Abstract
Chronic inflammation is the primary cause of gastric cancer (GC). NLRP3, as an important inflammasome component, has crucial roles in initiating inflammation. However, the potential roles of NLRP3 in GC is unknown. Here, we show that NLRP3 expression is markedly upregulated in GC, which promotes NLRP3 inflammasome activation and interleukin-1β (IL-1β) secretion in macrophages. In addition, NLRP3 binds to cyclin-D1 (CCND1) promoter and promotes its transcription in gastric epithelial cells. Consequently, NLRP3 enhances epithelial cells proliferation and GC tumorigenesis. Furthermore, we identify miR-22, which is constitutively expressed in gastric mucosa, as a suppressor of NLRP3. MiR-22 directly targets NLRP3 and attenuates its oncogenic effects in vitro and in vivo. However, Helicobacter pylori (H. pylori) infection suppresses miR-22 expression, while enhances NLRP3 expression, and that triggers uncontrolled proliferation of epithelial cells and the emergence of GC. Thus, our research describes a mechanism by which miR-22 suppresses NLRP3 and maintains homeostasis of gastric microenvironments and suggests miR-22 as a potential target for the intervention of GC.
Similar content being viewed by others

Introduction
Gastric cancer (GC) is the fifth most common cancer and the third leading cause of cancer-associated deaths worldwide.1 Although a number of factors (such as diet, smoking, chronic inflammation, and so on) have been reported to be associated with the growth and progression of GC, chronic inflammation is considered as the primary cause of GC.2, 3 Microbial pathogens take responsibility for the chronic inflammation in lots of cancers, particularly those originating in the gastrointestinal system, such as GC. Helicobacter pylori (H. pylori) infection is the most important factor for GC.2, 3, 4 H. pylori infection touches off acute inflammation initially and, in a subset of patients, gradually progresses to chronic inflammation, atrophic gastritis (AG), dysplasia, and finally GC, which is called as the ‘inflammation-carcinoma chain’.4, 5
Pattern-recognition receptors are critical to the recognition of invading microbial pathogens, generation of mature inflammatory cytokines (interleukin-1 beta (IL-1β), tumor necrosis factor-alpha (TNF-α), interleukin-6 (IL-6), and so on), and triggering inflammation. Given the association between chronic inflammation and GC is well established, the dysfunction of pattern-recognition receptors could modulate inflammation in gastric mucosa, and then subsequently influence the emergence and progression of GC.5 Multiple pattern-recognition receptors, including NLRs (nucleotide-binding oligomerization domain (NOD)-like receptors), TLRs (Toll-like receptors), and RLRs (retinoic acid-inducible gene (RIG)-I-like receptors), are involved in gastric carcinogenesis. H. pylori infection promotes development of GCs via TLR4.6 NOD1 expression level is markedly increased in GC tissues, and promotes IFN-γ signal pathway when H. pylori infect gastric epithelial and aggravates severity of disease.7 In H. pylori-infected human gastric mucosa overexpressed MDA5 (melanoma differentiation-associated gene 5, a RLR member) is associated with intestinal metaplasia and glandular atrophy.8
NLRP3 inflammasome is a multi-protein platform which contains NLRP3, Caspase-1 (CASP1) and ASC, which serves as platforms for the activation of caspase-1 (cysteine protease).9 Then pro-IL-1β was cleaved into mature IL-1β by activated caspase-1. NLRP3 inflammasome has taken part in many kinds of diseases, such as infectious diseases, gout, autoimmune disorders, atherosclerosis, type 2 diabetes, obesity and cancer.10, 11 NLRP3 inflammasome protects colon from tumor formation.12 The decreased expression of NLRP3 inflammasome contributes to hepatocellular carcinoma progression.13 In lung cancer, NLRP3 inhibits carcinogen-induced metastases and tumors mediated by NK cell.14 NLRP3 vitiates antitumor vaccine through promoting tumor-associated myeloid-derived suppressor cells accumulation.15 In addition, NLRP3 polymorphisms are associated with a higher susceptibility to melanoma and poor survival rates in colorectal cancer and myeloma patients.16, 17 NLRP3 polymorphism is also associated with GC risk.18 H. pylori infection can activate NLRP3 inflammasome and subsequent IL-1β secretion,19 indicating the potential roles of NLRP3 in H. pylori infection-associated diseases. However, the biological role played by NLRP3 in GC progression remains unknown.
MicroRNAs (miRNAs) suppress the expression of varieties of genes through binding with their 3'UTRs (3'-untranslated regions), and thus possibly regulate many steps of tumorigenesis and cancer progression, including GC.20, 21, 22 This report demonstrates that miR-22 is substantially decreased in GC and correlated with poor prognosis of GC patients. We show that miR-22 suppresses NLRP3 expression directly, attenuates NLRP3-driven GC cells proliferation and GC carcinogenesis both in vitro and in vivo. Furthermore, H. pylori infection markedly suppresses miR-22 expression, while enhances NLRP3 expression. Our studies reveal a new mechanism by which constitutively expressed miR-22 suppresses NLRP3 inflammasome activation and gastric epithelial cells proliferation, and thus maintains homeostasis in gastric mucosa. H. pylori infection can break the homeostasis in gastric mucosa and promotes the emergence and progression of GC.
Results
Enhanced NLRP3 level correlates with GC progression
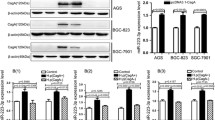
To explore the possible roles of NLRP3 in development of GC, we first searched the OncoMine database and found that NLRP3 was significantly upregulated in different types of GC (Figure 1a). However, the expression of key components of other inflammasomes, including AIM2, NLRC4, NLRP1 and NLRP6, was not enhanced (Figure 1a). NLRP3 expression in GC is higher than normal tissue in TCGA data set (Supplementary Figure 1a). In addition, two components of NLRP3 inflammasome, including ASC and CASP1, were also examined in the Oncomine data sets and TCGA data sets. Although ASC was overexpressed in most GC, CASP1 expression remained unchanged (Supplementary Figures 1b and c). Western blot confirmed that NLRP3 protein expression was markedly enhanced in four human GC and responding non-tumor samples, also in four GC cell lines compared with GES-1, a normal gastric mucous membrane epithelial cell line (Figures 1b and c). Immunohistochemistry (IHC) staining showed that NLRP3 expression in superficial gastritis (SG) was low, and its expression was slightly upregulated in atrophic gastritis (AG) and dysplasia (DYS) (Figures 1d and e). But, NLRP3 expression was markedly enhanced in GC samples (Figures 1d and e). In addition, NLRP3 expression was upregulated in both epithelium and the stromal components of GC (Figure 1d). RT–PCR analysis confirmed that NLRP3 expression was obviously upregulated in 30 human GC samples compared with 30 AG samples (Figure 1f). Taken together, above results strongly prove that NLRP3 is significantly upregulated in GC.
Enhanced NLRP3 levels correlates with GC progression. (a) The expression of NLRP3, AIM2, NLRC4, NLRP1 and NLRP6 in 80 normal and different types of GC. The data and P-values were obtained from the OncoMine database. (b) Western blot of NLRP3, p10 (cleaved caspase-1) and p17 (cleaved IL-1β) in four pairs of gastric tumor samples (T) and corresponding normal samples (N). (c) Western blot of NLRP3 in four GC cell lines (SGC-7901, BGC-823, HGC-27 and AGS) and normal gastric mucous membrane epithelial cell line (GES-1). (d) IHC staining for NLRP3 in SG (superficial gastritis), AG (atrophic gastritis), DYS (dysplasia) and GC samples. Scale bars, 200 μm (insets 50 μm). (e) IHC scores for NLRP3 in SG, AG, DYS and GC samples. (f) RT–PCR analysis of NLRP3 in 30 AG and 30 GC samples. The horizontal bars: mean value in each sample group. P<0.05 (Mann–Whitney U test). (g) Kaplan–Meier analysis of NLRP3 expression in survival of GC patients (P<0.05, log-rank test; n=90). (h) Kaplan–Meier analysis of NLRP3 (216016_at) expression in survival of GC patients (P<0.0001, log-rank test; n=876). The data and P-values were obtained from the Kaplan–Meier Plotter database. (i–k) Primary tumor gross appearance (i) and tumor growth curves (j) and the average tumor weight (k) of the GC tumor xenograft model. Each bar represents the mean±s.e.m. of six nude mice in (j) and (k). *P<0.05; ***P<0.001. (Student's t-test).
