Abstract
ALK mutations occur in 10% of primary neuroblastomas and represent a major target for precision treatment. In combination with MYCN amplification, ALK mutations infer an ultra-high-risk phenotype resulting in very poor patient prognosis. To open up opportunities for future precision drugging, a deeper understanding of the molecular consequences of constitutive ALK signaling and its relationship to MYCN activity in this aggressive pediatric tumor entity will be essential. We show that mutant ALK downregulates the ‘HMG-box transcription factor 1’ (HBP1) through the PI3K-AKT–FOXO3a signaling axis. HBP1 inhibits both the transcriptional activating and repressing activity of MYCN, the latter being mediated through PRC2 activity. HBP1 itself is under negative control of MYCN through miR-17~92. Combined targeting of HBP1 by PI3K antagonists and MYCN signaling by BET- or HDAC-inhibitors blocks MYCN activity and significantly reduces tumor growth, suggesting a novel targeted therapy option for high-risk neuroblastoma.
Similar content being viewed by others
Introduction
Neuroblastoma (NB) is a childhood tumor arising from the embryonic sympatho-adrenal lineage of the neural crest and represents the primary cause of cancer-related death in young children ages one to five [1]. These tumors are characterized by a heterogeneous clinical course, ranging from spontaneous regression to highly aggressive, metastatic disease refractory to therapy [2]. Sequencing efforts have resulted in a detailed molecular characterization of the neuroblastoma genomic landscape, exhibiting few recurrent driver mutations in a background of highly recurrent DNA copy number alterations [3]. MYCN amplification is observed in half of the high-risk tumors and more than 10% exhibit activating anaplastic lymphoma kinase (ALK) receptor mutations [2, 4,5,6,7]. These mutations are preferred targets for precision medicine and clinical trials using ALK-inhibitors have been initiated [8, 9]. However, as single compound approaches almost invariably lead to therapy resistance [10,11,12,13,14,15,16,17,18,19,20], a more detailed understanding of components implicated in ALK downstream signaling is warranted.
Previous studies have suggested genetic interaction between MYCN and ALK in neuroblastoma cells. We previously identified an ultra-high-risk patient subgroup with combined MYCN amplification and ALKF1174L mutation [21]. Subsequently, a mouse and zebrafish neuroblastoma model revealed accelerated tumor formation when both MYCN and ALKF1174L were expressed in sympathetic neuronal progenitor cells [22, 23]. Further, ALK was shown to control MYCN transcription levels and MYCN protein stabilization through the PI3K-AKT pathway [24,25,26,27], providing insight into the possible mechanism of mutant ALK mediated increased tumor aggressiveness. In this study, we further explored the interrelationship between ALK and MYCN based on our previously established ALK-driven 77-gene signature [28]. We identified consistent ALK controlled downregulation of HBP1 (‘high-mobility-group (HMG) box protein’), a previously established negative regulator of MYC(N) activity [29, 30]. Moreover, HBP1 is regulated by the PI3K-AKT–FOXO3 [31] and by phosphorylation by AKT [32], which is a major downstream pathway of ALK [4,5,6,7, 28, 33–35]. Therefore, we investigated the transcriptional and phenotypical effects of HBP1 modulation in ALK mutated and MYCN-amplified neuroblastoma cells. Finally, we also investigated the effects of different single and combined drug combinations on HBP1 levels.
Results
ALK downregulates HBP1 mRNA and protein expression levels
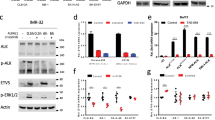
To further investigate the possible regulatory relationship between ALK and MYCN, we looked for hitherto unrecognized proteins implicated in MYCN regulation in neuroblastoma cells. To this end, we verified our previously published ALK-driven 77-gene signature [28] and identified the negative MYC regulator HBP1 as an ALK downregulated target. We first confirmed upregulation of HBP1 expression levels after pharmacological inhibition of ALK with TAE684 in six selected neuroblastoma cell lines. These included three ALK mutant cell lines with ALK amplification (NB-1), an ALKF1174L mutation (SK-N-SH) and an ALKR1275Q mutation (CLB-GA). Furthermore, we also selected two ALK wild-type non-responding to the inhibitor (SK-N-AS, IMR-32) and one ALK wild-type cell line (NGP) that responded to ALK inhibition (Supplementary Fig. 1A) [28, 35]. We confirmed increase in HBP1 mRNA expression levels in the ALK mutant cell lines and the ALK wild type responder NGP, while no notable effects were observed in wild type non-responders SK-N-AS and IMR-32 (Fig. 1a). We then analyzed the HBP1 expression over several time points in ALKR1275Q mutant CLB-GA cells following TAE684 treatment and observed HBP1 upregulation as early as two hours after drug exposure (Supplementary Fig. 1B). ALK-mediated HBP1 repression was confirmed by several next-generation ALK-inhibitors in CLB-GA cells [8, 33] (Fig. 1b, c). We could also confirm the effect of inhibition of mutant ALK signaling on HBP1 expression levels in vivo in mouse xenografted SH-SY5Y neuroblastoma cells treated with TAE684 and crizotinib (Fig. 1d).
ALK signaling downregulates HBP1 expression through PI3K/AKT–FOXO3a signaling in neuroblastoma and other ALKoma’s. a HBP1 mRNA expression in several ALK wild type (NGP, IMR-32, SK-N-AS) and ALK mutant (SK-N-SH, CLB-GA, NB-1) cell lines treated for 6 h with 0.3 μM TAE684 or DMSO relative to the levels in DMSO treated cells. b HBP1 mRNA levels in CLB-GA treated with 0.2 μM ceritinib and 0.5 μM crizotinib or DMSO for 6 h, relative to the DMSO control of each compound. Significant increase (p = 9.21e−05) of HBP1 expression was observed for the two treatments compared to the controls. c Western blot analysis showing HBP1 protein levels 24 h after treating the CLB-GA cell line with different ALK-inhibitors (0.2 μM ceritinib and 0.5 μM crizotinib or DMSO). d HBP1 mRNA levels in SH-SY5Y xenografted mice treated with TAE68, crizotinib or carrier solution, relative to the carrier solution. e HBP1 mRNA levels in SK-N-AS cell lines with TET-inducible ALKwt, ALKF1174L, or ALKR1275Q constructs treated with tetracycline or ethanol for 24 h, relative to the ethanol control of each cell line. f HBP1 expression in EML4/ALK fusion positive non-small-cell lung carcinoma cells (NSCLC)-xenografted mice treated with 4 or 20 mg/kg of the ALK-inhibitor alectinib. g HBP1 mRNA levels in two NB cell lines (one ALKamp and ALKR1275Q) treated with 8 μM MEK-inhibitor U0126, 0.5 μM PI3K/mTOR-inhibitor BEZ-235 or DMSO for 6 h, relative to the DMSO control of each cell line. Significant increase (P = 0.01) of HBP1 expression was observed for BEZ-235 treatment of the two cell lines compared to the control, while this was not the case for U0126 (P = 0.57). h Western blot analysis showing HBP1, phospho-AKT, and AKT protein levels 24 h after treating NB-1 and CLB-GA with the PI3K/mTOR-inhibitor (0.5 μM BEZ-235 or DMSO). i HBP1 mRNA levels in cells treated with or without 0.1 μg/ml doxycycline for 24 h to induce overexpression of HA-tagged FOXO3A and then treated with either 1 μM PI3K-inhibitor PI-103 or DMSO for 6 additional hours (ANOVA test followed by pairwise t-test for multiple comparisons on log normalized data). Error bars represents mean ± SD of, respectively, two technical replicates (a, b, g), two biological replicates (f), two biological replicates, each containing two technical replicates (d, e) or three biological replicates (i). Two-sided unpaired t-tests were performed on log normalized data with *P < 0.05, **P < 0.01, ***P < 0.001
The effects of ALK induction on HBP1 expression was tested in vitro using SK-N-AS (ALKwt) neuroblastoma cells transduced with tetracycline-inducible overexpression constructs for ALKwt, ALKF1174L, and ALKR1275Q (Fig. 1e). Furthermore, in a cohort of 283 primary human neuroblastoma tumors (GSE85047), a significant negative correlation between HBP1 and ALK gene expression was observed in keeping with the proposed negative regulatory effects of ALK on HBP1 (Supplementary Fig. 1C).
