Abstract
Background:
New screening methods that can add predictive diagnostic value for aggressive (high-grade, Gleason score ⩾7) prostate cancer (PCa) are needed to reduce unnecessary biopsies for patients with non-aggressive PCa. This is particularly important for men presenting for an initial biopsy with an equivocal PSA in the 2–10 ng ml−1 range. PCA3 and ERG are biomarkers that can add predictive value for PCa in urine; however, with a limited utility as a digital rectal exam (DRE) is required.
Methods:
First-catch urine samples were collected at six sites from men scheduled to undergo a prostate biopsy. Exosomal RNA was extracted, RNA copy numbers of ERG and PCA3 were measured by reverse transcription–quantitative PCR (RT–qPCR), and the EXO106 score (the sum of normalized PCA3 and ERG RNA levels) was computed. Performance was compared with standard of care (SOC; PSA, age, race or family history) parameters. Contingency table, logistic regression, receiver operating characteristics curve and box-plot analyses were performed.
Results:
In this cohort (N=195), a dichotomous EXO106 score demonstrated good clinical performance in predicting biopsy result for both any cancer and high-grade disease. For high-grade disease, the negative and positive predictive values were 97.5% and 34.5%, respectively. The discrimination between high-grade and Gleason score ⩽6 (including benign) biopsy results by a combination of EXO106 and SOC (area under the curve (AUC)=0.803) was significantly improved compared with SOC without EXO106 (AUC=0.6723, P=0.0009). The median EXO106 score correlated (P<0.001; Spearman’s rank order) with histologic grade.
Conclusions:
A novel molecular signature (EXO106 score) derived from non-DRE urine demonstrated independent, negative predictive value for the diagnosis of high-grade PCa from initial biopsy for men with ‘gray zone’ serum PSA levels. Its use in the biopsy decision process could result in fewer prostate biopsies for clinically insignificant disease.
Similar content being viewed by others
Introduction
Prostate cancer (PCa) is the second leading cause of cancer death among men in the United States, with an anticipated 233 000 new cases and nearly 29 480 deaths in 2014.1 The definitive diagnostic for PCa is the prostate needle biopsy, typically recommended for men with elevated serum PSA levels and/or a suspicious digital rectal exam (DRE) with added indication from family history, age and race. In addition to anxiety and pain associated with this invasive procedure, men may experience complications such as infection, bleeding, urinary obstruction,2, 3, 4 and, in some cases, even septic shock that results in death.5 The majority of PCa's remain indolent, infrequently resulting in death;6, 7, 8 thus, there is a major risk for detecting cancers that are clinically insignificant and do not require treatment. Unfortunately, due to the low positive predictive value of PSA and the high prevalence of low risk PCa, ~70–80% of men will undergo an unnecessary biopsy.9, 10
Even with the recently confirmed reduction in PCa mortality associated with PSA screening in the European Randomized Study of Screening for Prostate Cancer (ERSPC),11 the overdiagnosis and overtreatment resulting from current practices illustrates that improved methods are necessary to identify treatment-worthy disease. The strongest evidence of mortality reduction in PCa is in high-risk, generally high-grade, cancer with randomized trials showing benefit with radiotherapy or radical prostatectomy.12, 13 Thus, additional screening methods that can add diagnostic value for ruling out high-grade disease (Gleason score ⩾7) in particular are needed to reduce unnecessary biopsies not only for patients with benign results but also those with non-aggressive (Gleason score ⩽6) disease.
