Abstract
Androgen receptor (AR) activation is critical for prostate cancer (PCa) development and progression, including castration resistance. The nuclear export signal of AR (NESAR) has an important role in AR intracellular trafficking and proteasome-dependent degradation. Here, we identified the RNA helicase DHX15 as a novel AR co-activator using a yeast mutagenesis screen and revealed that DHX15 regulates AR activity by modulating E3 ligase Siah2-mediated AR ubiquitination independent of its ATPase activity. DHX15 and Siah2 form a complex with AR, through NESAR. DHX15 stabilized Siah2 and enhanced its E3 ubiquitin-ligase activity, resulting in AR activation. Importantly, DHX15 was upregulated in PCa specimens and its expression was correlated with Gleason scores and prostate-specific antigen recurrence. Furthermore, DHX15 immunostaining correlated with Siah2. Finally, DHX15 knockdown inhibited the growth of C4-2 prostate tumor xenografts in mice. Collectively, our data argue that DHX15 enhances AR transcriptional activity and contributes to PCa progression through Siah2.
Similar content being viewed by others
Introduction
Prostate cancer (PCa) is the most commonly diagnosed malignancy and the second leading cause of cancer-related mortality in American males, with an estimated 161 360 new cases and 26 730 deaths expected in 2017.1 Androgen-deprivation therapy remains the first-line therapy for men with metastatic PCa, however, most patients ultimately relapse with castration-resistant PCa (CRPC), which is presently incurable and accounts for most PCa mortality.2 The androgen receptor (AR), which plays a key role during all stages of prostate carcinogenesis including CRPC progression,3, 4 consists of an N-terminal domain, DNA-binding domain, hinge region and ligand-binding domain (LBD).5, 6 In the presence of androgens that bind to LBD, AR translocates from the cytoplasm to the nucleus and transactivates downstream genes. AR trafficking is regulated by multiple signals including two nuclear localization signals (NLs), NL1 in the DNA-binding domain and hinge region and NL2 in the androgen-bound LBD.7, 8, 9, 10 Our previous work identified one nuclear export signal (NESAR) within the androgen-free LBD and demonstrated NESAR (a.a. 743–817) is dominant over NLs in the absence of androgens and is necessary for the cytoplasmic localization of AR.9 Recently, we showed that NESAR can signal for polyubiquitination and proteasome-dependent degradation of NESAR-containing fusion proteins.11 NES is functionally conserved in the steroid receptor superfamily, and both GFP-tagged NESER and NESMR exhibit cytoplasmic localization and are targets of ubiquitination like NESAR, suggesting that this conserved region in the nuclear hormone receptor family mediates the cellular trafficking and ubiquitin-proteasome system-dependent degradation of these receptors. Unlike the leucine-rich NES, NESAR-mediated export is independent of the Crm-1/exporting-1 pathway12 and the mechanisms of NESAR function remain to be elucidated. Given that the aberrant nuclear localization and overexpression of AR represent two critical events in CRPC,13, 14 understanding the mechanisms underlying NESAR-mediated trafficking and degradation may provide new insights into mechanisms involved in castration resistance.
Siah2 is a RING finger type ubiquitin ligase consisting of a catalytic RING domain at its N terminus, two zinc fingers, and a C-terminal substrate-binding domain (SBD).15 A number of proteins have been identified as the substrates of Siah2, including N-CoR, PHD, Sprouty2, β-catenin and FIH.16, 17, 18, 19, 20 Accumulating evidence showed that Siah2 has an important role in tumorigenesis and metastasis in multiple cancers, including breast cancer, lung cancer, pancreatic cancer, melanoma and PCa (reviewed in reference 21). Recently, Siah2 was identified as an E3 ubiquitin ligase of AR specifically targeting a selective pool of NCOR1-bound, repressed AR chromatin complexes for degradation, promoting the expression of p300-bound AR downstream genes involved in lipid metabolism, cell motility and proliferation in PCa cells. Interestingly, Siah2 promotes AR activity without affecting the protein level of endogenous AR. Importantly, Siah2 was required for the growth of CRPC tumors in mice and analysis of human PCa specimens showed a significant upregulation of Siah2 in CRPCs.22 Thus, Siah2 is considered a critical player in the development of CRPC.23 Notably, both of the two major Siah2-binding sites on AR, a.a. 754 and a.a. 798, are located in the NESAR region,22 suggesting NESAR may have a role in Siah2-mediated ubiquitin-dependent degradation of AR.