Given the role of ALK activation in a subset of lung carcinomas, we analyzed a dataset of EML4/ALK fusion positive non-small-cell lung carcinoma cells (NSCLC) [34] and observed higher HBP1 expression levels upon ALK inhibition (Fig. 1f). Taken together, our data support that ALK leads to downregulation of HBP1 expression levels in neuroblastoma cells as well as in other cell types including NSCLC.
HBP1 levels are controlled through ALK-PI3K/AKT–FOXO3a
To test which of the two major ALK downstream pathways (MAPK versus PI3K/AKT) [4,5,6,7, 28, 36, 37] controls HBP1 expression, ALK mutant cells were treated with either a MEK-inhibitor (U0126) or a PI3K/mTOR-inhibitor (BEZ-235). In line with previous findings showing HBP1 transcriptional regulation through the PI3K-AKT–FOXO3 axis [31] as well as through phosphorylation by AKT [32], HBP1 upregulation was observed after exposure to the PI3K/mTOR-inhibitor, while MEK inhibition had no effect on HBP1 levels (Fig. 1g, h). Similar effects were obtained for NB-1 using the PI3K-inhibitor pictilisib (Supplementary Fig. 1D). In further support of these findings, the PI3K/AKT-inhibitor signature score [38] positively correlated with HBP1 gene levels in the cohort of 283 NB patients (GSE85047) (Supplementary Fig. 1E).
Based on previously reported observations showing negative regulation of FOXO3a by ALK through the PI3K-AKT pathway in neuroblastoma [28, 38] and anaplastic large cell lymphoma (ALCL) [39] and the control of HBP1 by FOXO3a in fibroblasts [31], we explored the role of FOXO3a in ALK controlled HBP1 regulation in neuroblastoma. First, we assessed a FOXO3a signature [38] in a cohort of 283 NB patients and observed positive correlation of the signature with HBP1 gene levels (GSE85047) (Supplementary Fig. 1F). Next, we looked into an available transcriptome dataset based on doxycycline-inducible FOXO3a overexpression in combination with the PI3K/mTOR-inhibitor PI-103 in the NB cells SH-SY5Y (ALKF1174L) [38]. As expected, PI-103 induces HBP1 levels through inhibition of PI3K activity. This results in the release of repression of FOXO3a and upregulation of its positively regulated target HBP1. Induction of FOXO3a expression only has no detectable effect on HBP1 expression levels due to sustained repression of FOXO3a by activated PI3K. Finally, combined activation of FOXO3a and PI3K inhibition through PI-103 causes a strong induction of HBP1 levels as PI-103 unleashes the repression of FOXO3a by PI3K, while FOXO3a itself is in addition ectopically overexpressed (Fig. 1i). Collectively, our data support that ALK negatively regulates HBP1 expression levels through the PI3K-AKT–FOXO3a pathway in neuroblastoma.
HBP1 is negatively regulated by MYCN through the miR-17~92 cluster
In view of the previously reported negative regulation of HBP1 through the miR-17~92 cluster in breast cancer, leukemia and lymphoma cells [40, 41] and the known positive regulation of miR-17~92 by MYCN in NB, leukemia and lymphoma cells [41,42,43], we decided to investigate this MYCN - miR-17~92 - HBP1 regulatory axis in more detail in the context of NB cells. To this end, we first evaluated HBP1 protein expression levels following shRNA-mediated MYCN knockdown in the neuroblastoma IMR-5/75 cell line and found elevated HBP1 protein levels upon MYCN knockdown (Fig. 2a). In line with this finding, we also show that in MYCN-driven mouse tumor formation, Hbp1 is transcriptionally downregulated (Supplementary Fig. 2A). The presumed MYCN control of HBP1 by the miR-17~92 cluster was then confirmed using the previously validated tetracycline-inducible miR-17~92 overexpression model in SH-EP neuroblastoma cells [42], showing HBP1 downregulation upon induction of miR-17~92 (Fig. 2b, c, Supplementary Fig. 2B, C). In summary, these data are in keeping with negative HBP1 regulation through MYCN induced elevated miR-17~92 levels.
HBP1 is negatively regulated by MYCN through the miR-17~92 miRNA cluster and represses MYCN activity. a Western blot showing HBP1 and MYCN protein levels in the neuroblastoma IMR-5/75 cells upon shRNA-mediated MYCN knockdown. b HBP1 mRNA levels in SH-EP cells treated with tetracycline (or ethanol as control) to induce the miR-17~92 cluster, expressed relative to the corresponding control. c Western blot analysis showing HBP1 protein levels at the same time points after inducing the miR-17~92 cluster in the SHEP cell line as in b. d Heatmap showing the gene sets of an in house compiled gene set collection containing all publically available MYC(N) activity or target signatures [44, 45, 52, 64,65,66,67,68], that are positively or negatively enriched upon HBP1 overexpression according to GSEA (with FDR < 0.25). e, f Spearman correlation between the HBP1 pathway signature and the MYCN gene levels (e) or the MYCN signature (f) in a cohort of 283 NB patients. g GSEA for the EZH2 down gene set in the HBP1 overexpression data and heatmap showing the leading edge (top 22 genes) of this gene set plotted in the HBP1 overexpression dataset. h, i Spearman correlation between HBP1 gene levels (h) or HBP1 pathway signature (i) and the EZH2 signature in a cohort of 283 NB patients. Error bars represents mean ± SD of three biological replicates, each containing two technical replicates. b Two-sided paired t-test was performed on log normalized data with *P < 0.05, **P < 0.01, ***P < 0.001
HBP1 is a suppressor of MYCN activity in neuroblastoma cells
Previous studies showed that HBP1 acts as a transcriptional repressor through direct interaction with other transcription factors, including MYC(N) [29, 30]. We therefore first evaluated the effects of HBP1 overexpression on MYCN mRNA and protein levels but observed no effect on transcriptional nor protein levels of MYCN thus excluding this possible mode of regulation (Supplementary Fig. 2D). To explore the role of HBP1 in regulation of MYCN activity in neuroblastoma, we analyzed the transcriptomes of the NGP-HBP1up versus NGP-parental cell line. Gene set enrichment analysis (GSEA [44]) of the genes upregulated upon HBP1 overexpression in NGP-HBP1up revealed enrichment (FDR < 0.25) for 11 out of 26 gene sets related to MYC(N) regulation and activity [45, 46] (Fig. 2d, Supplementary Fig. 2E, F).
To explore the functional relationship between HBP1 and MYCN in primary human tumors, we established a HBP1up pathway signature based on the differentially expressed genes in NGP-HBP1up cells as compared to the NGP-parental cells and tested this signature in a cohort of 283 primary human NB tumor samples (GSE85047), demonstrating strong inverse correlation between the HBP1up pathway signature and MYCN gene expression and MYCN activity score [45] (Fig. 2e, f). While these data point towards an inhibitory role of HBP1 on MYCN activity, we could not show direct interaction between both proteins, possibly due to the drastic effects on cell viability upon HBP1 overexpression in most neuroblastoma cell lines tested (not shown). Taken together, our data are in keeping with HBP1 acting as a suppressor of MYCN activity in neuroblastoma cells.
