Supplementary Fig. S3B). Since CTGF knockdown resulted in a decrease in focal adhesion and stress fiber formation, we measured activities of RhoA and Rac1, which are known to be responsible for focal adhesion and stress fiber assembly, using a pull-down assay [17]. Consistent with our previous findings, we found that RhoA and Rac1 activity was decreased in CTGF knockdown cells (Supplementary Fig. S3C).
CTGF induces Glut3 expression through integrin avβ3 signaling
Given our observation that CTGF knockdown cells had decreased focal adhesion, we next examined the activity of focal adhesion kinase (FAK) and Src, two major signaling components of focal adhesion [17]. Phosphorylation of both FAK and Src was decreased in CTGF knockdown cells (Fig. 4A). Among the downstream signaling molecules of Src, we focused on NF-κB p65 and Glut3, which is a transcriptional target of NF-κB p65, because proteins related to NF-κB signaling and glycolysis were downregulated in the secretome of CTGF knockdown cells (Supplementary Fig. S4A, B). Also, Glut3 was found to be upregulated in TNBC and Glut3 expression was positively correlated with CTGF expression in multiple datasets (Fig. 4B, C, Supplementary Fig. S5). Furthermore, TNBC patients with high Glut3 expression showed poorer RFS than patients with low Glut3 expression (Fig. 4C). Interestingly, we found that NF-κB p65 activity and Glut3 expression were decreased in CTGF knockdown cells (Fig. 4A). By treating the cells with specific inhibitors (PP2: Src inhibitor, 10 μM; celastrol: NF-κB p65 inhibitor, 0.5 μM), we found that Src induced NF-κB p65 phosphorylation, which led to transcriptional upregulation of Glut3, in accordance with previous reports indicating direct regulation of NF-κB p65 by Src and transcriptional upregulation of Glut3 by NF-κB p65 (Fig. 4D) [18,19,20,21,22]. Other known transcriptional targets of NF-κB p65 related to glycolysis were not altered by CTGF (Supplementary Fig. S6). Ectopic expression of CTGF in MCF7 cells, which showed low expression of CTGF, led to activation of FAK/Src/NF-κB p65 and transcriptional upregulation of Glut3, consistent with our findings (Supplementary Fig. S7).
A Upper, western blot analysis of total cell lysates with the indicated antibodies. Actin was used as a loading control. Middle, dual luciferase assays were performed using NF-κB-luc constructs. pRL-CMV was used as an internal control (n = 3, triplicates were repeated for three independent experiments). Lower, quantitative real-time PCR showing the transcript levels of Glut3 (n = 3, triplicates were repeated for three independent experiments). Values were normalized to GAPDH. B Upper, comparison of Glut3 mRNA expression levels using METABRIC (Luminal A, n = 1369; Luminal B, n = 109; HER2 + , n = 127; TNBC, n = 299) and TCGA (Luminal A, n = 171; Luminal B, n = 53; HER2 + , n = 19; TNBC, n = 42) cohort. Lower, Glut3 protein expression in breast cancer cell lines shown on Supplementary Fig. S5 was clustered into subtypes (Luminal, n = 3; HER2 + , n = 4; TNBC, n = 7). C Left, the correlation between CTGF and Glut3 mRNA expression levels was assessed using expression data from METABRIC (n = 1904) and TCGA (n = 421) cohort. Spearman correlations were calculated. Right, survival analysis of TNBC patients based on Glut3 expression levels (high, n = 86; low, n = 167). D Upper, western blot analysis of total cell lysates with the indicated antibodies. GAPDH was used as a loading control. Middle, dual luciferase assays were performed using NF-κB-luc constructs. pRL-CMV was used as an internal control (n = 3, triplicates were repeated for three independent experiments). Lower, quantitative real-time PCR showing the transcript levels of Glut3 (n = 3, triplicates were repeated for three independent experiments). Values were normalized to GAPDH. *P < 0.05; **P < 0.005; ***P < 0.0005. P values in A and D were calculated using one-way ANOVA, post hoc Dunnett’s multiple comparisons test; B, one-way ANOVA, post hoc Tukey’s multiple comparisons test; C left, two-tailed t test; C right, log-rank test.