We have developed a novel gene signature, the EXO106 score, derived from normalized PCA3 (PCa antigen 3) and ERG (V-ets erythroblastosis virus E26 oncogene homologs; often present as a gene fusion TMPRSS2:ERG) RNA expression in exosomes from urine that is predictive of initial biopsy results and does not require a DRE. Exosomes are small double lipid membrane vesicles (typically 30–200 nm in diameter, but herein considered all vesicles <800 nm in diameter) that are secreted from cells. Exosomes encapsulate a portion of the parent cell cytoplasm and are shed into various biofluids such as blood, cerebrospinal fluid and urine. They are a rich source of cellular protein and RNA and are particularly promising for profiling RNA expression from tumor cells as they are highly representative of their cell of origin14 and serve as a protective environment for mRNA during sample processing.14, 15, 16, 17, 18, 19 Exosomes in post-DRE urine collected from PCa subjects have been demonstrated to contain both PCA3 and TMPRSS2:ERG mRNA,15 and using advances in purification techniques we have been able to routinely isolate exosomal RNA from non-DRE urine that can be used to derive molecular signatures predictive of the diagnosis of PCa. The present study was designed to assess feasibility for isolating exosomes from prospectively collected urine samples at different sites and identify a stable gene signature that was potentially useful in discriminating higher Gleason disease on a subsequent biopsy.
Materials and methods
Study population
The study population consisted of men aged ⩾40 years scheduled for an initial or repeat prostate needle biopsy, due to a suspicious DRE and/or PSA levels, and met the eligibility criteria. Men were enrolled at six study sites (see Supplementary Table 1). The protocol was approved by the respective institutional review boards. All subjects provided written informed consent. The ‘intended use’ population was comprised only of men who were undergoing their initial biopsy and had equivocal ‘gray zone’ serum PSA levels (>2 and <10 ng ml−1).
Sample collection and handling
First-catch urines samples (25 to 120 ml) were collected and assigned a study ID as per standard protocols. Samples were stored in routine collection vessels (without preservatives) at 2–8 °C for up to 2 weeks until shipped on ice to a central laboratory (Exosome Diagnostic Laboratory, St Paul, MN, USA). The exosome isolation procedure has previously been extensively characterized by electron microscopy and nanoparticle tracking analysis.18, 19 A slightly modified version of this isolation platform has been since developed into a urine exosome clinical sample concentrator kit (Exosome Diagnostics, Cambridge, MA, USA). The Urine Clinical Sample Concentrator has gone through a rigorous design control in development to allow the method to be used in a clinical workflow. The samples were filtered through a 0.8-μm syringe filter and stored in 20 ml aliquots at −80 °C until further processing.
Sample processing, RNA extraction and RT–qPCR
A 20 ml aliquot of each sample (blinded to all lab personnel) was thawed in a water bath at 25 °C for ~20 min and processed with the Urine Clinical Sample Concentrator Kit (Exosome Diagnostics, St Paul, MN, USA). Included in the kit is an internal control that is added to each sample that co-isolates and co-extracts with exosomal RNA. From each urine sample, RNA was extracted and an aliquot was used for RT–qPCR. RNA extraction and complementary DNA preparation methods are presented in the Supplementary Information. For each batch of samples, an aliquot from a positive control urine pool was processed in parallel.
RT–qPCR data analysis
The qPCR reactions were performed using the Rotor-Gene Q System (QIAGEN, Venlo, Netherlands). For each reaction, a Ct (cycle threshold) value was derived for each biomarker and the internal control (Rotor-Gene Q User Manual, November 2012). Calibrators for each target gene plus the internal control were in each qPCR run and used to convert Ct-values to RNA copies. Calibration curves were generated for each qPCR plate to adjust for inter-run variability.
Determination of RNA levels
Samples were normalized for RNA levels with SPDEF (SAM-pointed domain-containing Ets transcription factor) to derive ‘ERG or PCA3 RNA copy number/SPDEF mRNA copy number.’
The performance of the EXO score using PSA RNA (encoded by kallikrein-related peptidase 3 (KLK3)) was also evaluated using receiver operating characteristics analysis. The area under the curve (AUC) resulting from the use of KLK3 as a normalizer was comparable, with that of SPDEF being slightly improved (data not shown).