Next, we investigated NLRP3 expression in 90 GC samples and their corresponding normal tissues. Univariate Cox regression analyses indicated that tumour, node and metastasis stage (P<0.001), tumor size (P=0.045), lymph node metastasis (P<0.001), invasion (P=0.006), distant metastasis (P=0.010) and NLRP3 expression (P=0.035) were significantly associated with GC patients’ survival. Furthermore, multivariate Cox regression analyses further confirmed the tumor size (P=0.042), lymph node metastasis (P<0.001) and NLRP3 expression (P=0.043) as independent predictors of the overall survival of GC patients (Supplementary Table 1). Survival curves were plotted to compare patient outcomes according to an analysis that showed difference in the expression levels of NLRP3. We found that survival times were lower in patients with high NLRP3 levels in different types of GC (Figure 1g). Kaplan–Meier survival plot shows that lower expression of NLRP3 results in a better overall survival (Figure 1h). In addition, survival analysis of 375 cases of GC samples in TCGA database also showed that lower expression of NLRP3 results in higher survival rate (Supplementary Figure 2). Collectively, above results hint a potential link between enhanced NLRP3 expression and GC progression.
To explore the biological function of NLRP3 in GC progression, a GC cell line BGC-823 was generated to stably overexpress NLRP3 (Supplementary Figure 3). And then, we subcutaneously injected BGC-823 cells into nude mice. The tumors formed by NLRP3-transduced BGC-823 cells were larger than the control tumors, both in size and weight (Figures 1i–k). Our data showed that NLRP3 expression contributed to in vivo tumor growth, indicating that the enhanced NLRP3 expression promotes GC tumorigenicity in vivo.
NLRP3 promotes GC cells proliferation via promoting CCND1 transcription
We then investigated the role of NLRP3 in GC progression. Colony formation assays and 5-ethynyl-2′-deoxyuridine (EdU) staining showed that NLRP3 overexpression markedly increased proliferation of GC cells (Figures 2a and b). Furthermore, NLRP3 greatly enhanced the amount of PCNA (proliferating cell nuclear antigen), a molecule related to proliferation, which confirmed the effects of NLRP3 on proliferation of GC cells (Figure 2c). To further elucidate the mechanisms by which NLRP3 promotes GC cells proliferation, we examined NLRP3 expression pattern in GC cells. NLRP3 located in both cytoplasm and nucleus (Figures 2d–f) and IL-1β greatly induced NLRP3 nucleic translocation in GC cells (Figures 2e and f). It has been reported that in the Th2 cells, nucleus NLRP3 bound to its binding sequence (5′-nGRRGGnRGRG-3′, where ‘R’ is any purine and ‘n’ is any nucleotide) of IL-4 promoter and transactivated it.23 We analyzed promoters of several proliferation-associated genes, including CCND1, CDK2, CDK4, CDK6 and CCNE1, and found two potential NLRP3-binding sequences within CCND1 promoter region (Figure 2g). By the analysis of public data sets from the GEO repository (GSE63089), we found that CCND1 expression was the most positively correlated gene with NLRP3 in 45 pairs of GC and corresponding normal tissues (Figure 2h). We also observed the positive correlation between CCND1 and NLRP3 in our own 30 AG and 30 GC samples in Figure1f (Supplementary Figure 4). Accordingly, NLRP3 overexpression significantly enhanced CCND1 mRNA expression in BGC-823 cells, while no effects on expression of other proliferation-associated genes, such as CDK2, CDK4, CDK6 and CCNE1 (Figure 2i). Consistently, NLRP3 overexpression also enhanced CCND1 protein expression and NLRP3 knockdown had the opposite effects (Figures 2j and k). Furthermore, siRNA-mediated knockdown of CCND1 blunted the proliferative effect of NLRP3 in BGC-823 cells (Figures 2l–n). Collectively, these results indicated that CCND1 is the functional downstream of NLRP3.
NLRP3 promotes GC cells proliferation via promoting CCND1 transcription. (a,b) Representative images (left) and quantification (right) of vector- or NLRP3 plasmid-transfected BGC-823 cells analyzed in a foci formation assay and EdU assays. Blue: DAPI, red: EdU-positive. (c) Western blot analysis of PCNA level in NLRP3 plasmid- or vector-transfected BGC-823 cells. (d) Immunofluorescence analysis of NLRP3 expression pattern in BGC-823 cells transfected with NLRP3 plasmid. (e) Western blot analysis of NLRP3 level in cytoplasm and nuclear extract of BGC-823 cells with IL-1β stimulation for 6 h. H3 served as a loading control for nuclear protein. GAPDH served as a loading control for cytoplasm protein. (f) Immunofluorescence analysis of NLRP3 expression pattern in BGC-823 cells after IL-1β stimulation for 6 h. (g) Predicted NLRP3 binding sequences in human CCND1 promoter. (h) Correlation of NLRP3 with CCND1 mRNA expression in 45 pairs of GC and respective normal samples. The data were obtained from the GEO database (GSE63089). (i) RT–PCR analysis of CCND1, CDK2, CDK4, CDK6 and CCNE1 mRNA expression in BGC-823 cells transfected with control or NLRP3 plasmid. (j) Western blot analysis of CCND1 level in BGC-823 cells that transfected with NLRP3 or control plasmid. (k) Western blot analysis of CCND1 level in BGC-823 cells that transfected with scramble control siRNA, NLRP3 siRNA 1 or NLRP3 siRNA 2, following IL-1β stimulation for 6 h. (l–n) Representative images (left) and quantification (right) of vector- or NLRP3 plasmid and Negative Control (NC)- or CCND1 siRNA-transfected BGC-823 cells analyzed in a foci formation assay (l) and EdU assays (m) and Western blot analysis of PCNA expression (n). Blue: DAPI, red: EdU-positive. (o) Diagram of CCND1 promoter luciferase reporters with different NLRP3 binding sites. (p) BGC-823 cells were transfected with different luciferase plasmids reporters, along with Vector or NLRP3 plasmid (NLRP3), and then luciferase assay was performed. (q) ChIP analysis of NLRP3 binds to CCND1 gene promoter region. Similar results were acquired in three independent experiments. Every bar represents the mean±s.e.m. of three independent experiments in (a), (b), (i), (l), (m), (p) and (q). *P<0.05; **P<0.01; ***P<0.001. (Student’s t-test). DAPI, 4-6-diamidino-2-phenylindole dihydrochloride.
We then explored whether NLRP3 increased CCND1 expression transcriptionally. A human CCND1 promoter plasmid (WT, -2079 to -1457, containing two potential NLRP3-binding sequences) was constructed and transiently transfected into BGC-823 cells. As shown in Figure 2p, NLRP3 overexpression greatly increased CCND1 promoter luciferase activity. To explore whether the potential NLRP3-binding sequences in CCND1 promoter were functional, three mutated CCND1 luciferase reporters with different NLRP3 binding site deletion were constructed (Figure 2o). NLRP3 overexpression lost its enhancement in CCND1 mt-1, mt-2 and mt-3 reporter (Figure 2p). Collectively, these results hinted that NLRP3 function relied on both of the two NLRP3-binding sequences.