Because integrin αvβ3 is known as the receptor for CTGF, we treated integrin αvβ3 neutralizing antibodies (2 μg/ml) and assessed alterations in signaling molecules to further elucidate upstream components involved in CTGF-mediated signaling [12]. We found that integrin αvβ3 antibody treatment decreased FAK/Src/NF-κB p65 phosphorylation and Glut3 expression to the same extent as CTGF neutralizing antibodies (2 μg/ml), indicating that extracellular CTGF signaling is relayed through integrin αvβ3 (Fig. 5A). Furthermore, treatment of CTGF knockdown cells with recombinant human CTGF (rhCTGF, 100 ng/ml) induced activation of the FAK/Src/NF-κB p65 pathway followed by Glut3 upregulation (Fig. 5B). Knockdown of integrin αv and integrin β3 diminished the effect of rhCTGF, further solidifying that CTGF-mediated signaling is relayed through integrin αvβ3 (Fig. 5B, Supplementary Fig. S8). Also, cell fractionation assays revealed that CTGF knockdown and neutralization inhibited NF-κB p65 nuclear translocation, while rhCTGF induced NF-κB p65 translocation to the nucleus (Supplementary Fig. S9). To directly determine if there was a physical association between integrin αvβ3 and CTGF, we performed co-immunoprecipitation assays and found that CTGF was bound to integrin αv and integrin β3 (Fig. 5C). Additionally, cell surface protein biotinylation assays revealed that cell surface-bound CTGF was diminished following integrin αv and integrin β3 knockdown (Fig. 5D).
A Upper, western blot analysis of total cell lysates with the indicated antibodies. GAPDH was used as a loading control. Middle, dual luciferase assays were performed using NF-κB-luc constructs. pRL-CMV was used as an internal control (n = 3, triplicates were repeated for three independent experiments). Lower, quantitative real-time PCR showing the transcript levels of Glut3 (n = 3, triplicates were repeated for three independent experiments). Values were normalized to GAPDH. B Upper, western blot analysis of total cell lysates with the indicated antibodies (MDA-MB-231 CTGF sh: clone 2; Hs578T CTGF sh: clone 1). Actin was used as a loading control. Middle, dual luciferase assays were performed using NF-κB-luc constructs. pRL-CMV was used as an internal control (n = 3, triplicates were repeated for three independent experiments). Lower, quantitative real-time PCR showing the transcript levels of Glut3 (n = 3, triplicates were repeated for three independent experiments). Values were normalized to GAPDH. C Co-immunoprecipitation assays were performed by immunoprecipitation using the indicated antibodies followed by western blot analysis with the indicated antibodies. D Cell surface biotinylation assays were performed with the cells using the indicated experimental conditions and analyzed by western blotting with the indicated antibodies. β-tubulin was used as a loading control. ***P < 0.0005. P values in A and B were calculated using one-way ANOVA, post hoc Dunnett’s multiple comparisons test.
CTGF/NF-κB p65/Glut3 signaling is required for enhanced proliferation of TNBC cells
Given our finding that extracellular CTGF-mediated signaling through the integrin αvβ3/FAK/Src/NF-κB p65 axis resulted in upregulation of Glut3, we next investigated whether Glut3 plays a functional role in TNBC. Glut3 knockdown (using two independent siRNAs) in control cells resulted in downregulation of glycolytic phenotypes while Glut3 overexpression (at similar expression levels with endogenous Glut3) in CTGF knockdown cells induced glycolytic phenotypes (Supplementary Figs. S10, S11A, B). Furthermore, neutralization of CTGF and integrin αvβ3 (Supplementary Fig. S11C) and treatment with PP2 and celastrol (Supplementary Fig. S11D) led to a decrease in glycolytic phenotypes while rhCTGF treatment increased glycolytic phenotypes through integrin αvβ3 (Supplementary Fig. S11E). These results indicate that aerobic glycolysis in TNBC cells, at least in part, relies on CTGF-mediated Glut3 expression. To further assess the functional consequences of CTGF-induction of Glut3 expression, we hypothesized that the CTGF/NF-κB p65/Glut3 axis may be required for TNBC cell growth and migration because aerobic glycolysis is known to be responsible for the aggressive growth and motility of TNBC cells. First, to test whether extracellular CTGF-mediated signaling directly affects cell proliferation, we treated cells with CTGF and integrin αvβ3 neutralizing antibodies and performed cell proliferation assays. We observed decreased cell proliferation following those treatments (Fig. 6A). Treatment with PP2 and celastrol both phenocopied the effects of CTGF downregulation on cell proliferation (Fig. 6B). Furthermore, rhCTGF treatment induced cell proliferation of CTGF knockdown cells and integrin αv or integrin β3 knockdown hindered the effect of rhCTGF (Fig. 6C). Furthermore, knockdown of Glut3 attenuated cell proliferation of control cells to a similar degree to CTGF knockdown cells while overexpression of Glut3 enhanced cell proliferation of CTGF knockdown cells to a similar degree to control cells (Fig. 6D, E). These results directly indicate that Glut3 is functionally responsible for CTGF-mediated upregulation of cell proliferation. Also, to further confirm the effect of CTGF/NF-κB p65/Glut3 axis on cell proliferation, we conducted colony formation assays and cell cycle analyses. CTGF and integrin αvβ3 neutralization, PP2 and celastrol treatment, and Glut3 knockdown decreased colony formation and induced G1 phase cell cycle arrest, while rhCTGF treatment and Glut3 overexpression showed the opposite effects, consistent with our previous results (Supplementary Figs. S12, S13, S14).
