Abstract
TET3 is a member of the ten-eleven translocation (TET) family of enzymes which oxidize 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC). Tet3 is highly expressed in the brain, where 5hmC levels are most abundant. In adult mice, we observed that TET3 is present in mature neurons and oligodendrocytes but is absent in astrocytes. To investigate the function of TET3 in adult postmitotic neurons, we crossed Tet3 floxed mice with a neuronal Cre-expressing mouse line, Camk2a-CreERT2, obtaining a Tet3 conditional KO (cKO) mouse line. Ablation of Tet3 in adult mature neurons resulted in increased anxiety-like behavior with concomitant hypercorticalism, and impaired hippocampal-dependent spatial orientation. Transcriptome and gene-specific expression analysis of the hippocampus showed dysregulation of genes involved in glucocorticoid signaling pathway (HPA axis) in the ventral hippocampus, whereas upregulation of immediate early genes was observed in both dorsal and ventral hippocampal areas. In addition, Tet3 cKO mice exhibit increased dendritic spine maturation in the ventral CA1 hippocampal subregion. Based on these observations, we suggest that TET3 is involved in molecular alterations that govern hippocampal-dependent functions. These results reveal a critical role for epigenetic modifications in modulating brain functions, opening new insights into the molecular basis of neurological disorders.
Similar content being viewed by others
Introduction
Neurons are long-lived cells, governed by a strict molecular regulation to maintain genomic stability, but also possess a remarkable plasticity to respond to external stimuli. The dynamic nature of neuronal function is precisely regulated by epigenetic changes [1]. DNA methylation is one of the most well-studied epigenetic marks and the discovery of the conversion of 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC), catalyzed by ten-eleven translocation (TET) family of dioxygenases [2], has contributed to a better understanding of the DNA demethylation process.
High 5hmC content is positively correlated with gene transcription and is a feature of postmitotic neurons, since very low levels were detected in immature neurons and nonneuronal cell types [3,4,5]. Tet genes have also been shown to be highly transcribed in the brain, with Tet3 being the most abundant. Moreover, Tet3 levels are similar across different forebrain regions, such as cortex, hippocampus, and cerebellum [6]. A putative link between neuronal TET protein function and cognitive processes gained more relevance following the discovery that (de)methylation of DNA in the brain appears to be relevant for learning and memory [7, 8]. Indeed, TET1 and TET2 were implicated in learning and memory processes in adult mice [9,10,11,12,13]. Concerning TET3, it was observed that Tet3 homozygous deletion in mice leads to neonatal lethality [14] and that Tet1/3 double knockout mice presented poor forebrain formation and abnormal facial structures resembling holoprosencephaly [15]. In addition, it was shown that fear extinction, a form of reversal learning, leads to TET3-mediated accumulation of 5hmC, and the knockdown of this enzyme in the infralimbic prefrontal cortex (PFC) leads to a significant impairment in fear extinction memory [16]. Kremer et al. further showed that Tet3 expression in hippocampal neurons, but not Tet1 or Tet2, is regulated in an activity-dependent manner and after formation of fear memory; moreover, expression of genes related with memory, such as Notch1, Creb1, Crebbp, and Gadd45b were regulated by Tet3 levels [17]. Importantly, Tet3 was described as a synaptic sensor, able to regulate neuronal activity, since Tet3 knockdown in hippocampal neuronal cultures increased excitatory glutamatergic synaptic transmission, whereas overexpression of Tet3 catalytic domain decreased it [18]. Concordantly, Tet3 deletion in young mice increases excitatory synaptic transmission, whereas GABAAR-mediated inhibitory synaptic transmission was significantly reduced [19]. Moreover, transcriptome analyses revealed a key role for TET3 in regulating gene expression in response to synaptic activity [18].
Here, we addressed the role of TET3 in adult behavior, namely in emotion and cognition, which are not independent behavioral dimensions. In fact most of neural regions related with modulation of emotional behavior are also involved in cognitive processes, such as the hippocampus and the PFC [20]. We mainly focused our study in hippocampal-dependent behaviors, since this structure presents a complex connectivity with numerous cortical and subcortical structures, integrating the neural circuitry of cognitive and emotional functions [21]. Indeed, the dorsal hippocampus is essential for spatial memory and navigation tasks. The ventral hippocampus establishes connections with structures, such as PFC, amygdala, and bed nucleus of stria terminalis (BNST), responsible for emotions-like anxiety and fear [22].
We specifically induced Tet3 deletion in mature forebrain neurons, at an adult stage, by crossing a Tet3 conditional knockout mouse line with calcium/calmodulin-dependent protein kinase II alpha-(Camk2a)-CreERT2-expressing transgenic line. Then, we performed behavioral analysis to assess anxiety- and depressive-like behaviors, as well as cognitive function. Our results showed that TET3 ablation in postmitotic neurons leads to increased anxiety and impaired spatial orientation, with concomitant hypercorticalism; transcriptomic analysis unveiled a deregulation of genes involved in glucocorticoid signaling pathway, controlled by the hypothalamic–pituitary–adrenal (HPA) axis, specifically in the ventral hippocampus. In addition, quantification of gene-specific transcript levels demonstrated that immediate early genes (IEGs) were upregulated in both dorsal and ventral hippocampus. Furthermore, Tet3 cKO mice showed increased synaptic maturation at the ventral CA1 hippocampal region. Thus, our study points to a role for TET3 as a regulator of HPA axis and neuronal activity-regulated genes, possibly associated with anxiety-like behavior and spatial orientation alterations in adult mice.