Aiming to examine whether NLRP3 can bind to its target binding sites in the CCND1 promoter, we performed ChIP (chromatin immunoprecipitation) assays. NLRP3 could interact with the CCND1 promoter region containing NLRP3-binding sequences (Figure 2q). Taken together, our data show that NLRP3 binds to the CCND1 promoter through two NLRP3 binding sites and facilitates CCND1 transcription, leading to the enhancement of GC cell proliferation.
MiR-22 directly targets and inhibits NLRP3 expression
The fact that NLRP3 promotes GC progression promoted us to investigate the underlying mechanisms of NLRP3 upregulation in GC. miRNAs are important post-transcriptional regulators of multiple molecules. We then performed miRNA microarray analysis using AG and GC samples, and identified 14 upregulated and 28 downregulated miRNAs (Figure 3a). By combining the microarray data with a bioinformatics search of Target-scan databases, we identified miR-22 was decreased in GC and suggested miR-22 as a potential NLRP3-targeting miRNA (Figure 3b). RT–PCR further confirmed that miR-22 was significantly reduced in GC samples compared with AG samples (Figure 3c). Analysis of clinical samples showed that the expression of miR-22 was negatively correlated with NLRP3 in 13 AG and 13 GC tissues (Supplementary Figure 5a). By the analysis of public data sets from the GEO repository (GSE54129), we found that miR-22 expression was negatively correlated with NLRP3 in 111 GC and 21 normal gastric tissues as well (Supplementary Figure 5b). In addition, the expression of miR-22 in four GC cell lines was lower compared with GES-1, a normal gastric mucous membrane epithelial cell line (Figure 3d).
miR-22 directly targets and inhibits NLRP3 expression. (a) MicroRNA expression profiles in tissues of AG and GC. (b) Predicted miR-22 target sequences in NLRP3 3′UTR (NLRP3 3′UTR WT) of different species. NLRP3 3′UTR Mut contains four mutated nucleotides in the seed sequence of miR-22. (c) RT–PCR analysis of miR-22 level in AG (n=13) and GC samples (n=13). The horizontal bars: mean value in each sample group. P=0.0046 (Mann–Whitney U test). (d) RT–PCR analysis of miR-22 expression in four GC cell lines (SGC-7901, BGC-823, HGC-27 and AGS) and a normal gastric mucous membrane epithelial cell (GES-1). (e) RT–PCR analysis of miR-22 level in BGC-823 cells that transfected with miR-22 mimics or inhibitor. (f, g) Western blot (f) and RT–PCR (g) analysis of NLRP3 level in BGC-823 or AGS cells that transfected with miR-22 mimics or inhibitors. (h) Western blot analysis of NLRP3, AIM2 and NLRC4 levels in THP-1 cells transfected with miR-22 inhibitors or mimics, following LPS stimulation for indicated time periods. (i, j) BGC-823 or AGS cells were transfected with miR-22 mimics (i) or inhibitors (j), along with wild-type NLRP3 3′UTR reporter plasmid (NLRP3-WT-3′UTR) or mutant NLRP3 3′UTR reporter plasmid (NLRP3-Mut-3′UTR) for 24 h, and then luciferase assay was performed. Every bar represents the mean±s.e.m. of three independent experiments in (d), (e), (g), (i) and (j). *P<0.05; **P<0.01; ***P<0.001. (Student’s t test).
In order to determine whether miR-22 affects NLRP3 expression, miR-22 mimics and inhibitors were synthesized and transfected in BGC-823 cells (Figure 3e). NLRP3 protein and mRNA expression were both dramatically decreased in miR-22-transfected BGC-23 and AGC cells, while miR-22 inhibitor had the opposite effects (Figures 3f and g). Moreover, miR-22 also markedly inhibited NLRP3 expression in THP-1 cells, with no inhibitory effects on AIM2 and NLRC4 expression (Figure 3h). To investigate whether NLRP3 is a direct target of miR-22, a luciferase reporter was used to measure translation driven by the 3′UTRs of NLRP3. MiR-22 overexpression significantly decreased the luciferase activity with NLRP3 3′UTR (Figures 3i and j), while miR-22 inhibitor greatly increased NLRP3-3′ UTR luciferase activity. However, miR-22 mimics and miR-22 inhibitor both did not affect the luciferase activity with mutated NLRP3 3′ UTR (Figures 3i and j). Taken together, above results show that miR-22 directly targets NLRP3.
NLRP3 promotes GC cells proliferation via inflammasome-dependent ways
IL-1β is crucial for the induction of GC.24 Colony formation assay and EdUstaining showed that IL-1β significantly increased colony formation and Edu-positive percent of BGC-823 and AGS cells (Figures 4a and b). Furthermore, IL-1β greatly enhanced the expression of PCNA, which confirmed the effects of IL-1β on proliferation of GC cells (Figure 4c). However, NLRP3 knockdown blunted the proliferation effect of IL-1β stimulation (Figure 4d). MCC950, a selective inhibitor of NLRP3 inflammasome activation, was used to block IL-1β production.25 MCC950 treatment blunted the proliferation effect of NLRP3 in vitro (Figure 4e). To explore the effects of IL-1β blockade in vivo, we performed a xenograft tumor model with NLRP3-overexpressed BGC-823 cells and MCC950 treatment. When tumor diameters reached 5–7 mm, MCC950 was injected into the tumor every 2 days for 2 weeks, and then the mice were killed. As shown in Figures 4f–h, the tumors after MCC950-treatment were smaller than the tumors treated with PBS, both in size and weight. Collectively, these results indicate that NLRP3 promotes GC cells proliferation, partially through promoting NLRP3 inflammasome activation and IL-1β in macrophage.
NLRP3 promotes GC cells proliferation via inflammasome dependent way. (a and b) Representative images (left) and quantification (right) of indicated cells that stimulated with IL-1β for 6 h analyzed in a foci formation assay (a) and EdU assays (b). Blue: DAPI, red: EdU-positive. (c) Western blot analysis of PCNA level in AGS and BGC-823 cells stimulated with IL-1β for indicated time periods. (d) Representative images (left) and quantification (right) of BGC-823 cells that stimulated with IL-1β for 6 h after Negative Control (NC)− or NLRP3 siRNA-transfection analyzed in a foci formation assay. (e) Representative images (left) and quantification (right) of vector- or NLRP3 plasmid-transfected BGC-823 cells that treated with MCC950 analyzed in a foci formation assay. (f–h) Primary tumor gross appearance (f) and tumor growth curves (g) and the average tumor weight (h) of the MCC950-treated GC tumor xenograft model. Similar results were acquired in three independent experiments. Every bar represents the mean±s.e.m. of three independent experiments in (a), (b), (d) and (e). Every bar represents the mean±s.e.m. of seven or eight nude mice in (g) and (h). *P<0.05; **P<0.01; ***P <0.001. (Student’s t-test). DAPI, 4-6-diamidino-2-phenylindole dihydrochloride.
MiR-22 inhibits NLRP3 inflammasome activation in macrophages
NLRP3 protein expression is crucial for the assembly and activation of NLRP3 inflammasome.26 We next evaluated the effects of miR-22 on NLRP3 inflammasome activation in macrophages. MiR-22 overexpression resulted in both decrease of cleaved caspase-1 p10 and NLRP3 expression in lipopolysaccharide (LPS)-primed and ATP-activated THP-1 cells (Figure 5a). Consistently, IL-1β secretion was dramatically decreased in miR-22-transfected THP-1 cells following NLRP3 inflammasome activation (Figure 5b). However, TNF-α expression was not changed with miR-22 overexpression (Figure 5c). Collectively, these results indicate that miR-22 selectively inhibit NLRP3 inflammasome activation and subsequent IL-1β secretion in macrophage.
miR-22 inhibits NLRP3 inflammasome activation in macrophages. THP-1 cells were transfected with control or miR-22 mimics, and then primed with LPS for indicated time periods and stimulated with ATP for 30 min at last. (a) Western blot analysis of NLRP3 and cleaved caspase-1 (p10) expression. (b,c) ELISA analysis of IL-1β (b) and TNF-α (c) secretion in supernatants. Similar results were obtained in three independent experiments. Every bar represents the mean±s.e.m. of three independent experiments in (b) and (c). *P<0.05; **P<0.01. (Student’s t-test).