Cell proliferation assays were performed with the indicated cell lines and experimental conditions (MDA-MB-231 CTGF sh: clone 2; Hs578T CTGF sh: clone 1). ***P < 0.0005. Data are presented as means ± SDs (n = 3, triplicates were repeated for three independent experiments). P values in A, B, C, and D were calculated using two-way ANOVA, post hoc Dunnett’s multiple comparisons test; E, two-way ANOVA, post hoc Tukey’s multiple comparisons test.
CTGF/NF-κB p65/Glut3 signaling confers cell migration and invasion
To examine whether the extracellular CTGF-mediated signaling axis plays a direct role in cell migration and invasion, we treated cells with CTGF and integrin αvβ3 neutralizing antibodies and performed transwell assays. Neutralization of CTGF and integrin αvβ3 resulted in decreased cell migration and invasion (Fig. 7A). Also, PP2 and celastrol treatment decreased cell migration and invasion, phenocopying the effects of CTGF inhibition (Fig. 7B). Decreased cell migration and invasion ability of CTGF knockdown cells were recovered by rhCTGF treatment, while integrin αv and β3 knockdown blocked the effect of rhCTGF (Fig. 7C). Furthermore, knockdown of Glut3 decreased cell migration and invasion of control cells, while overexpression of Glut3 increased cell migration and invasion of CTGF knockdown cells, indicating that CTGF-induced upregulation of Glut3 was responsible for the migration and invasion of control cells (Fig. 7D, E).
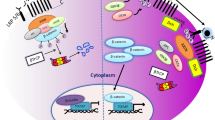
A, B, C, D, E Transwell migration and invasion assays were performed with the indicated cells and indicated experimental conditions (MDA-MB-231 CTGF sh: clone 2; Hs578T CTGF sh: clone 1). Quantified intensities of stained crystal violet using ImageJ is graphed below representative images. F A schematic diagram showing the CTGF-mediated signaling axis in TNBC. Data are presented as means ± SDs (n = 3, triplicates were repeated for three independent experiments). **P < 0.005; ***P < 0.0005. P values in A, B, C, and D were calculated using one-way ANOVA, post hoc Dunnett’s multiple comparisons test; E, two-tailed t test.
Since we previously found that RhoA and Rac1 activity was downregulated by CTGF knockdown, we sought to further clarify the role of RhoA and Rac1 in CTGF-mediated signaling (Supplementary Fig. S3C). RhoA and Rac1 knockdown downregulated proliferation, migration, and invasion but did not affect glycolysis (Supplementary Fig. S15). Also, RhoA and Rac1 knockdown had no effect on rhCTGF-mediated increase in proliferation, migration, invasion, glycolysis, and activation of the FAK/Src/NF-κB p65/Glut3 axis (Supplementary Figs. S16, S17). Since treatment with rhCTGF induced activation of RhoA and Rac1 but RhoA and Rac1 knockdown did not impair rhCTGF-mediated activation of the FAK/Src/NF-κB p65/Glut3 signaling and PP2 treatment downregulated RhoA and Rac1 activity, we concluded that RhoA and Rac1 was activated by CTGF, downstream of FAK/Src (Supplementary Fig. S17).
Also, our results indicate that TNBCs rely on Glut3 for enhanced glycolysis, leading to increased proliferation and migration. Since Glut1 is known as the ubiquitously expressed basal glucose transporter, we tried to compare the role of Glut1 and 3 on those phenotypes. Glut1 knockdown decreased glucose uptake in a higher degree compared to Glut3 knockdown (Supplementary Figs. S11A, S18A, B). However, the impact of Glut1 knockdown on cell proliferation, migration, and invasion was similar to Glut3 knockdown (Figs. 6D, 7D, Supplementary Figs. S12D, S14A, S18C–F).
