Abstract
Long noncoding RNAs (lncRNAs) are pervasive transcripts that play pivotal roles in regulating chromatin dynamics, gene and protein expression. Aberrant expression and mutations of lncRNAs represent a driving force behind tumor invasion and metastasis, making them attractive cancer targets. However, most of the lncRNAs are still being discovered and conclusive experimental evidence for their functional relevance is still lacking for most malignancies. In this study, a differentially expressed lncRNA, designated as lnc-CRCMSL, is identified by microarray-based screenings on non-metastatic and metastatic CRC specimens. Lnc-CRCMSL is verified as an anti-metastatic gene and negatively correlated with the poor prognosis of CRC patients. Lnc-CRCMSL overexpression restricts tumor growth and metastasis in vivo and in vitro. Instead, lnc-CRCMSL silencing accelerates CRC cell proliferation and migration. RNA-pulldown assay identifies high mobility group box 2 (HMGB2) as a downstream protein of lnc-CRCMSL. Mechanically, lnc-CRCMSL physically binds to HMGB2 and stabilizes the localization of HMGB2 in the cytoplasm. Notably, lnc-CRCMSL knockdown lead to the shift of HMGB2 into nuclear, in which it triggers epithelial to mesenchymal transition (EMT) programming. Importantly, lnc-CRCMSL controls the cytoplasmic retention of HMGB2 and attenuates the interaction between HMGB2 and OCT4 to suppress EMT. Treatment of leptomycin B (LMB), a potent and specific nuclear export inhibitor, counteracts lnc-CRCMSL-mediated suppression of aggressive phenotypes and EMT process by accumulating the nuclear HMGB2.
Conclusion
Our data highlight the anti-metastatic role of lnc-CRCMSL in stabilizing HMGB2 through lncRNA-protein interactions in the cytoplasm, and suggest that targeting lnc-CRCMSL may represent a therapeutic opportunity for managing metastatic CRC.
Similar content being viewed by others
Introduction
Long noncoding RNAs (lncRNAs) are a group of non-protein-coding transcripts that are longer than 200 nucleotides [1, 2]. Despite of fast turnover rates and low copy numbers, lncRNAs have been widely accepted as truly functional biomolecules [3]. LncRNAs carry out diverse functions in regulating development, differentiation, and metabolism. Especially, altered expression or mutations of lncRNA are closely linked to carcinogenesis, metastasis, and tumor staging [4]. The tremendously large number and tumor specific expression of certain lncRNAs have made lncRNAs as ideal biomarkers for diagnosis and attractive targets for tumor intervention [5, 6]. Emerging evidence points to an active role for lncRNAs in every stage of metastasis from cancer cell invasion, migration, and metastatic colonization in distant organs [7]. For example, colon-cancer associated transcript 2 (CCAT2) upregulated in microsatellite-stable CRC is correlated with migration and metastasis [8]. Oncogene c-myc directly induces the expression of the lncRNA H19 and drives the clonogenicity and anchorage-independent growth of cancer cells [9]. Although thousands of lncRNAs have been annotated, only a few lncRNAs have been functionally characterized.
Colorectal cancer (CRC) is a major public health problem, being the third most commonly diagnosed cancer and the fourth cause of cancer death worldwide. 20%–25% of CRC patients were diagnosed with metastatic foci, and additional 20%–25% of patients will develop metastases later. The overall mortality rate for metastatic CRC is 40%–45% [10]. Epithelial to mesenchymal transition (EMT) is widely regarded as a key step for CRC to acquire an invasive or metastatic phenotype. Our previous work shows that TGFβ triggers EMT process by dampening E-cadherin and upregulating vimentin in multiple CRC cell lines [11]. Recent studies also demonstrated that HOX transcript antisense intergenic RNA (HOTAIR) induces EMT through downregulating E-cadherin and augmenting vimentin and MMP9. Long noncoding RNA-activated by TGFβ (lncRNA-ATB), which is upregulated in serum of CRC patient, induces EMT by decreasing epithelial markers E-cadherin, ZO-1, and increasing expression of mesenchymal markers ZEB1 and N-cadherin (N-cad) [12]. Given the complex role of lncRNAs, their regulatory mechanisms underlying EMT and tumor metastasis are poorly clarified.
LncRNAs are pervasive transcripts that influence the complex metastasis process. However, many of which are still being discovered and are yet to be annotated. In the study, clinical specimens from CRC patients with or without metastasis were assayed to screen differentially expressed lncRNAs. We identify a candidate lncRNA that is negatively correlated with CRC metastasis, which is coined as lnc-CRCMSL (Colorectal Cancer Metastasis-Suppressed LncRNA). It is well known that lncRNA has a wide range of sources, including the UTR region. Lnc-CRCMSL, predicted as a cis-regulator of mitogen-activated protein kinase kinase 6 (MAP2K6) gene, overlaps the untranslated region (UTR) of a protein-coding gene. The lnc-CRCMSL has been previously reported, for example, TCGA database RNA-seq data analysis identified 83 lincRNAs showing the significant expression changes in breast tumors which lnc-CRCMSL included [13]. Lnc-CRCMSL were also downregulated after treatment with cisplatin in HepG2 cells and induced HepG2 cell cycle arrest with co-expression genes [14]. Our results further show that lnc-CRCMSL binds to high mobility group box 2 (HMGB2) and inhibits the EMT process in CRC cells. Our observation shows lnc-CRCMSL regulates a dynamic distribution of HMGB2 between cytoplasm and nucleus of CRC cells. Notably, lnc-CRCMSL regulates the nucleocytoplasmic shuttling of HMGB2 and functions as a CRC metastasis suppressor gene that has no translational potential.