Prp43 is an ATP-dependent DEAH-box helicase involved in the release of the intron lariat during pre-messenger RNA splicing and pre-ribosomal RNA processing.24, 25, 26 Most reported studies on Prp43 and its mammalian ortholog, DHX15, focused on their function as splicing factors in RNA metabolism.27, 28, 29 In the present study, using a yeast mutagenesis screen, we found that a point mutation in Prp43 inhibited the function of NESAR in a fusion protein. Further experiments showed that DHX15 forms a protein complex with AR and Siah2, regulates AR transcriptional activity independent of its ATPase activity and contributes PCa progression.
Results
Identification of PRP43 as a key regulator of NESAR in a yeast model system
We have previously reported that NESAR is functional in the yeast model12 and the fusion protein containing the NLSSV40 (2GFP- NLSSV40) localized strongly to the nucleus, whereas 2GFP-NLS SV40-NESAR showed cytoplasmic localization in yeast (Figure 1a). To identify factors required for NESAR to function, we performed a mutagenesis screen in yeast for mutants incapable of supporting NESAR function. In these yeast mutants, the 2GFP-NLSSV40-NESAR fusion protein should exhibit nuclear localization since NLSSV40 should not be suppressed by NESAR when a key factor required for NESAR function is mutated. Wild-type yeast cells expressing 2GFP-NLSSV40-NESAR were treated with ethyl methanesulfonate. Exposure to ethyl methanesulfonate typically results in point mutations, specifically a change of G→A. Over 6000 clones were visually assessed and several clones displaying nuclear localization of 2GFP-NLSSV40-NESAR were isolated. One of these mutants was chosen for further characterization and gene identification. The original 2GFP-NLSSV40-NESAR plasmid (exposed to mutagen) was removed and the mutant strain was retransformed with mutagen naive plasmids. When the mutant yeast was retransformed with 2GFP-NLSSV40 or 2GFP-NLSSV40-NESPKI, the localization of these fusion proteins was not different from those observed in the wild-type yeast (Figure 1a). However, when 2GFP-NLSSV40-NESAR was expressed in the mutant, GFP localization was nuclear, which was in contrast to the GFP cytoplasmic localization in wild-type cells (Figure 1a). As the mutation did not affect localization of 2GFP-NLSSV40, the mutation did not affect the nuclear import of 2GFP-NLSSV40-NESAR. Furthermore, it did not alter 2GFP-NLSSV40-NESPKI localization. Thus, the Crm-1 pathway, which mediates NESPKI-mediated export, should be intact in the mutant yeast. Collectively, these results suggested that the mutation specifically abrogated the ability of NESAR to localize the 2GFP-NLSSV40-NESAR fusion protein to the cytoplasm.
Mutation in Prp43 results in aberrant nuclear localization and upregulation of NESAR-containing fusion protein in yeast. (a) Localization of fusion protein 2GFP-NLSSV40, 2GFP-NLSSV40-NESPKI or 2GFP-NLSSV40–NESAR transformed into wildtype or mutant yeast. Liquid cultures were grown and cells were visualized under fluorescent microscopy. (b) Mutant was crossed to wildtype yeast and tetrad analysis was performed. GFP nuclear localization phenotype segregated in a ratio of nuclear:cytoplasmic of 2:2, indicating single-gene trait. Spore colony size also segregated 2:2 and small colony size was always associated with GFP nuclear phenotype. (c) Alignment of mouse, C. elegans, yeast and human amino acid mutated in yeast PRP43. Conserved amino acids in bold. Mutated glycine is underlined. (d) Mutant was transformed with empty vector, wild-type copy of PRP43 or another PRP gene, PRP8; 2GFP-NLSSV40–NESAR was visualized under fluorescent microscopy. (e) Western blot analysis of GFP in wild-type or prp43 mutant yeast cells transformed with 2GFP-NLSSV40-NESPKI (black arrow) or 2GFP-NLSSV40-GFP-NESAR (white arrow). Tpd3 served as control. Data are representative of three different experiments.