MiR-22 inhibits GC cells proliferation in vitro
IL-1β is crucial for the GC cells proliferation.27 We then examined the biological role of miR-22 in GC cells proliferation. As demonstrated in Figures 6a–d, colony formation assays and EdU staining showed that miR-22 markedly inhibited IL-1β-induced colony formation of BGC-823 and AGS cells. In addition, IL-1β treatment greatly enhanced NLRP3 expression and miR-22 overexpression dramatically reversed the effects (Figures 6e–f). Thus, miR-22 inhibits GC cells proliferation through inhibiting NLRP3 expression in GC cells and limiting IL-1β secretion.
miR-22 inhibits GC cells proliferation in vitro. (a–d) Representative images and quantification of control- or miR-22 mimics-transfected cells that were stimulated with IL-1β for 6 h analyzed in a foci formation assay (a, b) and EdU assays (c, d). (e) RT–PCR analysis of NLRP3 mRNA expression in BGC-823 and AGS cells stimulated with IL-1β for 6 h. (f) Western blot analysis of NLRP3 level in BGC-823 and AGS cells stimulated with IL-1β for indicated time periods. (g) Western blot analysis of NLRP3 and CCND1 levels in BGC-823 and AGS cells transfected with control or miR-22 mimics, following IL-1β stimulation for 6 h. Similar results were acquired in three independent experiments. Every bar represents the mean±s.e.m. of three independent experiments in (b), (d) and (e). *P<0.05; **P<0.01; ***P<0.001. (Student’s t-test).
MiR-22 agomir effectively suppresses NLRP3-driven tumorigenesis in vivo
Next, we further examined the inhibitory role of miR-22 in GC progression using an in vivo tumor model. MiR-22 agomir (cholesterol-conjugated miR-22 mimics) was transfected into NLRP3-overexpressed BGC-823 cells to enhance endogenous miR-22 expression and then a xenograft tumor model was generated (Figure 7a). When tumor diameters reached 5–7 mm, miR-22 was injected into the tumor every 4 days for 3 weeks, and then the mice were killed (Figure 7b). As shown in Figures 7c–e the tumors formed by miR-22-transfected cells were smaller than the tumors formed from control cells, both in size and weight. Moreover, IHC confirmed that both NLRP3 and CCND1 expressions were negatively correlated with miR-22 level (Figure 7f). Ki67 staining was nicely correlated with tumor growth status (Figure 7f). Taken together, these data indicate that miR-22 inhibits NLRP3-driven tumor growth in vivo and miR-22 might be an effective target for limiting GC tumorigenesis.
miR-22 agomir suppresses NLRP3-driven tumorigenesis in vivo. (a) RT–PCR analysis show miR-22 agomir efficacy. Every bar represents the mean±s.e.m. of three independent experiments. (b) Xenograft model in nude mice. Timeline of the miR-22 mimics therapeutic delivery experiment. (c) Representative fluorescence image of nude mice bearing xenograft tumors (upper panel) and images of tumors from all mice in each group (lower panel). (d) Tumor growth curves in each group. (e) Average tumor weight in each group. (f) IHC staining detecting NLRP3, CCND1 and Ki67 expression in the indicated tumors. Scale bars, 50 μm. Every bar represents the mean+s.e.m. of five nude mice. **P<0.01; ***P<0.001. (Student’s t-test).
H. pylori enhances NLRP3 expression and suppresses miR-22 expression
H. pylori infection is considered as the main cause of GC. RNA analysis of our clinical tissues positive/negative for H. pylori revealed that NLRP3 was induced in the gastric tissues with H. pylori infection, and CCND1 expression was positively correlated with NLRP3 (Figure 8a and Supplementary Figure 6a). Higher level of NLRP3 expression was observed both in epithelium and the stromal components in the H. pylori-positive gastric tissues (Figure 8b). We then investigated the relation between the infection of H. pylori and enhanced NLRP3 expression in gastric mucosa. AGS and BGC-823 cells were incubated with different H. pylori strains (Hp26695 or SS1). H. pylori infection greatly enhanced NLRP3 expression in these GC cell lines (Figures 8c–e). Moreover, NLRP3 expression and NLRP3 inflammasome activation were also enhanced following H. pylori infection in THP-1 cells (Figures 8f and g). In order to investigate the relation of H. pylori infection and NLRP3 expression in vivo, we performed H. pylori infection in MNU (N-methyl-N-nitrosourea)-treated mice.28 The gastric mucosa was normal and displayed no inflammatory cells infiltration in the control group. Low dose MNU induced chronic inflammation in gastric mucosa, and caused mild atrophic mucosa. However, a severe atrophic mucosa with intestinal metaplasia was observed following H. pylori infection in MNU-treated mice (Figure 8h). NLRP3 was slightly upregulated with MNU treatment, but its expression was markedly increased with MNU treatment and H. pylori infection (Figure 8h). Accordingly, Ki-67, a marker of cell proliferation, was greatly increased following H. pylori infection (Figure 8h).
Helicobacter pylori infection enhances NLRP3 expression and suppresses miR-22 expression. (a) RT–PCR analysis of NLRP3 expression in H. pylori-negative or -positive AG samples. The horizontal bars: mean value in each sample group. P=0.0032 (Mann–Whitney U test). (b) Representative images for IHC staining and IHC scores of NLRP3 in H. pylori-negative or -positive AG samples. Scale bars, 50 μm. (c) RT–PCR analysis of NLRP3 mRNA expression in H. pylori-infected BGC-823 and AGS. (d–f) Western blot analysis of NLRP3 level in H. pylori-infected AGS (d), BGC-823 (e) and THP-1 cells (f). (g) ELISA analysis of IL-1β in THP-1 cells infected with H. pylori for indicated time periods. (h) HE staining and IHC staining for NLRP3 and Ki-67 in control, MNU and MNU-Hp(SSI)-infected mice. Scale bars, 500 μm; 50 μm (insets). (i) RT–PCR analysis of miR-22 level in H. pylori-negative or -positive AG samples. The horizontal bars: mean value. P=0.0026 (Mann–Whitney U test). (j) RT–PCR analysis of miR-22 expression in H. pylori-infected BGC-823, AGS and THP-1 cells. (k, l) Western blot analysis of NLRP3 expression in control or miR-22 mimics transfected BGC-823, AGS and THP-1 cells, following H. pylori infection for 4 h. (m) RT–PCR analysis of miR-22 level in BGC-823, AGS and THP-1 cells, following IL-1β stimulation for 6 h. Similar results were acquired in three independent experiments. Every bar represents the mean±s.e.m. of three independent experiments in (c), (g), (j) and (m). *P<0.05; **P<0.01; ***P<0.001. (Student’s t-test).
As NLRP3 is a direct target of miR-22, we then explored whether H. pylori could regulate miR-22 expression and thus enhance NLRP3 expression. MiR-22 expression was significantly reduced in H. pylori-positive samples compared with the H. pylori-negative samples (Figure 8i). In addition, the level of miR-22 was negatively correlated with NLRP3 in samples with/without H. pylori infection (Supplementary Figure 6b). H. pylori infection greatly inhibited miR-22 in both GC cells and THP-1 cells (Figure 8j). Furthermore, miR-22 dramatically decreased H. pylori infection-induced NLRP3 expression in both GC cells and THP-1 cells (Figures 8k and l). Additionally, we found that IL-1β markedly inhibited miR-22 expression in GC cells and THP-1 cells (Figure 8m), indicating a feedback regulation loop between miR-22 and NLRP3 inflammasome activation in GC microenvironment. All together, our data indicate that H. pylori infection suppresses miR-22 expression, resulting in the enhancement of NLRP3 expression in both GC cells and macrophages during GC progression.