Material and methods
Animals and tamoxifen treatment
Tet3fl/fl mice on a C57BL/6N background [23, 24] were crossed with B6;129S6 mice (JAX stock #012362—Camk2a-CreERT2 [25]) expressing a tamoxifen-inducible Cre-recombinase under the control of the mouse Camk2a promoter region to generate mice heterozygous for the floxed Tet3 allele and Cre-recombinase. These mice were interbred with C57BL/6N mice homozygous for the floxed Tet3 allele to generate mice heterozygous for Cre-recombinase and homozygous for the floxed Tet3 allele, designated as Tet3 cKO mice. Mice homozygous for the Tet3 floxed allele, but not carrying Cre-recombinase, were used as controls. Animals were genotyped by PCR analysis using genomic DNA and primers specific to Cre-recombinase and the floxed Tet3 allele. Detection of the floxed transgene was done using a primer specific to the fragment, which allowed to detect the deleted or floxed allele (Supplementary Table S1a). To induce Tet3 deletion, 6-week-old male mice were administered tamoxifen (Sigma, St. Louis, MO; T-5648) dissolved in corn oil (Sigma; C-8267) at 20 mg/ml. Six-week-old mice were injected intraperitoneally with 50 mg/kg of tamoxifen twice a day for 5 consecutive days, with 7 days break followed by injections for 5 additional consecutive days. One month after tamoxifen administration, blood samples were collected from the animals, and corticosterone levels were measured using a commercial kit (Enzo Life Sciences, New York, USA). All procedures were carried out in accordance with EU Directive 2010/63/EU and NIH guidelines on animal care and experimentation and were approved by the Portuguese Government/Direção Geral de Alimentação e Veterinária (DGAV) with the project reference 0421/000/000/2017.
Behavioral analyses
We used standard behavioral tests to evaluate the following dimensions: motor and locomotor activity (OF—open field), emotional behavior—anxiety-like (OF and EPM—elevated plus maze) and antidepressant-like states (FST—forced-swim test and TST—tail suspension test)), and cognition (NOR—novel object recognition and MWM—Morris water maze).
Animals were submitted to behavioral testing 1 month after tamoxifen treatment. Mice were studied during the light phase and habituated to testing rooms for at least 30 min before each test. Behavioral assessment was performed following this order: OF, EPM, FST, TST, NOR, and MWM. These tests are described in detail in Supplementary information. The number of animals used for each test was: OF: n = 13 CTRL, 18 cKO; EPM: n = 14 CTRL, 16 cKO; TST and FST: n = 14 CTRL, 17 cKO; NOR: n = 14 CTRL, 15 cKO; MWM: n = 9 CTRL, 11 cKO. We performed all behavioral tests in two independent sets of animals, except the MWM that was performed only to a second set of animals. Animals were excluded from the analysis when they did not perform the test (exclusion of animals for each behavioral test is explained in the Supplementary experimental procedures). Behavioral experiments and analyses were conducted without randomization. The investigator was blinded to the genotype during the experiment and when assessing the outcome.
DNA/RNA extraction
After behavior assessment, the animals were first anesthetized with a mixture of ketamine (75 mg/kg, i.p.; Imalgene 1000, Merial, EUA) and medetomidine (1 mg/kg, i.p.; Dorbene Vet, Pfizer, EUA), and transcardially perfused with 0.9% saline. Brains were carefully removed and macrodissected, under a light microscope using anatomical landmarks by a single experimenter, and tissue samples were stored at −80 °C. The tissues were prepared by homogenization using Trizol® reagent (Invitrogen), and extracted according to the manufacturer’s instructions. The RNA was treated with DNase I (Thermo Scientific) and a total of 500 ng RNA was used for cDNA synthesis using the qScriptTM cDNA SuperMix (Quanta Biosciences, USA).
RNA sequencing analysis
RNA integrity was evaluated through chip-based capillary electrophoresis (Bioanalyzer, Agilent®), with all samples having a RQN (RNA Quality Number) >8.5. A total of 500 ng RNA was used for library construction, using the QuantSeq 3′mRNA-Seq Library Prep Kit (Lexogen) (n = 3 animals per group). The resulting library was then sequenced on Illumina NextSeq500. RNA-seq analysis was carried out on three independent technical replicates (GEO Accession number: GSE140850). Quality control was carried out on raw datasets using FastQC. Differential expression analysis was performed using edgeR [26], and genes were considered differentially expressed between conditions if FDR <5% and an absolute fold change >2. Gene ontology classification was carried out using Panther® and Ingenuity Pathway Analysis (IPA, Qiagen, Redwood City, CA, USA) software.
Immunofluorescence analysis of mouse brain sections
Mice were transcardially perfused with phosphate-buffered saline. Brains were then removed and fixed overnight in 4% PFA. Brain coronal cryosections (20 µm thick) collected onto slides were incubated O/N at 4 °C, with primary antibodies, sequentially. Primary antibodies were: NeuN (Cell signaling, rabbit, D4640, 1:100), Tet3 (Abcam, rabbit, ab153724, 1:100), GFAP (mouse, Thermo scientific, MA5-12023, 1:100), and CNPase (mouse, Millipore, MAB326, 1:200). Secondary antibodies—Alexa Fluor 488 and Alexa Fluor 594 (Molecular Probes)—were used for detection, and incubation was performed for 2 h at RT. Nuclei were counterstained using DAPI during 10 min at RT. Fluorescence images were acquired with the Olympus Fluoview FV1000 confocal microscope (Olympus, Hamburg, Germany) and the number of double-positive cells calculated using FIJI software (three sections per animal and three animals per group).
3D reconstruction of neurons
To assess the 3D dendritic morphology of hippocampal pyramidal neurons, we used the Golgi–Cox impregnation technique [27]. Dendritic arborization and spine numbers/density/types were analyzed in the dorsal and ventral CA1 of Control and Tet3 cKO mice, as described previously [28, 29]. More details are described in Supplementary information(n = 6 neurons for each animal; n = 3 animals per group).
Statistical analysis
The sample size was determined using G*Power [30]. The animals were divided in two groups: CTRL and Tet3 cKO; n = 45 per group (effect size = 0.25, α = 0.05, 1-β = 0.80, total sample size = 90). Although it was not possible to achieve this number of animals, statistical differences were observed in several behavioral tests. A confidence interval of 95% was assumed for hypothesis testing. Assumptions for all variables were validated prior to statistical testing (normality by the Shapiro–Wilk test and homoscedasticity by Levene’s test). For the comparison of two means, the two-tailed unpaired Student’s t test was carried out, followed by the Benjamini, Krieger, and Yekuteli correction when multiple hypotheses were tested. Analysis of variance (ANOVA) was used when two or more factors were tested, followed by Sidak’s post hoc test. Comparison of proportions was carried out using a two-tailed Chi-square statistical test. Appropriate effect size measures were reported for all statistical tests. All statistical analyses were carried out using SPSS 22.0® or GraphPad Prism 8.0®. Test details are described in figure captions, Supplementary information and Supplementary Table S2.