Results
Microarray analysis screens CRC metastasis-associated lncRNAs and mRNAs
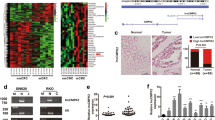
To identify transcripts that potentially drive CRC metastasis, lncRNA and messenger RNA (mRNA) expression profiles were determined by microarray analysis. The microarray data were deposited in NCBI Gene Expression Omnibus and are accessible through GEO series accession number GSE113296). A total of 134,870 lncRNA probes were detected, among which 65,174 genes were upregulated and 69,696 genes were downregulated. Screening conditions were set to reduce the number of candidate lncRNAs. Differential expressed intergenic genes (FC > 1.5 or < 0.67, and P-value < 0.01) that <3000 bp were selected and the repeated siftings have brought 105 candidate lncRNAs. Hierarchical clustering showed systematic variations in the expression of lncRNAs and protein-coding RNAs between metastatic CRC (mCRC) and non-metastatic CRC (nmCRC) tissues (Fig. 1a). We identified a candidate lncRNA as a colorectal cancer metastasis-suppressed lncRNA (referred to as lnc-CRCMSL hereafter), also named RP1193H18.2 (ENSG00000267194). As the locus of lnc-CRCMSL overlaps the UTR of MAP2K6 (Fig. 1b), we investigated the effect of lnc-CRCMSL introduction or knockdown on mRNA and protein expression of MAP2K6 in CRC cells. HCT116 cells were infected with the lentivirus containing the lnc-CRCMSL overexpression vector. On the contrary, SW620 cells were transfected with three small interfering RNA (siRNA) for lnc-CRCMSL to knock down lnc-CRCMSL expression and siRNA#2 was selected for further experiments (Fig. 1c). The QPCR and western blot results showed that no obvious changes of MA2PK6 were found after overexpression or RNA interference of lnc-CRCMSL. (Fig. 1c, d). Rapid amplification of cDNA ends (5’- and 3’-RACE) analysis was performed to determine the full-length transcript of lnc-CRCMSL. The sequence of the full-length lnc-CRCMSL clone is shown in Figure S1 and Text S1. To determine whether lnc-CRCMSL sequence represents a conserved protein-coding region, we checked Open Reading Frame (ORF) for lnc-CRCMSL using CPAT, which displayed no coding potential. We also subcloned the full length of lnc-CRCMSL into two eukaryotic expression vectors with 5-termianl or 3-terminal tag, respectively (Fig. S1). The frameshift design confirmed that lnc-CRCMSL had no protein-coding capability.
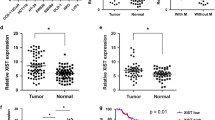
Lnc-CRCMSL is downregulated in aggressive CRC and ameliorates the prognosis of CRC patients. a Hierarchical clustering analysis of the top 105 lncRNAs that were differentially expressed (fold change > 2 & P < 0.05) between metastatic CRC (mCRC) and non-metastatic CRC (nmCRC) samples. Black arrowhead denotes lnc-CRCMSL. b Schematic annotation of lnc-CRCMSL genomic locus on chromosome 17. c Real-time PCR and western blot (d) analysis of MAP2K6 expression in indicated cell lines as well as the level of lnc-CRCMSL. The expression was normalized to that of GAPDH. Results are shown as means ± SD. *P < 0.05 and ***P < 0.001 by two-tailed Student’s t test. e CRCMSL expression was analyzed by qRT-PCR in CRC samples and adjacent non-tumor colorectal tissues (cohort 1, n = 20). Lnc-CRCMSL expression level was normalized to GAPDH. Results are shown as means ± SD by two-tailed Student’s t test. P < 0.001. f Lnc-CRCMSL expression in mCRC samples and nmCRC samples was measured by qRT-PCR (cohort 2, n = 64). Results are shown as means ± SD by two-tailed Student’s t test. P < 0.001. g Representative images of lnc-CRCMSL expression from different clinical pathological stages by ISH assays. Lnc-CRCMSL is significantly higher in stage I + II CRC than in stage III + IV CRC (cohort 3, n = 108). (H) a. Kaplan–Meier survival analysis of overall survival in all CRC patients, N0 subgroup, M0 subgroup and T3 + T4 subgroup based on CRCMSL expression, resprectively. Star *P < 0.05. Double stars **P < 0.01
Lnc-CRCMSL is downregulated in aggressive CRC and ameliorates the prognosis of CRC patients
qRT-PCR analysis confirmed that lnc-CRCMSL were downregulated in CRC tissues as compared with the corresponding paratumor mucosa tissues in a panel of 20 specimens (Fig. 1e, P < 0.001, cohort 1). Furthermore, lnc-CRCMSL was remarkably lower in additional panel of mCRC samples when compared with the nmCRC samples (Fig. 1f, P < 0.001, cohort 2). Using cytoplasmic and nuclear RNA fractions from HCT116 cells, we observed that lnc-CRCMSL is expressed in relative abundance mainly in the cytoplasm through qRT-PCR, which was consistent with FISH results in CRC cells (Fig. S3). We next detected the localization and expression of lnc-CRCMSL by in situ hybridization (ISH) and analyze its relationship with clinicopathological characteristics in 108 tumor tissue samples that had clinical information (Fig. 1e, Tables S2, cohort 3). Correlation and regression analysis showed that deceased expression of lnc-CRCMSL was significantly correlated with lymph node metastasis and clinical stages. Kaplan–Meier survival analysis of overall survival showed that CRC patients with low lnc-CRCMSL level had a significantly worse prognosis than those with high lnc-CRCMSL expression (Fig. 1f; P = 0.0304). The trend is more pronounced in subgroups of patients with M0 and T3/T4 stage. Multivariate Cox analysis was carried out to evaluate its predictive value in determining clinical outcomes of CRC patients. However, our current data did not support low expression of lnc-CRCMSL as an independent factor for poor prognosis in CRC patients (Table S3).