Discussion
Optimal NLRs expression and activation are crucial for host against invading pathogens and maintain immune homeostasis. Deregulation of NLRs function regulates the emergence and progression of multiple diseases, such as inflammatory disease and cancer. Nucleotide oligomerization domain 1 (NOD1) is significantly overexpressed in gastric tumor samples, promotes H. pylori-induced inflammatory responses and induces carcinogenic consequences in the gastric niche.7, 29 NOD2 protects against colorectal cancer via regulation of gut microbiota.30 NLR family CARD domain containing 3 (NLRC3) deficiency results in hyper-susceptible to colitis and colorectal tumorigenesis.31 NLRC5 expression was significantly associated with CD8+ T-cell mediated antitumor response and the elimination of multiple cancer cells, suggesting NLRC5 as a new prognostic biomarker and potential therapeutic target of cancers.32 NLRC4 inflammasome is critical to colonic inflammation-induced tumorigeneseis by regulating of epithelial cell response to injury in colorectal cancer.33
As for NLRP3, an important member of NLRs, also possesses vital regulatory roles in many types of tumors, such as colon tumor, liver tumor and lung tumor. However, NLRP3 plays beneficial or adverse effect in different cancer types. For example, NLRP3 inflammasome protect colon from tumor formation, and the deregulated NLRP3 inflammasome expression is associated with liver cancer progression.12, 13 On the other hand, NLRP3 inhibits carcinogen-induced metastases and tumors mediated by NK cell.14 But, the biological role of NLRP3 in GC progression remains unclear. In our study, we show that NLRP3 is upregulated in GC. NLRP3 acts as a transcriptional regulator by activating CCND1 transcription, and thus promotes cell proliferation. Second, enhanced NLRP3 expression facilitates NLRP3 inflammasome activation and subsequent secretion of IL-1β. Thus, NLRP3 enhances GC cells proliferation and GC progression, via both inflammasome-independent manner and inflammasome-dependent manner. Inhibition of NLRP3 expression may offer an interesting mechanism to alter the progression of GC.
Up to now, several mechanisms for negative regulation of NLRP3 expression have been established. The AhR (aryl hydrocarbon receptor) can inhibit NLRP3 transcription via binding to the XRE (xenobiotic response element) in the NLRP3 promoter.34 DRD1 (Dopamine D1 receptor) induces NLRP3 ubiquitination by E3 ubiquitin ligase MARCH7, leading to the autophagy-mediated degradation of NLRP3.35 E3 ubiquitin ligase TRIM31 inhibits NLRP3 expression by promoting proteasomal degradation of NLRP3.36 GPBAR1 (G-protein coupled bile acid receptor 1, also named TGR5)-induced PKA kinase activation leads to the ubiquitination of NLRP3 and thus blocks NLRP3 inflammasome activation.37 NLRP3 mRNA is suppressed by miR-223 by binding to the 3′UTR of NLRP3.38, 39 In this study, we identify another miRNA, miR-22, as a suppressor of NLRP3 inflammasome by directly targeting NLRP3 and inhibiting its expression.
As a tumor suppressor, miR-22 is downregulated in lots of cancers.40, 41 MiR-22 is involved in variety of cellular processes related to tumorigenesis by targeting multiple molecules, such as cyclin A2 (CCNA2), laminin gamma 1 (LAMC1), cyclin-dependent kinase 6 (CDK6), SIRT1 and Sp1.42, 43 As for GC, miR-22 suppressed cancer progression through directly targeting CD151, matrix metalloproteinase 14 (MMP14) and Snail.44, 45 In this study, we show that miR-22 is dramatically decreased in GC. The decreased miR-22 expression enhances NLRP3 expression in gastric epithelial cell and macrophages, and triggers uncontrolled cell proliferation and GC tumorigenesis. Consequently, miR-22 markedly inhibits tumor growth both in vivo and in vitro. Therefore, miR-22 may be a potential target to attenuate uncontrolled inflammation and gastric carcinogenesis.
Within tumor microenvironment, chronic inflammation and inflammation-like conditions contribute to cancer progression.23, 46 In gastric mucosa, H. pylori infection initiates chronic inflammation in gastric microenvironment, induces the release of several inflammatory cytokines (TNF-α, IL-6, IL-1β, etc.) by immune cells, such as macrophages.47 The proinflammatory cytokines bind to their corresponding receptors expressed in gastric epithelial cells, and then trigger uncontrolled proliferation of epithelial cells, leading to tumorigenesis eventually. It has been reported that a polymorphism of IL-1β gene is associated with GC risk and IL-1β is sufficient to induce gastric carcinoma.27, 48 IL-1β binds to IL-1R, and then activates NF-κB and JNK signaling to promote proliferation, angiogenesis, malignant transformation and tumor progression.49, 50, 51, 52
Here we describe a novel mechanism by which H. pylori infection alters the microenvironment of gastric mucosa and contributes to the emergence and progression of GC. Based on the experimental data, we propose a model to illustrate how miR-22 feedback sustains NLRP3 expression, maintains homeostasis in gastric mucosa and limits gastric carcinogenesis (Figure 9). In normal gastric mucosa, constitutively expressed miR-22 suppresses NLRP3 expression. Thus, CCND1 expression is relatively low in gastric epithelial cells. NLRP3 inflammasome assembly and activation is also attenuated, and IL-1β secretion is limited in macrophages (Figure 9a). Thus, NLRP3 and IL-1β-driven gastric epithelial cell proliferation is controlled at a normal status. Following H. pylori infection, miR-22 expression is markedly suppressed, while NLRP3 expression is dramatically enhanced. NLRP3 promotes CCND1 transcription directly in gastric epithelial cells, and then promotes cell proliferation. Furthermore, NLRP3 inflammasome assembly and activation are possibly induced, resulting in the increase of IL-1β secretion in macrophages (Figure 9b). IL-1β can restrict miR-22 expression and thus feedback enhances NLRP3 inflammasome activation. In addition, IL-1β activates NF-κB and JNK signal pathways to promote gastric epithelial cells proliferation. Therefore, H. pylori infection builds a cross-talk between macrophage and gastric epithelial cells in gastric mucosa through modulating miR-22 expression. H. pylori infection triggers uncontrolled cell proliferation, leading to the emergence and progression of GC.
Working model for MiR-22 attenuates H. pylori-induced gastric carcinogenesis. (a) In normal gastric mucosa, constitutively expressed miR-22 suppresses NLRP3 expression. Thus, CCND1 expression is limited in gastric epithelial cells. NLRP3 inflammasome assembly and activation is attenuated, and IL-1β secretion is limited in macrophages. In addition, CCND1 and IL-1β-driven gastric epithelial cell proliferation is controlled at a normal status. (b) Following H. pylori infection, miR-22 expression is markedly suppressed, while NLRP3 expression is dramatically enhanced. NLRP3 can promote CCND1 transcription directly in gastric epithelial cells, and then promote cell proliferation. Furthermore, NLRP3 inflammasome assembly and activation are possibly induced, resulting in the increase of IL-1β secretion in macrophages. IL-1β can restrict miR-22 expression and thus feedback enhances NLRP3 inflammasome activation. In additional, IL-1β activates NF-κB and JNK signal pathways in gastric epithelial cells to promote gastric epithelial cells proliferation. Thus, H. pylori infection triggers uncontrolled cell proliferation, leading to the emergence and progression of GC.
In summary, we identified miR-22 as a suppressor of NLRP3 expression and NLRP3 inflammasome activation. MiR-22 has critical roles in maintaining the homeostasis of gastric microenvironments by inhibiting NLRP3 expression in both gastric epithelial cells and macrophages. Our study suggested miR-22 as an attractive target for blocking the uncontrolled proliferation and gastric tumorigenesis induced by H. pylori infection. Modulating NLRP3 expression by miR-22 may has significant implications for the intervention of GC.
