Abstract
Objective
A causal obesity risk variant in the FTO locus was recently shown to inhibit adipocyte thermogenesis via increased adipose expression of the homeobox transcription factors IRX3 and IRX5. However, causal effects of IRX5 on fat storage remain to be shown in vivo, and discovery of downstream mediators may open new therapeutic avenues.
Methods
17 WT and 13 Irx5 knockout (KO) mice were fed low-fat control (Ctr) or high-fat (HF) diet for 10 weeks. Body weight, energy intake and fat mass were measured. Irx5-dependent gene expression was explored by transcriptome analysis of epididymal white adipose tissue (eWAT), confirmatory obesity-dependent expression in human adipocytes in vivo, and in vitro knock-down, overexpression and transcriptional activation assays.
Results
Irx5 knock-out mice weighed less, had diminished fat mass, and were protected from diet-induced fat accumulation. Key adipose mitochondrial genes Pparγ coactivator 1-alpha (Pgc-1α) and uncoupling protein 1 (Ucp1) were upregulated, and a gene network centered on amyloid precursor protein (App) was downregulated in adipose tissue of knock-out mice and in isolated mouse adipocytes with stable Irx5 knock-down. An APP-centered network was also enriched in isolated adipocytes from obese compared to lean humans. IRX5 overexpression increased APP promoter activity and both IRX5 and APP inhibited transactivation of PGC-1α and UCP1. Knock-down of Irx5 or App increased mitochondrial respiration in adipocytes.
Conclusion
Irx5-KO mice were protected from obesity and this can partially be attributed to reduced adipose App and improved mitochondrial respiration. This novel Irx5-App pathway in adipose tissue is a possible therapeutic entry point against obesity.
Similar content being viewed by others
Introduction
Obesity results from excess energy storage relative to expenditure through an interplay between environmental and genetic factors. Twin studies have estimated that heritability explains 40–80% of individual variability in body weight [1] and genome-wide association studies (GWAS) have pointed to several obesity-linked single-nucleotide variants (SNVs). Among these, the FTO locus has the strongest known GWAS association with body-mass index (BMI, kg/m2) in humans [2] with independent associations also with type 2 diabetes [3]. We recently reported that a causal cis-regulatory variant in intron 1 of the FTO gene, rs1421085, affects a molecular switch that controls the bioenergetic fate of individual adipocytes in a cell-autonomous manner [4]. Specifically, a risk allele-dependent increase in expression of two developmental homeoproteins, IRX3 and IRX5, promoted fat storage by inhibiting adipocyte browning and inducible thermogenesis [4]. We had previously reported differential expression of these and other homeoproteins in adipose tissue after profound fat loss in morbidly obese bariatric patients [5].
Homeoproteins, of which IRX5 is a family member, are key transcription factors in embryonic and adult development that share a common conserved DNA-binding domain, the homeodomain (HD) [6]. Despite their importance, knowledge of homeoprotein target genes and mechanistic action is limited [7], although bi-allelic mutations in IRX5 have been shown to impair craniofacial morphogenesis and heart, blood, bone and germ cell development in humans [8]. In mice, Irx5 has been found to be important for retinal [9] and cardiac [10, 11] development and function through correct temporal and spatial suppression of respective target genes. Our recent demonstration of the fat storing role of IRX5 in vitro raises the question of whether negatively targeting this factor in a metabolic context can be of therapeutic benefit. Of note, no obesity-related phenotypes have yet been reported after genetic manipulation of Irx5 in vivo. Furthermore, due to the pleiotropic impact of Irx5, unraveling downstream mediators of Irx5 action in adipose tissue is important to identify potentially more suitable therapeutic targets against obesity than Irx5 itself. We here report a severe loss of body weight and adipose tissue mass in Irx5 knock-out mice, linked to adipose gene networks centered around amyloid precursor protein (App) and improved mitochondrial respiration.
Experimental procedures
Animals and diets
The animal study was approved by the Norwegian State Board of Biological Experiments with Living Animals, and carried out in accordance with their guidelines. Generation of the CD1 Irx5-deficient mice is descried elsewhere [9]. Male heterozygote Irx5-KO mice were bred to acquire homozygote KO and WT mice and housed together with up to three littermates at 20 ± 3 °C. From the age of 8–10 weeks, the mice were subjected to a 10 week feeding experiment in which both WT and KO mice were randomized to ad libitum control (n = 7 for WT and n = 5 for KO) or high-fat (n = 10 for WT and n = 8 for KO) diet. Based on the strong anti-obesity phenotype of Irx3 whole-body knockout mice [12] and adipose-specific Irx3 dominant negative mice [4], we anticipated that 5–7 mice per group on control feed and 8–10 mice per group on high-fat diet would suffice to yield statistically significant obesity phenotypes. A lab technician randomly selected genotyped WT and KO mice, respectively, for distribution in new cages for control or high-fat feeding. The investigators were not blinded during the experiment or when assessing the outcome.
The control diet (D12450B, Brogaarden ApS, Hørsholm, Denmark, 3.85 kcal/g) consisted of 10 kcal% fat and 70 kcal% carbohydrate, whereas the HF diet (D12451, Brogaarden ApS, 4.72 kcal/g) comprised of 45 kcal% fat and 35 kcal% carbohydrate, with both diets containing 20 kcal% protein. Body weight and feed intake was measured and renal white adipose tissue (rWAT) was quantified by magnetic resonance (MR) in live animals as previously described [13]. The mice were euthanized after the 10th week of the feeding experiment and epididymal white adipose tissue (eWAT) was dissected out and weighed before RNA extraction.