The PRC2 complex cooperates with HBP1 in repression of gene activity
Gene set enrichment analysis (GSEA) of the NGP-HBP1up versus NGP-parental data using the ‘c6 Oncogenic Signatures v5.0’ from the Molecular Signatures Database (MSigDB), revealed positive enrichment for two PRC2 gene sets (Fig. 2g, Supplementary Fig. 2G) containing genes reported to be occupied by the Polycomb Repressive Complex 2 (PRC2) components SUZ12 or EZH2 [47], thus suggesting an upregulation by HBP1 of genes occupied by the PRC2 complex. Next, motif analysis using iRegulon [48] on the NGP-HBP1up versus NGP-parental differentially expressed genes revealed enrichments for SUZ12 motifs among the downregulated genes (Supplementary Fig. 2H). Moreover, a positive correlation between HBP1 gene levels as well as the HBP1up pathway signature scores and an EZH2-inhibitor signature score, generated upon EPZ6438 EZH2-inhibitor treatment in SK-N-BE(2c) cells [49], was observed in the cohort of 283 NB patients (GSE85047) (Fig. 2h, i).
In summary, we identified HBP1 as a negative regulator of MYCN activity and suggest that HBP1 interacts with the PRC2 complex in gene repression.
Increased HBP1 levels represses tumor aggressiveness
To elucidate the role of HBP1 on the neuroblastoma cellular phenotype, we evaluated the functional characteristics of the NGP-HBP1up cell line versus the NGP-parental cells. The overexpression of HBP1 was confirmed both at the mRNA level and at the protein level (Supplementary Fig. 3A). HBP1 overexpression increased the apoptotic response, negatively affected colony forming capacity and repressed cell growth both with normal and lower serum concentrations, as shown by a marked reduction in viability rate (Fig. 3a–c, Supplementary Fig. 3B). Furthermore, upon induction of miR-17~92 in the SHEP cell line in which we overexpressed HBP1, we observed a reduction of the attenuated growth observed upon HBP1 overexpression (Supplementary Fig. 3C). Since Li et al. [41]. showed through an elegant series of experiments that the ability of MYC(N) to sustain autonomous proliferation, self-renewal and survival is mainly miR-17~92-dependent, the observed reduction of attenuated growth suggests that at least a part of the functional effects observed upon HBP1 overexpression can be explained by the repressive effects of HBP1 on MYCN.
Increased HBP1 levels represses tumor aggressiveness. a, b Cell viability (a) and cell death (b) of NGP-parental cell line and the NGP cell line with stable HBP1 overexpression at the indicated time points (one out of 2 biological replicates shown). c Clonogenic survival assays in the parental and HBP1 overexpressing NGP cell lines, each consisting of 5 replicates (two out of three independent experiments shown). d NGP cell line with stable HBP1 overexpression forms a loose aggregate, while the NGP-parental cell line forms a dense spheroid on ultra-low-attachment plates. e, f Kaplan–Meier plot showing percentage of overall survival in patients with high or low HBP1 expression in tumors of all stages (e) and only stage 4 tumors (f) in the cohort of 283 NB patients. Error bars represents mean ± SD of three technical replicates (a, b)
Moreover, NGP-parental cells form compacted spheres within 3 days, an activity which is prevented by overexpression of HBP1 as indicated by the presence of loose aggregates where individual cells are recognizable (Fig. 3d).
Notably, HBP1 activity has previously been connected to cell differentiation in leukemic myeloid cells and in the cortical region of mouse brains [50, 51]. In keeping with this, we also observed higher scores for the differentiation signature of Frumm [52], in HBP1 overexpressing versus NGP-parental cells (Supplementary Fig. 3D).
Importantly, we also revealed that the lowest HBP1 mean expression was observed in the MYCN-amplified subgroup and in the stage 4 subgroup in a cohort of 283 neuroblastoma patients (GSE85047) (Supplementary Fig. 3E-F). Additionally, high levels of HBP1 expression significantly correlated with better event-free and overall survival in all tumor types and in stage 4 tumors only (Fig. 3e, f, Supplementary Fig. 3G, H). Moreover, HBP1 is a significant predictor of overall survival, independently of INSS stage, MYCN amplification and age (P = 0.02). To the best of our knowledge, no deletions nor inactivating mutations in HBP1 have been reported so far in NB primary tumors or cell lines.
Collectively, these data indicate that lower HBP1 levels mark tumor aggressiveness and that HBP1 could act as tumor suppressor in neuroblastoma.
Combined pharmacological upregulation of HBP1 and repression of MYCN induces synergistic effects
In view of the above, we assumed that HBP1 upregulation in combination with blocking of MYCN expression or activity in neuroblastoma cells could have an enhanced negative effect on tumor cell proliferation (Fig. 4a, b). This was further corroborated by the data obtained by combined HBP1 overexpression and shRNA knockdown of MYCN in IMR-5/75 cells (Supplementary Fig. 4A). In view of these data, we decided to explore possibilities for drugging this regulatory axis. We initially tested the green tea polyphenol epigallocatechin-3-gallate (EGCG) previously shown to reduce breast cancer cell proliferation through increase of HBP1 mRNA stability [53]. As a more clinically relevant compound, the PI3K/mTOR dual inhibitor BEZ-235 was found to induce HBP1 expression. First, we demonstrate HBP1 upregulation and decreased cell viability after EGCG treatment in the ALK mutant cell lines NB-1 and CLB-GA, while no effects were noted in the ALK wild type non-responder SK-N-AS cells (Supplementary Fig. 4B, Fig. 4c). Next, we performed combination drugging for EGCG and BEZ-235 with the BET-inhibitor JQ1, known to repress transcription elongation and MYCN expression in MYCN-amplified cell lines [54], and observed synergistic effects (combination index (CI) < 1) on cell growth and on MYCN protein levels as compared to treatment with the individual compounds in neuroblastoma cell lines (Fig. 4d, g; Supplementary Fig. 4C-D-E-F-G). Our findings confirm previous reports showing enhanced effects on MYC levels by combining a PI3K-AKT inhibitor with JQ1 [54,56,57]. We tested EGCG and JQ1 combination further in vivo in LSL-MYCN;dßh-iCre tumors engrafted in immunocompromised mice and showed significant effects on tumor growth, proliferation, apoptosis and survival of mice receiving this combination (Fig. 5a–d; Supplementary Fig. 4H-I).