Lnc-CRCMSL inhibits aggressive phenotypes and reverses EMT process of CRC cells in vitro
The expression of lnc-CRCMSL was analyzed in 8 CRC cell lines and a normal human intestinal mucosal cell line NCM460. As shown in Fig. 2a, relative lower expression of lnc-CRCMSL was found in all CRC cell lines [15], except SW620 cells, in comparison with NCM460. To evaluate the effects of lnc-CRCMSL on cell biological behaviors, gain and loss function in vitro were performed. HCT116 and RKO were infected with the lentivirus containing the lnc-CRCMSL overexpression vector. On the contrary, SW620 cells were transfected with siRNA for lnc-CRCMSL to knock down lnc-CRCMSL expression (Fig. 1c and Fig. S4A). Cell-counting kit-8 (Fig. 2b and Fig. S4B) and colony forming assays (Fig. 2c and Fig. S4C) indicated that lnc-CRCMSL significantly reduced cell viability and colony forming ability. Meanwhile, transwell assay (Fig. 2d and Fig. S4D), matrigel assay (Fig. 2e and Fig. S4E) and wound healing assay (Fig. 2f) showed that lnc-CRCMSL strikingly decreased the potential of cell migration, invasion and motility, respectively. The initiation of EMT is the drive force for tumor invasion and metastasis. Lnc-CRCMSL overexpression upregulated mRNA expression of epithelial markers E-cadherin and ZO-1. In contrast, lnc-CRCMSL silencing downregulated their mRNA expression as expected (Fig. 3a). The protein expression of epithelial markers E-cadherin, ZO-1 and mesenchymal marker fibronectin was further confirmed in HCT116, RKO and SW620 cells (Fig. 3b and Fig. S4F).
Lnc-CRCMSL inhibits aggressive phenotypes and reverses EMT process of CRC cells in vitro. a Expression levels of CRCMSL in 8 CRC cell lines. NCM460 was used as a normal control. Data are shown as means ± SD. Data are representative of three independent experiments. b CCK8 assay was used to assess the ability of cell proliferation in indicated cells. Data represent mean ± SD, *P < 0.01 and ***P < 0.001 as determined by one-way ANOVA. c Colony formation assay of CRC cells transfected with lentivirus or siRNA for lnc-CRCMSL. Bars on the right panel represent the number of formed clones. Results are shown as means ± SD. **P < 0.01 and ***P < 0.001 by two-tailed Student’s t test. d Transwell assay in indicated CRC cells. Bars on the right panel represent the number of migrated cells. Results are shown as means ± SD. **P < 0.01 and ***P < 0.001 by two-tailed Student’s t test. e Matrigel assay in indicated CRC cells. Bars on the right panel represent the number of invaded cells. Results are shown as means ± SD. **P < 0.01 and ***P < 0.001 by two-tailed Student’s t test. f Wound-healing assay of indicated cells. Bars on the right panel represent the migration index of wound-healing assay. The distance migrated by treated cells was relative to that migrated by control cells. Representative figures are shown. The results are from 3 independent experiments. Results are shown as means ± SD. **P < 0.01 and ***P < 0.001 by two-tailed Student’s t test
Lnc-CRCMSL overexpression inhibits tumorigenic and metastasis of CRC in vivo. a Real-time PCR analysis of EMT markers expression. Error bars represent mean ± SD based on 3 independent experiments. Results are shown as means ± SD. **P < 0.01 by two-tailed Student’s t test. b Western blot analysis was performed to detect EMT markers. The immunosignal was quantified using quantity one software, and the relative protein abundance was determined by normalization with β-tubulin. c Tumor cells were injected subcutaneously into the back of nude mice to evaluate tumorigenesis. Representative figure of tumors formed is shown. Data represent mean ± SD, *P < 0.01 as determined by one-way ANOVA. d Tumor volumes and weights in the back of nude mice injected with indicated cells were measured. The data of all primary tumors are expressed as mean ± SD. Scatter plots summarize tumor weights (P = 0.016) and volumes (P = 0.010) derived from indicated cells at 18 d after subcutaneous implantation. Results are shown as means ± SD by two-tailed Student’s t test. e Tumor cells were injected into nude mice through the tail vein to evaluate the lung homing potential of cells. The number of metastatic lung nodules in individual mice was counted under the microscope. The magnification areas indicated metastatic nodes in the lung. f Kaplan–Meier survival curves and univariate analyses (log-rank) for the mice with CRCMSL overexpressing cells versus control cells were performed. Representative figures are shown. The results are from 3 independent experiments
Lnc-CRCMSL overexpression inhibits tumorigenic and metastasis of CRC in vivo
We evaluated the effect of lnc-CRCMSL on tumor growth in vivo. Growth of lnc-CRCMSL-overexpressing xenograft tumors was significantly slower than that of control group (Fig. 3c). As showed in Fig. 3d, tumor weight and volume was reduced by lnc-CRCMSL overexpression at the end of the experiment. To assess the effect of lnc-CRCMSL on lung homing capacity of tumor cells in vivo, the number of nodules was counted in mice injected with tumor cells through tail vein. Lnc-CRCMSL overexpression significantly reduced the lung homing potential of HCT116 cells (Fig. 3e) and ameliorated the survival of tumor-bearing mice (Fig. 3f). Collectively, these results demonstrate that lnc-CRCMSL overexpression decreased tumor growth and lung colonization.
Lnc-CRCMSL physically interacts with HMGB2 proteins and regulates its nucleocytoplamic shuttling
To seek proteins that are interacted with lncRNA-CRCMSL, RNA-pulldown assay was conducted in SW620 cells. RNA-associated proteins were resolved by SDS-PAGE, and the bands were excised and subjected to mass spectrometry (Fig. 4a, Table S4). Among all of the proteins identified by mass spectrometry, HMGB2 was successfully validated by western blot from RNA pull-down assays. A significant enrichment of HMGB2 was detected after pull-down preformed with the sense sequence of lnc-CRCMSL (Fig. 4b), but no enrichment was observed with the antisense lnc-CRCMSL as blotted by HMGB2 antibody. We further implemented HMGB2 antibody as a bait to carry out RNA immunoprecipitation (RIP) with cell extracts from the HCT116 tumor cell lines. We observed the lnc-CRCMSL enrichment, with GAPDH mRNA remains unchanged, using the HMGB2 antibody versus a nonspecific antibody (IgG control) (Fig. 4c).