We then tested whether the mutant phenotype was resulted from a mutation in a single gene. The tetrad analysis showed a 2:2 ratio of mutant:wild-type phenotype, suggesting that the mutant phenotype was the result of the mutation of a single gene. Interestingly, tetrad analysis also revealed a 2:2 colony size phenotype (Figure 1b). Further analysis showed that the GFP nuclear localization phenotype always segregated with the small colony size phenotype (Figure 1b). In addition, when cells were incubated at the restrictive temperature of 39°C, mutant cells were unable to grow, indicating that in addition to having a role in NESAR-mediated localization, this gene was important for cell growth.
To identify the mutated gene, we performed a rescue screen. The mutant strain was transformed with a wild-type yeast genomic DNA library and then, taking advantage of the fact that the mutant displayed temperature sensitive lethality, we looked for those clones that were able to grow at the restrictive temperature of 39°C. We identified a clone that rescued both temperature sensitive lethality and mislocalization of 2GFP-NLSSV40-NESAR. The mutation was subsequently identified as a G→A transition at bp 911, resulting in a change of Glu→Asp in aa 304 of PRP43 (PRP43G304D), an RNA helicase in the DEAH-box family. PRP43 is a conserved protein with ~65% shared identity between yeast PRP43 and its human ortholog, DHX15 (Figure 1c). To confirm PRP43 being the gene responsible for the mutant phenotypes, we transformed the mutant yeast with a wild-type copy of PRP43 or PRP8, another gene involved in RNA splicing.30 PRP43, but not PRP8, rescued the localization phenotype (Figure 1d) and the growth defect.
NESAR not only mediates the localization of AR, but also plays a key role in AR ubiquitination and degradation.11 Thus, the mutation in PRP43 may also influence the abundance of the fusion protein containing NESAR. The protein levels of 2GFP-NLSSV40-NESPKI were similar in wild-type yeast and the PRP43G304D mutant (Figure 1e). In wild-type cells, 2GFP-NLSSV40-NESAR protein levels were much lower than that of 2GFP-NLSSV40-NESPKI (Figure 1e). Notably, 2GFP-NLSSV40-NESAR protein level in the PRP43G304D mutant was significantly increased as compared with its level in the wildtype (Figure 1e). These data suggest that PRP43 is required for NESAR to modulate AR localization and degradation.
DHX15 is a co-activator of AR in PCa cells
Given that NESAR has an important role in AR export and ubiquitination, we tested whether knockdown of the mammalian ortholog of PRP43, DHX15, influenced AR activity in PCa cells. We used siRNA to inhibit DHX15 expression both in androgen-dependent LNCaP cells and castration-resistant C4-2 cells. Knockdown of DHX15 led to a decrease in prostate-specific antigen (PSA) transcripts, whereas no obvious changes were detected in terms of AR mRNA either in the absence or the presence of R1881 (Figures 2a and b). Consistent with RT-PCR results, knockdown of DHX15 using two different siRNAs led to a decrease in PSA, but not AR protein levels (Figure 2c). DHX15 knockdown reduced the transcripts of AR downstream genes: PSA, TMPRSS2, NKX3.1 and SCL45A3, but not FKBP5 and MASPIN (Figure 2d), which indicated an important role for DHX15 in regulating the expression of a subset of AR downstream genes.