Combined pharmacological upregulation of HBP1 and repression of MYCN induces synergistic effects on tumor growth. a ALK regulates MYCN activity through several mechanisms. It enhances MYCN protein stability by blocking GSK3β through PI3K, while it simultaneously phosphorylates FOXO3 through this same pathway. In this way, FOXO3 stays cytoplasmatic and can not activate HBP1, which is a negative regulator of MYCN activity. MYCN itself inhibits HBP1 through induction of the miR-17–92 cluster, thereby forming a negative feedback loop. To block these pathways in order to upregulate HBP1 while negatively affecting MYCN activity, there are several nodes that can be targeted by compounds: ALK-inhibitors (TAE684, crizotinib, ceritinib, alectinib), GSK3β-inhibitors, PI3K-inhibitors (BEZ-235, PI-103, pictilisib), HDAC-inhibitors (SAHA, panobinostat), JQ1 which inhibits BRD3 and in this way MYCN indirectly, while the green tea component EGCG upregulates HBP1 expression directly. b Hypothetical scheme showing how HBP1 has an impact on the dual role of MYCN: HBP1 causes downregulation of genes involved in cell cycle that are positively regulated by MYCN, while it upregulates differentiation genes that are repressed by MYCN. c Cell viability assay in SK-N-AS, NB-1, and CLB-GA, showing the EGCG dose–response curves 48 h after treatment. d Cell viability of the NGP cell line, showing the dose–response curves of EGCG, JQ1, and the combination 48 h after treatment. e Combination index (CI)-values at IC-50 showing synergistic effect in a panel of 7 NB cell lines with different ALK and MYCN status. Synergy: CI < 1.0, additive: CI = 1.0, antagonism: CI > 1.0. f, Cell viability of the NGP cell line, showing the dose–response curves of BEZ-235, JQ1 and the combination 48 h after treatment. g Western blot analysis showing MYCN protein levels 48 h after treating the CLB-GA cells with DMSO, 0.405 μM BEZ-235, 1.215 μM JQ1, or the combination. c, d, f Mean cell viability measurements of three biological replicates
BEZ-235 in combination with JQ1 or HDACi synergistically upregulates HBP1 levels with concomitant repression of MYCN activity a, Kaplan–Meier plot showing percentage survival of LSL-MYCN;dßh-iCre grafted mice treated with DMSO, EGCG (20 mg/kg), JQ1 (50 mg/kg), or the combination. Black stars: survival DMSO versus combination, blue stars: survival EGCG versus combination, n = 7 mice per group. b, c Graphs showing Ki-67 positive cells (b) and cleaved caspase-3 positive cells (c) in immunohistochemistry (IHC) staining in tumor sections of the forced treatment group of mice. d IHC staining for the proliferative cells (Ki-67) and apoptotic cells (cleaved caspase-3) mentioned in b and c. Scale bar = 200 µm. e Cell viability of the NGP cell line, showing the dose–response curves of BEZ-235, SAHA and the combination 48 h after treatment. f, g MYCN expression (f) and MYCN activity score (g) in NB-1 cells treated with DMSO, 0.5 μM BEZ-235, 0.5 μM SAHA, or the combination. h Cell viability of the NGP cell line, showing the dose–response curves of BEZ-235, panobinostat, and the combination 48 h after treatment. Error bars represents mean ± SD of seven biological replicates (b, c), or at least two biological replicates, each consisting of two technical replicates (e, h) or of three biological replicates (f, g). ANOVA test followed by pairwise t-test for multiple comparisons (b, c, f) with *P < 0.05, **P < 0.01, ***P < 0.001
Given the reported effect of HDAC-inhibitors on MYCN protein levels [58], we also tested the effects of combined BEZ-235 and HDAC-inhibitor SAHA (vorinostat) treatment in a panel of 8 cell lines and in the patient-derived xenograft (PDX)-derived cell line (LU-NB-2) [59, 60], showing synergistic effects on cell survival (Fig. 5e; Supplementary Fig. 5A-B-C-D) and a synergistic effect on HBP1 and MYCN levels in the CLB-GA cell line (Supplementary Fig. 5E). Moreover, analysis of transcriptomes of the treated cells demonstrates downregulation of MYCN expression and more importantly, the MYCN activity score [45], thus indicating the expected reduction in MYCN activity (Fig. 5f, g). Finally, we also tested the more recently developed potent HDAC-inhibitor panobinostat [61], also demonstrating the expected synergism on cell viability in the tested neuroblastoma cell lines (Fig. 5h; Supplementary Fig. 5F-G) and the patient-derived xenograft (PDX)-derived cell line (LU-NB-2) [59, 60] (Supplementary Fig. 5H) and upregulation of HBP1 mRNA in the CLB-GA cell line upon the BEZ-235 and panobinostat combination (Supplementary Fig. 5I).
Discussion
We provide in vitro and in vivo evidence for the existence of another mechanism for activation of MYCN activity through ALK-PI3K–FOXO3a controlled downregulation of the negative regulator of MYC(N) activity HBP1. Together with the previously described ALK-ERK5-driven transcriptional induction of MYCN and regulation of the oncogenic activity of MYCN through increased mRNA levels and protein phosphorylation by the ALK-PI3K/mTOR-GSK3β axis [24,25,26,27, 62, 63], this represents a third mechanism of ALK controlled MYCN activation. We also show that in neuroblastoma cells MYCN itself indirectly represses HBP1 expression levels through its downstream upregulated miRNA cluster miR-17∼92 that targets HBP1. As such, a complex inter-regulatory network emerges where ALK regulates MYCN through three distinct mechanisms [24,25,26,27, 56, 57], MYCN transcriptionally activates ALK [64] and both ALK and MYCN repress HBP1 expression levels. Given the proven role of MYCN as driver oncogene in neuroblastoma oncogenesis [2], our data provide a further mechanistic explanation for the previously reported increased tumor aggressiveness in patients with combined MYCN amplification and ALKF1174L mutations and mouse and zebrafish modeling which demonstrated a key role for mutated ALK in accelerated MYCN-driven neuroblastoma formation [21,22,23].
In this study, we investigated for the first time the functional interaction of HBP1 with MYCN in the context of neuroblastoma. We showed evidence for physical interaction between MYCN and HBP1, while HBP1 overexpression revealed significant enrichment for gene sets related to MYC(N) regulation and activity. Furthermore, in a cohort of 283 primary human NB tumor samples, we demonstrated strong inverse correlation between the HBP1 upregulated pathway signature and MYCN gene expression and MYCN activity score. Taken together, these data support the role of HBP1 as a suppressor of MYCN activity in neuroblastoma cells.
Unexpectedly, the data mining of the transcriptome alterations upon HBP1 overexpression and EZH2-inhibitor signature scores suggest that HBP1 abrogates the MYCN/PRC2 controlled repression of pro-differentiation genes. Our findings provide further evidence for a role for EZH2 and SUZ12 as components of the PRC2 complex in the MYCN-mediated gene repression [49, 65], and for the first time assigns a putative function to HBP1 as regulatory factor mediating the release of transcriptional repression by the MYCN/EZH2 complex. Further, HBP1 upregulation also leads to repression of MYCN activated genes, possibly through recruitment of HDAC and SIN3B as described for MYC [41]. Taken together, while the exact mode-of-action of HBP1 in relation to MYCN and PRC2 remains to be resolved, our data support previous findings that HBP1 affects both the MYC(N) transcriptional activating and repressing activity [29]. As such, our findings also support the recently suggested role for PRC2 in MYCN controlled gene repression [49, 65].
Unraveling of the ALK signaling cascade has previously provided novel putative drugging approaches as illustrated by the finding of RET and ERK5 as druggable downstream ALK targets [28, 62]. In this study, we explored several drug combinations targeting HBP1 and/or MYCN to explore possible synergistic interactions. We tested a PI3K/mTOR-inhibitor aimed to activate (among others) HBP1 expression in combination with BET-inhibitor JQ1, a known negative regulator of MYC(N) activity in MYCN-amplified cell lines [54], in order to further decrease cell viability as well as MYC(N) activity and observed strong synergistic effects in neuroblastoma cells. Secondly, we combined the same PI3K/mTOR-inhibitor with different HDAC-inhibitors mediating MYCN suppression [58] and previously shown to give promising effects on MYC-driven medulloblastoma [61] and Burkitt lymphoma [66] and also observed synergistic effects, both on conventional neuroblastoma as well as PDX-derived cell lines [59]. In addition to these combinations (Fig. 4a, b), other approaches can be envisioned to target MYCN or MYCN activity, thus offering the potential for consecutive series of different drug combinations to achieve sustained blocking of MYCN activity in neuroblastoma. Such approaches and other potent drug combinations as well as novel immunotherapeutic approaches may ultimately lead to accomplish the final goal to achieve better and long-term survival for NB patients.
Material and methods
Cell lines and reagents
Human neuroblastoma cells (Supplementary Table I: cell line information) were cultured in RPMI-1640 medium (Invitrogen), supplemented with fetal bovine serum (10%), kanamycin (100 μg/ml), penicillin/streptomycin (100 IU/ml), l-glutamin (2 mM), and HEPES (25 mM) (Life Technologies), while the mouse mNB-A1 cells were cultured in this medium supplemented with N-2 Supplement and B-27 Serum-Free Supplement (Life Technologies). The PDX-derived LU-NB-2 cell line, derived from a MYCN-amplified neuroblastoma, orthotopically xenografted in NSG mice forming metastasizing tumors, was cultured under serum-free, stem-cell promoting culture conditions [59, 60]. Cells were maintained at 37 °C in a 5% CO2-humidified environment. Mycoplasma testing was performed every 2 months.