Patient samples and adipocyte isolation
The human study was approved by the Western Norway Regional Committee for Medical Research Ethics (REK) (Norway), and each of the subjects gave written informed consent. Subcutaneous adipose tissue was collected from severely obese patients undergoing bariatric surgery (average BMI of 45.5 kg/h2, n = 12, thereof 4 men) and healthy lean patients undergoing hernia repairs (average BMI of 24.2 kg/h2, n = 12, thereof 7 men) between 27 and 56 years of age. Adipocytes were liberated from the adipose tissue by collagenase treatment and filtering before RNA isolation.
Cell cultures
Primary white preadipocyte cells were obtained by collagenase digestion of WAT from C57BL/6 mice and immortalization by the SV40 virus. The cells were grown in DMEM containing L-glutamine and 4.5 g/L glucose (Lonza, Basel, Switzerland), supplemented with 10% calf serum (CS) (GE healthcare, Little Chalfont, Great Britain) and 1% penicillin-streptomycin (Sigma, St. Louis, MO, USA). Stable knock-down (KD) of Irx5 in the WAT cells was performed by CRISPR-Cas9 in line with [14]. Briefly, cells were transfected with a single plasmid containing both guide RNA (TTATGGGGACCCCGCGTACCGG) and Cas9 fused to GFP (Sigma), using the TransIT-LT1 transfection reagent (Mirus Bio LLC, Madison, WI, USA). A Crispr construct without guide RNA (Sigma) was used as negative control. The day after transfection, cells were trypsinated, sorted by fluorescent activated cell sorting (FACS) on the Sony SH800 flow cytometer (Sony, Minato, Tokyo, Japan) and single live cells positive for GFP were automatedly seeded in each well of 96-well plates. Cells were incubated in proliferation medium supplemented with 0.01 µg/µL FGF (Sigma) and 0.001 µg/µL EGF (Sigma) for one week. During this period, growing colonies were trypsinated and re-seeded in 96-well plates to keep cells sub-confluent. After one week, the medium was changed to regular proliferation medium without supplemented growth factors and colonies were expanded and DNA-sequenced. Heterozygous knock-out of Irx5 was confirmed by Sanger Sequencing using the BigDye 3.1 protocol (Thermo Fisher Scientific) according to manufacturer’s instructions. The CRISPR-edited WAT cells were employed for gene expression and oxygen consumption rate analyses. COS-1 monkey kidney cells (ATCC, Manassas, Virginia, USA) were grown in DMEM containing 1 g/L glucose (Lonza) supplemented with 10% FBS, 1% penicillin-streptomycin and 2 mM l-glutamine. These cells were used for transactivation assays. Mouse embryonic fibroblast Rb−/− Line 3 (ME3) cells, a model of beige pre-adipocytes [15,16,17], were grown in AmnioMAX -C100 (Thermo Fisher Scientific, Waltham, Massachusetts, USA) supplemented with 7.5% FBS, 7.5% C100 (Thermo Fisher Scientific), 1% penicillin-streptomycin and 2 mM l-glutamine. These cells were used for transactivation assays. Cells were tested for mycoplasma.
Transactivation assays
COS-1 and ME3 cells were grown for 24 h prior to transient co-transfection of indicated overexpression plasmids (40 ng each) with the firefly luciferase reporter (50 ng) under control of the relevant promoter sequences. The total amount of DNA per well was adjusted to 130 ng with empty plasmid. The reporter constructs employed were pGL4-phAPP-luc (RIKEN cat.no RDB07692, −1447/+110 of APP_1 promoter), pGL3-hUCP1–3.1kb-luc [18] and pGL3-mPgc-1α-luc (Addgene plasmid #8887) [19]. The following overexpression plasmids were utilized: pCMV6-hIRX5 (Origene, Rockville, MD, USA), pCDNA-mPgc-1a (Origene), pCMV6-mApp-myk-ddk (Origene) and pCDNA-empty. ME3 and COS-1 cells were transfected using the TransIT-LT1 (Mirus Bio LLC) and Superfect (Qiagen) reagents, respectively, according to manufacturers’ instructions. Cells were lysed 48 h after transfection with 25 mM Tris-Acetate-EDTA (pH 7.8), 2 mM dithiothreitol (DTT), 1 mM EDTA, 10% glycerol and 1% Triton X-100 and analyzed with the BioThema luciferase assay (BioThema, Handen, Sweden) on a FLUOStar Optima (BMG Labtech, Ortenberg, Germany) luminescence plate reader.
RNA isolation, cDNA synthesis, and Real-time qPCR analysis
RNA isolation of tissue and isolated adipocytes was performed by lysis in Qiazol (Qiagen, Hilden, Germany) and homogenization in the TissueLyser (Qiagen) followed by chloroform extraction according to manufacturer’s instructions. RNA isolation from cell cultures was performed using the RNeasy Mini kit (Qiagen) on the QIAcube instrument (Qiagen) according to manufacturer’s instructions. RNA integrity was verified using the Agilent RNA 6000 Nano kit on the Agilent 2100 BioAnalyzer instrument (Agilent, Santa Clara, USA). cDNA was synthesized using 500 ng RNA input with the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Waltham, MA, USA) and diluted 1:10 in PCR-grade water (Roche, Basel, Switzerland) before quantification. Real-time qPCR was performed in the LightCycler 480 system (Roche) using 4 pmol of each primer and 1.25 μL cDNA template with 5 μL SYBR I Green in a total volume of 10 μL. Quantification of target genes was calculated using the delta-delta Ct method relative to reference gene Rps13. Primers were designed using either the Universal ProbeLibrary Assay Design Center (Roche) or the Primer-BLAST [20] softwares. Irx5, forward 5′-CGAGGAGGAGGAGGAGAACAT-3′ and reverse 5′-CCTTAAAATCCGAGTCGCTGAG-3′; Pgc-1a, forward 5′-AATTTTTCAAGTCTAACTATGCAGACC-3′ and reverse 5′-AAAATCCAGAGAGTCATACTTGCTC-3′; App, forward 5′-AGGACTGACCACTCGACCAG-3′ and reverse 5′-CTTCCGAGATCTCTTCCGTCT-3′; Ucp1, forward 5′-GGGCATTCAGAGGCAAATCAG-3′ and reverse 5′-TTTCCGAGAGAGGCAGGTGTTT-3′.