CRCMSL interacts with HMGB2 and suppress it into the nucleus. a RNA pull-downs were performed with SW620 cells using full-length CRCMSL transcript (sense) and antisense. The candidate bands were identified by mass spectrometry. HMGB2 (band #1) was identified as the band unique to lnc-CRCMSL. b HMGB2 was confirmed by immunoblotting. c The interaction of CRCMSL with HMGB2 was verified by an RNA immunoprecipitation (RIP) assay. Results are shown as means ± SD. **P < 0.01 by two-tailed Student’s t test. d The expression of HMGB2 in nucleus and cytoplasm of CRCMSL-overexpressing HCT116 cells and CRCMSL-knockdown SW620 cells were shown by western blot. The immunosignal was quantified using quantity one software, Nuclear segregation is assayed by Histone H3. Cytoplasmic segregation is assayed by β-actin. e ISH and IHC were used to detect the localization and correlation of CRCMSL and HMGB2 in the same fields of non-metastatic CRC samples and metastatic CRC samples (Left panel). IHC analysis of HMGB2 in subcutaneous tumor tissues (Right panel). 100 × for all, scale bar = 100 μm. 400 × for all, scale bar = 20 μ. f immunofluorescence assay was performed to observe the localization of HMGB2 in indicated cells. ×1000 for all, scale bar = 10 μ
HMGB2 is a nuclear protein that binds to DNA and functions in chromatin remodeling. Lnc-CRCMSL obviously reduced its expression in the nucleus, but slightly reduced the expression of HMGB2 in the cytoplasm (Fig. 4d). In contrast, lnc-CRCMSL silencing led to the enrichment of HMGB2 in the nuclear. Then, the expression pattern of lnc-CRCMSL and HMGB2 was carefully checked in specimens from CRC patients. ISH assays showed that the positive signal of lnc-CRCMSL was mainly localized in the cytoplasm of tumor cells. The lower expression of lnc-CRCMSL in mCRC specimens was frequently accompanied with the nuclear enrichment of HMGB2 proteins (Fig. 4e). IHC assay in xenograft tumors further confirmed that lnc-CRCMSL overexpression induced the translocation of HMGB2 to the cytosol from the mucleus (Fig. 4e). Immunofluorescence provided direct evidence that lnc-CRCMSL upregulated the cytoplasmic retention of HMGB2 and siRNA for lnc-CRCMSL drived shift of HMGB2 into nucleus (Fig. 4f).
HMGB2 is overexpressed in CRC and contributes CRC progression
To clarify the effects of HMGB2 on biological and clinical characters of CRC, its expression and functional assay were performed in both cell lines and clinical specimens. The mRNA and protein expression of HMGB2 was significantly higher in all CRC cell lines than that of NCM460 (Fig. 5a, left). Western blot from fresh clinical samples showed that higher expression of HMGB2 was also observed in CRC tissues as compared to the paired adjacent non-tumor tissues (Fig. 5b). Transwell and wound-healing assay indicated that overexpression of HMGB2 increased the potential of migration and motility in HCT116 cells and vice versa for HMGB2 silencing in SW620 cells (Fig. 5c and Fig. S5). To further confirm the effects of HMGB2 on tumor metastasis, the number of metastatic lung nodules was observed in mice injected with SW620 cells. As showed in Fig. 5d, HMGB2 shRNA significantly reduced the number and size of nodule in lungs as compared to the control. It also prolonged the survival time of mice as monitored consecutively for 10 weeks. We further evaluated the role of HMGB2 on EMT phenotype in CRC cells. HMGB2 overexpression downregulated the mRNA expression levels of epithelial markers E-cadherin and ZO-1. Similarly, HMGB2 overexpression decreased protein expression of epithelial markers E-cadherin, ZO-1 and increased protein expression of mesenchymal markers vimentin in HCT116 cells and vice versa for HMGB2 silencing in SW620 cells (Fig. 5e).
HMGB2 is overexpression in CRC and contributes CRC progression. a Real-time PCR and western blot analysis of HMGB2 expression in CRC cell lines. The expression of HMGB2 was increased in all night CRC cell lines compared with the normal human colon epithelial cell line NCM460. Error bars represent mean ± SD based on three independent experiments. The immunosignal was quantified using quantity one software, and the relative protein abundance was determined by normalization with GAPDH. b Western blot analysis of HMGB2 in CRC tissues (T) and adjacent non-tumor tissues (N). The immunosignal was quantified using quantity one software, and the relative protein abundance was determined by normalization with GAPDH. c Transwell assay of CRC cells transfected with HMGB2 Vector or siRNA-1009 for HMGB2. Bars on the right panel represent the number of migrated cells. Representative figures are shown. The results are from three independent experiments. **P < 0.01; ***P < 0.001. d Tumor cells were injected into nude mice through the tail vein to evaluate the lung homing potential of cells. The number of metastatic lung nodules in individual mice was counted under the microscope. The magnification areas indicated metastatic nodes in the lung. Kaplan–Meier survival curves and univariate analyses (log-rank) for the mice with shRNA transfected cells versus control cells were performed. e Real-time PCR and western blot analyses were performed in indicated cells to detect EMT markers. Results are shown as means ± SD. *P < 0.05 and **P < 0.01 by two-tailed Student’s t test. The immunosignal was quantified using quantity one software, and the relative protein abundance was determined by normalization with GAPDH
Downregulated lnc-CRCMSL increases nuclear HMGB2 from cytoplasm and triggers OCT4-mediated EMT programming
To investigate the role underlying nuclear enrichment of HMBG2, the interaction between HMGB2 and key EMT-associated transcription factors, such as octamer-binding transcription factor 4 (Oct4), ZEB1, c-Myc, STAT3, slug and snail, was probed in CRC cells. Co-IP assays only validated the interaction of HMGB2 and Oct4, but not ZEB1, c-Myc, STAT3, slug and snail (Fig. 6a and Fig. S6). Immunofluorescence assays further confirmed the nuclear co-localization of Oct4 and HMGB2 (Fig. 6b). Interestingly, lnc-CRCMSL cannot affect the expression of HMGB2 and OCT4 at both the transcriptional and post-transcriptional level (Fig. S7). So, lnc-CRCMSL affects the interaction between HMGB2 and OCT4 rather than the stability of them. Lnc-CRCMSL introduction dissociated the interaction. As expected, lnc-CRCMSL knockdown increased their binding (Fig. 6a). To further explore the effects of lnc-CRCMSL on nucleoplasmic shuttle of HMGB2, leptomycin B (LMB) was adopted to suppress the export of nuclear proteins. HMGB2 was pulled out of the nuclear and prefer to stay in the cytoplasm in the presence of lnc-CRCMSL. However, LMB suppressed the export of HMGB2 and induced the nuclear accumulation of HMGB2 (Fig. 6c). The upregulated mRNA and protein expression of epithelial markers E-cadherin and ZO-1 by lncRNA-CRCMSL were significantly abrogated by LMB pretreatment (Fig. 6d). Transwell assay was also performed to further confirm our conclusion (Fig. 6e). Finally, we transfected HMGB2 plasmid and nuclear exporting signal (NES)-HMGB2 plasmid respectively in HMGB2-KD SW620 cell line. As shown in Figure S8, the invasiveness of transfected HMGB2 cells was enhanced, while the invasiveness of NES-HMGB2 group was weakened compared with that of HMGB2 group, but stronger than that of NC group. Additionally, the interaction between HMGB2 and OCT4 was significantly decrased in HMGB2-knock down cells treated with exogenous NES-HMGB2 vector than that of HMGB2 vector using Co-IP assays.