Results
TET3 is present in mature neurons and oligodendrocytes, but not in astrocytes, in adult mouse cortex and hippocampus brain regions
To elucidate in which cell types TET3 is present in the adult brain cortex and hippocampus, we performed double immunofluorescence staining for TET3 and typical markers for postmitotic neurons (NeuN, Neuronal nuclear protein), astrocytes (GFAP, Glial fibrillary acidic protein), or oligodendrocytes (CNPase, 2′,3′-Cyclic-nucleotide 3′-phosphodiesterase). We observed a strong co-localization of TET3 and NeuN in the cortical and hippocampal brain regions, pointing out neurons as the main source of TET3 protein (Fig. 1a). We did not observe GFAP and TET3-double-positive cells, and only a few number of cells co-expressed CNPase and TET3, suggesting that TET3 is absent in astrocytes and lowly expressed in oligodendrocytes (Fig. 1a). In addition, in order to investigate if forebrain regions presented a distinct pattern of expression of Tet3, we measured Tet3 levels in the PFC, hippocampus, amygdala, and BNST. We observed similar Tet3 transcript levels in these brain regions (Supplementary Fig. S1a).
a Representative double immunostaining for NeuN and TET3 proteins, showing strong expression of TET3 in postmitotic neurons in the cortex and dorsal CA1 brain regions. TET3 expression was detected in some oligodendrocytes stained with CNPase marker. No TET3 staining was found in GFAP positive cells. Scale bars, 50 and 25 μm. b–e Reduction in Tet3 mRNA levels and maintenance of Tet1 and Tet2 levels was observed in Tet3 cKO animals. mRNA expression in forebrain regions was measured by qRT-PCR in control and Tet3 cKO animals in (b) prefrontal cortex; (c) dorsal hippocampus; (d) ventral hippocampus; and (e) amygdala (n = 3–5 per group). Two-tailed Student’s t test; *p < 0.05, **p < 0.01. f Representative double immunostaining for NeuN and TET3 proteins in the CA1, CA3, and DG hippocampal regions, showing reduction in TET3 expression in the Tet3 cKO animals. Scale bars, 50 and 25 μm. g The percentage of TET3 positive cells in NeuN-positive cells (postmitotic neurons) was quantified in Tet3 cKO animals, relative to controls (n = 3 per group). Quantifications are presented as the mean ± SEM. Adjusted two-tailed Student’s t test; *p < 0.05, **p < 0.01. h No alterations were found in the global 5hmC levels in forebrain regions analyzed by ELISA (n = 3 per group). Quantifications are presented as the mean ± SEM.
Adult Tet3 conditional knockout mice show significant reduction of Tet3 levels, but not Tet1 or Tet2, in different forebrain regions
In order to determine the function of TET3 in mature neurons at an adult stage, we generated a Tet3 conditional knockout mouse model, by crossing Tet3 floxed mice, in which the exon 7 (exon 5 of the coding sequence) of Tet3 gene is flanked by LoxP sites (for Cre-induced site-specific recombination) [23, 24] with a Camk2a-CreERT2 inducible line to specifically delete Tet3 in mature forebrain neurons, after tamoxifen administration. Tet3 was disrupted by deletion of the targeted exon resulting in a truncated protein lacking the catalytic domain. Tet3 deletion was confirmed at the DNA and mRNA levels, by PCR and RT-PCR, respectively (Supplementary Fig. S2a, b). Tet3 cKO mice showed a significant reduction of Tet3 mRNA levels in several forebrain regions—PFC, amygdala, dorsal, and ventral hippocampus (Fig. 1b–e) (t-test, p < 0.05). We also analyzed the BNST region but, no reduction of Tet3 transcripts was observed (Supplementary Fig. S1b). Importantly, decreased levels of Tet3 did not interfere with transcript levels of Tet1 or Tet2 (Fig. 1b–e), suggesting the absence of a compensatory effect in the brain. To better determine the effectiveness of the conditional knockout strategy in neuronal cells, we quantified the number of postmitotic neurons (NeuN-positive cells) showing TET3 staining in the hippocampus of control and Tet3 cKO mice (Fig. 1f), and observed a significant reduction in TET3/NeuN-positive cells (Adjusted t-test, p < 0.05, ω2p = 0.841; Fig. 1g). We further assessed whether the conditional deletion of Tet3 affected global 5hmC levels in forebrain regions using an ELISA-based assay, and observed no changes in the PFC, hippocampus, and amygdala of Tet3 cKO mice when comparing to the control group (Fig. 1h).
Tet3 deletion in neurons results in increased anxiety-like behavior and hypercorticalism, and impaired spatial orientation
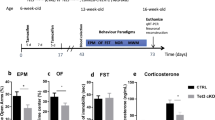
In order to ascertain if Tet3 ablation in adult postmitotic neurons has an effect in the behavioral performance of mice, we performed a plethora of behavioral tests assessing various paradigms related to emotional and cognitive domains (Fig. 2a). We used two behavioral tests to detect anxiety-like behavior, namely the OF test and the EPM, and the FST and TST to assess antidepressant-like behavior. No differences were found in the average velocity and vertical counts in the OF test (Supplementary Fig. S3a, b). In addition, analysis of the total distance traveled in the open arena and EPM arms revealed no differences between groups, as they presented similar locomotor activity (Supplementary Fig. S3c, d). Regarding the anxiety-like behavioral dimension, Tet3 cKO mice spent less time in the center of the OF (Fig. 2b). No statistical differences were found in the time spent in the open arms of the EPM when compared with the control group (Fig. 2c), but the magnitude effect, represented by Cohen’s d [31], is medium–large (t-test, OF, p < 0.05, d = 0.810; EPM, p = 0.114, d = 0.684). Moreover, in the EPM test, we found that Tet3 cKO mice showed significantly lower frequency of head dips in the open arms (Fig. 2d, p < 0.01, d = 1.400) and higher latency to enter for the first time in the open arms when compared with control animals (Fig. 2e, p < 0.05, d = 0.881). In addition, we discovered a significant positive correlation between the performance in the OF and EPM behavioral tests (r = 0.652; p < 0.01) (Fig. 2f). Altogether, these parameters are indicative of increased anxiety-like behavior in Tet3 cKO animals.