Methods
Human clinical specimens
RNA was obtained from 109 atrophic gastritis (AG) and 30 GC samples, which were obtained from Jinan Central Hospital and Qilu Hospital (Shandong, China). The tissues were stored in RNAlater (Qiagen, Hilden, Germany) at −80 °C before RNA extraction. A 13C urea breath test was used to test H. pylori positive or negative in AG patients. For IHC, 36 samples of SG, AG and dysplasia respectively were obtained from Bengbu Medical University, Anhui province; 69 AG were acquired from Jinan Central Hospital. Twenty-eight GC were acquired from Qilu Hospital; 105 GC and corresponding non-cancer samples were acquired from Shanghai Outdo Biotech Co., Ltd. (Shanghai, China). The specimen were collected immediately after surgery and stored at formalin. For western blot analysis, eight freshly collected GC samples and corresponding non-tumor gastric samples were stored in liquid nitrogen before protein extraction. All involved patients provided signed written informed consent. The histological types and clinical stages of the tumors were defined according to WHO classification criteria.
Reagents
Recombinant human IL-1β was obtained from Peprotech (Rocky Hill, NJ, USA). MNU, MCC950, anti-β-actin, LPS (Escherichia coli, 055:B5) were obtained from Sigma-Aldrich (St Louis, MO, USA). The stimuli concentrations were as follows: IL-1β, 100 ng/ml; LPS, 100 ng/ml; MCC950: 1 and 10 μM. Anti-CagA, anti-AIM2,anti-NLRC4, anti-Ki67 were from Abcam (Cambridge, MA, USA). Anti-NLRP3 was obtained from Abcam and AdipoGen (San Diego, CA, USA). Anti-ASC was from Enzo Life Sciences (Farmingdale, NY, USA). Anti-PCNA, anti-IL-1β, anti-CCND1 were obtained from Cell Signaling Technology (Danvers, MA, USA). Anti-caspase-1 and anti-caspase1-p10 were from Santa Cruz Biotechnology (Dallas, TX, USA).
Transfection
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) were used for stable transfection of NLRP3 plasmid into BGC-823 cells. For transient silencing, siRNA was transfected into cells with INTERFERin Reagent (Polyplus-transfection, Illkirch, France) as per the manufacturer’s instructions. For NLRP3 transient silencing, target sequences were as follows: 5′-GAAGTGAAAGCCAAAGCTA-3′ (siRNA1) and 5′-CCTACTTGGAGAAGAAATT-3′ (siRNA2). For CCND1 transient silencing, target sequences were as follows: 5′-CCACAGATGTGAAGTTCAT-3′ (siRNA1) and 5′-GTAAGAATAGGCATTAACA-3′ (siRNA2). Lipofectamine 2000 (Invitrogen) were used for MiR-22 mimic, inhibitor and agomir (Ribobio, Guangzhou, China) transfection.
IHC and quantification
FFPE (formalin-fixed, paraffin-embedded) sections acquired from patient or mouse tissues were subjected to deparaffination and dehydration. After antigen retrieval and H2O2 treatment, 3% bovine serum albumin were used for blocking for 30 min and then specific antibodies were incubated for one night at 4 °C. Finally, the secondary antibodies were used for incubating the sections for 50 min and a diaminobenzidine staining kit were used for detecting the immune complexes. A scale from 0 to 3 was used to score the intensity of positive staining: 0, no staining; 1, light brown; 2, medium brown and 3, dark brown. The proportion of positively stained cells was scored as follows: 0 (0%), 1 (<25%), 2 (25–75%) and 3 (>75%). The results that reported as the expression score were the multiply of above two scores. Samples with an IHC score ⩾5 were high expression, while those with an IHC score <5 were low expression.
Cell culture
AGS cells were cultured in F12 (HyClone, Logan, UT, USA) added with 10% FBS (Gibco, Carlsbad, CA, USA) as well as penicillin-streptomycin (Gibco). THP-1, BGC-823, GES-1, SGC-7901 and HGC-27 cells were cultured in RPMI-1640 (Gibco) added with 10% FBS. BGC-823 cells stably expressing NLRP3 were selected using 2 μg/ml puromycin (Gibco).
Luciferase assays
The promoter region human CCND1 (WT) was subcloned into the pGL3 basic reporter vector (Promega, Madison, WI, USA). All the NLRP3 binding site deleted CCND1 promoter were generated based on CCND1 WT plasmid with KOD-Plus-Mutagenesis kit (Toyobo, Osaka, Japan). The 3′UTR regions of human NLRP3 and corresponding mutants were cloned into the pMIR-GLO basic reporter vector (Promega). DNA sequencing was used to confirm all constructs. GC cells were seeded 24 h before transfection with the internal control vector pRL-TK and the promoter or 3′UTR reporters using Roche Transfection Reagent (Roche, Basel, Switzerland) as per the manufacturer’s instructions. After 48 h of transfection, luciferase assays were performed with a Luciferase Assay System (Promega) as per the manufacturer’s instructions.33, 35, 52 Renilla luciferase activity was used to normalize firefly luciferase activity.
Western blot
Protein lysis buffer supplemented with phosphatase and protease inhibitors was used to lyse cells. Same amounts of extracts were separated through SDS–PAGE. The proteins were transferred onto polyvinylidene fluoride membranes, and then the membranes were blocked with 5% nonfat dry milk. Subsequently the membranes were incubated first with specific primary antibodies and then with the respective secondary antibodies. Immunoblot signals in western blot were detected using Millipore ECL regents.34, 36, 53
RT–PCR
TRIzol reagent (Invitrogen) was used for extracting total RNA as per the manufacturer’s instructions. cDNA was synthesized using mRNA as a template with random primers and MMLV reverse transcriptase (Fermentas, Burlington, Canada). SYBR-Green RT-PCR assays (TaKaRa, Shiga, Japan) was used for RT–PCR analysis. β-actin expression was used to normalize data. The expression of miR-22 (RiboBio) was normalized to tU6 RNA (RiboBio). Specific primers used for RT–PCR assays are shown as below: 5′-AGTTGCGTTACACCCTTTCTTG-3′ and 5′-CACCTTCACCGTTCCAGTTTT-3′ for β-actin; 5′-AGCCTCAACAAACGCTACAC-3′ and 5′-GGTCAAACAGCAACTCCATC-3′ for NLRP3; 5′-ATGGAACACCAGCTCCTGTG-3′ and 5′-ACCTCCAGCATCCAGGTGGC-3′ for CCND1; 5′-CAGTACTGCCATCCGAGAGA-3′ and 5′-GAATCTCCAGGGAATAGGGC- 3′ for CDK2; 5′-TTTGTGGCCCTCAAGAGTGT-3′ and 5′-TCCTTGATCGTTTCGGCTGG-3′ for CDK4; 5′-GCTGACCAGCAGTACGAATG-3′ and 5′-GTCTGTTCGTGACACTGTGC-3′ for CDK6; 5′-AGAGGAAGGCAAACGTGACC-3′ and 5′-TGGTGCAATAATCCGAGGCT-3′ for CCNE1.
ChIP
SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling) was used to perform chromatin immunoprecipitation (ChIP) assays as per the manufacturer’s instructions. Chromatin fragments derived from GC cells were immunoprecipitated using 2 μg of an NLRP3 antibody.
H. pylori cultures and H. pylori-infected mouse model
Microaerophilic cultures (5% O2, 10% CO2 and 85% N2) were performed for the H. pylori strains 26695 and SSI in Brucella broth supplemented with 5% FBS at 37 °C. The bacteria were harvested through centrifugation, quantified according to their OD600 and immediately added to cell cultures at a bacteria-to-cell ratio of 100:1. 48 C57BL/6 mice (male) were separated into three groups. The control group consisted of 12 mice named as group 1. Groups 2 and 3 were given MNU (30 ppm) in their drinking water for 70 days and were allowed to drink distilled water for 14 days after the administration of MNU was stopped. Group 3 was then inoculated with the SS1 strain (1 × 109 colony-forming U/ml) every 2 days, a total of three times. These mice were provided with distilled water that did not contain MNU or SSI. All of the mice were killed for further analyses after 350 days.