Microarray gene expression analyses
Epididymal white adipose tissue (eWAT) from the mice was analyzed with Illumina iScan and MouseWG-6 v2.0 Expression BeadChips (6 WT and 6 Irx5-KO mice), and the isolated human adipocytes were analyzed with the Illumina iScan and HumanHT-12 v.3 BeadChip. Raw data files were imported into J-Express. Before further analysis, missing values were replaced by the LSimpute_adaptive method [21] and signal intensity values were quantile normalized [22] and log transformed (base 2). The mouse and human datasets were combined based on ENSEMBL gene IDs. The genes identified in both datasets were then sorted based on similar or divergent expression in obesity. Ingenuity Pathway Analysis (IPA) was used to identify globally predominant gene networks (accessed on March 21, 2016).
Oxygen consumption rate
Cellular oxygen consumption rate (OCR) was measured using the Seahorse XF Cell Mito Stress Test kit (Agilent, Santa Clara, USA) and the Seahorse XFe96 Analyzer (Agilent) with 12 replicates. ME3 cells were transfected the day after seeding and left to grow for 48 h in 1 μM isoproterenol before measurement of OCR. Wild-type WAT and Crispr-WAT cells were induced to differentiate two days post confluence (day 0) by addition of fresh culture media containing CS instead of FBS, and supplemented with 172 nM insulin, 0.5 mM IBMX, 1 µM dexamethasone and 1 µM rosiglitazone (all components from Sigma). Cells were transfected on day 0 with either 25 nM siRNA (Dharmacon, Lafayette, USA) using Hiperfect (Qiagen), or 40 ng expression plasmid encoding either Irx5 or App as indicated. Cells were either assayed on day 2, or further cultured to day 7 by addition of fresh medium supplemented with insulin only for two more days and culture medium only for the last three days before assaying. 1 µM isoproterenol was added 48 h before measuring OCR.
On the day of assay, culture media was replaced with DMEM D5030 (Agilent) supplemented with 2 mM l-glut (Sigma), 2 mM sodium pyruvate (Sigma) and 10 mM glucose (Sigma) and cells were incubated at 37 °C in an incubator without CO2 infusion for 1–2 h before running the OCR assay. After establishing baseline OCR, the following inhibitors of mitochondrial respiration were added in successive order: oligomycin (3 µM) to block ATP production, carbonyl cyanide m-chlorophenylhydrazone (CCCP, 1.5 µM) to uncouple oxygen consumption from ATP production and rotenone (1 µM) with antimycin (1 µM) to inhibit complexes I and III (all inhibitors from Sigma). Three measurement cycles were performed between the addition of each inhibitor.
Statistical analyses
Analysis of significance was performed by unpaired Student’s t-test, one-way ANOVA, two-way ANOVA and repeated measures two-way ANOVA with Tukey or Sidak’s post hoc test as indicated. Data are shown as either mean ± standard deviation or boxplots with Tukey whiskers (1.5 times interquartile range) with outliers shown as filled circles, as indicated. Data were tested for normality using Shapiro-Wilk test and for homogeneity of variances using Lavene’s test. Outliers, as detected by the ROUT test (Q = 1%) were plotted, but omitted from calculations of statistical significance.
Data deposition
Microarray data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-6726 (Irx5-KO mice) and E-MTAB-6728 (Human adipocytes).
Results
Diminished fat mass in Irx5 −/− mice
To determine the effect of Irx5-KO on body weight and fat mass, we performed a 10-week feeding experiment in adult mice, where wild-type (WT) and KO mice were randomized to either a low-fat control (Ctr) or a high-fat (HF) diet (Fig. 1a). One week before intervention, the Irx5-KO mice weighed on average 42% less (p < 0.001) than WT littermates (Figure S1A). The significant genotype-dependent difference in body weight persisted throughout the experiment regardless of feed (Fig. 1a). Furthermore, whereas the WT mice weighed significantly (p < 0.05) more after 10 weeks on HF compared to the control diet, the Irx5-KO mice were completely protected from weight gain induced by the HF diet. Despite the great differences in body weight between WT and KO mice, no significant difference in absolute or relative energy intake was observed (Figure S1B). Quantification of the renal and epididymal white adipose tissue depots (rWAT and eWAT, respectively) revealed that Irx5-KO mice on HF diet possessed on average 47% less rWAT (p < 0.001) and 79% less eWAT (p < 0.001) compared to WT littermates (Fig. 1b-d). Moreover, whereas the eWAT from WT mice fed a HF diet weighed roughly 2-fold that of mice given the control diet (p < 0.01), no difference in eWAT mass was observed in the Irx5 KO on the two diets. Taken together, mice devoid of Irx5 have a profound anti-obesity phenotype.