To further confirm our results, we reproduced rhCTGF-mediated activation of the integrin αvβ3/FAK/Src/NF-κB p65/Glut3 axis leading to increased cell proliferation, migration, and invasion using other CTGF knockdown clones (Supplementary Figs. S19, S20). Also, shRNA-resistant CTGF expression restored CTGF-mediated signaling in the CTGF knockdown cells (Supplementary Fig. S21).
Discussion
TNBCs do not express ER, PR, or HER2, and thus do not respond to currently available targeted therapeutics in contrast with hormone receptor and HER2-positive tumors [8]. In this study, we identified a novel function of CTGF in TNBC, reinforcing the therapeutic potential of neutralizing extracellular CTGF for TNBC treatment. We found that CTGF is overexpressed in TNBC using a breast cancer patient tissue array, public cancer cell line database, and directly measuring the expression levels of this protein in breast cancer cell lines. High CTGF expression was correlated with advanced breast cancer stages and with poorer TNBC patient survival. Knockdown of CTGF abolished tumorigenesis in vivo and led to decreased proliferation, migration, invasion, and glycolysis in vitro. We demonstrated a mechanistic link between CTGF and Glut3, whereby CTGF induces Glut3 at the transcriptional level, leading to increased glycolytic phenotypes followed by upregulation of cell growth and motility (Fig. 7F).
Several studies have demonstrated an oncogenic function for CTGF in numerous types of cancers including breast cancer, chondrosarcoma, colon cancer, gallbladder carcinoma, glioma, leukemia, melanoma, pancreatic cancer, prostate cancer, and thyroid carcinoma [23,24,25,26,27,28,29,30,31,32]. Shimo et al. demonstrated that secreted CTGF from breast cancer and fibrosarcoma cells led to increased neovascularization, while neutralization of secreted CTGF using CTGF-specific antibodies diminished the pro-angiogenic effects of CTGF [33]. Also, the same research group reported that neutralization of CTGF decreased bone metastasis of breast cancer cells in mice [34]. CTGF stimulated migration and angiogenesis of breast cancer through the C-terminal CT domain [35]. Knockdown of CTGF in glioma cells resulted in decreased cell migration through downregulation of FAK, p-FAK, and SPARC [36]. Welch et al. found that CTGF was overexpressed in 75% of precursor B-cell acute lymphoblastic leukemias and was correlated with a poor prognosis [30]. Immunohistochemical analysis of 131 thyroid tissue samples revealed that CTGF level was increased in papillary thyroid carcinoma tissues compared to benign thyroid tumor tissues. Also, CTGF level was positively correlated with advanced stages and lymph node metastasis [29]. In pancreatic cancer, FG-3019, a humanized CTGF neutralizing antibody, inhibited tumor growth and metastasis and enhanced gemcitabine efficacy in vivo [37, 38]. By contrast, CTGF was downregulated in ovarian cancer through promoter hypermethylation and CTGF expression was lower in primary lung cancer specimens than normal lung samples [39, 40]. Interestingly, CTGF was found to be overexpressed in MDA-MB-231, a TNBC cell line, and conferred cell migration and invasion through integrin αvβ3/ERK signaling-mediated upregulation of S100A4 and induced chemoresistance through upregulation of Bcl-xL and cIAP1 through the same signaling axis [12, 13]. We also found that CTGF knockdown cells had decreased levels of ERK phosphorylation (data not shown). Because ERK is known to be associated with diverse biological phenotypes of cancers such as cell proliferation, metastasis, and survival, we assume that CTGF-mediated carcinogenesis does not rely solely on the linear signaling axis that results in Glut3 upregulation [41]. Rather, we propose that our novel finding that CTGF induces Glut3 through activation of the FAK/Src/NF-κB p65 axis represents an additional dimension of CTGF-mediated signaling in TNBC. Also, since we found decreased RhoA and Rac1 activity in CTGF knockdown cells, we tried to further determine the role of RhoA and Rac1 in CTGF-mediated signaling (Supplementary Fig. S3C). RhoA and Rac1 knockdown resulted in decreased cell proliferation, migration, and invasion and alteration of the cell cycle, consistent with previous reports (Supplementary Fig. S15) [42, 43]. However, RhoA and Rac1 knockdown had no effect on glycolytic phenotypes of TNBC cells and rhCTGF-mediated upregulation of cell proliferation, migration, invasion, glycolysis, and FAK/Src/NF-κB p65/Glut3 signaling (Supplementary Figs. S15E, S16, S17B). While rhCTGF treatment activated RhoA and Rac1, RhoA and Rac1 knockdown did not alter rhCTGF-mediated activation of the FAK/Src/NF-κB p65/Glut3 axis and treatment with PP2 decreased RhoA and Rac1 activity (Supplementary Fig. S17). Collectively, these results indicate that RhoA and Rac1 activity was modulated by CTGF, downstream of the FAK/Src signaling axis.
