Abstract
Predicting antidepressant response has been a clinical challenge for mood disorder. Although several genome-wide association studies have suggested a number of genetic variants to be associated with antidepressant response, the sample sizes are small and the results are difficult to replicate. Previous animal studies have shown that knockout of the serotonin receptor 7 gene (HTR7) resulted in an antidepressant-like phenotype, suggesting it was important to antidepressant action. In this report, in the first stage, we used a cost-effective pooled-sequencing strategy to sequence the entire HTR7 gene and its regulatory regions to investigate the association of common variants in HTR7 and clinical response to four selective serotonin reuptake inhibitors (SSRIs: citalopram, paroxetine, fluoxetine and sertraline) in a retrospective cohort mainly consisting of subjects with bipolar disorder (n = 359). We found 80 single-nucleotide polymorphisms (SNPs) with false discovery rate < 0.05 associated with response to paroxetine. Among the significant SNPs, rs7905446 (T/G), which is located at the promoter region, also showed nominal significance (P < 0.05) in fluoxetine group. GG/TG genotypes for rs7905446 and female gender were associated with better response to two SSRIs (paroxetine and fluoxetine). In the second stage, we replicated this association in two independent prospective samples of SSRI-treated patients with major depressive disorder: the MARS (n = 253, P = 0.0169) and GENDEP studies (n = 432, P = 0.008). The GG/TG genotypes were consistently associated with response in all three samples. Functional study of rs7905446 showed greater activity of the G allele in regulating expression of HTR7. The G allele displayed higher luciferase activity in two neuronal-related cell lines, and estrogen treatment decreased the activity of only the G allele. Electrophoretic mobility shift assay suggested that the G allele interacted with CCAAT/enhancer-binding protein beta transcription factor (TF), while the T allele did not show any interaction with any TFs. Our results provided novel pharmacogenomic evidence to support the role of HTR7 in association with antidepressant response.
Similar content being viewed by others
Introduction
Serotonin (5-HT) is a monoamine neurotransmitter with a broad range of physiological functions including sleep, mood, cardiovascular function, circadian rhythms, body temperature, food intake and endocrine regulation. These effects are mediated by a large number of 5-HT receptors, comprising seven families (HTR1 to HTR7) and at least 14 subtypes, among which HTR7 displays the highest affinity for 5-HT [1,2,3]. HTR7 is a G protein-coupled receptor that links to adenylate cyclase and transduces signals mainly through the cyclic adenosine monophosphate pathway [3, 4]. HTR7 has been shown to be expressed abundantly in both peripheral tissues like smooth muscle and intestine and in brain regions including the forebrain, hippocampus, hypothalamus, brainstem and cerebellum [4,5,6,7].
A growing body of evidence has indicated that HTR7 plays a role in the pathophysiology of psychiatric disorders. Genome-wide association studies (GWASs) have suggested a relationship between HTR7 genetic polymorphisms and schizophrenia and the development of alcohol dependence [8,9,10]. HTR7 was also shown to influence behaviors in rodents that mimic obsessive–compulsive disorder and substance abuse [11, 12]. Much attention has been devoted to the possible role of HTR7 in depression. HTR7 knockout mice or mice with pharmacological blockade of HTR7 showed antidepressant-like behavior [13,14,15,16]. A recent study showed genetic polymorphisms in HTR7 were associated with hypocortisolism in a gender-specific manner in African-American subjects, suggesting HTR7 may contribute to stress system dysregulations [17]. Emerging preclinical evidence have suggested that HTR7 is involved in the action of antidepressants. Several antidepressants, both tricyclics and selective serotonin reuptake inhibitors (SSRIs), induce c-fos expression in a fashion that is similar to HTR7 activation, while chronic treatment by fluoxetine downregulates HTR7 expression [18, 19]. In addition, blockade of HTR7 by SB-269970, a highly selective HTR7 antagonist, was found to potentiate the effects of SSRI and norepinephrine reuptake inhibitors (NARIs) [14]. Indeed, several antidepressant and antipsychotic drugs with clinically established antidepressant efficacy showed high affinity for HTR7, such as amitriptyline, amoxapine, amisulpride, clozapine, aripiprazole, lurasidone, risperidone and perospirone [20,21,22,23]. Thus, the above evidence suggests HTR7 could play an important role in SSRI action and may serve as a potential target for the treatment of depression.