Irx5 knock-out mice are protected against diet-induced obesity. a WT and Irx5-KO mice on Ctr diet (n = 7 for WT and n = 5 for KO) or HF (n = 10 for WT and n = 8 for KO) diet for 10 weeks. Mean body weight ± SD shown. Statistical analysis by two-way repeated measures ANOVA with Sidak’s post hoc test. *p < 0.05 between Ctr and HF diet at week 10; n.s, not significant; †p < 0.05, ††p < 0.01, †††p < 0.001 between WT and KO group on control diet; §§§p < 0.001 between WT and KO group on HF diet. b Magnetic resonance imaging (MRI)-quantification of renal white adipose tissue (rWAT) depots (n = 22). Percent rWAT of the quantified area of the mice shown. Data displayed as Boxplots with Tukey whiskers. Statistical analysis by regular two-way ANOVA with Sidak’s post hoc test. *p < 0.05, ***p < 0.001. c Weight of epididymal white adipose tissue (eWAT) as % of total body weight. Statistical analysis by regular two-way ANOVA with Sidak’s post hoc test. **p < 0.01, ***p < 0.001. d Representative MRI images of rWAT in WT and Irx5-KO mice fed a control or HF-diet, transverse (top) and coronal view (bottom). See also Figure S1
Adipose gene networks suppressed via Irx5 center around App
To elucidate mechanisms underlying the diminished lipid accumulation of the Irx5-KO mice, we systematically searched for the most affected genes and gene networks in eWAT of Irx5-KO compared to WT mice fed a HF diet. Through genome-wide expression analysis we identified 438 down-regulated and 238 up-regulated genes in the Irx5-KO mice compared to the WT mice (fold change > 1.5 and q-value < 0.25) (Fig. 2a). When interrogating the differentially expressed genes with Ingenuity Pathway Analysis, the top-scoring network for the 238 upregulated genes in the Irx5-KO mice centered around the down-regulated gene amyloid precursor protein (App) and included genes important for mitochondrial biogenesis and function, i.e., Pgc-1α and Ucp1 (Fig. 2b). These microarray results were validated by qPCR (Fig. 2c). To investigate whether Pgc-1α and Ucp1 levels could be altered by Irx5 manipulation in a cell-autonomous manner in adipose tissue, we generated heterozygous Irx5-KO (overall 50% knock-down) mouse primary white adipocytes by CRISPR-Cas9, and measured the respective mRNA levels by qPCR (Fig. 2d). Cells were treated with and without 1 μM isoproterenol (iso), a β-adrenergic agonist that enhances transcription of Ucp1 and stimulates lipolysis which activates Ucp1-mediated thermogenesis. The Pgc-1α expression was approximately 12-fold higher (p < 0.001) in Irx5 knock-down (KD) cells compared to controls, regardless of iso treatment. Ucp1 could not be detected in the unstimulated control cells, but was expressed in the Irx5 KD-cells (p = 0.0131), as well as in both cell lines stimulated with iso. In the stimulated state, however, no significant difference in Ucp1 expression between control and KD cell lines was observed (Fig. 2d). Taken together, these data suggest a suppressive role of Irx5 on transcription of Pgc-1α and Ucp1.
Irx5 knock-out mice show increased adipose expression of genes involved in energy metabolism. a WT (n = 6) and Irx5-KO (n = 6) mice fed HF diet for 10 weeks and global gene expression in epididymal white adipose tissue (eWAT) measured by microarrays. Fold change > 1.5 and q-value cut-off < 0.25. b Upregulated genes centered around the down-regulated amyloid precursor protein (App) gene. The color scale indicates fold change in Irx5-KO mice gene expression compared to WT, with reduction shown in red and increase in green. Dotted lines, gene co-expression in previous studies; solid lines, gene-protein or protein-protein interactions. c Pgc-1α and Ucp1 expression relative to Rps13 in the same eWAT dissected from the mice, measured by RT-qPCR. Boxplots with Tukey shown, black dots denote outliers and these were omitted from calculations of statistical significance. Statistical analysis by two-way ANOVA with Sidak’s post hoc test. *p < 0,05. d Pgc-1α and Ucp1 expression relative to Rps13 in response to stable Irx5 knock-down in primary white adipocyte cells isolated from WT C57BL/6 mice. Iso: 1 µM isoproterenol for 24 h. Statistical analyses by two-way ANOVA with Sidak’s post hoc test. *p < 0.05, **p < 0.01,***p < 0.001. e, f Luciferase reporter under control of the mouse Pgc-1α (e) or the human UCP1 promoter (f), with and without overexpression of Irx5 and/or Pgc-1α. Data are presented as mean ± SD. Statistical analyses by one-way ANOVA with Tukey’s post hoc test. **p < 0.01; ***p < 0.001. Representative of three independent experiments
To further investigate whether Irx5 inhibits the Pgc-1α and Ucp1 expression, we overexpressed Irx5 in COS-1 kidney and ME3 preadipocyte cells together with respective luciferase reporter constructs under control of either the mouse Pgc-1α promoter (Fig. 2e) or the human UCP1 promoter (Fig. 2f). Since Pgc-1α can coactivate the expression of itself as well as Ucp1, we performed the luciferase assays with and without overexpression of Pgc-1α. In both cell lines, Irx5 significantly suppressed basal Pgc-1α reporter activity and in COS-1 cells abolished any stimulatory effect of Pgc-1α on activating its own promoter (Fig. 2e). Of note, Irx5 did not affect the basal activity of the UCP1 reporter, but strongly suppressed Pgc-1α-mediated activation of UCP1 (Fig. 2f).