Downregulated lnc-CRCMSL increases nuclear HMGB2 from cytoplasm and triggers OCT4-mediated EMT programming. a The interaction between HMGB2 and OCT4 was detected in lnc-CRCMSL-overexpressing HCT116 cells and lnc-CRCMSL-knockdown SW620 cells using Co-IP assays. b The colocalization between HMGB2 and OCT4 was detected by immunofluorescence staining. c Immunofluorescence assays of HMGB2 in the treated cells as indicated. Representative figures are shown. d Real-time PCR and western blot analyses of EMT markers after treatment with Leptomycin B (LMB) in indicated cells. Results are shown as means ± SD. **P < 0.01 by two-tailed Student’s t test. The immunosignal was quantified using quantity one software, and the relative protein abundance was determined by normalization with β-tubulin. e Transwell assay of the indicated cells treated with LMB. Representative figures are shown. The results are from three independent experiments. **P < 0.01 by two-tailed Student’s t test. f A hypothetical model illustrating that lncRNA-CRCMSL suppresses Oct4-induced reprogramming in colorectal cancer progression through nucleocytoplasmic shuttling of HMGB2
Discussion
Aberrantly expressed lncRNAs play vital roles in CRC metastasis that accounts for most cancer-related deaths in CRC patients. Using the lncRNAs microarray scanning and bioinformatics analyses, we identified a potential tumor metastasis suppressor in CRC, designated as lnc-CRCMSL. As lnc-CRCMSL is located in the 3’UTR region of MAP2K6 gene, there is a very real possibility that the biological behaviors induced by lnc-CRCMSL wholly or partially arise through off-target effects on the MAP2K26 gene. Our data showed that the expression of MAP2K6 was not significantly altered by introduction or knockdown of lnc-CRCMSL, suggesting that the observed phenotypes are not the result of directly manipulating MAP2K6 gene through its UTR. In previous studies, however, it has been demonstrated that some transcripts annotated as lncRNAs are actually encode for small proteins [16]. The full-length cDNA of lnc-CRCMSL was subcloned into two eukaryotic expression vectors with N-terminal or C-terminal tag. No protein expression was observed in all three coding patterns with frameshift designing. The expression of lnc-CRCMSL was not only associated with good survival outcome but also negatively correlated with advanced clinical staging in CRC patients. Then, a series of in vivo and in vitro assays provided consolidated data that lnc-CRCMSL suppress the growth and metastasis of CRC, suggested that lnc-CRCMSL serves as a potential CRC metastasis suppressor.
High mobility group proteins are a family of non-histone chromatin-bound proteins that modulate transcription by altering the architecture of chromatin [17, 18]. The functions of HMGB proteins are determined by their relocation from the nucleus to the cytoplasm or secretion outside of the cell [18,19,20]. Although HMGB2 has scarcely been found relocating to extranuclear or extracellular compartment, HMGB1 has been reported to locate out of nucleus and function as a potent proinflammatory cytokine [19, 21]. Abnormal expression and localization of HMGB proteins has been found in various kinds of tumor. Elevated expression of HMGB2 significantly correlated with shorter overall survival time in patients with hepatocellular carcinoma [22]. HMGB2 was highly expressed in the nuclei of breast cancer cells, compared with the adjacent normal breast tissues [23], which are consistent with our results. In our study, lnc-CRCMSL is mainly distributed in cytoplasm of the cells. It tethers with HMGB2 and forms an inhibitory complex which stabilize the cytoplamic location of HMGB2. Therefore, lnc-CRCMSL normally promotes the retention of HMGB2 in the cytoplasm. Our work shows that decreased expression of lnc-CRCMSL will allow HMGB2 shifting to the nuclear in multiple CRC cells. The liberation of HMGB2 from lnc-CRCMSL back to the nuclear will cause invasion and metastasis phenotype of CRC both in clinical data and in vivo experiments. However, LMB treatment significantly reduces the cytoplasmic localization of HMGB2, suggested that lnc-CRCMSL prompts the nuclear export of HMGB2 and holds it in the cytosol. Lnc-CRCMSL might act as a key regulator of HMGB2 that fine-tunes the nucleocytoplasmic shuttling of HMGB2 [2, 24].