SSRIs (e.g., paroxetine and fluoxetine) are the most widely used antidepressants for the treatment of major depressive disorder (MDD); however, around half of the patients show poor response to SSRIs [24]. Treatment resistance in MDD is common and evidence shows that a substantial portion of the treatment-resistant MDD patients may later be diagnosed as bipolar disorder (BD) [25]. BD is a complex and chronic psychiatric condition affecting 1–2% of the population and characterized by shifts in mood between manic and depressive states [26]. Although mania is the most dramatic manifestation of BD, in reality patients spend most of their time depressed when ill [27]. Though there are many effective treatments for mania, treating bipolar depression remains a considerable clinical challenge [28]. The primary dilemma is the use of antidepressants; there is a risk of inducing a manic episode or rapid cycling, though the larger question is one of efficacy. Despite widespread safe and seemingly effective use in the community, many controlled trials have failed to show efficacy for antidepressants in BD [28]. This suggests heterogeneity in drug response and possibly disease mechanism. Several large-scale GWASs have examined the association between genetic markers and antidepressant response, but the results are difficult to replicate and only a limited number of single-nucleotide polymorphisms (SNPs) in HTR7 have been covered [29,30,31]. The overall goal of this study is to identify genes that influence SSRI response in BD. In this report, in the first stage, we utilized a cost-effective pooled-sequencing strategy to sequence the entire HTR7 gene and its regulatory regions in a retrospectively characterized cohort mainly consisting of subjects with BD, aimed to investigate the genetic association of HTR7 and SSRI response. In the second stage, we replicated the findings from stage one in two independent prospective cohorts consisting of patients with MDD (MARS and GENDEP).
Methods
Pooled-sequencing of HTR7 gene in a retrospective cohort
Subjects
All subjects (n = 359) were ascertained as part of several cohorts collected for genetic studies of BD. All subjects were selected because they had a BD type 1 (BD-I) diagnosis, or they had major depression and a first-degree relative with BD-I, or schizoaffective disorder, bipolar type. Subjects were identified through Department of Veterans Affairs (VA) and University of California, San Diego (UCSD) clinics, as well as advertisement and patient support groups. All subjects provided written informed consent according to UCSD Institutional Review Board-approved procedures and consent form.
Assessment of SSRI response
All subjects were directly interviewed using the Diagnostic Interview for Genetic Studies (DIGS) [32] which had been modified to collect information regarding past drug trials. Interviewers underwent a training course, reliability was tested regularly and was consistently high. Information from the modified DIGS was reviewed by a panel of experienced clinicians along with medical records and information from family informants. Patients were queried regarding all their past medication trials including a past history of SSRI treatment. Subject’s response to medications over their lifetime was assessed based on self-reporting. Blind raters considered all information about all medication trials over the patient’s life in order to assess response. Good responders were those who were estimated to have 50% reduction in symptoms or episode frequency during entire illness. Subject demographic information classified by treatment groups is shown in Table 1.
Pooled-DNA sequencing
DNA was quantified with PicoGreen and equal quantities from each subject were combined into 32 pools (ranging from 11 to 24 subjects per pool) grouped by medication (citalopram, paroxetine, fluoxetine and sertraline) and type of response (good and poor). The entire HTR7 gene, promoter and 5’- and 3’-untranslated region were covered and amplified by 13 long-range polymerase chain reactions, generating DNA fragments from 10 to 13 kb covering the region of Chr10: 92499978–92623668. We performed 2 × 150 bp paired-end, multiplexed sequencing on an Illumina MiSeq sequencer (Illumina, San Diego, CA). The quality of raw reads were examined using FastQC [33] and were aligned to human reference genome (GRCh37/hg19) using BWA [34]. We used CRISP (v0.7) [35] with the default setting as the variant caller and filtered the variants in the VCF files that showed EMpass, quality value > 100 and minor allele frequency > 0.05. The variants were annotated by ANNOVAR [36].
Replication study I in the Munich Antidepressant Response Signature (MARS) project
The MARS project is a prospective naturalistic study of adult inpatients with depression in Germany [30, 37]. Diagnoses were based on diagnostic and statistical manual of mental diseases (DSM-IV) criteria of a major depressive episode, including first-episode MDD, recurrent MDD and BD. The severity of the depressive symptoms was assessed weekly based on the 21-item Hamilton Depression Rating Scale (HDRS-21) [38]. In this study, we included samples with only unipolar depression diagnosis and Caucasian ancestry (n = 837) and evaluated the treatment response at week 6. We defined remission as HDRS-21 < 10. For further details about the MARS project, see Hennings et al. [37].
Replication study II in the Genome-based Therapeutic Drugs for Depression (GENDEP) study
GENDEP is a multicenter part-randomized open-label pharmacogenomic study of patients with moderate to severe unipolar depression diagnosed according to DSM-IV and established in the semi-structured SCAN (Schedules for Clinical Assessment in Neuropsychiatry) interview [39]. Patients with personal and family history of schizophrenia or bipolar affective disorder and current dependence on alcohol or drugs were excluded from the study. Response was assessed weekly by three established measures of depression severity: the clinician-rated 10-item Montgomery–Åsberg Depression Rating Scale (MADRS) [40], the HDRS-17 [41] and the self-report 21-item Beck Depression Inventory [42]. In this study, we included patients with European ancestry, and evaluated the treatment response at week 12. We defined remission as HDRS-17 ≤ 7 [43]. For further details about the GENDEP project, see Uher et al. [39, 44].