Metabolic reprogramming from OXPHOS to glycolysis is a well-known characteristic of cancer, first described by Otto Warburg [10]. Genes related to the glycolysis pathway are upregulated in more than 70% of cancer cases [44]. Increased glycolysis, even in the presence of oxygen, supports the growth of cancer through rapid ATP production and providing the building blocks required for biosynthesis. The Warburg effect also promotes tumor migration and invasion through diverse mechanisms such as creating an acidic microenvironment, which activates proteases that induce ECM degradation [45]. Recent studies have demonstrated that the altered expression of genes that cause this metabolic shift in cancer is due to transcription factors such as NF-κB, hypoxia-inducible factor-1, and loss of p53. Interestingly, Kawauchi et al. showed that NF-κB p65 is activated in p53 deficient cancer cells, inducing Glut3 upregulation followed by enhanced glucose metabolism. Furthermore, exogenous Glut3 expression induced glucose uptake and oncogenic cell transformation [20]. The cell lines we used in our study are both known to have a mutant form of p53; the CTGF-mediated activation of NF-κB p65/Glut3 is therefore consistent with the findings of previous studies regarding the oncogenic function of NF-κB p65-induced Glut3 expression. Since NF-κB p65 is also known to regulate Glut1, hexokinase 2, pyruvate kinase isozyme M2, and synthesis of cytochrome c oxidase 2, which are genes related to glucose metabolism, we checked whether the expression of those genes were altered by CTGF [46,47,48,49]. We found that the expression levels of those genes were not altered while Glut3 was significantly downregulated in CTGF knockdown cells (Supplementary Fig. S6). Also, rhCTGF treatment to the CTGF knockdown cells restored Glut3 expression while the expression of other NF-κB p65 targets were not altered (Supplementary Fig. S6).
Glut3 has not been extensively studied in breast cancer since Glut1 was first characterized as the major glucose transporter in diverse tissues [50]. However, recent reports have indicated that both Glut1 and Glut3 are associated with breast cancer progression [51]. Expression analysis of 70 breast cancer patient samples revealed that Glut1 and Glut3 were expressed in 50 and 40% of samples, respectively. Glut3 expression was also significantly higher in breast tumors with advanced grades [52]. Interestingly, Kuo et al. showed that Glut3 is upregulated in brain metastatic breast cancer and that Glut3 conferred cell malignancy through enhanced glucose metabolism [53]. Our results suggest that CTGF induces the growth, migration, and invasion of TNBC cells in a Glut3-dependent manner. Since Glut1 is known as the basal glucose transporter in various cell types, to compare the dependency of TNBC on Glut1 and Glut3, we transfected Glut1 siRNA to MDA-MB-231 and Hs578T cells to knockdown Glut1 and performed proliferation, migration, invasion, and metabolism assays (Supplementary Fig. S18). While Glut1 knockdown resulted in 64–69% decreased glucose uptake, Glut3 knockdown resulted in 48–59% decrease, indicating that glucose uptake of TNBCs relies more on Glut1 than Glut3 (Supplementary Figs. S11A, S18B). However, ATP level, lactate production, proliferation, migration, and invasion rates were similar in Glut1 and Glut3 siRNA transfected cells, suggesting that Glut1 and Glut3 both contributes to enhanced glycolysis followed by increased proliferation, migration, and invasion in a similar degree (Figs. 6D, 7D, Supplementary Figs. S11A, S18). Extracellular CTGF induced Glut3 expression through the FAK/Src/NF-κB p65 signaling axis; blocking extracellular CTGF binding to integrin αvβ3 using CTGF-specific antibodies resulted in significant downregulation of Glut3, followed by diminished oncogenic properties. This novel mechanistic link between CTGF, an ECM component, and glucose metabolism in TNBC furthers our understanding of how outside-in signaling can enhance cancer progression through upregulation of glycolysis.