The compounds NVP-TAE684 (ALK-inhibitor, S1108), BEZ-235 (PI3K/mTOR-inhibitor, S1009), pictilisib (PI3K-inhibitor, S1065), U0126 (MEK-inhibitor, S1102), EGCG (S2250), and panobinostat (HDAC-inhibitor, S1030) were purchased from SelleckChem, crizotinib (ALK-inhibitor, PZ0191) and SAHA (vorinostat, HDAC-inhibitor, SML0061) from Sigma-Aldrich, ceritinib (ALK-inhibitor, A-1189–5, LDK378) from Hoelzel Biotech and JQ1 (BET-inhibitor, 27401) from BPS Bioscience. Compounds were dissolved in sterile DMSO, stored at −20 °C and further diluted to an appropriate final concentration in culture medium at the time of use. DMSO was used as solvent control for every treatment.
Evaluation of HBP1 expression upon ALK and downstream pathway inhibition
HBP1 expression levels were evaluated in (1) 6 NB cell lines with different ALK status (ALKwt, ALKF1174L, ALKR1275Q, ALKamp) treated with 0.32 μM TAE684 for 6 h, (2) the CLB-GA cell line (ALKR1275Q) treated with 0.32 μM TAE684 and harvested at different time points (10, 30, 60, 120, 240, 360 min) (E-MTAB-3205 [28]), (3) CLB-GA cells treated with 0.2 μM ceritinib (LDK378), 0.32 μM TAE684, 0.06 μM X396, 0.5 μM crizotinib, 0.05 μM trametinib, 0.5 μM BEZ-235 and 9.5 μM vandetanib for 6 h (E-MTAB-3206 [28]), (4) SK-N-AS cell lines with tetracycline-inducible ALKwt, ALKF1174L or ALKR1275Q constructs treated with 2 μg/ml tetracycline or ethanol for 24 h (E-MTAB-3207 [28]), (5) NB-1 and CLB-GA treated with 0.5 μM BEZ-235, 0.5 μM pictilisib or 8 μM U0126 for 6 h for RNA and protein, (6) mice with subcutaneously xenografted SH-SY-5Y neuroblastoma cells, treated with TAE684 and crizotinib, (7) publically available data of NSCLC treated with Alectinib (GSE25118 [34]), (8) a published dataset of a FOXO3-inducible SH-SY5Y cell line (GSE42762 [38]) and (9) a cohort of 283 neuroblastoma tumor samples (GSE85047)
Testing synergism of drug combinations in vitro
Neuroblastoma cells were seeded in 96-well tissue culture plates in triplicate at 30% confluency, allowed to recover overnight and subsequently treated with a range of concentrations of the two inhibitors while keeping the final concentration of DMSO constant. Forty-eight hours after treatment, cell viability was measured using Cell-Titer Glo (Promega), according to the manufacturer’s protocol and luminescence was measured with the GloMax®-Multi Detection System (Promega). To evaluate possible synergism, combination indexes (CI) for each combination were calculated using the CalcuSyn software (Biosoft, Ferguson, MO), which uses the Median Effect method [67]. Synergism is defined as a CI-value < 1.0, while additive effects result in a CI-value equal to 1.0.
Synergistic effects of EGCG and JQ1 in vivo
LSL-MYCN;dßh-iCre tumors were re-grafted in the flank of immuno-deficient mice (nu/nu female mice of 6 weeks old). The sample size calculation for means was used to determine the number of animals in the groups. With an estimated difference in means (treated-to-control ratio) of 50% (estimated tumor volume of 1.2 cm3 in the control group and 0.6 cm3 in the treated group), standard deviation of 30%, p-value of 0.05 and power of 80%, you need at least 6 mice per group. The short-term treatment experiment consisted of 4 groups each containing 3 mice: vehicle (12,5% DMSO in 5% glucose), EGCG (20 mg/kg), JQ1 (50 mg/kg), or the combination (EGCG + JQ1). When tumors reached a volume of around 500 mm3, mice were treated twice daily (100 μl i.p.) during 3 days. Animals were therefore first sorted by tumor volume. Mice with order number 1, 5, 9 were allocated to cohort a, mice 2, 6, 10 to cohort b, mice 3, 7, 11 to cohort c, and mice 4, 8, 12 to cohort d. Hereafter, mice were killed by cervical dislocation. Tumors were excised, formalin-fixed, analyzed for histology (H&E) and immunohistochemically stained for Ki-67 and cleaved caspase-3. To monitor the effects on tumor growth and survival, a long-term experiment was performed. After tumors reached a volume of around 100–150 mm3, 28 mice were randomly assigned to the 4 groups, according to the system earlier mentioned. Mice were treated daily (100 μl i.p.) for a period of maximal 23 days. When tumor sizes exceeded 3000 mm3, mice were killed by cervical translocation. For both experiments, the person, who measured the tumors did not knew which cohort the present animal belonged to. All experiments were performed in accordance with the council of Europe guidelines for housing and care of laboratory animals, and protocols were approved by the animal facility of the University Hospital Essen.
SHEP-miR-17~92 system
The miR-17∼92 cluster was induced with 2 μg/ml tetracycline or ethanol (control) in the previously described SHEP-TR-miR-17~92 model system [42] and cells were harvested at different time points (0, 24, 48, 72 h).
For the rescue experiment, these cells were nucleofected (Neon™ Transfection System 100 µl Kit, Cat No MPK10025) with either SHC002-GFP (control) or a Precision LentiORF Human HBP1 overexpression construct (ThermoScientific) for HBP1 overexpression. The cell lines with the HBP1 construct were thereafter sorted on GFP to select for HBP1 overexpression. The four conditions (miR-17~92 off & HBP1 off, miR-17~92 on & HBP1 off, miR-17~92 off & HBP1 on and miR-17~92 on & HBP1 on) were seeded in complete medium in 96-well tissue culture plates in triplicate at 30% confluency. Cell proliferation was assessed continuously by using the IncuCyte® Live Cell imaging system (Essen BioScience, Cat No 4641), by taking photos every 2–3 h of the whole well. Masking was done using the IncuCyte® ZOOM Software.
IMR-5/75 system
IMR-5/75 cell line was nucleofected (Neon™ Transfection System 100 µL Kit, Cat No MPK10025) with either SHC002-GFP (control) or a Precision LentiORF Human HBP1 overexpression construct (ThermoScientific) for HBP1 overexpression, and treated with 1 µg/ml doxycylin or not for the induction of shMYCN. 24 h after seeding, cells were refreshed with complete RPMI medium, and subsequently 48 h later cell proliferation was assessed continuously by using the IncuCyte® Live Cell Imaging System (Essen BioScience, Cat No 4641), by taking photos every 2–3 h of the whole well. Masking was done using the IncuCyte® ZOOM Software.
RNA isolation, cDNA synthesis, and RT-qPCR
RNA isolation, cDNA synthesis and RT-qPCR of the generated samples was performed as we described earlier [28]. The Cq-values for target gene expression were normalized with at least three reference genes (primer sequences: Supplementary Table II) and qBasePlus software (Biogazelle) was used to analyze the results [68].
Establishment of stabile HBP1 overexpressing cell lines
NGP cells were transduced with Precision LentiORF Human HBP1 viral particles (ThermoScientific). Transduced cells were selected using blasticidin (20 µg/ml), subsequently maintained and harvested for further experiments.