a Scheme illustrating the protocol used to induce Tet3 deletion and the behavior paradigm timeline. Six-week-old mice were injected intraperitoneally with 50 mg/kg of tamoxifen twice a day for 5 consecutive days, with 7 days break, followed by injections for 5 additional consecutive days. Animals were submitted to behavioral testing 1 month after the last tamoxifen injection and euthanized after this assessment. b–f Anxiety-like behavior was tested both in the open-field test (OF) (b) and elevated plus maze (EPM) (c–e). b Time spent in the center of the OF; c time spent in the open arms of the EPM; d The frequency number of head dips (EPM); e Latency to enter open arms (EPM); f Correlation between OF and EPM performances (n = 13–18 per group). The presence of depressive-like behavior was assessed in the tail suspension test (TST) (g) and forced swimming test (FST) (h) (n = 13–17 per group). i Basal serum concentration of corticosteroids in control and Tet3 cKO mice, both in the morning and at night, revealed a significant increased production by Tet3 cKO mice (n = 8 per group). Quantifications are presented as the mean ± SEM. b–e; g–h Two-tailed Student’s t test; *p < 0.05; f correlation Pearson; p < 0.01; i Adjusted two-tailed Student’s t test; **p < 0.01.
In contrast, we did not observe differences in immobility times between the two groups in the TST and FST (t-test, TST, p = 0.270, d = 0.511; FST, p = 0.316, d = 0.353) (Fig. 2g, h). Considering the involvement of the HPA axis in the modulation of behavior, we further determined the serum basal levels of corticosterone in control and Tet3 cKO mice. Both in nadir and zenith time points, Tet3 cKO mice presented an overactivation of the HPA axis, as suggested by the increased levels of corticosterone (Adjusted t-test, p < 0.01, ω2p = 0.417; Fig. 2i).
Next, we assessed the possible effect of Tet3 neuronal ablation in different cognitive domains, such as learning and memory. We tested Tet3 cKO mice in the MWM test to assess reference memory, a task that relies on hippocampal activity. Both Tet3 cKO and control mice were able to successfully learn the spatial reference memory task, as confirmed by the decreasing latencies during the trials (Fig. 3a; mixed ANOVA, genotype, p = 0.274, ω2p = 0.015) and by their performance in the probe trial, assessed by the same preference (percentage of time swum) for the goal quadrant, where the platform was located during the acquisition phase (t-test, p = 0.417, d = 0.417) (Fig. 3b). However, analysis of the strategies adopted by the mice to achieve the escape platform in the MWM task, divided in random searching/scanning (nonhippocampal strategies) or directed strategies (hippocampal strategies) [32], revealed that Tet3 cKO mice adopted significantly less hippocampal-dependent strategies when compared with control mice (Chi-square test, p < 0.05, φ = 0.466), indicating a poor spatial orientation (Fig. 3c).
a–c Morris Water Maze test. Spatial acquisition performances were recorded during 4-day training. a Escape latency. b Time in the target quadrant. c Cognitive strategies during water maze learning. A schematic representation and color code for each group of strategy and the average prevalence by trial number are shown. Representation of the percentage of mice using directed strategies (hippocampal-dependent strategies) (n = 8–11 per group). d–f Novel object recognition test. Animals were allowed to explore two identical objects for 10 min. d After an interval of 1 h in their home cages, the novel object was displaced to the opposite side of the box and mice were allowed to explore this new configuration, evaluating spatial recognition memory (displacement). e After 24 h, mice were returned to the arena, where one of the familiar objects was replaced with a novel one, evaluating long-term memory. f After 24 h, two new objects were placed in the box and mice were allowed to explore them. One hour after, one object was replaced by a novel one, and the animals placed in the arena, evaluating short-term memory (n = 14–18 per group). Quantifications are presented as mean ± SEM. a Mixed ANOVA, genotype; b Two-tailed Student’s t test; c Chi-square test; *p < 0.05; d–f two-tailed Student’s t test.
To evaluate recognition memory, we performed the NOR test. In this task, Tet3 cKO and control mice showed similar time exploring the object displaced after a short period of time (1 h) (Fig. 3d; t-test, p = 0.553, d = 0.379) indicating normal object location memory. Tet3 cKO and control mice dedicated similar percentages of time exploring the novel object displayed 24 h after habituation to familiar objects, indicating no deficits in long-term object recognition memory (Fig. 3e; t-test, p = 0.462, d = 0.267). Moreover, when short-term memory was evaluated, Tet3 cKO displayed identical discrimination index, indicating no deficits of short-term memory as well (Fig. 3f; t-test, p = 0.504, d = 0.235).
Transcriptomic analysis revealed that Tet3 deletion affects gene expression mainly in the ventral hippocampus
To determine which genes were affected by the loss of Tet3 and considering the impairment of hippocampal-dependent function described above, we performed a transcriptomic analysis, using QuantSeq 3′ mRNA Sequencing [33], in RNA extracted from dorsal and ventral hippocampus of Tet3 cKO and control mice. Notably, only 20 transcripts were found to be differentially expressed in the dorsal hippocampus of Tet3 cKO mice, with only seven being protein coding and apparently with no particular biological relevance for this study (Fig. 4a, b and Supplementary Table S3); on the other hand, in the ventral hippocampus, the number of differentially expressed genes was higher—143 (Fig. 4a, c and Supplementary Table S4), with 90 being downregulated and 53 upregulated. This reveals a greater sensitivity of the ventral hippocampus to deletion of neuronal Tet3.
Gene expression analysis showed an increase in the expression of neuronal activity-regulated genes in both regions. a Number of differentially expressed genes (up and downregulated) in the dorsal and ventral regions of the hippocampus. b, c Volcano plot of all transcripts identified in the dorsal (29,407 targets) and ventral hippocampus (29,146 targets). The log fold change represents control versus Tet3 cKO mice. d, e Gene ontology classification of the molecular function and protein class of differentially expressed targets in the ventral hippocampus of Tet3 cKO mice, based on the PANTHER database. f, g Enriched level 4 and 5 gene ontology classes and pathways of differentially expressed targets in the ventral hippocampus of Tet3 cKO mice, based on the ingenuity pathway analysis (IPA). h Expression of selected genes from the IPA analysis was evaluated by qRT-PCR, confirming the QuantSeq results (n = 3 per group). i, j Expression of immediate early genes (IEGs) in the dorsal and ventral hippocampus (n = 4–6 per group). Fold change relative to controls was calculated using the 2−ΔΔCT relative gene expression analysis. Quantifications are presented as the mean ± SEM. h–j Two-tailed Student’s t test; *p < 0.05, **p < 0.01.