Materials and methods
Cell culture
MDA-MB-231, Hs578T, and MCF7 human breast cancer cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). All cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM, 10-013-CVR, Corning, NY, USA) supplemented with 10% fetal bovine serum (FBS, Youngin Frontier, Seoul, Korea), 100 U/ml penicillin (15140-122, Corning), and 100 µg/ml streptomycin (15140-122, Corning). Cells were incubated at 37 °C in a humidified atmosphere with 5% CO2.
Detailed descriptions of the materials and methods, including the list of antibodies (Supplementary Table S1) and oligonucleotide sequences used for shRNA generation (Supplementary Table S2), cloning (Supplementary Table S3), mutagenesis (Supplementary Table S3), and qRT-PCR (Supplementary Table S4), used in this study are provided in the Supplementary Materials and Methods.
References
Bradham DM, Igarashi A, Potter RL, Grotendorst GR. Connective tissue growth factor: a cysteine-rich mitogen secreted by human vascular endothelial cells is related to the SRC-induced immediate early gene product CEF-10. J Cell Biol. 1991;114:1285–94.
Leask A, Abraham DJ. All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J Cell Sci. 2006;119:4803–10.
Hall-Glenn F, Lyons KM. Roles for CCN2 in normal physiological processes. Cell Mol Life Sci. 2011;68:3209–17.
Lipson KE, Wong C, Teng Y, Spong S. CTGF is a central mediator of tissue remodeling and fibrosis and its inhibition can reverse the process of fibrosis. Fibrogenes Tissue Repair. 2012;5:S24.
Jun JI, Lau LF. Taking aim at the extracellular matrix: CCN proteins as emerging therapeutic targets. Nat Rev Drug Discov. 2011;10:945–63.
Kim H, Son S, Shin I. Role of the CCN protein family in cancer. BMB Rep. 2018;51:486–92.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Bosch A, Eroles P, Zaragoza R, Vina JR, Lluch A. Triple-negative breast cancer: molecular features, pathogenesis, treatment and current lines of research. Cancer Treat Rev. 2010;36:206–15.
Aysola K, Desai A, Welch C, Xu J, Qin Y, Reddy V. et al. Triple negative breast cancer - an overview. Hereditary Genet. 2013;2013:001.
Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41:211–8.
Xie D, Nakachi K, Wang H, Elashoff R, Koeffler HP. Elevated levels of connective tissue growth factor, WISP-1, and CYR61 in primary breast cancers associated with more advanced features. Cancer Res. 2001;61:8917–23.
Chen PS, Wang MY, Wu SN, Su JL, Hong CC, Chuang SE, et al. CTGF enhances the motility of breast cancer cells via an integrin-alphavbeta3-ERK1/2-dependent S100A4-upregulated pathway. J Cell Sci. 2007;120:2053–65.
Wang MY, Chen PS, Prakash E, Hsu HC, Huang HY, Lin MT, et al. Connective tissue growth factor confers drug resistance in breast cancer through concomitant up-regulation of Bcl-xL and cIAP1. Cancer Res. 2009;69:3482–91.
Sun X, Wang M, Wang M, Yu X, Guo J, Sun T, et al. Metabolic reprogramming in triple-negative breast cancer. Front Oncol. 2020;10:428.
Geiger B, Bershadsky A, Pankov R, Yamada KM. Transmembrane crosstalk between the extracellular matrix-cytoskeleton crosstalk. Nat Rev Mol Cell Biol. 2001;2:793–805.
Tai YL, Chen LC, Shen TL. Emerging roles of focal adhesion kinase in cancer. Biomed Res Int. 2015;2015:690690.
Wozniak MA, Modzelewska K, Kwong L, Keely PJ. Focal adhesion regulation of cell behavior. Biochim Biophys Acta. 2004;1692:103–19.
Lee HS, Moon C, Lee HW, Park EM, Cho MS, Kang JL. Src tyrosine kinases mediate activations of NF-kappaB and integrin signal during lipopolysaccharide-induced acute lung injury. J Immunol. 2007;179:7001–11.
Lluis JM, Buricchi F, Chiarugi P, Morales A, Fernandez-Checa JC. Dual role of mitochondrial reactive oxygen species in hypoxia signaling: activation of nuclear factor-{kappa}B via c-SRC and oxidant-dependent cell death. Cancer Res. 2007;67:7368–77.
Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat Cell Biol. 2008;10:611–8.