Adipose gene networks dependent on Irx5 also center around App
Amyloid precursor protein (App) was also the center of the top scoring network of the 438 downregulated genes in eWAT of the Irx5-KO mice (Fig. 3a). qPCR confirmed absence of Irx5 in the KO mice (Fig. 3b) and significantly lower App expression levels (p = 0.0012) in Irx5-KO mice compared to WT littermates after overfeeding (Fig. 3b). Furthermore, App levels were significantly (p = 0.0466) higher in WT mice fed HF diet compared to control diet, and this effect was blunted in KO mice. To investigate whether Irx5 manipulation could alter App levels in a cell-autonomous manner in adipose tissue, we analyzed the primary adipocytes with stable Irx5 knock-down. Indeed, knock-down cells showed 60% reduction (p < 0.001) in App levels and this effect was independent of iso treatment (Fig. 3c). Conversely, overexpression of Irx5 increased App expression in ME3 preadipocytes (Fig. 3d), and in transactivation assays we moreover found that Irx5 overexpression strongly induced the App promoter activity in COS-1 and ME3 cells (p = 0.0016 and p = 0.0006 respectively) (Fig. 3e). Taken together, these experiments demonstrate that Irx5 promotes expression of App.
Downregulation of an APP-related gene network in adipocytes from Irx5-KO mice. WT (n = 6) and Irx5-KO (n = 6) mice fed HF diet for 10 weeks and global gene expression in eWAT measured by microarrays. Fold change > 1.5 and q-value cut-off < 0.25. a Downregulated genes centered around the amyloid precursor protein (App) gene. Dotted lines: gene co-expression in previous studies; solid lines: gene–protein or protein–protein interactions. b Irx5 and App expression relative to the housekeeping gene Rps13 in eWAT measured by RT-qPCR. Boxplots with Tukey shown. n.d: not detectable. Statistical analyses by two-way ANOVA with Sidak’s post hoc test. *p < 0,05, **p < 0.01,***p < 0.001. c App expression relative to Rps13 in response to stable Irx5 knock-down in primary white adipocyte cells isolated from WT C57BL/6 mice. Iso: 1 µM isoproterenol. Statistical analyses by two-way ANOVA with Sidak’s post hoc test. ***p < 0.001. d App expression relative to Rps13 in response to over-expression of Irx5 in ME3 preadipocytes. Statistical analysis by student’s t-test. *p < 0.05. e Luciferase reporter under control of human amyloid precursor protein (APP) promoter with and without overexpression of Irx5. Data are presented as mean ± SD. Statistical analyses by Student’s t-test. **p < 0.01,***p < 0.001. Representative of three independent experiments
Validation of obesity-linked genes in human adipocytes
To test the relevance of the identified Irx5-dependent genes for human obesity, we searched for consistently regulated genes in mature adipocytes isolated from subcutaneous white adipose tissue of lean and obese patients. Consistent with conserved obesity-related gene networks, we found a highly significant enrichment of Irx5-dependent mouse eWAT genes among the most differentially expressed genes in human adipocytes (Fig. 4a), thereof 18 consistently upregulated and 84 downregulated genes in both the lean Irx5-KD mice and lean human patients (Fig. 4b) (q-value < 0.05 in both datasets, genes with fold difference > 1.5 are shown in Table S1). Submitting the 84 genes with consistently reduced expression in lean mice and people (i.e., upregulation in obesity) to Ingenuity Pathway Analysis, again the top-scoring network centered on APP (Fig. 4c).
Validation of Irx5 knockout-dependent obesity genes in human adipocytes. a Gene Set Enrichment Analysis (GSEA) of differentially expressed genes (based on Significance Analysis of Microarrays), showing a significant enrichment of Irx5-dependent genes identified in mice adipose tissue among the most differentially expressed genes in adipocytes from obese compared to lean humans. Only genes showing the same direction of regulation in the mouse and human datasets were included, leaving 84 downregulated and 18 upregulated genes in lean subjects within the q-value < 0.05 significance cut-off used for both datasets. b Heatmap showing relative gene expression (Illumina microarray signal intensities) of these genes, eWAT from wild-type and Irx5 knock-out mice fed obesogenic diet for 10 weeks (left) and mature adipocytes isolated from subcutaneous adipose tissue of obese and lean humans (average body-mass index (BMI) of 45.5 and 24.2 kg/h2, respectively) (right). eWAT, epididymal white adipose tissue; FDR, false discovery rate. c Top-scoring networks for Irx5-dependent genes with consistent upregulation in human obesity. Ingenuity pathway analysis was performed for 84 genes with a consistent increase in adipose expression in obesity in mouse adipose tissue and human adipocytes (Significance analysis of microarrays q-value < 0.05, shown in Fig. 4). The color scale indicates the fold differential expression in mature human adipocytes from lean and obese people (n = 10 per group). Dotted lines indicate gene co-expression in previous studies, and solid lines indicate gene–protein or protein–protein interactions
Repression of App increases mitochondrial respiration
Given the inhibitory effect of Irx5 on key mitochondrial genes and the central position of App in the network of these genes, we sought to investigate the functional outcome of reduced Irx5 and App expression levels on adipocyte mitochondrial function. In primary white adipocytes, transient knock-down of App early in differentiation significantly increased OCR (p < 0.001) in fully differentiated white adipocytes in response to CCCP (Fig. 5a), with significant increases (p < 0.001) in maximal respiration and spare capacity (Fig. 5b). In the beige-like ME3 cells, the effect of siRNA against App on maximal respiration and spare capacity was borderline significant (Figure S2) whereas transient knock-down of Irx5 resulted in significant increase in maximal respiration (p = 0.0011) and spare capacity p < 0.001) (Figure S2).