Technical sample exclusion criteria
Samples were excluded based on qPCR data if either of the following were not met: (1) Internal controls were within ±3 s.d. from the mean of all internal control values in the cohort and 2) SPDEF detected at >30 copies per reaction (minimum expression to ensure appropriate normalization).
EXO106 test algorithm
Utilizing RNA copy numbers of ERG, PCA3 and SPDEF, the EXO106 test algorithm was developed to provide a quantitative number, the EXO106 score, per subject. The linear score, transformed to generate values between 0 and 30, is defined to be proportional to the probability of a prostate biopsy to be positive for cancer. The score is defined as sum log-transformed ERG and PCA3 RNA copy number relative to SPDEF RNA copy number. Multiple binary cutoffs yielded a negative predictive value (NPV) of >95% with the optimal cutoff of 10 selected based on an NPV of >95% a maximized Youden’s J-statistic.
ERG and PCA3 imputation
To compute an EXO106 score for samples that did not have adequate levels of ERG or PCA3 RNA to yield a Ct-value (that is, less than one RNA copy per reaction) an imputed value of one copy was assigned. Noteworthy, the absence of a detectable ERG or PCA3 expression level is diagnostically significant and imputation allows for individual subject inclusion in model development.
Statistical analysis
A summary of the statistical analysis is presented in the Supplementary Information. All data processing and statistical analysis were carried out using R statistical computing environment version 3.0.2 (http://www.r-project.org) along with packages from Bioconductor Open Source Software (http://www.bioconductor.org).
Results
Subject and sample characteristics
Non-DRE urine samples were collected from 453 subjects enrolled between June 6 2013 and November 7 2013. For each subject, exosomal RNA was extracted and the RNA copy numbers of ERG (including wild-type and most fusion partners such as TMPRSS2:ERG), PCA3 and SPDEF were determined. Samples from 107 subjects who had incomplete subject data and/or were >40 ml were excluded. Among the remaining 346 subjects, 212 met the criteria for the ‘intended use population’: no prior biopsies and gray zone serum PSA levels (>2 and <10 ng ml−1). Of all, 195 had sufficient qPCR data: SPDEF expression>30 copies and appropriate urine volume. ERG expression was detected in 90 (46%) study patients which had >1 copy of ERG RNA; PCA3 expression was detected in all subjects. Absence of ERG expression was encoded through imputation of one copy as ERG expression value for subjects with inadequate ERG levels (that is, ERG absence=one copy; ERG presence=actual copy number). The number of patients with a prior negative biopsy was insufficient to effectively evaluate performance of the Exo106 assay (data not provided).
The median age was 62 years and median pre-biopsy serum PSA was 5.1 ng ml−1 (Table 1). Most subjects had a non-suspicious DRE (70%) and no family history of PCa (76%). Of all, 83% were Caucasian, and 13% were African-American. The positive biopsy prevalence was 46%, with 22% having a Gleason score ⩾7.
EXO106 score for prediction of initial biopsy results
For each subject, exosomal RNA copy numbers of ERG, PCA3 and SPDEF were used to derive an EXO106 score (see Materials and methods). For both any cancer and high-grade (Gleason score ⩾7) disease, a dichotomous EXO106 score alone (cutoff value of 10) demonstrated good clinical performance in predicting initial biopsy result (Table 2). The EXO106 score demonstrated relatively greater sensitivity (95.2 vs 75.3%) and NPV (97.5 vs 72.2%) for high-grade disease.
The predictive accuracy of the Exo106 binary cut point of 10 was further evaluated in predicting biopsy Gleason scores (Table 3). Among the 42 biopsies with Gleason score ⩾7, the EXO106 score correctly identified 100% of Gleason scores ⩾4+3 and only missed 2 positive Gleason 3+4 patients (8%).