SNP genotyping
Genotyping of rs7905446 (T/G) in the retrospective cohort was performed using a TaqMan SNP genotyping assay (Thermo Fisher Scientific, Waltham, MA, USA) as previously described [45]. The genotyping success rate was >95%. Twenty percent of the samples were genotyped in duplicate, with 100% reproducibility. For SNP imputation for the MARS and GENDEP cohorts see supplementary materials/methods.
Transfection and luciferase reporter assay
HTR7 promoter containing rs7905446 (T/G) SNP was amplified followed by ligation into pGL4.26 luciferase reporter vector (Promega, Madison, WI, USA). HT-22 and SK-N-MC cell lines were transfected with rs79054446-T or rs7905446-G vectors together with pGL4.74 Renilla Luciferase control vector (Promega) using Lipofectamine 3000 reagent (Thermo Fisher Scientific). Cells were assayed for luciferase and Renilla luciferase activity using Dual-Glo Luciferase Assay System (Promega) according to the manufacturer’s instruction. For details see supplementary materials/methods.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed using the LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. In brief, Hela cell nuclear extracts and biotin-labeled probes spanning rs7905446 (T/G) region were incubated at room temperature for 40 min followed by electrophoresis separation and transferring to the nylon membrane. The competition reaction was performed using 200-fold molar excess of unlabeled probe. For supershift analysis, 1 µg anti-CCAAT/enhancer-binding protein beta (CEBPB) antibody (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) was added to the nuclear extract prior to the binding reaction. The DNA-protein complexes were detected using chemiluminescence. For details see supplementary materials/methods.
Statistical analysis
The association between drug response and allelic SNPs identified from pooled-sequencing were performed using logistic regression (PLINK version 1.9) [46]. In this analysis, because of the pooling, Caucasians and a small portion of other ethnicities were included. However, the association between drug response and rs7905446 genotype was performed using logistic regression within the Caucasian population, adjusted for age and sex. χ2 tests were used to compare the sex distribution between responders and non-responders. Group differences were analyzed using Student’s t-test. A P value of < 0.05 was considered nominally statistically significant.
Results
Common SNPs in HTR7 are associated with SSRI response in BD
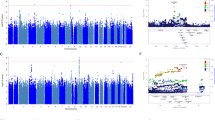
In the retrospective cohort, we performed pooled-sequencing of HTR7 gene in a total of 359 subjects (Table 1) and examined the association between SSRI treatment response and common SNP variations based on an allelic model. We found that 80 out of 169 common SNPs survived false discovery rate (FDR) < 0.05 in the paroxetine group and 95% (n = 76) of the significant SNPs were located in intronic regions (for the full list see Supplementary file). We were particularly interested in the SNP rs7905446 (FDR = 0.0387, Table 2) that was located at the promoter region, because several validated transcription factors (TFs) from the Encyclopedia of DNA Elements (ENCODE) database showed binding signals around this region, implicating a functional SNP. Further, rs7905446 also showed nominal significance (P = 0.047) in the fluoxetine group (Supplementary file) and is in high linkage disequilibrium with the other two top SNPs in the 5’ upstream, rs6583737 and rs12254390 (Fig. 1). We validated rs7905446 in Caucasian subjects using a TaqMan SNP genotyping assay in both paroxetine and fluoxetine groups (n = 266). The genotype distribution was significantly different between responders and non-responders of these two SSRIs (responders: TT vs GT vs GG = 29.7% vs 54.9% vs 15.4%; non-responders: TT vs GT vs GG = 46.0% vs 41.5% vs 12.5%; Pearson χ2 = 6.697, P = 0.035). Next, using logistic regression we found that TT genotype was significantly associated with poor paroxetine response compared with TG/GG genotypes, when controlled for gender and age (TT vs TG/GG: P = 0.005, odds ratio (OR) = 5.250; Table 3). When combining both paroxetine and fluoxetine groups, TT genotype was again shown to be associated with poor response in two SSRIs (TT vs TG/GG: P = 0.008, OR = 2.135; Table 3). Gender seemed to influence SSRI response in the BD samples and, specifically, men were more likely to be poor responders (P < 0.001, OR = 2.623; Table 3 and Fig. 2). No gender × rs7905446 interaction was found in either the paroxetine group or paroxetine+fluoxetine groups. Four SNPs including rs7905446 in the fluoxetine showed nominal P < 0.05 (Supplementary file). No SNPs with nominal P < 0.05 were detected in citalopram and sertraline groups.