Irx5 and App repress mitochondrial respiration. Real-time whole-cell oxygen consumption rate (OCR). Data represented as mean ± SD of 12 wells per treatment, adjusted for cell number. Representative of two independent experiments. a White preadipocytes were treated with 25 nM siRNA against App on day 0 of differentiation and OCR was measured on day 7. Statistical analysis by two-way repeated measures ANOVA with Sidak’s post hoc test. ***p < 0.001. b Calculated basal- and maximal respiration, spare capacity, ATP production and proton leak. Statistical analysis by multiple t-tests with FDR correction, Q = 1%. ***p < 0.001. c Over-expression of mouse App in white preadipocytes with or without stable knock-down of Irx5 on day 0 of differentiation, with OCR measured on day 2. Statistical analysis by two-way repeated measures ANOVA with Tukey’s post hoc test. ***p < 0.001, Ctr vs. Irx5-KD; ††p < 0.01, †††p < 0.001, empty vs. over-expression (oex) of App in control cells; ‡‡‡p < 0.001, empty vs oex of App in KD-Irx5 cells; §§p < 0.001, §§§p < 0.001, Ctr oex-App vs KD-Irx5 empty. d Calculated basal- and maximal respiration, spare capacity, ATP production and proton leak. Statistical analysis by one-way ANOVA with Tukey’s post hoc test. *p < 0.05, **p < 0.01, ***p < 0.001. Oligo: oligomycin; am + rot: antimycin and rotenone
We next measured OCR in the primary adipocytes with stable CRISPR-Cas9-mediated Irx5 knock-down, and assessed whether overexpression of App could counteract the phenotype of the knock-down cells. As hypothesized, stable knock-down of Irx5 significantly elevated OCR in response to CCCP (p < 0.001), and this effect was partially reversed by overexpression of App (p < 0.001) (Fig. 5c). Both basal and maximal respiration as well as spare capacity, ATP production and proton leak were significantly elevated in the Irx5-KD cells compared to controls, and overexpression of App partially reversed the effect of Irx5-knock down on all measures except proton leak (Fig. 5d).
App inhibits transactivation of Pgc-1α and Ucp1
To determine whether App could affect the transactivation of Pgc-1α or Ucp1, we overexpressed App together with the same luciferase constructs as before, containing the promoter of either Pgc-1α or Ucp1 (Fig. 6). Consistent with the effects of overexpressed Irx5, App strongly suppressed both promoters (p < 0.001).
App represses transactivation of Pgc-1α and Ucp1 Luciferase reporter under control of the mouse Pgc-1α (a) or the human UCP1 promoter (b), with and without overexpression of App. Representative of three independent experiments in each of the two cell lines COS-1 and ME3. Data are presented as mean ± SD. Statistical analyses by Student’s t-test. *p < 0.05, ***p < 0.001
Discussion
We and others previously showed that a risk-variant in the FTO obesity locus increases mRNA expression of both IRX3 and IRX5 through a common enhancer in adipocytes, and that adipose-selective Irx3-KO in mice prevents diet-induced obesity [4]. The present study reports effects of global Irx5-KO on body weight, fat storage and adipose gene expression in mice.
The Irx5 knock-out mice exhibit a profound anti-obesity phenotype with a dramatic loss of fat mass. Genes vital to mitochondrial biogenesis and thermogenesis (e.g., Pgc-1α, Ucp1) were upregulated in the eWAT of KO mice in vivo and in isolated adipocytes with transient or stable knock-down of Irx5 in vitro. Moreover, our transactivation data demonstrated that Irx5 suppresses these genes by inhibiting transcription of Pgc-1α as well as by impairing the coactivator function of the Pgc-1α protein on the Ucp1 promoter. Stable knock-down of Irx5 in isolated white adipocytes elevated oxygen consumption rate and proton uncoupling, in agreement with our previous study [4]. Taken together, these data point to a potential for increased thermogenic capacity in the Irx5-KO mice. However, because thermogenesis was not measured in vivo in this study, the overall contribution of thermogenesis to the observed anti-obesity phenotype in the Irx5-KO mice was not determined.
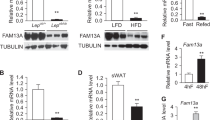
In search of other factors that could further contribute to the lean Irx5-KO phenotype, our attention turned to the downregulated amyloid precursor protein (App) gene that centered the top-scoring networks of both the downregulated and upregulated genes in eWAT of the KO mice and also in the differentially expressed genes between obese and lean human adipocytes. Intriguingly, App-KO mice were recently shown to be resistant to diet-induced obesity, displaying reduced body weight, adipocyte size and visceral fat content [23] comparable to our Irx5-KO mice with reduced App-levels. Moreover, multiple studies have associated APP or its cleavage products with obesity or adipose tissue function [24,25,26,27,28,29,30]. These data point to a central role for adipose App in obesity development. Because insulin has anti-inflammatory effects [31] and has been reported to suppress APP and other Alzheimer’s disease related genes in peripheral mononuclear blood cells [32], it is conceivable that insulin resistance in obese individuals may lead to elevated App expression in adipocytes. However, our in vitro data clearly show a causal dependency of App expression on Irx5 in adipocytes. The reduced adipose App levels in Irx5-KO versus WT mice therefore likely primarily result from reduced Irx5 levels rather than higher insulin sensitivity. Taken together, these data support the anti-obesity effect observed in Irx5-KO mice to be at least partially mediated through reduced App levels.
Exactly how App may contribute to obesity is not clear, although it is well established that App or its cleavage products impair mitochondrial function in neurons through a number of different mechanisms [33,34,35,36]. Our data show that App has negative effects on mitochondria also in adipocytes, i.e., a strong repression of Pgc-1α by App in luciferase assays and the improved oxygen consumption rate and respiratory capacity of adipocytes following siRNA-mediated knock-down of App. Of note, several different mouse models have revealed reduced respiratory capacity in white adipose tissue as a characteristic of obesity [37].