In vitro assessment of HBP1 effect
NGP-parental and NGP HBP1 cells were seeded in complete medium and a 1:200 dilution of IncuCyte® Annexin V Red Reagent for apoptosis (Essen Bioscience Cat No 4641) in 96-well tissue culture plates in triplicate at 30% confluency. Cell proliferation and cell death (Annexin V positivity) was assessed continuously by using the IncuCyte® Live Cell imaging system (Essen BioScience, Cat No 4641), by taking photos every 2–3 h of the whole well. Masking was done using the IncuCyte® ZOOM Software. Additionally, NGP-HBP1up and the NGP-parental cell lines were seeded at 7000 cells/well in sextuple in 96-well tissue culture plates in the presence of 10, 5, 1, or 0.1% serum and monitored and quantified using IncuCyte® Live Cell imaging system (Essen BioScience, Cat No 4641) and the IncuCyte® ZOOM Software.
To assess the colony formation capacity, cells were seeded in 5-fold at a concentration of 2000 cells per 6-cm dish. Cells were allowed to recover and to form colonies during a period of at least one week, followed by fixation of the cells by adding 0,5 ml 4% formaldehyde to each dish. After 1 h, dishes were washed and colonies were colored with 0.005% crystal violet. After washing and air-drying, the dishes were scanned and evaluated with ImageJ to quantify the differences. For the spheroid formation, NGP-parental and NGP-HBP1up cells were seeded at 4000 cells/well in 48-fold in hydrogel coated 96-well plates, termed ultra-low attachment plates (ULA, Corning 7007) and monitored and quantified using IncuCyte ZOOM technology (Essen BioScience).
Protein isolation, antibodies, and western blotting
Protein isolation and western blotting was performed as we described earlier [28]. AKT (9272, 1:1000), phosphorylated AKT (S473) (3787, 1:1000), MYCN (9405, 1:1000), SUZ12 (3737, 1:1000), secondary anti-rabbit (7074, 1:50,000) and anti-mouse (7076, 1:50,000) antibodies were obtained from Cell Signaling, while antibodies against the loading proteins vinculin (V9141, 1:10,000) and α-tubulin (T4026, 1:10,000) from Sigma Aldrich and the HBP1 (A-5) antibody (sc-376831, 1:400) from Santa Cruz Biotechnology.
Microarray-based gene expression profiling
RNA quality was analyzed using Experion (Bio-Rad). Samples from the parental and NGP HBP1 cell lines were labeled and hybridized to the Sureprint G3 human GE 8 × 60 K microarrays (Agilent Technologies), according to the manufacturer’s guidelines and starting from 200 ng RNA. The data were normalized with the vsn method, using the vsn and Limma packages in R.
Signature score generation and analysis and GSEA
Using the limma R-package, differential expression analysis was performed comparing the parental and NGP HBP1 samples. The established signatures consist of the differentially expressed genes with adjusted P-value (False Discovery Rate (FDR)) < 0.05. Next, we generated an EZH2-inhibitor signature, based on public data of EHZ2 inhibition with EPZ6438 in the SK-N-BE(2c) cell line [49] and a differentiation signature, based on public data of treating SK-N-BE(2c) and SH-SY5Y with pro-differentiating agents [52]. Signature score analyses were conducted using a rank-scoring algorithm [28]. Gene set enrichment analysis (GSEA [44]) (Supplementary Table III) was performed using the MSigDB ‘c6 Oncogenic Signatures v5.0’ gene sets (software.broadinstitute.org/gsea/msigdb) and an in house compiled gene set catalog containing all MYC target gene sets from the MSigDB ‘Hallmark v5.0’ catalog as well as publically available MYC(N) activity or target signatures [45, 46, 54, 68,70,71,72,73]. The gene sets, showing positively or negatively enrichment and with a FDR < 0.25 are plotted in a heatmap.
RNA sequencing after SAHA and BEZ
RNA quality was evaluated using the Fragment Analyzer (Advanced Analytics). Library prep was performed with the TruSeq Stranded mRNA Library Prep Kit LT (Illumina), following manufacturer’s instructions. Quality of the library was assessed with BioAnalyzer (Agilent) and concentrations were checked with the Kapa Library Quantification Kit (Kapa Biosystems). After pooling the samples, RNA sequencing was performed with the NextSeq 500 High Output kit, V2, 75 cycli, single-end (Illumina), following manufacturer’s instructions, on the NextSeq 500 (Illumina). Quality of the data was checked by fastQC. Thereafter, the data was mapped to Hg38 by STAR and count data was generated using RSEM. Differential expression analysis was performed with Limma-voom in R.
Statistical analyses
Statistical significance was calculated with GraphPad Prism7 by unpaired one-way ANOVA with Bonferroni correction when comparing more than two unmatched groups, while unpaired or paired t-test was chosen when comparing two groups. Parametric t-tests and ANOVA tests were performed on log-transformed expression data, which are considered to have a normal distribution. Spearman was used to evaluate correlation between datasets, which does not require normal distribution of the data. For survival curves, we choose the survival analysis within GraphPad Prism7, which makes use of the LogRank test.
Accession number
The data have been deposited in NCBI’s Gene Expression Omnibus and is accessible through GEO Series accession number GSE95193.
References
Louis CU, Shohet JM. Neuroblastoma: molecular pathogenesis and therapy. Annu Rev Med. 2015;66:49–63.
Cheung N-K, Dyer M. Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nat Rev Cancer Nat. 2013;13:397–411.
Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–93.
Mossé Y, Laudenslager M, Longo L, Cole K, Wood A, Attiyeh E, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–5.
George R, Sanda T, Hanna M, Fröhling SW II, Zhang J, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Cah Rev The. 2008;455:975–8.
Chen Y, Takita J, Choi Y, Kato M, Ohira M, Sanada M, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Cah Rev The. 2008;455:971–4.
Janoueix-Lerosey I, Lequin D, Brugières L, Ribeiro A, Pontual L, Combaret V, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Cah Rev The. 2008;455:967–70.
Carpenter EL, Mossé YP. Targeting ALK in neuroblastoma—preclinical and clinical advancements. Nat Rev Clin Oncol. 2012;9:391–9.
Guan Tucker, Wan Chand, Danielson Ruuth, et al. The ALK inhibitor PF-06463922 is effective as a single agent in neuroblastoma driven by expression of ALK and MYCN. Dis Model Mech Highwire. 2016;9:941–52.
Amin A, Li L, Rajan S, Gokhale V, Groysman M, Pongtornpipat P, et al. TKI sensitivity patterns of novel kinase-domain mutations suggest therapeutic opportunities for patients with resistant ALK+tumors. Oncotarget. 2016;7:23715–29.
Pall G. The next-generation ALK inhibitors. Curr Opin Oncol. 2015;27:118.
Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, et al. Dynamic reprogramming of the kinome in response to targeted MEK Inhibition in triple-negative breast. Cancer Cell. 2012;149:307–21.
Toyokawa Seto. Updated evidence on the mechanisms of resistance to ALK inhibitors and strategies to overcome such resistance: clinical and preclinical data. Oncol Res Treat. 2015;0:291–8.
Bresler SC, Wood AC, Haglund EA, Courtright J, Belcastro LT, Plegaria JS, et al. Differential inhibitor sensitivity of anaplastic lymphoma kinase variants found in neuroblastoma. Sci Transl Med. 2011;3:108ra114.
Bresler SC, Weiser DA, Huwe PJ, Park JH, Krytska K, Ryles H, et al. ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer Cell. 2014;26:682–94.
Heuckmann J, Hölzel M, Sos M, Heynck S, Balke-Want H, Koker M, et al. ALK mutations conferring differential resistance to structurally diverse ALK inhibitors. Clin Cancer Res. 2011;17:7394–401.