Rs7905446 is in high linkage disequilibrium with two top single-nucleotide polymorphisms (SNPs; rs6583737 and rs12254390) in the 5’ upstream region of HTR7 gene. A number of transcription factors such as CCAAT/enhancer-binding protein beta (CEBPB) in the Encyclopedia of DNA Elements (ENCODE) database showed binding signals around rs7905446, implicating a functional SNP
Rs7905446 is associated with antidepressant response in unipolar depression in MARS and GENDEP cohorts
We next investigated if rs7905446 was associated with antidepressant response in MDD in two larger-scale prospective cohorts. The treatment in MARS cohort is naturalistic, selected by clinician, which includes a variety of antidepressants such as SSRIs, serotonin and norepinephrine reuptake inhibitors (SNRIs) and tricyclics etc. We first examined if rs7905446 can predict antidepressant response in general, i.e., including all antidepressant drugs. We found TT genotype was significantly associated with non-remission status, while TG/GG genotypes predicted treatment remission at week 6, when controlling for gender and age (TT vs TG/GG: P = 0.032, OR = 1.385; Table 3). Next, we found similar results in patients who underwent SSRI or SNRI treatments (P = 0.044, Table 3) or were only treated with SSRI (P = 0.017, Table 3). Other top SNPs (rs6583737 and rs12254390), which are in high linkage disequilibrium with rs7905446, showed similar predictive effects. In the GENDEP cohort, two antidepressants (escitalopram and nortriptyline) that represent the two most common mechanisms of action of antidepressants were administered in a part-randomized manner. Interestingly, we found TG/GG genotypes predicted remission only in the escitalopram-treated group, escitalopram being an SSRI (P = 0.008, Table 3) but not nortriptyline which acts like NARI (P = 0.154, Table 3). There was no significant gender effect on response to antidepressants in the MARS and GENDEP cohorts.
Functional validation of rs7905446
We used a luciferase reporter assay to test if rs7905446 was a functional SNP in two neuronal-related cell lines, SK-N-MC (neuroblastoma cell line) and HT-22 (mice hippocampal cell line). In both cell lines, we observed the rs7905446-G allele, associated with better antidepressant response, exhibited stronger luciferase signals compared with the T allele, suggesting a higher promoter activity (SK-N-MC: P < 0.01; HT-22: P < 0.001; Fig. 3). Gender seemed to play a role in modulating antidepressant response: men were more than twofold more likely to become non-responders in the BP retrospective samples (Table 3), suggesting estrogen may enhance the effect of antidepressant efficacy. We treated the HT-22 cell line with different concentrations of estrogen, and found the high activity of the rs7905446-G allele was decreased after estrogen treatment at a concentration of 1 µM, while the activity of the rs7905446-T allele was not influenced at any concentration tested (Fig. 3). The ENCODE database suggests rs7905446 position overlaps with the binding sites of several potential TFs, including CEBPB, which can recruit both activators like EP300 and repressors like the estrogen receptor 1 (ESR1) [47, 48]. EMSA showed the rs7905446-G allele was able to generate a shift, and when adding an anti-CEBPB antibody, a supershift was observed. In contrast, biotin-probe spanning the T allele did not show binding potentials of any TFs in the nuclear extract (Fig. 4).
Electrophoretic mobility shift assay showed biotin-labeled probe containing the rs7905446-G can produce a shift (arrow) when incubated with the HeLa cell nuclear extract, suggesting an interaction with transcription factors. An anti-CCAAT/enhancer-binding protein beta (CEBPB) antibody generated a supershift (asterisk), suggesting an interaction with CEBPB transcription factor
Discussion
To our knowledge, this is the first study showing a consistent association between a functional variant, rs7905446, in HTR7 gene and SSRI response in three independent clinical cohorts. We also showed that the rs7905446-G allele, which was associated with better antidepressants response, displayed higher promoter activity than the T allele, and estrogen treatment decreased the promoter activity in only the G allele.
Rs7905446 is associated with response to drugs with different mechanisms of action
SSRIs are chemically diverse and therefore are different from each other in pharmacological profiles and clinical efficacy. For example, citalopram is a racemic mixture and escitalopram is its S-enantiomer; the latter was shown to have superior efficacy [49]. Paroxetine and fluoxetine have a high potential to interact with other drugs compared to citalopram and sertraline [50]. In addition, paroxetine exhibits relatively high affinity to muscarinic receptors and fluoxetine shows high affinity to HTR2A/2C receptors. Whether these additional actions of SSRIs will influence HTR7 function awaits further investigation. We did not find that rs7905446 was associated with response to citalopram or sertraline, suggesting poor power, or that HTR7 is not as prominent in the mechanism of action for these two drugs. In the GENDEP cohort, we noticed that rs7905446 can predict remission only in patients treated with escitalopram but not with nortriptyline; the latter is a tricyclic antidepressant with a 100 times higher affinity to norepinephrine transporter than to the serotonin transporter [39]. Consistently, in the MARS cohort, rs7905446 in predicting remission to SSRI exhibited a much lower P value compared to the P value predicting SSRI+SNRI together. Our result suggested HTR7 polymorphisms were strongly associated with response to SSRIs but not inhibitors of norepinephrine reuptake.