Gene ontology analysis of differentially expressed transcripts in the ventral hippocampus, using the Panther® classification system [34], revealed that the most common molecular function was binding activity, and the most represented protein classes were transporters, hydrolases, and enzyme modulators (Fig. 4d, e). Moreover, we performed a level 4 and 5 gene ontology classification using the Consensus Pathway Database [35]; while none of the top 10 categories were specifically related with neuronal activity (Fig. 4f), pathway enrichment analysis revealed an impact in the glucocorticoid signaling pathway (HPA axis) and FOXA1 transcription factor network, amongst others (Fig. 4g). Regarding HPA axis, we confirmed by qRT-PCR the downregulation of corticotropin releasing hormone receptor type 2 (Crhr2), involved in stress-related disorders, such as anxiety and depression [36] (Fig. 4h). For FOXA1 transcription factor-related genes, we confirmed the downregulation of Poua2af1, Col8a1, and Lmx1a (Fig. 4h). In addition, we obtained information from the Ingenuity Pathway Analysis software [37], namely in canonical pathways (where glucocorticoid receptor signaling was newly identified), upstream regulators (where Dopamine Receptor D2 appears) or diseases and disorders (being the most enriched category, cancer) (Supplementary Fig. S4a–c).
Tet3 cKO mice displayed increased expression of neuronal activity-regulated genes in the hippocampus
A plethora of neuronal genes is involved in neural plasticity related to learning and memory processes, and transcriptional activity of these genes is crucial to control these cognitive processes. The QuantSeq RNA results allowed to identify c-fos (Fos), as an upregulated gene in Tet3 cKO animals (Supplementary Table S4). Hence, we explored whether Tet3 deletion in neurons leads to dysregulation of other activity-induced genes involved in synaptic plasticity in the hippocampus (Fig. 4i, j) and observed an increase in transcript levels of some of these genes. The IEGs Npas4 and c-fos were the most significantly upregulated in the dorsal hippocampus of Tet3 cKO mice (Fig. 4i; adjusted t-test, c-fos p < 0.05, d = 1.534; Npas4 p < 0.01, d = 2.321). On the other hand, in the ventral part, only Npas4 was significantly increased (Fig. 4j; adjusted t-test: Npas4, p < 0.05, d = 0.520; c-fos, p = 0.248, d = 0.523). These results suggest that TET3 regulates these IEG’s levels.
Tet3 cKO mice harbor increased dendritic spine maturation in ventral hippocampal pyramidal neurons
To further correlate the observed behavioral and neuronal activity-regulated gene expression changes in Tet3 cKO mice with putative alterations in neural plasticity mechanisms, we analyzed neuronal and spines morphology of pyramidal neurons in the adult dorsal and ventral hippocampus CA1. We did not observe alterations in dendritic length and complexity of their arborization in the dorsal and ventral regions of Tet3 cKO (Fig. 5a, b, e, f). However, and although Tet3 deletion in neurons did not impact on spines density (Fig. 5c, g), Tet3 cKO mice displayed a decrease in the proportion of immature thin spines and an increase in the mature mushroom type of spines specifically in the ventral, but not in the dorsal, region of the hippocampus (Fig. 5d, h; factorial ANOVA, genotype, p < 0.001, ω2p < 0), suggesting that TET3 regulates spine maturation in the ventral CA1 region.
Tet3 cKO presented no alterations in neuronal morphology, in dorsal and ventral hippocampus, assessed by total length and Sholl analysis (a–b; e–f). Regarding spines analysis, no differences were found at the dorsal part (c, d), but a robust increase in spine differentiation (mushroom type) and a decrease in immature spines (thin type) of the apical dendrites spines was found in the ventral region in Tet3 cKO mice (h), without alterations in spine density (g). n = 13–20 neurons; three mice per group. Quantifications are presented as the mean ± SEM. a, b, e, f Factorial ANOVA; c, g Two-tailed Student’s t test; d, h Factorial ANOVA, genotype; ***p < 0.001.
Discussion
Despite recent advances, the role of TET enzymes in the brain, particularly of Tet3 which is highly transcribed, is largely unknown. Here we show that, in forebrain regions of the adult brain, TET3 is present in neurons, as observed by others [16, 38], but sparsely expressed in oligodendrocytes and absent in astrocytes.
Since TET3 is highly expressed in mature neurons in forebrain regions of the adult brain, we used a conditional and inducible knockout mouse model to ablate Tet3 in Camk2a-positive mature neurons allowing to study the function of TET3 in these cells at an adult stage. Importantly, Tet3 deletion did not affect Tet1 and Tet2 expression, as we observed previously in neural precursor cells [39]; in fact, this deletion did not influence the global genomic levels of 5hmC, and this might reflect action of Tet1 and Tet2, which also convert 5mC into 5hmC.
Our results show that neuronal Tet3 deletion increases anxiety-like behavior and impairs hippocampal spatial orientation. Regarding TET enzymes and the control of anxiety-like behaviors, only the role of Tet1 was previously addressed. Although TET1 KO mice showed normal anxiety and depression-related behaviors [11], Feng et al. showed that neuronal Tet1 deletion in neurons of nucleus accumbens produced antidepressant-like effects in several behavioral assays [40]. In contrast, in our model in which we deleted Tet3 in mature forebrain neurons, we observed an anxiety-like phenotype. Nevertheless, the deletion of Tet1 and Tet3 in these two studies was performed in distinct brain regions, possibly influencing the observed results. On the other hand, the distinct behavioral phenotype triggered by deletion of Tet1 and Tet3 might suggest a different regulatory activity for these enzymes in the anxiety regulation of the adult brain.