Tumor xenograft model
Thymus-null BALB/c nude mice were obtained from QING ZI LAN Animal Company (Nanjing, China). BGC-823 cells were transfected with vector or NLRP3 plasmid stably. We subcutaneously injected transfected BGC-823 cells (2 × 105) into the right or left flank of six nude mice. We monitored the tumor growth every 2 days, and the mice were killed after 26 days. In the second experiments, BGC-823 cells (3 × 105) that stably transfected with vector or NLRP3 plasmid were subcutaneously injected into 30 nude mice, respectively. On day 12 after injection, when palpable tumors were present, both of vector group (14 nude mice) or NLRP3 group (16 nude mice) mice were divided into two groups. MCC950 (20 mg/kg) was intratumorally injected into one group. PBS was intratumorally injected into another group, and this group served as the negative control. Tumor growth was monitored every 2 days, and the mice were killed after 26 days. In the third experiments, BGC-823 cells that stably transfected with NLRP3 plasmid were transfected with miR-22 or mock agomirs (RiboBio), and 24 h later, 3 × 105 of these cells were collected and subcutaneously injected into 10 nude mice, respectively. On day 9 after injection, when palpable tumors were present, the mice that had received the miR-22 agomir received an intratumoral injection of the miR-22 agomir. The other group (five mice) received the mock agomir; this group served as the negative control. After 26 days, fluorescence images of the tumors were acquired, and the mice were then killed. The mice were anesthetized with isoflurane for 5 min and subsequently photographed using a Xenogen IVIS Spectrum imaging system.
ELISA
ELISA kits (eBioscience, San Diego, CA, USA) were used to evaluate the concentration of IL-1β and TNF-α according to the manufacturer’s protocol.
EdU staining
DNA synthesis was analyzed using the Cell-Light EdU Apollo488 In Vitro Imaging Kit as per the manufacturer’s instructions (RiboBio).
Colony formation assay
Methanol and Giemsa were used to fix and stain cells. The colonies numbers more than 100 were counted for subsequent analysis.
Immunofluorescence and confocal imaging
Sterile coverslips were put into six-well plates, and then cells were seeded on it before being exposed to the appropriate treatments. PBS and fresh 4% paraformaldehyde were used to wash and fix the cells. Then cells probed overnight with a specific primary antibody against NLRP3 in a humidified chamber at 4 °C. On the second day, anti-mouse IgG (H+L) and F(ab′)2 fragment (Cell Signaling) was used to incubate the slides, and 4-6-diamidino-2-phenylindole dihydrochloride was used to stain nuclei. Olympus BX51 fluorescence microscope was used to visualize the cells. Confocal images were acquired with Zeiss LSM780 and ZEN 2010 software.
Study approval
All clinical samples were approved for analysis by Shandong University Research Ethics Committee. All animal experiments were reviewed and approved by Shandong University Research Ethics Committee.
Statistical analysis
Unless otherwise indicated, the significant differences between any two groups were determined by two-tailed Student’s t-tests. Values of P<0.05 were considered as statistical significance. Survivals between experimental groups were compared by a log-rank (Mantel–Cox) test in the Kaplan–Meier plots. In the univariate and multivariate analysis, the effects of the clinicopathological variables and NLRP3 expression on the patients’ survival were determined by a Cox proportional hazard regression model. The univariate and multivariate analysis were performed using SPSS version 23.0. All other statistical analyses were performed using GraphPad PRISM version 6.01.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A . Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108.
Fox JG, Wang TC . Inflammation, atrophy, and gastric cancer. J Clin Invest 2007; 117: 60–69.
Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ . Gastric cancer. Lancet 2009; 374: 477–490.
Amieva M, Peek RM Jr. . Pathobiology of Helicobacter pylori-induced gastric cancer. Gastroenterology 2016; 150: 64–78.
Castano-Rodriguez N, Kaakoush NO, Mitchell HM . Pattern-recognition receptors and gastric cancer. Front Immunol 2014; 5: 336.
Chochi K, Ichikura T, Kinoshita M, Majima T, Shinomiya N, Tsujimoto H et al. Helicobacter pylori augments growth of gastric cancers via the lipopolysaccharide-toll-like receptor 4 pathway whereas its lipopolysaccharide attenuates antitumor activities of human mononuclear cells. Clin Cancer Res 2008; 14: 2909–2917.
Allison CC, Ferrand J, McLeod L, Hassan M, Kaparakis-Liaskos M, Grubman A et al. Nucleotide oligomerization domain 1 enhances IFN-gamma signaling in gastric epithelial cells during Helicobacter pylori infection and exacerbates disease severity. J Immunol 2013; 190: 3706–3715.
Tatsuta T, Imaizumi T, Shimoyama T, Sawaya M, Kunikazu T, Matsumiya T et al. Expression of melanoma differentiation associated gene 5 is increased in human gastric mucosa infected with Helicobacter pylori. J Clin Pathol 2012; 65: 839–843.
Davis BK, Wen H, Ting JP . The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol 2011; 29: 707–735.
Strowig T, Henao-Mejia J, Elinav E, Flavell RI . Inflammasomes in health and disease. Nature 2012; 481: 278–286.
Petrilli V . The multifaceted roles of inflammasome proteins in cancer. Curr Opin Oncol 2017; 29: 35–40.
Allen IC, TeKippe EM, Woodford RM, Uronis JM, Holl EK, Rogers AB et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med 2010; 207: 1045–1056.
Wei Q, Mu K, Li T, Zhang Y, Yang Z, Jia X et al. Deregulation of the NLRP3 inflammasome in hepatic parenchymal cells during liver cancer progression. Lab Invest 2014; 94: 52–62.
Chow MT, Sceneay J, Paget C, Wong CS, Duret H, Tschopp J et al. NLRP3 suppresses NK cell-mediated responses to carcinogen-induced tumors and metastases. Cancer Res 2012; 72: 5721–5732.
van Deventer HW, Burgents JE, Wu QP, Woodford RM, Brickey WJ, Allen IC et al. The inflammasome component NLRP3 impairs antitumor vaccine by enhancing the accumulation of tumor-associated myeloid-derived suppressor cells. Cancer Res 2010; 70: 10161–10169.
Verma D, Bivik C, Farahani E, Synnerstad I, Fredrikson M, Enerback C et al. Inflammasome polymorphisms confer susceptibility to sporadic malignant melanoma. Pigment Cell Melanoma Res 2012; 25: 506–513.
Ungerback J, Belenki D, Jawad ul-Hassan A, Fredrikson M, Fransen K, Elander N et al. Genetic variation and alterations of genes involved in NFkappaB/TNFAIP3- and NLRP3-inflammasome signaling affect susceptibility and outcome of colorectal cancer. Carcinogenesis 2012; 33: 2126–2134.
Castano-Rodriguez N, Kaakoush NO, Goh KL, Fock KM, Mitchell HM . The NOD-like receptor signalling pathway in Helicobacter pylori infection and related gastric cancer: a case-control study and gene expression analyses. PloS One 2014; 9: e98899.
Semper RP, Mejias-Luque R, Gross C, Anderl F, Muller A, Vieth M et al. Helicobacter pylori-induced IL-1beta secretion in innate immune cells is regulated by the NLRP3 inflammasome and requires the cag pathogenicity island. J Immunol 2014; 193: 3566–3576.
Bartel DP . MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297.
Calin GA, Croce CM . MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6: 857–866.
Song JH, Meltzer SJ . MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology 2012; 143: 35–47 e32.