Estrogen plays a role in antidepressant action
In accordance with our findings in the BD cohort, there are reports suggesting SSRI are more effective in women than in men [51, 52]. In contrast, the effect of gender on antidepressant response was not observed in the two depression cohorts, suggesting gender may play different roles in BD and unipolar depression. While most studies showed an almost equal gender ratio in lifetime prevalence in BD, women were twice as likely than men to suffer unipolar depression [52, 53]. A number of studies have suggested estrogen as antidepressant or as co-adjuvant to facilitate the effect of antidepressants like fluoxetine [54]. Our work showed a novel mechanism for estrogen’s antidepressant effect: via reduced HTR7 expression. We speculate that CEBPB will predominantly recruit activators (e.g., EP300) when in conditions of absent or low levels of estrogen, and thus we observed a high promoter activity in the G allele. In contrast, high levels of estrogen will trigger ESR1 (a repressor) competing with other activators to interact with CEBPB, since we observed a significant decrease of promoter activity with 1 µM β-estradiol treatment but not with 10 nM.
Multiple roles of HTR7
HTR7 has been shown to promote neurite outgrowth [55], dendritic spines and synaptogenesis [56], suggesting responders may receive more 5-HT input during neurodevelopment or in learning and memory formation. HTR7 may also play a role in mediating inflammatory response. Casas-Engel et al. [57] showed that serotonin inhibited lipopolysaccharide-stimulated proinflammatory cytokine production (e.g., interleukin-12 and tumor necrosis factor-α) in macrophages. This effect was blocked by a highly selective HTR7 antagonist SB-269970 [57], suggesting an increased HTR7 expression may be associated with lower inflammatory cytokine levels. Interestingly, a recent meta-analysis showed that a heightened inflammatory profile may underly the treatment resistance in depression [58]. Besides, HTR7 seems to have a dual role in regulating γ-aminobutyric acid (GABA) synaptic transmission. Activation of HTR7 in raphe nuclei reduces GABA-mediated inhibition of serotonergic neurons and consequently enhances 5-HT release. However in the hippocampus, HTR7 activation was shown to stimulate GABAergic interneuron activity [59]. HTR7 can form heterodimer with HTR1A, which will inhibit HTR1A-mediated activation of Gi protein and G protein-gated potassium channels while accelerating agonist-mediated internalization of HTR1A receptor, initiating G protein-independent signaling pathways such as mitogen-activated protein kinases [60]. It has been suggested that HTR1A/7 heterodimers are more prevalent in postsynaptic populations in depression condition than in physiological condition, leading to an increased internalization of postsynaptic HTR1A and neuronal hyper-excitability [61]. Decreasing HTR7 activity may inhibit HTR1A/7 dimerization-induced neuronal hyper-excitability which may enhance the treatment effect of SSRIs. Thus, whether HTR7 expression level can predict SSRI response remains elusive but a decrease of HTR7 level seems to be associated with a reduction in severity of depressive symptoms.
Limitations
The evaluation of SSRI response in the bipolar cohort was retrospective, and thus recall bias may be present. However, we previously compared retrospective ratings with prospective response on the same patients (n = 40) who completed the prospective arm of a lithium study. The patients’ records were then retrospectively and blindly rated using the Alda scale [62]. We demonstrated a strong correlation between the Alda score and prospectively measured response (r = 0.67, P < 0.001) supporting the validity of our retrospective assessment. These results are being reported separately; due to the retrospective assessment of the bipolar cohort, we were unable to distinguish if there was a risk for mania/rapid cycling after SSRI treatment and the activity of different concomitant medications such as mood stabilizer; in the MARS study, we cannot examine the association between rs7905440 genotype and specific type of SSRI due to a lack of detailed drug information. The genetic association analyses in MARS and GENDEP studies included imputed data. We cannot provide haplotype analysis regarding the SNPs that showed significance since we used pooled-sequencing method.
Conclusion remark
Heterogeneity in drug response has been a great challenge in treating mood disorder, which may be related to different pathophysiology of the disease and metabolism of the drug, both factors thought to be influenced by an individual’s genetic background [63]. Understanding the relationship between genetic factors and treatment response may allow for the clinical implementation of pharmacogenetic tests and the development of personalized treatment in patients. Our study showed a functional SNP, rs7905446 in the HTR7 gene was associated with response to antidepressants in both bipolar and unipolar depression, which warrants further investigation as a potential novel pharmacogenetic diagnostic marker.
References
Hannon J, Hoyer D. Molecular biology of 5-HT receptors. Behav Brain Res. 2008;195:198–213.
Nichols DE, Nichols CD. Serotonin receptors. Chem Rev. 2008;108:1614–41.
Ruat M, Traiffort E, Leurs R, Tardivel-Lacombe J, Diaz J, Arrang JM, et al. Molecular cloning, characterization, and localization of a high-affinity serotonin receptor (5-HT7) activating cAMP formation. Proc Natl Acad Sci USA. 1993;90:8547–51.
Bard JA, Zgombick J, Adham N, Vaysse P, Branchek TA, Weinshank RL. Cloning of a novel human serotonin receptor (5-HT7) positively linked to adenylate cyclase. J Biol Chem. 1993;268:23422–6.