Our previous work shows that TGFβ1 significantly induces EMT process in CRC [25]. TGFβ1 activates Sox2, Nanog, and Oct4 to trigger EMT and switch to a more stem cell like phenotype in in bladder cancer [26]. It has been reported that HMGB2 can bind to DNA without sequence specificity and promotes transcription by expanding the accessibility of the chromatin to transcription factors [27, 28]. HMGB2 interacts with OCT4 and maintains the pluripotent gene expression signature in murine embryonic stem cells [29]. Oct4 is a homeodomain transcription factor that upregulates EMT and cell migratory ability [30]. As we all know, the lncRNAs frequently play a role as a regulator of transcription and/or protein stabilization. In the present scenario, we confirmed that lnc-CRCMSL cannot affect the stability of HMGB2 and OCT4 at both the transcriptional and post-transcriptional level. Further, lnc-CRCMSL silencing unleashes HMGB2 and relocates the latter to nucleus. HMGB2 then directly binds to Oct4, which in turn, upregulates EMT-associated gene expression and increases migratory and invasive ability [30]. Nuclear HMGB2 thus catalyze the EMT process by binding to Oct4 to promote the assembly of specific transcriptional complex and their accessibility to chromatins [30]. The increased expression of Oct4 correlates with decreased E-cadherin and increased expression of vimentin [31]. Therefore, Oct4 is a potential target of nuclear HMGB2 through physical interaction. Both of them cooperatively switch EMT and commit metastasis in CRC patients [31, 32].
Lnc-CRCMSL is a potent tumor suppressor that correlates with better prognosis in CRC patients. It exerts anti-metastatic effects by stabilizing cytoplasmic localization of HMGB2. LncRNAs can be functionally classified as signals, decoys, guides, and scaffolds [33]. Lnc-CRCMSL functions as a guide that associates with HMGB2 and directs the cytoplasmic retention of HMGB2, which subsequently inhibits the downstream EMT signaling and metastatic events [34]. Taken together, lnc-CRCMSL suppresses EMT and metastasis of CRC by keeping cytoplamic localization of HMGB2. In the condition of lower level lnc-CRCMSL, HMGB2 translocates into nuclear and associates with Oct4 to trigger EMT (Fig. 6f). In conclusion, lnc-CRCMSL holds HMGB2 in the cytoplasm to suppress EMT and metastasis by disassociating the interaction between HMGB2 and Oct4. Our findings indicate that lnc-CRCMSL may be a promising therapeutic target and serve as a prognostic biomarker of CRC metastasis.
Materials and methods
Patients and Microarray analysis
The samples of colorectal cancer in non-metastatic (N = 0) and metastatic (N > 1) were obtained from Department of Gastrointestinal Surgery, Guangdong General Hospital (Guangzhou, china). Total RNA was isolated using Trizol reagent (Cat#15596-018, Life technologies, US). The gene expression patterns of non-metastatic (N0) and metastatic (N1 + N2) were analyzed by microarray analysis using Gene Expression Hybridization Kit (Cat#5188-5242, Agilent technologies Santa Clara, CA, USA) and Agilent Human lncRNA 4 × 180 K chips (design ID: 045142) in Shanghai Biotechnology Corporation (Shanghai, China). Each group has eight biological replicates. Differential gene expression was determined using the Gene Spring Software (Santa Clara, CA, USA), and gene ontology analysis was performed. The gene expression data has been assigned an accession ID as GSE113296. The standard selection criteria to identify differentially expressed genes are as follows: (i) log2 |Fold change| ≥ 1.5 or < 0.5. (ii) P < 0.05. (iii) log2 ratios = “NA” and the higher expression group had a signal value greater than 4.
Cell culture
CRC cell lines HCT116, SW480, SW620, LS174T, RKO, HT29 and LOVO were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and maintained as previously described [15]. All cells were authenticated by short tandem repeat (STR) profiling before receipt and were propagated for less than 6 months after resuscitation. Additionally, a human CRC cell subline SW480/M5 with unique liver metastatic potential was established in our laboratory [35]. And the normal human colon epithelial cell line NCM460 was provided by Incell Corporation (San Antonio, TX). All of the cells were cultured in RPMI 1640 (Gibco, Thermo Fisher Science, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco-BRL, Invitrogen, Paisley, UK) at 37oC with a humidity of 90–95% and 5% CO2.
Animals
Five-week-old female BALB/c nude mice were purchased from the Experimental Animal Centre of Southern Medical University, which is certified by the Guangdong Provincial Bureau of Science. All animal experiments were carried out with the approval of the Southern Medical University Animal Care and Use Committee in accordance with the guidelines for the ethical treatment of animals. Nude nu/nu mice were maintained in a barrier facility in racks filtered with high-efficiency particulate air filter. The animals were fed with an autoclaved laboratory rodent diet. All animal experiments involved ethical and humane treatment under a license from the Guangdong Provincial Bureau of Science. Animals were randomly allocated and the investigators were blinded to the group allocation during the experiment and when assessing the outcome.
For the tumor growth assay, 1 × 106 LV-Ctrl-transfected cells and LV-CRCMSL-transfected cells were independently injected subcutaneously into the back of nude mice (n = 6/group). The tumor volume was calculated using the following formula: V = 0.5 × D × d2, where V represents volume, D represents the longitudinal diameter, and d represents the latitudinal diameter.
For tumor metastasis assay, we injected 5 × 106 indicated cells into nude mice (n = 7/group) through the tail vein to determine the lung homing potential of cancer cells in vivo. Whole-body optical images were obtained to monitor primary tumor growth and the formation of metastatic lesions. After all the mice were sacrificed, individual organs were removed and metastatic tissue was analyzed using hematoxylin and eosin (H&E) and immunohistochemical stains.
Transfection of lnc-CRCMSL siRNA and CRCMSL overexpression lentiviral vector
siRNAs and lentiviral vectors were all purchased from GenePharma (Shanghai, China). siRNA sequences used in this study were shown in Table S1. Lipofectamine 2000 reagent was purchased from Invitrogen (Carlsbad, California, USA). CRC cells at exponential growth phase were plated into 6-well plates for 24 h at a density of 0.5 × 105 cells/mL, and transfected according to the manufacturer’s protocol.