Based on the whole-body knock-out model we cannot completely rule out the possibility that loss of Irx5 exerted anti-obesity effects via organs other than adipose tissue, such as the brain which shows a notable expression of Irx5. IRX5 expression may also partly depend on additional regulatory elements and mechanisms unrelated to the identified causal FTO obesity variant rs1421085 [4], and it remains to be determined if altered IRX5 expression can influence fat storage via additional cell types. However, the profound protection against obesity we have observed in Irx5 knock-out mice, as in adipose-selective aP2-Irx3 dominant negative mice [4], may likely in large part be ascribed to Irx5 action in the adipose tissue itself. Firstly, the highly conserved cis-regulatory module (CRM) found to regulate IRX5 expression in the FTO obesity locus showed potential for gene regulation particularly in the adipocyte lineage [4]. Secondly, eQTL analysis has suggested that variants in the FTO locus explain only around 1% of individual differences in IRX5 in the human brain tissue, while they explain around 200% in isolated primary human adipose cells [4]. Thirdly, the present study confirms a causal effect of Irx5 on expression of App, Pgc-1α and Ucp1 as well as mitochondrial respiration in isolated primary adipocytes with Irx5-KD.
In conclusion, our study is the first to reveal how Irx5 impacts energy metabolism and fat mass in vivo. We identified a profound loss of body weight and fat mass in Irx5-KO mice with increased adipose expression of thermogenic regulators Pgc-1a and Ucp1 and reduced expression of App. Our in vitro data demonstrate an Irx5-dependent transcriptional regulation of these genes, and an Irx5-mediated and App-mediated repression of adipocyte mitochondrial respiration. Collectively, these findings unravel an Irx5-App pathway in adipose tissue as a possible therapeutic avenue to mitigate obesity.
References
Allison DB, Kaprio J, Korkeila M, Koskenvuo M, Neale MC, Hayakawa K. The heritability of body mass index among an international sample of monozygotic twins reared apart. Int J Obes Relat Metab Disord. 1996;20:501–6.
Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–48.
Hertel JK, Johansson S, Sonestedt E, Jonsson A, Lie RT, Platou CGP, et al. FTO, type 2 diabetes, and weight gain throughout adult life: a meta-analysis of 41,504 subjects from the scandinavian HUNT, MDC, and MPP studies. Diabetes. 2011;60:1637–44.
Claussnitzer M, Dankel SN, Kim K-H, Quon G, Meuleman W, Haugen C, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373:895–907.
Dankel SN, Fadnes DJ, Stavrum AK, Stansberg C, Holdhus R, Hoang T, et al. Switch from stress response to homeobox transcription factors in adipose tissue after profound fat loss. PLoS ONE 2010;5.
Bürglin TR, Affolter M. Homeodomain proteins: an update. Chromosoma. 2016;125:497–521.
Hueber SD, Lohmann I. Shaping segments: Hox gene function in the genomic age. Bioessays. 2008;30:965–79.
Bonnard C, Strobl AC, Shboul M, Lee H, Merriman B, Nelson SF, et al. Mutations in IRX5 impair craniofacial development and germ cell migration via SDF1. Nat Genet. 2012;44:709–13.
Cheng CW, Chow RL, Lebel M, Sakuma R, Cheung HOL, Thanabalasingham V, et al. The Iroquois homeobox gene, Irx5, is required for retinal cone bipolar cell development. Dev Biol. 2005;287:48–60.
Costantini DL, Arruda EP, Agarwal P, Kim KH, Zhu Y, Zhu W, et al. The homeodomain transcription factor Irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell. 2005;123:347–58.
Gaborit N, Sakuma R, Wylie JN, Kim K-H, Zhang S-S, Hui C-C, et al. Cooperative and antagonistic roles for Irx3 and Irx5 in cardiac morphogenesis and postnatal physiology. Development. 2012;139:4007–19.
Smemo S, Tena JJ, Kim K-H, Gamazon ER, Sakabe NJ, Gómez-Marín C, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–5.
Thomas EL, Saeed N, Hajnal JV, Brynes A, Goldstone AP, Frost G. et al. Magnetic resonance imaging of total body fat. J Appl Physiol. 1998;85:1778–85.
Lonowski LA, Narimatsu Y, Riaz A, Delay CE, Yang Z, Niola F, et al. Genome editing using FACS enrichment of nuclease-expressing cells and indel detection by amplicon analysis. Nat Protoc. 2017;12:581.
Lukas J, Bartkova J, Rohde M, Strauss M, Bartek J. Cyclin D1 is dispensable for G1 control in retinoblastoma gene-deficient cells independently of cdk4 activity. Mol Cell Biol. 1995;15:2600–11.
Hansen JB, Jørgensen C, Petersen RK, Hallenborg P, De Matteis R, Bøye HA, et al. Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proc Natl Acad Sci USA. 2004;101:4112–7.
Hakim-Weber R, Krogsdam A-M, Jørgensen C, Fischer M, Prokesch A, Bogner-Strauss JG, et al. Transcriptional regulatory program in wild-type and retinoblastoma gene-deficient mouse embryonic fibroblasts during adipocyte differentiation. BMC Res Notes. 2011;4:157.
Rim JS, Kozak LP. Regulatory motifs for CREB-binding protein and Nfe2l2 transcription factors in the upstream enhancer of the mitochondrial uncoupling protein 1 gene. J Biol Chem. 2002;277:34589–600.
Handschin CC, Rhee JJ, Lin JJ, PTPT Tarr, BMBM. Spiegelman. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Pnas. 2003;100:7111–6.
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012;13:134.
Bø TH, Dysvik B, Jonassen I. LSimpute: accurate estimation of missing values in microarray data with least squares methods. Nucleic Acids Res. 2004;32:e34–e34.
Bolstad BM, Irizarry Ra, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93.
Puig KL, Brose SA, Zhou X, Sens MA, Combs GF, Jensen MD, et al. Amyloid precursor protein modulates macrophage phenotype and diet-dependent weight gain. Sci Rep. 2017;7:43725.