Yan X, Kennedy CR, Tilkens SB, Wiedemeier O, Guan H, Park J-II, et al. Cooperative cross-talk between neuroblastoma subtypes confers resistance to anaplastic lymphoma kinase inhibition. Genes Cancer. 2011;2:538–49.
Debruyne Bhatnagar, Sharma Luther, Moore CheungN-K, et al. ALK inhibitor resistance in ALK(F1174L)-driven neuroblastoma is associated with AXL activation and induction of EMT. Oncogene. 2016;35:3681–91.
Isozaki H, Ichihara E, Takigawa N, Ohashi K, Ochi N, Yasugi M, et al. Non-small cell lung cancer cells acquire resistance to the ALK inhibitor alectinib by activating alternative receptor tyrosine kinases. Cancer Res Highwire. 2015;76:1506–16.
Lovly CM, Shaw AT. Molecular pathways: resistance to kinase inhibitors and implications for therapeutic strategies. Clin Cancer Res. 2014;20:2249–56.
De Brouwer S, De Preter K, Kumps C, Zabrocki P, Porcu M, Westerhout E, et al. Meta-analysis of neuroblastomas reveals a skewed ALK mutation spectrum in tumors with MYCN amplification. Clin Cancer Res. 2010;16:4353–62.
Zhu S, Lee J-S, Guo F, Shin J, Perez-Atayde A, Kutok J, et al. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell. 2012;21:362–73.
Berry T, Luther W, Bhatnagar N, Jamin Y, Poon E, Sanda T, et al. The ALKF1174L mutation potentiates the oncogenic activity of MYCN in neuroblastoma. Cancer Cell. 2012;22:117–30.
Schönherr C, Ruuth K, Kamaraj S, Wang C-L, Yang H-L, Combaret V, et al. Anaplastic lymphoma kinase (ALK) regulates initiation of transcription of MYCN in neuroblastoma cells. Oncogene. 2012;31:5193–200.
Chesler L, Schlieve C, Goldenberg DD, Kenney A, Kim G, McMillan A, et al. Inhibition of phosphatidylinositol 3-kinase destabilizes Mycn protein and blocks malignant progression in neuroblastoma. Cancer Res. 2006;66:8139–46.
Cage TA, Chanthery Y, Chesler L, Grimmer M, Knight Z, Shokat K, et al. Downregulation of MYCN through PI3K inhibition in mouse models of pediatric neural cancer. Front Oncol. 2015;5:111. Jan 4
Vaughan L, Clarke P, Barker K, Chanthery Y, Gustafson C, Tucker E, et al. Inhibition of mTOR-kinase destabilizes MYCN and is a potential therapy for MYCN-dependent tumors. Oncotarget. 2016;7:57525–44.
Lambertz I, Kumps C, Claeys S, Lindner S, Beckers A, Janssens E, et al. Upregulation of MAPK negative feedback regulators and RET in mutant ALK neuroblastoma: implications for targeted treatment. Clin Cancer Res. 2015;21:3327–39.
Escamilla-Powers J, Daniel C, Farrell A, Taylor K, Zhang X, Byers S, et al. The tumor suppressor protein HBP1 is a novel c-Myc-binding protein that negatively regulates c-Myc transcriptional activity. J Biol Chem Highwire. 2010;285:4847–58.
Tevosian Shih, Mendelson Sheppard, Paulson Yee. HBP1: a HMG box transcriptional repressor that is targeted by the retinoblastoma family. Genes Dev. 1997;11:383–96.
Coomans de Brachène A, Bollaert E, Eijkelenboom A, de Serra A, van der Vos KE, Burgering BM, et al. The expression of the tumour suppressor HBP1 is down-regulated by growth factors via the PI3K/PKB/FOXO pathway. Biochem J. 2014;460:25–34.
Bollaert E, Johanns M, Herinckx G, Serra A, Vandewalle V, Havelange V, et al. HBP1 phosphorylation by AKT regulates its transcriptional activity and glioblastoma cell proliferation. Cell Signal. 2018;44:158–70.
Mossé Y, Lim M, Voss S, Wilner K, Ruffner K, Laliberte J, et al. Safety and activity of crizotinib for paediatric patients with refractory solid tumours or anaplastic large-cell lymphoma: a Children’s Oncology Group phase 1 consortium study. Lancet Oncol. 2013;14:472–80.
Sakamoto H, Tsukaguchi T, Hiroshima S, Kodama T, Kobayashi T, Fukami T, et al. CH5424802, a Selective ALK inhibitor capable of blocking the resistant gatekeeper mutant. Cancer Cell Sci. 2011;19:679–90.
Passoni L, Longo L, Collini P, Coluccia A, Bozzi F, Podda M, et al. Mutation-independent anaplastic lymphoma kinase overexpression in poor prognosis neuroblastoma patients. Cancer Res. 2009;69:7338–46.
Heukamp L, Thor T, Schramm A, Preter K, Kumps C, Wilde B, et al. Targeted expression of mutated ALK induces neuroblastoma in transgenic mice. Sci TranslMedicine. 2012;4:141ra91–141ra91.
Moore NF, Azarova AM, Bhatnagar N, Ross KN, Drake LE, Frumm S, et al. Molecular rationale for the use of PI3K/AKT/mTOR pathway inhibitors in combination with crizotinib in ALK-mutated neuroblastoma. Oncotarget. 2014;5:8737–49.
Santo E, Stroeken P, Sluis P, Koster J, Versteeg R, Westerhout E. FOXO3a is a major target of inactivation by PI3K/AKT signaling in aggressive neuroblastoma. Cancer Res. 2013;73:2189–98.
Gu T-LL, Tothova Z, Scheijen B, Griffin JD, Gilliland DG, Sternberg DW. NPM-ALK fusion kinase of anaplastic large-cell lymphoma regulates survival and proliferative signaling through modulation of FOXO3a. Blood. 2004;103:4622–9.
Li H, Bian C, Liao L, Li J, Zhao R. miR-17-5p promotes human breast cancer cell migration and invasion through suppression of HBP1. Breast Cancer Res Treat. 2011;126:565–75.
Li Y, Choi PS, Casey SC, Dill DL, Felsher DW. MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell. 2014;26:262–72.
Mestdagh Fredlund, Pattyn Schulte, Muth Vermeulen, et al. MYCN/c-MYC-induced microRNAs repress coding gene networks associated with poor outcome in MYCN/c-MYC-activated tumors. Oncogene. 2009;29:1394–404.
Althoff Beckers, Bell Nortmeyer, Thor Sprüssel, et al. A Cre-conditional MYCN-driven neuroblastoma mouse model as an improved tool for preclinical studies. Oncogene. 2015;34:3357–68.
Subramanian A, Tamayo P, Mootha V, Mukherjee S, Ebert B, Gillette M, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50.
Valentijn LJ, Koster J, Haneveld F, Aissa RA, van Sluis P, Broekmans ME, et al. Functional MYCN signature predicts outcome of neuroblastoma irrespective of MYCN amplification. Proc Natl Acad Sci USA. 2012;109:19190–5.
Zeller K, Jegga A, Aronow B, O’Donnell K, Dang C. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:1–10.
Bracken A, Dietrich N, Pasini D, Hansen K, Helin K. Genome-wide mapping of polycomb target genes unravels their roles in cell fate transitions. Genes Dev Highwire. 2006;20:1123–36.
Janky R, Verfaillie A, Imrichová H, Sande B, Standaert L, Christiaens V, et al. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol. 2014;10:e1003731.
Henrich K-O, Bender S, Saadati M, Dreidax D, Gartlgruber M, Shao C, et al. Integrative genome-scale analysis identifies epigenetic mechanisms of transcriptional deregulation in unfavorable neuroblastomas. Cancer Res. 2016;76:5523–37.
Watanabe N, Kageyama R, Ohtsuka T. Hbp1 regulates the timing of neuronal differentiation during cortical development by controlling cell cycle progression. Development. 2015;142:2278–90.