We further showed that Tet3 cKO mice displayed a specific impairment of hippocampal-dependent spatial orientation; however, other hippocampal-dependent tasks, such as object location and long- and short-term recognition memories, remained unaffected. Importantly, Tet1 and Tet2 were shown to be relevant to control spatial learning [9, 10], but Tet3 seems to be only critical to control spatial orientation, since the spatial learning remained unaltered in our model. This observation, linked to the finding of role of the ventral hippocampus in spatial navigation [41], supports the impact of Tet3 in the function of ventral hippocampus. In addition, the impairment in the use of directed strategies reflects alteration of the goal-directed behavior, and can be correlated with the anxiety-like behavior, since the ability to demonstrate goal-directed behavior requires a suppression of emotional states, namely anxiety [42, 43].
The molecular mechanisms through which TET3 controls anxiety and spatial orientation remained to be explained. Hence, we studied whether Tet3 deletion could impact the expression of gene networks known to be involved in the dysregulation of behavioral domains. We performed a transcriptomic analysis of the hippocampus of Tet3 cKO mice that showed few dysregulated genes in the dorsal region, but several networks were altered in the ventral region. The ventral hippocampus has been shown to be involved in the modulation of emotional behavior, namely anxiety [22]. Strikingly, an impact of neuronal Tet3 deletion in key networks involved in the regulation of anxiety-like behavior was observed in the ventral hippocampal region.
Enrichment pathway analysis of differentially expressed targets identified the glucocorticoid signaling pathway (HPA axis), specifically in this hippocampal subregion. In fact, dysfunction of the HPA axis has been implicated in the pathogenesis of psychiatric disorders, including anxiety disorders [44]. The persistent activation of the HPA system results in a sustained increase of cortisol (humans) or corticosterone (rodents) levels, and is one of the most consistent findings in psychiatry diseases, namely anxiety and depression [44]. Importantly, Tet3 cKO animals presented increased levels of corticosterone in the blood serum suggesting an overactivation of the HPA axis, which can be correlated with the increased anxiety-like behavior. Remarkably, we also found Crhr2 to be downregulated in the ventral hippocampus of Tet3 cKO mice; and CRHR2-deficient mice display increased anxiety-like behavior [45, 46], suggesting that the observed decrease of Crhr2 levels in the ventral hippocampus of Tet3 cKO mice can be linked to the development of this altered emotional behavior. However, Kishimoto et al. suggest that Crhr2 predominantly mediates a central anxiolytic effect independent of the HPA axis activity, since they did not observe hypercorticalism in Crhr2−/+ and Crhr2−/−mice at basal levels. Nevertheless, hypercorticalism was observed in Crhr2−/− mice upon acute stress in Kishimoto et al. work [46], and in various other studies [47, 48]. Yet, the direct or indirect causal relationship between Crhr2 decrease, corticosterone increases, and stress has not been established. Importantly, it is relevant to consider that Kishimoto et al. used a full KO mouse model, which can develop compensatory effects, hiding alterations in the basal corticosterone levels.
Neuronal activity-regulated genes are known to play important roles in diverse cellular processes such as neurotransmission, neuronal plasticity, learning and memory [49, 50]. Notably, and contrary to TET3 function in our model, TET1 was shown to be a positive regulator of IEGs expression [11,12,13]. Amongst all the IEGs analyzed in Tet3 cKO, we showed that the key upstream regulatory gene Npas4 displayed the highest magnitude of change amongst all the IEGs analyzed in Tet3 cKO. Npas4 controls a transcriptional program involving neural activity-regulated genes and is essential for cognitive function [50, 51]. Moreover, this gene is involved in neural circuitry plasticity, maintaining circuit homeostasis [51, 52]. Thus, we speculate that the aberrant increase in Npas4 and c-fos transcript levels in the dorsal hippocampus of Tet3 cKO mice might lead to a dysregulation of neuronal activity and possibly explain the spatial orientation impairment observed. Despite the vast amount of data supporting changes in IEGs expression in cognitive processes, in psychiatric conditions, only few studies have evaluated the direct implication of IEGs. However, it was already shown that Npas4 KO mice have less anxiety when compared with control animals [53]. Accordingly, Tet3 cKO mice showed increased anxiety-like behavior and increased expression of Npas4 mRNA transcript in the ventral hippocampus. Based on our results, we hypothesize that upregulation of Npas4 can lead to an increase in the expression of its targets, namely c-fos, promoting increased hippocampal neuronal activity. This dysfunctional hippocampal neuronal activity may alter the capacity of neurons to correctly respond to stimulus and culminate in behavioral dysfunction. Our hypothesis is in agreement with findings that TET3 is a negative regulator of synaptic activity [18, 19]. In accordance, we herein observed that Tet3 deletion triggered a shift of thin immature spines to the mushroom mature type in the ventral CA1 hippocampus. This increased synaptic complexity [54] fits with the observations of increased glutamatergic transmission in Tet3 deficient animals [19] and with the role of glutamate in spine maturation [55, 56]. Hence, our data first suggest that dendritic spine morphology is modified by Tet3 deletion specifically in mature neurons. This overactivation of the ventral hippocampus is suggestive of an anxiogenic phenotype in these animals and may reveal to be an interesting therapeutic target.
In summary, we here suggest that Tet3 plays an important role in modulation of anxiety-like behavior, as well as in spatial orientation tasks. The epigenetic control of behavior and neurophysiology is a topical subject currently and very relevant to various neurological and psychiatric conditions [57]. However, TET3 has been poorly studied in the context of psychiatric disorders, and to our knowledge, there is only one report showing no alterations in Tet3 levels in the parietal cortex of psychotic patients [58]. Future research on TET3 function, as well as on other members of TET family, may contribute to our understanding of the impact of epigenetic regulation on behavioral performance. Particularly, TET3 can represent a potential therapeutic target in pathologies related with anxiety spectrum.
References
Feng J, Fouse S, Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatr Res. 2007;61:58r–63r.
Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in Mammalian DNA by MLL partner TET1. Science. 2009;324:930–5.
Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14:1607–16.
Cadena-del-Castillo C, Valdes-Quezada C, Carmona-Aldana F, Arias C, Bermudez-Rattoni F, Recillas-Targa F. Age-dependent increment of hydroxymethylation in the brain cortex in the triple-transgenic mouse model of Alzheimer’s disease. J Alzheimer’s Dis. 2014;41:845–54.