Balakrishnan K, Verdile G, Mehta PD, Beilby J, Nolan D, Galvão DA, et al. Plasma Aβ42 correlates positively with increased body fat in healthy individuals. J Alzheimers Dis. 2005;8:269–82.
Lee Y-H, Tharp WG, Maple RL, Nair S, Permana PA, Pratley RE. Amyloid precursor protein expression is upregulated in adipocytes in obesity. Obesity. 2008;16:1493–500.
Ghanim H, Monte SV, Sia CL, Abuaysheh S, Green K, Caruana Ja, et al. Reduction in inflammation and the expression of amyloid precursor protein and other proteins related to Alzheimer’s disease following gastric bypass surgery. J Clin Endocrinol Metab. 2012;97:E1197–201.
Lee Y-H, Martin JM, Maple RL, Tharp WG, Pratley RE. Plasma amyloid-beta peptide levels correlate with adipocyte amyloid precursor protein gene expression in obese individuals. Neuroendocrinology. 2009;90:383–90.
Puig KL, Floden AM, Adhikari R, Golovko MY, Combs CK. Amyloid precursor protein and proinflammatory changes are regulated in brain and adipose tissue in a murine model of high fat diet-induced obesity. PLoS ONE. 2012;7.
Freeman LR, Zhang L, Dasuri K, Fernandez-Kim SO, Bruce-Keller AJ, Keller JN. Mutant amyloid precursor protein differentially alters adipose biology under obesogenic and non-obesogenic conditions. PLoS ONE. 2012;7.
Wang J, Gu BJ, Masters CL, Wang YJ. A systemic view of Alzheimer disease—insights from amyloid-β metabolism beyond the brain. Nat Rev Neurol. 2017;13:612–23.
Sun Q, Li J, Gao F. New insights into insulin: the anti-inflammatory effect and its clinical relevance. World J Diabetes. 2014;5:89–96.
Dandona P, Mohamed I, Ghanim H, Sia CL, Dhindsa S, Dandona S, et al. Insulin suppresses the expression of amyloid precursor protein, presenilins, and glycogen synthase kinase-3beta in peripheral blood mononuclear cells. J Clin Endocrinol Metab. 2011;96:1783–8.
Mattson MP. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430:631–9.
Anandatheerthavarada HK, Devi L. Amyloid precursor protein and mitochondrial dysfunction in Alzheimer’s disease. Neurosci. 2007;13:626–38.
Chen JX, Yan SDu. Amyloid-beta-induced mitochondrial dysfunction. J Alzheimers Dis. 2007;12:177–84.
Shi C, Zhu XM, Wang J, Long D. Intromitochondrial IκB/NF-κB signaling pathway is involved in amyloid β peptide-induced mitochondrial dysfunction. J Bioenerg Biomembr. 2014;46:371–6.
Schöttl T, Kappler L, Fromme T, Klingenspor M. Limited OXPHOS capacity in white adipocytes is a hallmark of obesity in laboratory mice irrespective of the glucose tolerance status. Mol Metab. 2015;4:631–42.
Acknowledgements
We thank Carol Cook, Margit Solsvik, Rita Holdhus, Alba Kaci, Regine Åsen Jersin, André Madsen, Mona Nilsen and Tone Flølo for excellent technical assistance. We thank the patients and hospital staff at Voss and Haraldsplass Hospitals for the adipose tissue biopsies. The Irx5-KO mice were a gift from Drs. Kyoung-Han Kim and Chi-chung Hui. The pGL4-phAPP-luc reporter construct was provided by the RIKEN BRC through the National Bio-Resource Project of the MEXT, Japan. The mPgc-1α-luc construct was a gift from Dr. Bruce Spiegelman and the pGL3-UCP1-luc construct was a gift from Dr. Leslie P. Kozak. The ME3 cells were a gift from Dr. Karsten Kristiansen.
Author contributions
SD and GM conceived the study and designed the experiments. JIB, CH, and OG designed and conducted the experiments. HJN and VV obtained the human adipose tissue biopsies and performed the bariatric surgery. OPN made the CRISPR-cells. SD and JIB performed the data analyses. JIB, CH and SD drafted the manuscript. JIB, CH, OG, OPN, HJN, VV, PRN, JVS, SD and GM contributed to data interpretation, discussion and reviewed and edited the manuscript.
Funding
Funding for this project was provided by Persontilpasset medisin for barn og voksne med diabetes mellitus (PERSON-MED-DIA), the Western Norway Regional Health Authority and Meltzerfondet. PRN was supported by the European Research Council (AdG #293574).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic Information
Rights and permissions
About this article
Cite this article
Bjune, JI., Haugen, C., Gudbrandsen, O. et al. IRX5 regulates adipocyte amyloid precursor protein and mitochondrial respiration in obesity. Int J Obes 43, 2151–2162 (2019). https://doi.org/10.1038/s41366-018-0275-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41366-018-0275-y
- Springer Nature Limited
This article is cited by
-
MRI adipose tissue segmentation and quantification in R (RAdipoSeg)
Diabetology & Metabolic Syndrome (2022)
-
IRX5 promotes adipogenesis of hMSCs by repressing glycolysis
Cell Death Discovery (2022)
-
Deficiency of Irx5 protects mice from obesity and associated metabolic abnormalities
International Journal of Obesity (2022)
-
Irx3 and Irx5 in Ins2-Cre+ cells regulate hypothalamic postnatal neurogenesis and leptin response
Nature Metabolism (2021)
-
Dysregulation of amyloid precursor protein impairs adipose tissue mitochondrial function and promotes obesity
Nature Metabolism (2019)