Neumaier JF, Sexton TJ, Yracheta J, Diaz AM, Brownfield M. Localization of 5-HT(7) receptors in rat brain by immunocytochemistry, in situ hybridization, and agonist stimulated cFos expression. J Chem Neuroanat. 2001;21:63–73.
Varnas K, Thomas DR, Tupala E, Tiihonen J, Hall H. Distribution of 5-HT7 receptors in the human brain: a preliminary autoradiographic study using [3H]SB-269970. Neurosci Lett. 2004;367:313–6.
Beattie DT, Smith JA. Serotonin pharmacology in the gastrointestinal tract: a review. Naunyn Schmiede Arch Pharmacol. 2008;377:181–203.
East SZ, Burnet PW, Kerwin RW, Harrison PJ. An RT-PCR study of 5-HT(6) and 5-HT(7) receptor mRNAs in the hippocampal formation and prefrontal cortex in schizophrenia. Schizophr Res. 2002;57:15–26.
Mowry BJ, Ewen KR, Nancarrow DJ, Lennon DP, Nertney DA, Jones HL, et al. Second stage of a genome scan of schizophrenia: study of five positive regions in an expanded sample. Am J Med Genet. 2000;96:864–9.
Ikeda M, Iwata N, Kitajima T, Suzuki T, Yamanouchi Y, Kinoshita Y, et al. Positive association of the serotonin 5-HT7 receptor gene with schizophrenia in a Japanese population. Neuropsychopharmacology. 2006;31:866–71.
Hedlund PB, Sutcliffe JG. The 5-HT7 receptor influences stereotypic behavior in a model of obsessive-compulsive disorder. Neurosci Lett. 2007;414:247–51.
Ballaz SJ, Akil H, Watson SJ. Analysis of 5-HT6 and 5-HT7 receptor gene expression in rats showing differences in novelty-seeking behavior. Neuroscience. 2007;147:428–38.
Guscott M, Bristow LJ, Hadingham K, Rosahl TW, Beer MS, Stanton JA, et al. Genetic knockout and pharmacological blockade studies of the 5-HT7 receptor suggest therapeutic potential in depression. Neuropharmacology. 2005;48:492–502.
Sarkisyan G, Roberts AJ, Hedlund PB. The 5-HT(7) receptor as a mediator and modulator of antidepressant-like behavior. Behav Brain Res. 2010;209:99–108.
Hedlund PB, Huitron-Resendiz S, Henriksen SJ, Sutcliffe JG. 5-HT7 receptor inhibition and inactivation induce antidepressantlike behavior and sleep pattern. Biol Psychiatry. 2005;58:831–7.
Wesolowska A, Nikiforuk A, Stachowicz K, Tatarczynska E. Effect of the selective 5-HT7 receptor antagonist SB 269970 in animal models of anxiety and depression. Neuropharmacology. 2006;51:578–86.
Swanson G, Miller S, Alyahyawi A, Wilson B, Saadatmand F, Lee C. et al. Genetic polymorphisms in the serotonin receptor 7 (HTR7) gene are associated with cortisol levels in African American young adults [version 1; referees: 1 not approved]. F1000Research. 2017;6:19
Mullins UL, Gianutsos G, Eison AS. Effects of antidepressants on 5-HT7 receptor regulation in the rat hypothalamus. Neuropsychopharmacology. 1999;21:352–67.
Sleight AJ, Carolo C, Petit N, Zwingelstein C, Bourson A. Identification of 5-hydroxytryptamine7 receptor binding sites in rat hypothalamus: sensitivity to chronic antidepressant treatment. Mol Pharmacol. 1995;47:99–103.
Roth BL, Craigo SC, Choudhary MS, Uluer A, Monsma FJ Jr., et al. Binding of typical and atypical antipsychotic agents to 5-hydroxytryptamine-6 and 5-hydroxytryptamine-7 receptors. J Pharmacol Exp Ther. 1994;268:1403–10.
Monsma FJ Jr., Shen Y, Ward RP, Hamblin MW, Sibley DR. Cloning and expression of a novel serotonin receptor with high affinity for tricyclic psychotropic drugs. Mol Pharmacol. 1993;43:320–7.
Abbas AI, Hedlund PB, Huang XP, Tran TB, Meltzer HY, Roth BL. Amisulpride is a potent 5-HT7 antagonist: relevance for antidepressant actions in vivo. Psychopharmacology (Berl). 2009;205:119–28.
Ishibashi T, Horisawa T, Tokuda K, Ishiyama T, Ogasa M, Tagashira R, et al. Pharmacological profile of lurasidone, a novel antipsychotic agent with potent 5-hydroxytryptamine 7 (5-HT7) and 5-HT1A receptor activity. J Pharmacol Exp Ther. 2010;334:171–81.
Rush AJ, Trivedi MH, Wisniewski SR, Nierenberg AA, Stewart JW, Warden D, et al. Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: a STAR*D report. Am J Psychiatry. 2006;163:1905–17.
Sharma V, Khan M, Smith A. A closer look at treatment resistant depression: is it due to a bipolar diathesis? J Affect Disord. 2005;84:251–7.