Santiago M, Antunes C, Guedes M, Sousa N, Marques CJ. TET enzymes and DNA hydroxymethylation in neural development and function—how critical are they? Genomics. 2014;104:334–40.
Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181.
Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–69.
Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem. 2008;89:599–603.
Zhang RR, Cui QY, Murai K, Lim YC, Smith ZD, Jin S, et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell. 2013;13:237–45.
Gontier G, Iyer M, Shea JM, Bieri G, Wheatley EG, Ramalho-Santos M, et al. Tet2 rescues age-related regenerative decline and enhances cognitive function in the adult mouse brain. Cell Rep. 2018;22:1974–81.
Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, Le T, et al. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109–22.
Kumar D, Aggarwal M, Kaas GA, Lewis J, Wang J, Ross DL, et al. Tet1 oxidase regulates neuronal gene transcription, active DNA hydroxy-methylation, object location memory, and threat recognition memory. Neuroepigenetics. 2015;4:12–27.
Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming GL, et al. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79:1086–93.
Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–10.
Kang J, Lienhard M, Pastor WA, Chawla A, Novotny M, Tsagaratou A, et al. Simultaneous deletion of the methylcytosine oxidases Tet1 and Tet3 increases transcriptome variability in early embryogenesis. Proc Natl Acad Sci USA. 2015;112:E4236–45.
Li X, Wei W, Zhao QY, Widagdo J, Baker-Andresen D, Flavell CR, et al. Neocortical Tet3-mediated accumulation of 5-hydroxymethylcytosine promotes rapid behavioral adaptation. Proc Natl Acad Sci USA. 2014;111:7120–5.
Kremer EA, Gaur N, Lee MA, Engmann O, Bohacek J, Mansuy IM. Interplay between TETs and microRNAs in the adult brain for memory formation. Sci Rep. 2018;8:1678.
Yu H, Su Y, Shin J, Zhong C, Guo JU, Weng YL, et al. Tet3 regulates synaptic transmission and homeostatic plasticity via DNA oxidation and repair. Nat Neurosci. 2015;18:836–43.
Wang L, Li MY, Qu C, Miao WY, Yin Q, Liao J, et al. CRISPR-Cas9-mediated genome editing in one blastomere of two-cell embryos reveals a novel Tet3 function in regulating neocortical development. Cell Res. 2017;27:815–29.
Liu Y, Fu QF, Fu XL. The interaction between cognition and emotion. Chinese Sci Bull. 2009;54:4102–16.
Strange BA, Witter MP, Lein ES, Moser EI. Functional organization of the hippocampal longitudinal axis. Nat Rev Neurosci. 2014;15:655–69.
Fanselow MS, Dong H-W. Are the dorsal and ventral hippocampus functionally distinct structures? Neuron. 2010;65:7–19.
Peat JR, Dean W, Clark SJ, Krueger F, Smallwood SA, Ficz G, et al. Genome-wide bisulfite sequencing in zygotes identifies demethylation targets and maps the contribution of TET3 oxidation. Cell Rep. 2014;9:1990–2000.
Santos F, Peat J, Burgess H, Rada C, Reik W, Dean W. Active demethylation in mouse zygotes involves cytosine deamination and base excision repair. Epigenet Chromatin. 2013;6:39.
Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–40.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40.
Gibb R, Kolb B. A method for vibratome sectioning of Golgi-Cox stained whole rat brain. J Neurosci Methods. 1998;79:1–4.
Bessa JM, Ferreira D, Melo I, Marques F, Cerqueira JJ, Palha JA, et al. The mood-improving actions of antidepressants do not depend on neurogenesis but are associated with neuronal remodeling. Mol Psychiatr. 2008;14:764.
Mateus-Pinheiro A, Alves ND, Patrício P, Machado-Santos AR, Loureiro-Campos E, Silva JM, et al. AP2γ controls adult hippocampal neurogenesis and modulates cognitive, but not anxiety or depressive-like behavior. Mol Psychiatr. 2016;22:1725.
Erdfelder E, Faul F, Buchner A. GPOWER: a general power analysis program. Behav Res Methods, Instrum Comput. 1996;28:1–11.
Cohen J, (ed). Statistical power analysis for the behavioral sciences. New York: Academic Press; 1977.
Graziano A, Petrosini L, Bartoletti A. Automatic recognition of explorative strategies in the Morris water maze. J Neurosci Methods. 2003;130:33–44.
Moll P, Ante M, Seitz A, Reda T. QuantSeq 3’ mRNA sequencing for RNA quantification. Nat Methods. 2014;11: i–iii.
Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–386.
Kamburov A, Stelzl U, Lehrach H, Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013;41:D793–800.
Reul JMHM, Holsboer F. On the role of corticotropin-releasing hormone receptors in anxiety and depression. Dialogues Clin Neurosci. 2002;4:31–46.
Kramer A, Green J, Pollard J Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–30.
Montalban-Loro R, Lozano-Urena A, Ito M, Krueger C, Reik W, Ferguson-Smith AC, et al. TET3 prevents terminal differentiation of adult NSCs by a non-catalytic action at Snrpn. Nat Commun. 2019;10:1726.
Santiago M, Antunes C, Guedes M, Iacovino M, Kyba M, Reik W, et al. Tet3 regulates cellular identity and DNA methylation in neural progenitor cells. Cell Mol Life Sci. 2019. https://doi.org/10.1007/s00018-019-03335-7.
Feng J, Pena CJ, Purushothaman I, Engmann O, Walker D, Brown AN, et al. Tet1 in nucleus accumbens opposes depression- and anxiety-like behaviors. Neuropsychopharmacology. 2017;42:1657–69.
McDonald RJ, Balog RJ, Lee JQ, Stuart EE, Carrels BB, Hong NS. Rats with ventral hippocampal damage are impaired at various forms of learning including conditioned inhibition, spatial navigation, and discriminative fear conditioning to similar contexts. Behav Brain Res. 2018;351:138–51.
Jimenez JC, Su K, Goldberg AR, Luna VM, Biane JS, Ordek G, et al. Anxiety cells in a hippocampal-hypothalamic circuit. Neuron. 2018;97:670–.e676.
Yoshida K, Drew MR, Mimura M, Tanaka KF. Serotonin-mediated inhibition of ventral hippocampus is required for sustained goal-directed behavior. Nature Neurosci. 2019;22:770–7.