Bruchard M, Rebe C, Derangere V, Togbe D, Ryffel B, Boidot R et al. The receptor NLRP3 is a transcriptional regulator of TH2 differentiation. Nat Immunol 2015; 16: 859–870.
Grivennikov SI, Greten FR, Karin M . Immunity, inflammation, and cancer. Cell 2010; 140: 883–899.
Coll RC, Robertson AA, Chae JJ, Higgins SC, Muñoz-Planillo R, Inserra MC et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med 2015; 21: 248–255.
Dowling JK, O’Neill LA . Biochemical regulation of the inflammasome. Crit Rev Biochem Mol Biol 2012; 47: 424–443.
Tu S, Bhagat G, Cui G, Takaishi S, Kurt-Jones EA, Rickman B et al. Overexpression of interleukin-1beta induces gastric inflammation and cancer and mobilizes myeloid-derived suppressor cells in mice. Cancer Cell 2008; 14: 408–419.
Nam KT, Hahm KB, Oh SY, Yeo M, Han SU, Ahn B et al. The selective cyclooxygenase-2 inhibitor nimesulide prevents Helicobacter pylori-associated gastric cancer development in a mouse model. Clin Cancer Res 2004; 10: 8105–8113.
Suarez G, Romero-Gallo J, Piazuelo MB, Wang G, Maier RJ, Forsberg LS et al. Modification of Helicobacter pylori peptidoglycan enhances NOD1 activation and promotes cancer of the stomach. Cancer Res 2015; 75: 1749–1759.
Couturier-Maillard A, Secher T, Rehman A, Normand S, De Arcangelis A, Haesler R et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest 2013; 123: 700–711.
Karki R, Man SM, Malireddi RK, Kesavardhana S, Zhu Q, Burton AR et al. NLRC3 is an inhibitory sensor of PI3K-mTOR pathways in cancer. Nature 2016. PMID: 27951586.
Yoshihama S, Roszik J, Downs I, Meissner TB, Vijayan S, Chapuy B et al. NLRC5/MHC class I transactivator is a target for immune evasion in cancer. Proc Natl Acad Sci USA 2016; 113: 5999–6004.
Hu B, Elinav E, Huber S, Booth CJ, Strowig T, Jin C et al. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. Proc Natl Acad Sci USA 2010; 107: 21635–21640.
Huai W, Zhao R, Song H, Zhao J, Zhang L, Zhang L et al. Aryl hydrocarbon receptor negatively regulates NLRP3 inflammasome activity by inhibiting NLRP3 transcription. Nat Commun 2014; 5: 4738.
Yan Y, Jiang W, Liu L, Wang X, Ding C, Tian Z et al. Dopamine controls systemic inflammation through inhibition of NLRP3 inflammasome. Cell 2015; 160: 62–73.
Song H, Liu B, Huai W, Yu Z, Wang W, Zhao J et al. The E3 ubiquitin ligase TRIM31 attenuates NLRP3 inflammasome activation by promoting proteasomal degradation of NLRP3. Nat Commun 2016; 7: 13727.
Guo C, Xie S, Chi Z, Zhang J, Liu Y, Zhang L et al. Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity 2016; 45: 802–816.
Bauernfeind F, Rieger A, Schildberg FA, Knolle PA, Schmid-Burgk JL, Hornung V . NLRP3 inflammasome activity is negatively controlled by miR-223. J Immunol 2012; 189: 4175–4181.
Haneklaus M, Gerlic M, Kurowska-Stolarska M, Rainey AA, Pich D, McInnes IB et al. Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1beta production. J Immunol 2012; 189: 3795–3799.
Yang F, Hu Y, Liu HX, Wan YJ . MiR-22-silenced cyclin A expression in colon and liver cancer cells is regulated by bile acid receptor. J Biol Chem 2015; 290: 6507–6515.
Wan WN, Zhang YQ, Wang XM, Liu YJ, Zhang YX, Que YH et al. Down-regulated miR-22 as predictive biomarkers for prognosis of epithelial ovarian cancer. Diagn Pathol 2014; 9: 178.
Xu D, Takeshita F, Hino Y, Fukunaga S, Kudo Y, Tamaki A et al. miR-22 represses cancer progression by inducing cellular senescence. J Cell Biol 2011; 193: 409–424.
Pasqualini L, Bu H, Puhr M, Narisu N, Rainer J, Schlick B et al. miR-22 and miR-29a are members of the androgen receptor cistrome modulating LAMC1 and Mcl-1 in prostate cancer. Mol Endocrinol 2015; 29: 1037–1054.
Zuo QF, Cao LY, Yu T, Gong L, Wang LN, Zhao YL et al. MicroRNA-22 inhibits tumor growth and metastasis in gastric cancer by directly targeting MMP14 and Snail. Cell Death Dis 2015; 6: e2000.
Wang X, Yu H, Lu X, Zhang P, Wang M, Hu Y . MiR-22 suppresses the proliferation and invasion of gastric cancer cells by inhibiting CD151. Biochem Biophys Res Commun 2014; 445: 175–179.
Lu H, Ouyang W, Huang C . Inflammation, a key event in cancer development. Mol Cancer Res 2006; 4: 221–233.
Lamb A, Chen LF . Role of the Helicobacter pylori-induced inflammatory response in the development of gastric cancer. J Cell Biochem 2013; 114: 491–497.
El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature 2000; 404: 398–402.
Hai Ping P, Feng BoT, Li L, Nan Hui Y, Hong Z . IL-1beta/NF-kb signaling promotes colorectal cancer cell growth through miR-181a/PTEN axis. Arch Biochem Biophys 2016; 604: 20–26.
Qin Y, Milton DR, Oba J, Ding Z, Lizee G, Ekmekcioglu S et al. Inflammatory IL-1beta-driven JNK activation in stage III melanoma. Pigment Cell Melanoma Res 2015; 28: 236–239.
Karin M . Nuclear factor-kappaB in cancer development and progression. Nature 2006; 441: 431–436.
Wagner EF, Nebreda AR . Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer 2009; 9: 537–549.
Huai W, Song H, Yu Z, Wang W, Han L, Sakamoto T et al. Mint3 potentiates TLR3/4- and RIG-I-induced IFN-beta expression and antiviral immune responses. Proc Natl Acad Sci USA 2016; 113: 11925–11930.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Nos. 81772151, 81571960, 81371781, 81670146, 81470318, 81372680, 81471991 and 81501720), the Science Foundation of Shandong Province (2017GSF18194, 2017GSF18109, 2014GSF118114 and ZR2014HQ070), and the Independent Innovation Foundation of Shandong University.
Author contributions
JJ conceived of the study and provided overall direction; SL, WZ and JJ designed the experiments; SL, XL, LM, LS, TL, LZ, AS, WS and CC performed the experiments; SL, WZ and JJ analyzed the data and wrote the paper.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Li, S., Liang, X., Ma, L. et al. MiR-22 sustains NLRP3 expression and attenuates H. pylori-induced gastric carcinogenesis. Oncogene 37, 884–896 (2018). https://doi.org/10.1038/onc.2017.381
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.381
- Springer Nature Limited
This article is cited by
-
Britannilactone 1-O-acetate induced ubiquitination of NLRP3 inflammasome through TRIM31 as a protective mechanism against reflux esophagitis-induced esophageal injury
Chinese Medicine (2024)
-
Mechanisms and effects of NLRP3 in digestive cancers
Cell Death Discovery (2024)
-
Harnessing innate immune pathways for therapeutic advancement in cancer
Signal Transduction and Targeted Therapy (2024)
-
The ‘speck’-tacular oversight of the NLRP3-pyroptosis pathway on gastrointestinal inflammatory diseases and tumorigenesis
Journal of Biomedical Science (2023)
-
The E3 ubiquitin ligase TRIM31 attenuates NLRP3 inflammasome activation in Helicobacter pylori-associated gastritis by regulating ROS and autophagy
Cell Communication and Signaling (2023)