Relationship of the EXO106 score and tumor histologic grade
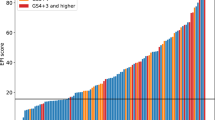
Consistent with the predictive accuracy for high-grade disease, a statistically significant direct correlation (P<0.001; Spearman’s rank order, although with a wide s.d.) between the EXO106 score and Gleason score was observed (Figure 1). Compared with biopsy-negative samples (N=106), the median EXO106 score was significantly higher in biopsy-positive samples, with the score increasing from low-grade (Gleason score 6, N=47) to high-grade (Gleason score ⩾7, N=42).
EXO106 also provided improvement in the binary predictive performance of standard of care (SOC) parameters (that is, serum PSA levels, age, race and family history of PCa) in logistic regression models with a fixed sensitivity of 90%. Binary performance for predicting high-grade disease increased by 33.3% in specificity, 12.2% in positive predictive value and 5.4% in NPV, when EXO106 was included as coefficient to a multivariable logistic regression model (Table 4). The increase in binary performance for predicting any cancer is less pronounced.
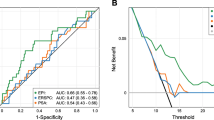
The performance of the EXO106 signature to discriminate between biopsy-positive and -negative samples was also evaluated by receiver operating characteristic analyses and compared with SOC parameters. This assessment demonstrated that the EXO106 alone has significant accuracy in predicting both any cancer and high-grade disease, independent of the SOC parameters (Figure 2). The AUC of EXO106 alone was 0.715 and 0.764 for predicting any cancer (Figure 2a) and high-grade disease (Figure 2b), respectively. As illustrated, the AUCs of SOC were significantly increased when evaluated in combination with EXO106 (P<0.0003 and P<0.0009) for any cancer (Figure 2a) and high-grade disease (Figure 2b), respectively. The AUCs for SOC alone were 0.563 and 0.672 for any cancer and high-grade disease, respectively, compared with 0.732 and 0.803 when combined with EXO106.
EXO106 is predictive of biopsy result for any cancer (a) and high-grade disease (b) and adds significant predictive value to that of SOC alone. For each cohort, the AUC was determined for ROC analyses of multivariable models that included SOC parameters with EXO106 as an additional predictor, as well as for SOC parameters alone vs EXO106 alone. AUC, area under the curve; ROC, receiver operating characteristics; SOC, standard of care parameter.
Discussion
We identified a urine exosome RNA signature, the EXO106 score, predictive of high-grade (Gleason score ⩾7) PCa in men presenting for an initial biopsy with serum PSA levels within an expanded gray zone (>2 and <10 ng ml−1).20, 21 The EXO106 signature, derived from first-catch, non-DRE urine samples shipped without preservatives to a central lab for processing, demonstrated predictive performance that was independent and improved compared with SOC parameters, as assessed by using logistic regression models.
This novel gene signature is derived from ERG and PCA3 RNA normalized with SPDEF RNA. Both ERG (often present as a gene fusion, most commonly TMPRSS2:ERG) and PCA3 RNA are known to be overexpressed in PCa tissue22, 23 and have been identified as biomarkers in post-DRE urine.24, 25, 26, 27, 28, 29, 30, 31, 32 The ETS transcription factor SPDEF, although not prostate or PCa specific, interacts with the androgen receptor to induce PSA expression and appears to function as a tumor suppressor gene in PCa.31 Since PSA and SPDEF are both expressed (although not exclusively) in benign prostate epithelial cells and dysregulated in cancer, they each represent appropriate normalizing genes reflecting prostate epithelial content and disease potential. In this cohort, normalization with SPDEF RNA outperformed PSA RNA (encoded by KLK3), particularly in identifying high-grade disease. Of note, the EXO106 score maximizes the detection of ERG+ tumors as it represents total ERG RNA (that is, both wild type and fusions). There is increasing evidence that suggests total ERG RNA levels may actually be more accurate for predicting not only the presence of PCa but also overall disease progression than the TMPRSS2:ERG fusion alone.34, 35, 36
In our population of initial biopsy subjects, the EXO106 score accurately predicted both any cancer (AUC=0.715, NPV=72.22%) and, with relatively improved performance, high-grade disease (Gleason score ⩾7; AUC=0.764, NPV=97.5%). Of particular clinical significance, among biopsies with a Gleason score ⩾7, a dichotomous EXO106 score correctly identified all biopsies with a Gleason score ⩾4+3 and only 2 of 25 (8%, all Gleason score 3+4) were predicted negative. These results are comparable to a recently published first-biopsy post-DRE PCA3 clinical nomogram, which missed 8% of Gleason score ⩾7 biopsies (specifically, 10 Gleason score 3+4 and 1 Gleason score 4+4),37 reaffirming a role for the EXO106 score in early detection as it does not require a DRE.