Zha X, Hu Z, Ji S, Jin F, Jiang K, Li C, et al. NFkappaB up-regulation of glucose transporter 3 is essential for hyperactive mammalian target of rapamycin-induced aerobic glycolysis and tumor growth. Cancer Lett. 2015;359:97–106.
Watanabe M, Abe N, Oshikiri Y, Stanbridge EJ, Kitagawa T. Selective growth inhibition by glycogen synthase kinase-3 inhibitors in tumorigenic HeLa hybrid cells is mediated through NF-kappaB-dependent GLUT3 expression. Oncogenesis. 2012;1:e21.
Bennewith KL, Huang X, Ham CM, Graves EE, Erler JT, Kambham N, et al. The role of tumor cell-derived connective tissue growth factor (CTGF/CCN2) in pancreatic tumor growth. Cancer Res. 2009;69:775–84.
Braig S, Wallner S, Junglas B, Fuchshofer R, Bosserhoff AK. CTGF is overexpressed in malignant melanoma and promotes cell invasion and migration. Br J Cancer. 2011;105:231–8.
Garcia P, Leal P, Ili C, Brebi P, Alvarez H, Roa JC. Inhibition of connective tissue growth factor (CTGF/CCN2) in gallbladder cancer cells leads to decreased growth in vitro. Int J Exp Pathol. 2013;94:195–202.
Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–49.
Ladwa R, Pringle H, Kumar R, West K. Expression of CTGF and Cyr61 in colorectal cancer. J Clin Pathol. 2011;64:58–64.
Shakunaga T, Ozaki T, Ohara N, Asaumi K, Doi T, Nishida K, et al. Expression of connective tissue growth factor in cartilaginous tumors. Cancer. 2000;89:1466–73.
Wang G, Zhang W, Meng W, Liu J, Wang P, Lin S, et al. Expression and clinical significance of connective tissue growth factor in thyroid carcinomas. J Int Med Res. 2013;41:1214–20.
Welch MD, Greene WK, Kees UR. Hypomethylation of the CTGF gene locus is a common feature of paediatric pre-B acute lymphoblastic leukaemia. Br J Haematol. 2013;162:537–41.
Yang F, Tuxhorn JA, Ressler SJ, McAlhany SJ, Dang TD, Rowley DR. Stromal expression of connective tissue growth factor promotes angiogenesis and prostate cancer tumorigenesis. Cancer Res. 2005;65:8887–95.
Yin D, Chen W, O’Kelly J, Lu D, Ham M, Doan NB, et al. Connective tissue growth factor associated with oncogenic activities and drug resistance in glioblastoma multiforme. Int J Cancer. 2010;127:2257–67.
Shimo T, Nakanishi T, Nishida T, Asano M, Sasaki A, Kanyama M, et al. Involvement of CTGF, a hypertrophic chondrocyte-specific gene product, in tumor angiogenesis. Oncology. 2001;61:315–22.
Shimo T, Kubota S, Yoshioka N, Ibaragi S, Isowa S, Eguchi T, et al. Pathogenic role of connective tissue growth factor (CTGF/CCN2) in osteolytic metastasis of breast cancer. J Bone Min Res. 2006;21:1045–59.
Chien W, O’Kelly J, Lu D, Leiter A, Sohn J, Yin D, et al. Expression of connective tissue growth factor (CTGF/CCN2) in breast cancer cells is associated with increased migration and angiogenesis. Int J Oncol. 2011;38:1741–7.
Lee HK, Bier A, Cazacu S, Finniss S, Xiang C, Twito H, et al. MicroRNA-145 is downregulated in glial tumors and regulates glioma cell migration by targeting connective tissue growth factor. PLoS ONE. 2013;8:e54652.
Neesse A, Frese KK, Bapiro TE, Nakagawa T, Sternlicht MD, Seeley TW, et al. CTGF antagonism with mAb FG-3019 enhances chemotherapy response without increasing drug delivery in murine ductal pancreas cancer. Proc Natl Acad Sci USA. 2013;110:12325–30.
Dornhofer N, Spong S, Bennewith K, Salim A, Klaus S, Kambham N, et al. Connective tissue growth factor-specific monoclonal antibody therapy inhibits pancreatic tumor growth and metastasis. Cancer Res. 2006;66:5816–27.
Kikuchi R, Tsuda H, Kanai Y, Kasamatsu T, Sengoku K, Hirohashi S, et al. Promoter hypermethylation contributes to frequent inactivation of a putative conditional tumor suppressor gene connective tissue growth factor in ovarian cancer. Cancer Res. 2007;67:7095–105.