Martinsson L, Wei Y, Xu D, Melas PA, Mathe AA, Schalling M, et al. Long-term lithium treatment in bipolar disorder is associated with longer leukocyte telomeres. Transl Psychiatry. 2013;3:e261.
Post RM, Leverich GS, Nolen WA, Kupka RW, Altshuler LL, Frye MA, et al. A re-evaluation of the role of antidepressants in the treatment of bipolar depression: data from the Stanley Foundation Bipolar Network. Bipolar Disord. 2003;5:396–406.
Vazquez GH, Tondo L, Undurraga J, Baldessarini RJ. Overview of antidepressant treatment of bipolar depression. Int J Neuropsychopharmacol. 2013;16:1673–85.
Uher R, Perroud N, Ng MY, Hauser J, Henigsberg N, Maier W, et al. Genome-wide pharmacogenetics of antidepressant response in the GENDEP project. Am J Psychiatry. 2010;167:555–64.
Ising M, Lucae S, Binder EB, Bettecken T, Uhr M, Ripke S, et al. A genomewide association study points to multiple loci that predict antidepressant drug treatment outcome in depression. Arch Gen Psychiatry. 2009;66:966–75.
Garriock HA, Kraft JB, Shyn SI, Peters EJ, Yokoyama JS, Jenkins GD, et al. A genomewide association study of citalopram response in major depressive disorder. Biol Psychiatry. 2010;67:133–8.
Nurnberger JI Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, et al. Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry. 1994;51:849–59. discussion 863-4
Andrews S. FastQC: a quality control tool for high throughput sequence data. 2015. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Bansal V. A statistical method for the detection of variants from next-generation resequencing of DNA pools. Bioinformatics. 2010;26:i318–24.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164.
Hennings JM, Owashi T, Binder EB, Horstmann S, Menke A, Kloiber S, et al. Clinical characteristics and treatment outcome in a representative sample of depressed inpatients - findings from the Munich Antidepressant Response Signature (MARS) project. J Psychiatr Res. 2009;43:215–29.
Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry. 1960;23:56–62.
Uher R, Huezo-Diaz P, Perroud N, Smith R, Rietschel M, Mors O, et al. Genetic predictors of response to antidepressants in the GENDEP project. Pharm J. 2009;9:225–33.
Montgomery SA, Asberg M. A new depression scale designed to be sensitive to change. Br J Psychiatry. 1979;134:382–9.
Hamilton M. Development of a rating scale for primary depressive illness. Br J Soc Clin Psychol. 1967;6:278–96.
Beck AT, Ward CH, Mendelson M, Mock J, Erbaugh J. An inventory for measuring depression. Arch Gen Psychiatry. 1961;4:561–71.
Fabbri C, Tansey KE, Perlis RH, Hauser J, Henigsberg N, Maier W, et al. Effect of cytochrome CYP2C19 metabolizing activity on antidepressant response and side effects: Meta-analysis of data from genome-wide association studies. Eur Neuropsychopharmacol. 2018;28:945–54.
Uher R, Maier W, Hauser J, Marusic A, Schmael C, Mors O, et al. Differential efficacy of escitalopram and nortriptyline on dimensional measures of depression. Br J Psychiatry. 2009;194:252–9.
Wei YB, Martinsson L, Liu JJ, Forsell Y, Schalling M, Backlund L, et al. hTERT genetic variation in depression. J Affect Disord. 2016;189:62–9.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Mink S, Haenig B, Klempnauer KH. Interaction and functional collaboration of p300 and C/EBPbeta. Mol Cell Biol. 1997;17:6609–17.
Stein B, Yang MX. Repression of the interleukin-6 promoter by estrogen receptor is mediated by NF-kappa B and C/EBP beta. Mol Cell Biol. 1995;15:4971–9.
Montgomery S, Hansen T, Kasper S. Efficacy of escitalopram compared to citalopram: a meta-analysis. Int J Neuropsychopharmacol. 2011;14:261–8.
Marken PA, Munro JS. Selecting a selective serotonin reuptake inhibitor: clinically important distinguishing features. Prim Care Companion J Clin Psychiatry. 2000;2:205–10.
Martenyi F, Dossenbach M, Mraz K, Metcalfe S. Gender differences in the efficacy of fluoxetine and maprotiline in depressed patients: a double-blind trial of antidepressants with serotonergic or norepinephrinergic reuptake inhibition profile. Eur Neuropsychopharmacol. 2001;11:227–32.
Keers R, Aitchison KJ. Gender differences in antidepressant drug response. Int Rev Psychiatry. 2010;22:485–500.
Diflorio A, Jones I. Is sex important? Gender differences in bipolar disorder. Int Rev Psychiatry. 2010;22:437–52.
Estrada-Camarena E, Lopez-Rubalcava C, Vega-Rivera N, Recamier-Carballo S, Fernandez-Guasti A. Antidepressant effects of estrogens: a basic approximation. Behav Pharmacol. 2010;21:451–64.