Despite these very promising results there are a few limitations to this study. First, the lack of a central pathology review likely introduced some variability, specifically when reporting small volume cancers and on the breakdown of Gleason 7 (that is, 3+4 vs 4+3). However, all participating centers represent large urology group practices with highly experienced uropathologists. Another point of concern is that the exosomal ERG RNA copy number is low (average of 1 copy/reaction) and suggests that performance of this gene may be impacted by urine volume, shipping conditions and properties of the urine sample including acidity, protein content and so on. To address the potential impact of urine volume and a dilutional effect on exosome isolation, analyses were limited to samples of ⩽40 ml and a standard reduced volume urine collection vessel has been introduced for ongoing studies. Finally, given the variety of assays currently available in the liquid clinical space including the prostate health index (Food and Drug Administration approved), 4 K score and the post-DRE urine combination of PCA3 with TMPRSS2:ERG, additional studies are planned to compare performance of the current exosome test with these approaches, as well as examine cost-effectiveness and clinical adoption models.
Conclusion
In summary, this study presents a novel noninvasive, molecular assay utilizing a first-catch, non-DRE urine, exosomal RNA signature as a prognostic tool for predicting high-grade PCa among men presenting for an initial biopsy with serum PSA levels within an expanded gray zone (>2 and <10 ng ml−1). Once validated in a larger cohort, we anticipate that this assay could be used along with PSA and other prognostic factors to inform and guide current biopsy decision models and reduce unnecessary biopsies.
References
Siegel R, Ma J, Zou Z, Jemal A . Cancer statistics, 2014. CA J Clin 2014; 64: 9–29.
Bjurlin MA, Wysock JS, Taneja SS . Optimization of prostate biopsy: review of technique and complications. Urol Clin North Am 2014; 41: 299–313.
Lundstrom KJ, Drevin L, Carlsson S, Garmo H, Loeb S, Stattin P et al. Nationwide population based study of infections after transrectal ultrasound guided prostate biopsy. J Urol 2014; 192: 1116–1122.
Nam RK, Saskin R, Lee Y, Liu Y, Law C, Klotz LH et al. Increasing hospital admission rates for urological complications after transrectal ultrasound guided prostate biopsy. J Urol 2010; 183: 963–968.
Bruyere F, Malavaud S, Bertrand P, Decock A, Cariou G, Doublet JD et al. Prosbiotate: a multicenter, prospective analysis of infectious complications after prostate biopsy. J Urol 2014; 193: 145–150.
Abouassaly R, Thompson IM Jr, Platz EA, Klein EA Epidemiology, etiology, and prevention of prostate cancer. In: McDougal WS, Wein A, Kavoussi L, Novick AC, Partin AW, Peters CA et al (eds). Campbell-Walsh Urology, 10th edn. Elsevier Saunders: Philadelphia, PA, USA, 2012.
Berman DM, Epstein JI . When is prostate cancer really cancer? Urol Clin North Am 2014; 41: 339–346.
Klotz L . Active surveillance versus radical treatment for favorable-risk localized prostate cancer. Curr Treat Opt Oncol 2006; 7: 355–362.