Chen PP, Li WJ, Wang Y, Zhao S, Li DY, Feng LY, et al. Expression of Cyr61, CTGF, and WISP-1 correlates with clinical features of lung cancer. PLoS ONE. 2007;2:e534.
Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–90.
Pille JY, Denoyelle C, Varet J, Bertrand JR, Soria J, Opolon P, et al. Anti-RhoA and anti-RhoC siRNAs inhibit the proliferation and invasiveness of MDA-MB-231 breast cancer cells in vitro and in vivo. Mol Ther. 2005;11:267–74.
Yoshida T, Zhang Y, Rivera Rosado LA, Chen J, Khan T, Moon SY, et al. Blockade of Rac1 activity induces G1 cell cycle arrest or apoptosis in breast cancer cells through downregulation of cyclin D1, survivin, and X-linked inhibitor of apoptosis protein. Mol Cancer Ther. 2010;9:1657–68.
Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84:1014–20.
Han T, Kang D, Ji D, Wang X, Zhan W, Fu M, et al. How does cancer cell metabolism affect tumor migration and invasion? Cell Adh Migr. 2013;7:395–403.
Wang X, Liu R, Qu X, Yu H, Chu H, Zhang Y, et al. alpha-Ketoglutarate-activated NF-kappaB signaling promotes compensatory glucose uptake and brain tumor development. Mol Cell. 2019;76:148–62.e147.
Londhe P, Yu PY, Ijiri Y, Ladner KJ, Fenger JM, London C, et al. Classical NF-kappaB metabolically reprograms sarcoma cells through regulation of hexokinase 2. Front Oncol. 2018;8:104.
Han D, Wei W, Chen X, Zhang Y, Wang Y, Zhang J, et al. NF-kappaB/RelA-PKM2 mediates inhibition of glycolysis by fenofibrate in glioblastoma cells. Oncotarget. 2015;6:26119–28.
Mauro C, Leow SC, Anso E, Rocha S, Thotakura AK, Tornatore L, et al. NF-kappaB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nat Cell Biol. 2011;13:1272–9.
Adekola K, Rosen ST, Shanmugam M. Glucose transporters in cancer metabolism. Curr Opin Oncol. 2012;24:650–4.
Ancey PB, Contat C, Meylan E. Glucose transporters in cancer - from tumor cells to the tumor microenvironment. FEBS J. 2018;285:2926–43.
Krzeslak A, Wojcik-Krowiranda K, Forma E, Jozwiak P, Romanowicz H, Bienkiewicz A, et al. Expression of GLUT1 and GLUT3 glucose transporters in endometrial and breast cancers. Pathol Oncol Res. 2012;18:721–8.
Kuo M-H, Chang W-W, Yeh B-W, Chu Y-S, Lee Y-C, Lee H-T. Glucose transporter 3 is essential for the survival of breast cancer cells in the brain. Cells. 2019;8:1568.
Acknowledgements
This work was supported by National Research Foundation of Korea (NRF) grants funded by the Korea government (MIST) (2019R1H1A2079999, 2020R1F1A1048616, 2020R1A6A3A13074546).
Financial Support
Grant source, National Research Foundation of Korea (NRF); Grant numbers (grant recipient), 2019R1H1A2079999 (Incheol Shin), 2020R1F1A1048616 (Incheol Shin), 2020R1A6A3A13074546 (Hyungjoo Kim).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, H., Son, S., Ko, Y. et al. CTGF regulates cell proliferation, migration, and glucose metabolism through activation of FAK signaling in triple-negative breast cancer. Oncogene 40, 2667–2681 (2021). https://doi.org/10.1038/s41388-021-01731-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-021-01731-7
- Springer Nature Limited
This article is cited by
-
Application of color doppler ultrasound and US shear wave elastography with connective tissue growth factor in the risk assessment of papillary thyroid carcinoma
BMC Medical Imaging (2024)
-
The effect of selenium on the proliferation of bovine endometrial epithelial cells in a lipopolysaccharide-induced damage model
BMC Veterinary Research (2024)
-
Expected and unexpected effects after systemic inhibition of Hippo transcriptional output in cancer
Nature Communications (2024)
-
Trophoblast fusion in fetal growth restriction is inhibited by CTGF in a cell-cycle-dependent manner
Journal of Molecular Histology (2024)
-
GPER deletion triggers inhibitory effects in triple negative breast cancer (TNBC) cells through the JNK/c-Jun/p53/Noxa transduction pathway
Cell Death Discovery (2023)