Speranza L, Chambery A, Di Domenico M, Crispino M, Severino V, Volpicelli F, et al. The serotonin receptor 7 promotes neurite outgrowth via ERK and Cdk5 signaling pathways. Neuropharmacology. 2013;67:155–67.
Speranza L, Labus J, Volpicelli F, Guseva D, Lacivita E, Leopoldo M, et al. Serotonin 5-HT7 receptor increases the density of dendritic spines and facilitates synaptogenesis in forebrain neurons. J Neurochem. 2017;141:647–61.
de las Casas-Engel M, Dominguez-Soto A, Sierra-Filardi E, Bragado R, Nieto C, Puig-Kroger A, et al. Serotonin skews human macrophage polarization through HTR2B and HTR7. J Immunol. 2013;190:2301–10.
Strawbridge R, Arnone D, Danese A, Papadopoulos A, Herane Vives A, Cleare AJ. Inflammation and clinical response to treatment in depression: a meta-analysis. Eur Neuropsychopharmacol. 2015;25:1532–43.
Ciranna L, Catania MV. 5-HT7 receptors as modulators of neuronal excitability, synaptic transmission and plasticity: physiological role and possible implications in autism spectrum disorders. Front Cell Neurosci. 2014;8:250.
Renner U, Zeug A, Woehler A, Niebert M, Dityatev A, Dityateva G, et al. Heterodimerization of serotonin receptors 5-HT1A and 5-HT7 differentially regulates receptor signalling and trafficking. J Cell Sci. 2012;125(Pt 10):2486–99.
Naumenko VS, Popova NK, Lacivita E, Leopoldo M, Ponimaskin EG. Interplay between serotonin 5-HT1A and 5-HT7 receptors in depressive disorders. CNS Neurosci Ther. 2014;20:582–90.
Schulze TG, Alda M, Adli M, Akula N, Ardau R, Bui ET, et al. The International Consortium on Lithium Genetics (ConLiGen): an initiative by the NIMH and IGSLI to study the genetic basis of response to lithium treatment. Neuropsychobiology. 2010;62:72–8.
Salloum NC, McCarthy MJ, Leckband SG, Kelsoe JR. Towards the clinical implementation of pharmacogenetics in bipolar disorder. BMC Med. 2014;12:90.
Acknowledgements
This work was supported by grants to JRK from the NIMH (U01 MH92758) and the Department of Veterans Affairs and UCSD CTRI Pilot Project Grant (to MM and JRK). YBW was supported by the Swedish Research Council (Reg no. 2015–06372). HR was supported by the Alberta Centennial Addiction and Mental Health Research Chair held by KJA. GENDEP was funded by a European Commission Framework 6 grant (Contract Ref: LSHB-CT-2003503428). Lundbeck provided both nortriptyline and escitalopram free of charge. GlaxoSmithKline, the Medical Research Council and the Biomedical Research Centre for Mental Health at the Institute of Psychiatry, King’s College London and South London and Maudsley NHS Foundation Trust (funded by the National Institute for Health Research, Department of Health, UK) contributed by funding add-on projects in the London center. A joint grant from the Medical Research Council, UK, and GlaxoSmithKline (G0701420) provided additional funding for the array genotyping. The funders had no role in the design and conduct of the study, in data collection, analysis, interpretation or writing the report. MARS samples were supported by the German Federal Ministry of Education and Research (BMBF) through the Integrated Network IntegraMent (Integrated Understanding of Causes and Mechanisms in Mental Disorders), under the auspices of the e: Med Programme (grant 01ZX1314J to EBB).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
KJA has been a member of various advisory boards, received consultancy fees and honoraria, and received research grants from companies including Johnson and Johnson Pharmaceuticals Research and Development, Bristol-Myers Squibb Pharmaceuticals Limited, and Janssen Inc., Canada. MM serves as scientific consultant to Janssen Pharmaceuticals. EBB receives a research grant from Böhringer Ingelheim.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Wei, Y.B., McCarthy, M., Ren, H. et al. A functional variant in the serotonin receptor 7 gene (HTR7), rs7905446, is associated with good response to SSRIs in bipolar and unipolar depression. Mol Psychiatry 25, 1312–1322 (2020). https://doi.org/10.1038/s41380-019-0397-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41380-019-0397-1
- Springer Nature Limited
This article is cited by
-
Progress and Implications from Genetic Studies of Bipolar Disorder
Neuroscience Bulletin (2024)
-
Role of estrogen in sex differences in memory, emotion and neuropsychiatric disorders
Molecular Biology Reports (2024)
-
Regulatory T cells in ischemic stroke
Acta Pharmacologica Sinica (2022)
-
ERICH3: vesicular association and antidepressant treatment response
Molecular Psychiatry (2021)
-
Implication of 5-HT7 receptor in prefrontal circuit assembly and detrimental emotional effects of SSRIs during development
Neuropsychopharmacology (2020)