Western blot
Total protein was harvested by RIPA lysis buffer with Protease Inhibitor Cocktail, and quantified using the BCA protein assay kit (Beyotime Biotechnology). This protein was separated by SDS-PAGE gel and transferred onto PVDF membrane (Millipore). The membranes were incubated with rabbit antibodies to β-actin, β-tubulin, GAPDH (1:1000; Santa Cruz), rabbit antibodies to Histone H3, E-cadherin, ZO-1, Fibronectin, Vimentin, MAP2K6 (1:1000; CST), rabbit antibody to HMGB2 (1:1000; Abcam), mouse antibody to OCT4 (1:1000; Abcam) overnight, and followed by HRP-conjugated secondary antibody (1:10000; CST), respectively. The signal was detected using enhanced chemiluminescence detection system (Pierce, Rockford, IL) as described by the manufacturer.
RNA preparation, reverse transcription and quantitative real-time PCR
Fresh primary CRC specimens and paired noncancerous colorectal tissue were provided by Guangdong General Hospital. The study was approved by the Ethics Committee of Guangdong General Hospital and all aspects of the study comply with the Declaration of Helsinki. All reagents for RNA preparation, reverse transcription and quantitative real-time PCR were purchased from Takara (Japan) and following the manufacturer’s protocol. PCR primers used in this study was shown in Table S1.
Rapid amplification of cDNA ends (RACE)
The full-length cDNA sequence of lnc-CRCMSL gene was determined using 3′RACE and 5′RACE. The 3′-Full RACE Core Set with PrimeScript RTase was purchased from Takara (Japan) and following the manufacturer’s protocol. The supplied Oligo dT-3 sites Adaptor Primer is specially designed for efficient cDNA synthesis from 3’end of ploy(A) + RNA (3′ RACE Outer primer: TACCGTCGTTCCACTAGTGATTT, 3′ RACE Inner primer: CGCGGATCCTCCACTAGTGATTTCACTATAGG, 3′ RACE specially primer: firstly, GTTCCACGGTTCATCTGGCTAC; secondly, GTGGGATTCCCATCACATC). 5′ RACE was done by Sangon Biotech(Shanghai, China) (5′ RACE Outer primer: GGCCACGCGTCGACTAGTAC, 5′ RACE Inner primer: GCCACGCGTCGACTAGTACGGGGGGGGGG, 5′ RACE specially primer: firstly, AAACAGTTGCTGTGAAACCCCAGCTATT; secondly, ACCTCCTCAAGTGGCTTTTATGCAGATT and primer of reverse transcription: GCACAAAGAAAGCTGAAGAAATCCACCTC).
RNA in situ hybridization analysis
Paraffin sections of patients that had undergone elective surgery for CRC between 2007 and 2010 were provided by Nanfang Hospital. The probe was provided by Redland (Guangzhou, China). After deparaffinization and hydration, slides were air dried and treated with pepsin solution for 10 min at 37 °C. Slides were hybridized with target Botin-labeled probes for 3 h at 50°C, followed by a series of washing steps (2 × SSC, 0.5 × SSC, 0.2 × SSC). Blocking buffer was applied for 15 min at 30 °C followed by Streptomycin-Alkaline phosphatase (Boster Biological Technology, Wuhan, China) for 1 h at 37 °C and then NBT/BCIP (Roche, American) for 3 h at 37 °C. After dehydration, slides were mounted neutral gum and examined by microscopy.
RNA pull-down assay and RIP assay
In vitro biotin-labeled RNAs (CRCMSL, its antisense RNA, and control RNAs) were transcribed with Biotin RNA Labeling Mix and DIG RNA Labeling Kit (SP6. T7) (Roche) and purified with RNeasy Mini Kit (QIAGEN). Biotinylated RNAs were incubated with CRC cell nuclear extracts, and then added Streptavidin Magnetic Beads (Pierce™, Invitrogen). And pull-down proteins were run on SDS-PAGE gels, and then gels were stained by silver staining, and differential bands were choosed for mass spectroscope. RIP assay was performed using RIP™ RNA-Binding Protein Immunoprecipitation Kit (Merck Millipore) and following the manufacturer’s protocol.
Statistical analysis
Data were analyzed using SPSS 19.0 software (SPSS; Chicago, USA) and GraphPad Prism 5. Differences between two comparisons were evaluated using Student’s t test. Analysis of variance method was performed to analyze the differences in multiple comparisons. Log-rank analysis was applied in survival comparisons. Statistical significance was established at P < 0.05.
References
Lagarde J, Uszczynska-Ratajczak B, Carbonell S, Perez-Lluch S, Abad A, Davis C, et al. High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat Genet. 2017;49:1731–40.
Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4.
Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407.
Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–81.
Weidle UH, Birzele F, Kollmorgen G, Ruger R. Long Non-coding RNAs and their role in metastasis. Cancer Genom Proteom. 2017;14:143–60.
Cao C, Sun J, Zhang D, Guo X, Xie L, Li X, et al. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of beta-catenin in HCC cells. Gastroenterology. 2015;148:415–26.
Huang Q, Yan J, Agami R. Long non-coding RNAs in metastasis. Cancer Metastas Rev. 2018;37:75–81.
Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–61.
Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330–7.
Schmoll HJ, Van Cutsem E, Stein A, Valentini V, Glimelius B, Haustermans K, et al. ESMO Consensus Guidelines for management of patients with colon and rectal cancer. a personalized approach to clinical decision making. Ann Oncol. 2012;23:2479–516.
Wang H, Shi J, Luo Y, Liao Q, Niu Y, Zhang F, et al. LIM and SH3 protein 1 induces TGFbeta-mediated epithelial-mesenchymal transition in human colorectal cancer by regulating S100A4 expression. Clin Cancer Res. 2014;20:5835–47.
Yue B, Qiu S, Zhao S, Liu C, Zhang D, Yu F, et al. LncRNA-ATB mediated E-cadherin repression promotes the progression of colon cancer and predicts poor prognosis. J Gastroenterol Hepatol. 2016;31:595–603.
Zhang Y, Wagner EK, Guo X, May I, Cai Q, Zheng W, et al. Long intergenic non-coding RNA expression signature in human breast cancer. Sci Rep. 2016;6:37821.