Getzenberg RH, Partin AW. Prostate cancer tumor markers. In: Wein AJ, Kavoussi LR, Novick AC, Partin AW, Peters CA (eds). Campbell-Walsh Urology, 10th edn. Elsevier Saunders: Philadelphia, PA, USA, 2012.
Loeb S, Vellekoop A, Ahmed HU, Catto J, Emberton M, Nam R et al. Systematic review of complications of prostate biopsy. Eur Urol 2013; 64: 876–892.
Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Zappa M, Nelen V et al. Screening and prostate cancer mortality: results of the european randomised study of screening for prostate cancer (ERSPC) at 13 years of follow-up. Lancet 2014; 384: 2027–2035.
Bill-Axelson A, Holmberg L, Garmo H, Rider JR, Taari K, Busch C et al. Radical prostatectomy or watchful waiting in early prostate cancer. N Engl J Med 2014; 370: 932–942.
Wilt TJ, Brawer MK, Jones KM, Barry MJ, Aronson WJ, Fox S et al. Radical prostatectomy versus observation for localized prostate cancer. N Engl J Med 2012; 367: 203–213.
van der Vos KE, Balaj L, Skog J, Breakefield XO . Brain tumor microvesicles: insights into intercellular communication in the nervous system. Cell Mol Neurobiol 2011; 31: 949–959.
Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer 2009; 100: 1603–1607.
Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008; 10: 1470–1476.
Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO . Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9: 654–659.
Miranda KC, Bond DT, McKee M, Skog J, Paunescu TG, Da Silva N et al. Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int 2010; 78: 191–199.
Miranda KC, Bond DT, Levin JZ, Adiconis X, Sivachenko A, Russ C . Massively parallel sequencing of human urinary exosome/microvesicle RNA reveals a predominance of non-coding RNA. PLoS One 2014; 9: e96094.
Thompson IM, Ankerst DP, Chi C, Goodman PJ, Tangen CM, Lucia MS et al. Assessing prostate cancer risk: results from the Prostate Cancer Prevention Trial. J Nati Cancer Inst 2006; 98: 529–534.
Thompson IM, Pauler DK, Goodman PJ, Tangen CM, Lucia MS, Parnes HL et al. Prevalence of prostate cancer among men with a prostate-specific antigen level < or =4.0 ng per milliliter. N Engl J Med 2004; 350: 2239–2246.
Hessels D, Klein Gunnewiek JM, van Oort I, Karthaus HF, van Leenders GJ, van Balken B et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol 2003; 44: 8–15; discussion -6.
St John J, Powell K, Conley-Lacomb MK, Chinni SR . TMPRSS2-ERG fusion gene expression in prostate tumor cells and its clinical and biological significance in prostate cancer progression. J Cancer Sci Ther 2012; 4: 94–101.
Gittelman MC, Hertzman B, Bailen J, Williams T, Koziol I, Henderson RJ et al. PCA3 molecular urine test as a predictor of repeat prostate biopsy outcome in men with previous negative biopsies: a prospective multicenter clinical study. J Urol 2013; 190: 64–69.
Laxman B, Morris DS, Yu J, Siddiqui J, Cao J, Mehra R et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res 2008; 68: 645–649.
Laxman B, Tomlins SA, Mehra R, Morris DS, Wang L, Helgeson BE et al. Noninvasive detection of TMPRSS2:ERG fusion transcripts in the urine of men with prostate cancer. Neoplasia 2006; 8: 885–888.
Leyten GH, Hessels D, Jannink SA, Smit FP, de Jong H, Cornel EB et al. Prospective multicentre evaluation of PCA3 and TMPRSS2-ERG gene fusions as diagnostic and prognostic urinary biomarkers for prostate cancer. Eur Urol 2014; 65: 534–542.
Rice KR, Chen Y, Ali A, Whitman EJ, Blase A, Ibrahim M et al. Evaluation of the ETS-related gene mRNA in urine for the detection of prostate cancer. Clin Cancer Res 2010; 16: 1572–1576.