Faravelli C, Lo Sauro C, Lelli L, Pietrini F, Lazzeretti L, Godini L, et al. The role of life events and HPA axis in anxiety disorders: a review. Current Pharm Des. 2012;18:5663–74.
Bale TL, Contarino A, Smith GW, Chan R, Gold LH, Sawchenko PE, et al. Mice deficient for corticotropin-releasing hormone receptor-2 display anxiety-like behaviour and are hypersensitive to stress. Nat Genet. 2000;24:410–4.
Kishimoto T, Radulovic J, Radulovic M, Lin CR, Schrick C, Hooshmand F, et al. Deletion of crhr2 reveals an anxiolytic role for corticotropin-releasing hormone receptor-2. Nat Genet. 2000;24:415–9.
Coste SC, Kesterson RA, Heldwein KA, Stevens SL, Heard AD, Hollis JH, et al. Abnormal adaptations to stress and impaired cardiovascular function in mice lacking corticotropin-releasing hormone receptor-2. Nat Genet. 2000;24:403–9.
Preil J, Müller MB, Gesing A, Reul JMHM, Sillaber I, van Gaalen MM, et al. Regulation of the hypothalamic-pituitary-adrenocortical system in mice deficient for CRH receptors 1 and 2. Endocrinology. 2001;142:4946–55.
Loebrich S, Nedivi E. The function of activity-regulated genes in the nervous system. Physiol Rev. 2009;89:1079–103.
Coutellier L, Beraki S, Ardestani PM, Saw NL, Shamloo M. Npas4: a neuronal transcription factor with a key role in social and cognitive functions relevant to developmental disorders. PloS ONE. 2012;7:e46604.
Ramamoorthi K, Fropf R, Belfort GM, Fitzmaurice HL, McKinney RM, Neve RL, et al. Npas4 regulates a transcriptional program in CA3 required for contextual memory formation. Science. 2011;334:1669–75.
Ploski JE, Monsey MS, Nguyen T, DiLeone RJ, Schafe GE. The neuronal PAS domain protein 4 (Npas4) is required for new and reactivated fear memories. PloS ONE. 2011;6:e23760.
Jaehne EJ, Klaric TS, Koblar SA, Baune BT, Lewis MD. Effects of Npas4 deficiency on anxiety, depression-like, cognition and sociability behaviour. Behav Brain Res. 2015;281:276–82.
Berry KP, Nedivi E. Spine dynamics: are they all the same? Neuron. 2017;96:43–55.
Mattison HA, Popovkina D, Kao JPY, Thompson SM. The role of glutamate in the morphological and physiological development of dendritic spines. Eur J Neurosci. 2014;39:1761–70.
McKinney RA. Excitatory amino acid involvement in dendritic spine formation, maintenance and remodelling. J Physiol. 2010;588(Pt 1):107–16.
Antunes C, Sousa N, Pinto L, Marques CJ. TET enzymes in neurophysiology and brain function. Neurosci Biobehav Rev. 2019;102:337–44.
Dong E, Gavin DP, Chen Y, Davis J. Upregulation of TET1 and downregulation of APOBEC3A and APOBEC3C in the parietal cortex of psychotic patients. Transl Psychiatry. 2012;2:e159.
Acknowledgements
We thank Julian Peat and Christel Krueger (Babraham Institute, UK) for help with the Tet3 floxed mice, João Sobral (Genomics Unit, IGC, Portugal) for RNA-Seq analysis, and Daniel Neves and Daniel Sobral (Bioinformatics Unit, IGC, Portugal) for Bioinformtaic analysis. This work was supported by National Funds through Foundation for Science and Technology (FCT) fellowships (PD/BD/106049/2015 to CA, PD/BD/128074/2016 to JDS, IF/01079/2014 to LP, SFRH/BD/101298/2014 to SGG, SFRH/BD/131278/2017 to ELC, and IF/00047/2012 and CEECIND/00371/2017 to CJM); FCT project grant (PTDC/BIA-BCM/121276/2010) to CJM; EpiGeneSys Small Collaborative project to LP and MRB; BIAL Foundation Grant 427/14 to LP; Northern Portugal Regional Operational Programme (NORTE 2020), under the Portugal 2020 Partnership Agreement, through the European Regional Development Fund (FEDER; NORTE-01-0145-FEDER-000013); FEDER funds, through the Competitiveness Factors Operational Programme (COMPETE), and National Funds, through the FCT (POCI-01-0145-FEDER-007038).
Author information
Authors and Affiliations
Contributions
CA designed the study, performed the experiments, analyzed the data, and wrote the manuscript; JDS performed gene ontology analysis of QuantSeq results, statistical analysis, and wrote the manuscript. SGG and NDA helped with the behavioral tests and respective analysis. ELC helped with the behavioral tests. FF helped with neuronal morphology analysis. MRB helped with RNA-Seq data. NS organized and wrote the manuscript; WR contributed with the Tet3 conditional mouse strain; LP and CJM designed the study, organized, and wrote the manuscript. All authors revised and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Antunes, C., Da Silva, J.D., Guerra-Gomes, S. et al. Tet3 ablation in adult brain neurons increases anxiety-like behavior and regulates cognitive function in mice. Mol Psychiatry 26, 1445–1457 (2021). https://doi.org/10.1038/s41380-020-0695-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-020-0695-7
- Springer Nature Limited
This article is cited by
-
Tet1-mediated 5hmC regulates hippocampal neuroinflammation via wnt signaling as a novel mechanism in obstructive sleep apnoea leads to cognitive deficit
Journal of Neuroinflammation (2024)
-
Recent Advances and Therapeutic Implications of 2-Oxoglutarate-Dependent Dioxygenases in Ischemic Stroke
Molecular Neurobiology (2024)
-
TET2 deficiency promotes anxiety and depression-like behaviors by activating NLRP3/IL-1β pathway in microglia of allergic rhinitis mice
Molecular Medicine (2023)
-
Epigenetics of cognition and behavior: insights from Mendelian disorders of epigenetic machinery
Journal of Neurodevelopmental Disorders (2023)
-
Post-stroke depression: epigenetic and epitranscriptomic modifications and their interplay with gut microbiota
Molecular Psychiatry (2023)