Wang P, Cui J, Wen J, Guo Y, Zhang L, Chen X. Cisplatin induces HepG2 cell cycle arrest through targeting specific long noncoding RNAs and the p53 signaling pathway. Oncol Lett. 2016;12:4605–12.
Zhao L, Wang H, Liu C, Liu Y, Wang X, Wang S, et al. Promotion of colorectal cancer growth and metastasis by the LIM and SH3 domain protein 1. Gut. 2010;59:1226–35.
Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21.
Pennisi E. Long noncoding RNAs may alter chromosome’s 3D structure. Science. 2013;340:910.
Reeves R. High mobility group (HMG) proteins: Modulators of chromatin structure and DNA repair in mammalian cells. DNA Repair. 2015;36:122–36.
Takeuchi T, Sakazume K, Tonooka A, Zaitsu M, Takeshima Y, Mikami K, et al. Cytosolic HMGB1 expression in human renal clear cell cancer indicates higher pathological T classifications and tumor grades. Urol J. 2013;10:960–5.
Ueda T, Yoshida M. HMGB proteins and transcriptional regulation. Biochim Biophys Acta. 2010;1799:114–8.
Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–5.
Kwon JH, Kim J, Park JY, Hong SM, Park CW, Hong SJ, et al. Overexpression of high-mobility group box 2 is associated with tumor aggressiveness and prognosis of hepatocellular carcinoma. Clin Cancer Res. 2010;16:5511–21.
Fu D, Li J, Wei J, Zhang Z, Luo Y, Tan H, et al. HMGB2 is associated with malignancy and regulates Warburg effect by targeting LDHB and FBP1 in breast cancer. Cell Commun Signal. 2018;16:8.
Castillo-Quan JI, Blackwell TK. Metformin: restraining nucleocytoplasmic shuttling to fight cancer and aging. Cell. 2016;167:1670–1.
Zhang F, Luo Y, Shao Z, Xu L, Liu X, Niu Y, et al. MicroRNA-187, a downstream effector of TGFbeta pathway, suppresses Smad-mediated epithelial-mesenchymal transition in colorectal cancer. Cancer Lett. 2016;373:203–13.
Islam SS, Mokhtari RB, Noman AS, Uddin M, Rahman MZ, Azadi MA, et al. Sonic hedgehog (Shh) signaling promotes tumorigenicity and stemness via activation of epithelial-to-mesenchymal transition (EMT) in bladder cancer. Mol Carcinog. 2016;55:537–51.
Aird KM, Iwasaki O, Kossenkov AV, Tanizawa H, Fatkhutdinov N, Bitler BG, et al. HMGB2 orchestrates the chromatin landscape of senescence-associated secretory phenotype gene loci. J Cell Biol. 2016;215:325–34.
Ugrinova I, Pashev IG, Pasheva EA. Nucleosome binding properties and Co-remodeling activities of native and in vivo acetylated HMGB-1 and HMGB-2 proteins. Biochemistry. 2009;48:6502–7.
Campbell PA, Rudnicki MA. Oct4 interaction with Hmgb2 regulates Akt signaling and pluripotency. Stem Cells. 2013;31:1107–20.
Mandal G, Biswas S, Chowdhury SR, Chatterjee A, Purohit S, Khamaru P, et al. Heterodimer formation by Oct4 and Smad3 differentially regulates epithelial-to-mesenchymal transition-associated factors in breast cancer progression. Biochim Biophys Acta. 2018;1864:2053–66.
Zhi X, Lin L, Yang S, Bhuvaneshwar K, Wang H, Gusev Y, et al. betaII-Spectrin (SPTBN1) suppresses progression of hepatocellular carcinoma and Wnt signaling by regulation of Wnt inhibitor kallistatin. Hepatology. 2015;61:598–612.
Zhao Y, Yang Z, Wu J, Wu R, Keshipeddy SK, Wright D, et al. High-mobility-group protein 2 regulated by microRNA-127 and small heterodimer partner modulates pluripotency of mouse embryonic stem cells and liver tumor initiating cells. Hepatol Commun. 2017;1:816–30.
Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14.
Ulitsky I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat Rev Genet. 2016;17:601–14.
Zhang YF, Liu L, Ding YQ. [Isolation and characterization of human colorectal cancer cell subline with unique metastatic potential in the liver]. Nan Fang Yi Ke Da Xue Xue Bao. 2007;27:126–30.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Nos. 81572813, 81773082, 81573848, 81774172), Guangdong Natural Science Foundation (2015A030313274), Science and Technology Program of Guangzhou (1563000235) and Special Funds for the Cultivation of Guangdong College Students’ Scientific and Technological Innovation (pdjhb0105).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Precis: Findings identify and report a CRC metastasis suppressor lnc-CRCMSL with potential therapeutic applications.
Supplementary information
Rights and permissions
About this article
Cite this article
Han, Q., Xu, L., Lin, W. et al. Long noncoding RNA CRCMSL suppresses tumor invasive and metastasis in colorectal carcinoma through nucleocytoplasmic shuttling of HMGB2. Oncogene 38, 3019–3032 (2019). https://doi.org/10.1038/s41388-018-0614-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0614-4
- Springer Nature Limited
This article is cited by
-
Exploring the interplay of natural products and long non-coding RNAs in colorectal cancer: pathogenesis, diagnosis, and overcoming drug resistance
Naunyn-Schmiedeberg's Archives of Pharmacology (2024)
-
Sirt3 restricts tumor initiation via promoting LONP1 deacetylation and K63 ubiquitination
Journal of Translational Medicine (2023)
-
Long non-coding RNAs as the critical regulators of epithelial mesenchymal transition in colorectal tumor cells: an overview
Cancer Cell International (2022)
-
The LncRNA RP11-301G19.1/miR-582-5p/HMGB2 axis modulates the proliferation and apoptosis of multiple myeloma cancer cells via the PI3K/AKT signalling pathway
Cancer Gene Therapy (2022)
-
Long Non-coding RNAs (lncRNAs), A New Target in Stroke
Cellular and Molecular Neurobiology (2022)