Yao Works, Romagnoli Austin. Effects of overexpression of HBP1 upon growth and differentiation of leukemic myeloid cells. Leukemia. 2005;19:1958–68.
Frumm S, Fan Z, Ross K, Duvall J, Gupta S, VerPlank L, et al. Selective HDAC1/HDAC2 inhibitors induce neuroblastoma differentiation. Chem Biol. 2013;20:713–25.
Kim J, Zhang X, Rieger-Christ KM, Summerhayes IC, Wazer DE, Paulson KE.Yee AS, Suppression of Wnt signaling by the green tea compound (-)-epigallocatechin 3-gallate (EGCG) in invasive breast cancer cells. Requirement of the transcriptional repressor HBP1. J Biol Chem. 2006;281:10865–75. https://www.ncbi.nlm.nih.gov/pubmed/16495219.
Puissant A, Frumm S, Alexe G, Bassil C, Qi J, Chanthery Y, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3:308–23.
Schubbert S, Cardenas A, Chen H, Garcia C, Guo W, Bradner J, et al. Targeting the MYC and PI3K pathways eliminates leukemia-initiating cells in T-cell acute lymphoblastic leukemia. Cancer Res. 2014;74:7048–59.
Stratikopoulos EE, Dendy M, Szabolcs M, Khaykin AJ, Lefebvre C, Zhou M-M, et al. Kinase and BET inhibitors together clamp inhibition of PI3K signaling and overcome resistance to therapy. Cancer Cell. 2015;27:837–51.
Stratikopoulos E, Parsons R. Molecular pathways: targeting the PI3K pathway in cancer—BET inhibitors to the rescue. Clin Cancer Res. 2016;22:2605–10.
Cortés C, Kozma S, Tauler A, Ambrosio S. MYCN concurrence with SAHA-induced cell death in human neuroblastoma cells. Cell Oncol. 2015;38:341–52.
Braekeveldt N, Wigerup C, Gisselsson D, Mohlin S, Merselius M, Beckman S, et al. Neuroblastoma patient‐derived orthotopic xenografts retain metastatic patterns and geno‐ and phenotypes of patient tumours. Int J Cancer. 2015;136:E252–E261.
Persson CU, von Stedingk K, Bexell D, Merselius M, Braekeveldt N, Gisselsson D, et al. Neuroblastoma patient-derived xenograft cells cultured in stem-cell promoting medium retain tumorigenic and metastatic capacities but differentiate in serum. Sci Rep. 2017;7:10274.
Pei Y, Liu K-W, Wang J, Garancher A, Tao R, Esparza LA, et al. HDAC and PI3K antagonists cooperate to inhibit growth of MYC-driven medulloblastoma. Cancer Cell. 2016;29:311–23.
Umapathy G, Wakil A, Witek B, Chesler L, Danielson L, Deng X, et al. The kinase ALK stimulates the kinase ERK5 to promote the expression of the oncogene MYCN in neuroblastoma. Sci Signal. 2014;7:ra102–ra102.
Duffy D, Krstic A, Schwarzl T, Higgins D, Kolch W. GSK3 inhibitors regulate MYCN mRNA levels and reduce neuroblastoma cell viability through multiple mechanisms, including p53 and Wnt signaling. Mol Cancer Ther. 2014;13:454–67.
Hasan K, Nafady A, Takatori A, Kishida S, Ohira M, Suenaga Y, et al. ALK is a MYCN target gene and regulates cell migration and invasion in neuroblastoma. Sci Rep Nat. 2013;3:3450.
Corvetta D, Chayka O, Gherardi S, D’Acunto C, Cantilena S, Valli E, et al. Physical interaction between MYCN oncogene and polycomb repressive complex 2 (PRC2) in neuroblastoma functional and therapeutic implications. J Biol Chem. 2013;288:8332–41.
Ferreira A, de-Freitas-Junior J, Morgado-Díaz J, Ridley A, Klumb C. Dual inhibition of histone deacetylases and phosphoinositide 3-kinases: effects on Burkitt lymphoma cell growth and migration. J Leukoc Biol. 2016;99:569–78.
Chou T-C, Talalay P. Analysis of combined drug effects: a new look at a very old problem. Trends Pharmacol Sci Sci. 1983;4:450–4.
Hellemans J, Mortier G, Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:1–14.
Coller H, Grandori C, Tamayo P, Colbert T, Lander E, Eisenman R, et al. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci. 2000;97:3260–5.
Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–38.
Fredlund E, Ringnér M, Maris JM, Påhlman S. High Myc pathway activity and low stage of neuronal differentiation associate with poor outcome in neuroblastoma. Proc Natl Acad Sci USA. 2008;105:14094–9.
Yu D, Cozma D, Park A, Thomas-Tikhonenko A. Functional validation of genes implicated in lymphomagenesis: an in vivo selection assay using a Myc-induced B-cell tumor. Ann N Y Acad Sci. 2005;1059:145–59.
Kim Y, Girard L, Giacomini C, Wang P, Hernandez-Boussard T, Tibshirani R, et al. Combined microarray analysis of small cell lung cancer reveals altered apoptotic balance and distinct expression signatures of MYC family gene amplification. Oncogene. 2005;25:130–8.
Acknowledgements
We thank Jeroen Schacht, Jolien Van Laere, Els De Smet, Fanny De Vloed, Givani Dewyn, and Aline Eggermont from our lab as well as Glenn Wagemans from the LECR for their outstanding technical assistance. Furthermore, we thank Els Janssens, Annelies Fieuw, Sara De Brouwer, and Irina Lambertz for their guidance during this project.
Funding
S.C. is supported by a pre-doctoral fellowship of the Research Foundation—Flanders (FWO; 11J8313N) and an Emmanuel van der Schueren grant (‘Kom op tegen Kanker’). S.V. is funded by the VLK (Flemish League against cancer) and ‘Stichting Villa Joep’. B.D. and S.L. are supported by a pre-doctoral fellowship of the FWO Research Foundation—Flanders (FWO). K.D. is supported by Ghent University (BOF; BOF16/PDO/043). C.V., B.D.W. and T.V.M. are senior clinical investigators of the Research Foundation—Flanders (FWO; 18B1716N (B.D.W.), 12N6917N (C.V.), 1803115 N (T.V.M.)). We would further like to thank the following funding agencies: the Belgian Foundation against Cancer (project 2014–175) to F.S., Ghent University (BOF10/GOA/019, BOF16/GOA/23) to F.S., the Belgian Program of Interuniversity Poles of Attraction (IUAP Phase VII–P7/03) to F.S., the fund for Scientific Research Flanders (Research projects G053012N, G050712N, G051516N to F.S) and ‘Stichting Villa Joep’ to F.S.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Claeys, S., Denecker, G., Durinck, K. et al. ALK positively regulates MYCN activity through repression of HBP1 expression. Oncogene 38, 2690–2705 (2019). https://doi.org/10.1038/s41388-018-0595-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0595-3
- Springer Nature Limited
This article is cited by
-
Integrated systematic functional screen and fine-mapping decipher the role and genetic regulation of RPS19 in colorectal cancer development
Archives of Toxicology (2024)
-
The transcriptional co-repressor Runx1t1 is essential for MYCN-driven neuroblastoma tumorigenesis
Nature Communications (2024)
-
Methylation of HBP1 by PRMT1 promotes tumor progression by regulating actin cytoskeleton remodeling
Oncogenesis (2022)
-
The ETS transcription factor ETV5 is a target of activated ALK in neuroblastoma contributing to increased tumour aggressiveness
Scientific Reports (2020)
-
The HMG box transcription factor HBP1: a cell cycle inhibitor at the crossroads of cancer signaling pathways
Cellular and Molecular Life Sciences (2019)