Salami SS, Schmidt F, Laxman B, Regan MM, Rickman DS, Scherr D et al. Combining urinary detection of TMPRSS2:ERG and PCA3 with serum PSA to predict diagnosis of prostate cancer. Urol Oncol 2013; 31: 566–571.
Dijkstra WS, Birker IL, Smit FP, Leyten GH, de Reijke TM, van Oort IM et al. Prostate cancer biomarker profiles in urinary sediments and exosomes. J Urol 2014; 191: 1132–1138.
Cheng XH, Black M, Ustiyan V, Le T, Fulford L, Sridharan A et al. SPDEF inhibits prostate carcinogenesis by disrupting a positive feedback loop in regulation of the Foxm1 oncogene. PLoS Genet 2014; 10: e1004656.
Haller AC, Tan W, Payne-Ondracek R, Underwood W, Tian L, Morrison C et al. High SPDEF may identify patients who will have a prolonged response to androgen deprivation therapy. Prostate 2014; 74: 509–519.
Oettgen P, Finger E, Sun Z, Akbarali Y, Thamrongsak U, Boltax J et al. PDEF, a novel prostate epithelium-specific ets transcription factor, interacts with the androgen receptor and activates prostate-specific antigen gene expression. J Biol Chem 2000; 275: 1216–1225.
Hagen RM, Adamo P, Karamat S, Oxley J, Aning JJ, Gillatt D et al. Quantitative analysis of ERG expression and its splice isoforms in formalin-fixed, paraffin-embedded prostate cancer samples: association with seminal vesicle invasion and biochemical recurrence. Am J Clin Pathol 2014; 142: 533–540.
Svensson MA, Perner S, Ohlson AL, Day JR, Groskopf J, Kirsten R et al. A comparative study of ERG status assessment on DNA, mRNA, and protein levels using unique samples from a Swedish biopsy cohort. Appl Immunohistochem Mol Morphol 2014; 22: 136–141.
He J, Schepmoes AA, Shi T, Wu C, Fillmore TL, Gao Y et al. Analytical platform evaluation for quantification of ERG in prostate cancer using proteins and mRNA detection methods. J Transl Med 2015; 54: 1–14.
Hansen J, Rink M, Bianchi M, Kluth LA, Tian Z, Ahyai SA et al. External validation of the updated Briganti nomogram to predict lymph node invasion in prostate cancer patients undergoing extended lymph node dissection. Prostate 2013; 73: 211–218.
Acknowledgements
Technical and scientific writing was done by Dr Julie Deardorff, Pacifica Scientific Consulting.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
MN, SB, SB, JS and VO'N are employees of Exosome Diagnostics; MJD is a consultant to Exosome Diagnostics. The remaining authors declare no conflict of interest
Additional information
Supplementary Information accompanies the paper on the Prostate Cancer and Prostatic Diseases website
Supplementary information
Rights and permissions
About this article
Cite this article
Donovan, M., Noerholm, M., Bentink, S. et al. A molecular signature of PCA3 and ERG exosomal RNA from non-DRE urine is predictive of initial prostate biopsy result. Prostate Cancer Prostatic Dis 18, 370–375 (2015). https://doi.org/10.1038/pcan.2015.40
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/pcan.2015.40
- Springer Nature Limited
This article is cited by
-
PSA-Test und moderne Biomarker zur Detektion von Prostatakrebs
Urologie in der Praxis (2024)
-
Urinary marker panels for aggressive prostate cancer detection
Scientific Reports (2022)
-
Predicting high-grade prostate cancer at initial biopsy: clinical performance of the ExoDx (EPI) Prostate Intelliscore test in three independent prospective studies
Prostate Cancer and Prostatic Diseases (2022)
-
Prostate cancer and the role of biomarkers
Abdominal Radiology (2020)
-
How should radiologists incorporate non-imaging prostate cancer biomarkers into daily practice?
Abdominal Radiology (2020)