DHX15 regulates AR transcriptional activity. (a, b) LNCaP or C4-2 cells were transfected with siRNAs targeting DHX15 or control for 24 h, subsequently cultured in CS for 24 h, and then treated with 1 nm R1881 or vehicle for another 24 h. The mRNA expression of DHX15, AR and PSA were detected by qPCR. (c) LNCaP or C4-2 cells were transfected with two different siRNAs targeting DHX15 or control for 72 h and subjected to western blotting for detecting the expression of DHX15, AR and PSA. (d) DHX15 was knocked down with siRNA in LNCaP cells for 72 h, and the mRNA expression of different AR downstream genes were detected by qPCR. (e, f) LNCaP and C4-2 cells were transfected with siRNAs targeting DHX15 or control for 24 h, and then were transfected with PSA-Luc plasmid and pRL Renilla plasmid. 24 h later, cells were cultured in CS with or without 1 nm R1881 as indicated for another 24 h, and luciferase activity were analyzed in a luminometer. (g) DHX15, PSA-Luc and pRL Renilla plasmids were transfected into C4-2 cells for 24 h, subsequently cultured in CS with or without 1 nm R1881 for another 24 h. The luciferase activity was analyzed. (h) DHX15-WT, DHX15-K166A or DHX15-D260A were co-transfected with PSA-Luc and pRL Renilla plasmids into C4-2 cells for 24 h, subsequently cultured in CS with or without 1 nm R1881 for another 24 h. The luciferase activity was analyzed. (i) C4-2 cells stably transfected with control or DHX15 shRNA were cultured in CS for 48 h, then treated with 10 nm DHT for 2 h, and collected for ChIP assays using AR antibody. Purified chromatin was analyzed by qPCR for the ARE regions of PSA enhancer (PSA-EN), PSA promoter (PSA-PR) and TMPRSS2. Data represent average of a minimum of three technical replicates representative of three different experiments. (*P<0.05, **P<0.01, ***P<0.001).
To further determine whether DHX15 regulates PSA expression at the transcriptional level, the PSA promoter/enhancer-driven luciferase assay coupled with DHX15 knockdown was performed in LNCaP and C4-2 cells. DHX15 knockdown decreased luciferase activity by 30% in the presence of R1881 and by 50% in the absence of R1881 (Figures 2e and f). These results indicated that DHX15 was required for PSA transcription under conditions of both very low and normal concentrations of androgens (1 nm R1881). As expected, overexpression of DHX15 resulted in increased PSA-luciferase activity (Figure 2g). To assess whether the ATPase activity of DHX15 was required for the activation of AR transcription, we used the alanine substitution mutants of DHX15, DHX15-K166A and DHX15-D260A, which lack ATPase activities.31 Overexpression of ATPase-deficient mutants of DHX15 resulted in activation of luciferase reporter similarly to that of wild-type DHX15 (Figure 2h), suggesting that DHX15 activates AR signaling in an ATPase-independent manner. DHX15 knockdown or overexpression did not affect AR splice variants expression in both C4-2 and 22Rv1 PCa cell lines (Supplementary Figure S1), also suggesting that ATP-dependent RNA helicase or RNA splicing function of DHX15 is not involved in its role in AR stimulation. To further address the role of DHX15 in AR activity regulation, we determined whether DHX15-modulated AR binding to AREs on target genes using ChIP assay. C4-2 cells stably expressing DHX15 short hairpin RNA (shRNA) were established using a lentiviral shRNA expression system. Four different shRNAs elicited 60%~90% knockdown of DHX15, and consistent with results with siRNAs knockdown, all shDHX15s led to a decrease of PSA expression (Supplementary Figure S2). We used shDHX15.1 to perform the following experiments. Our ChIP assay showed knockdown of DHX15 reduced the binding of AR to the AREs of PSA and TMPRSS2 by 40% and 67% separately (Figure 2h).
To evaluate if the LBD is required for DHX15 modulation of AR transcriptional activity, we tested the effect of DHX15 knockdown or overexpression on GFP-NAR(AR lacking LBD) transactivation of PSA-luciferase reporter in C4-2 cells. The results showed that DHX15 knockdown or overexpression did not affect GFP-NAR transactivation of PSA-luciferase reporter in C4-2 cells (Supplementary Figure S3).
DHX15 regulates AR transcriptional activity by stimulating Siah2-mediated ubiquitination
To further understand DHX15 regulation of AR activity, first we assessed whether DHX15 influenced AR trafficking in PCa cells. Knockdown of DHX15 retarded, but did not completely inhibit AR nuclear translocation response to androgen (Supplementary Figures S4A and B). Because NESAR locates in LBD domain, we also tested whether loss of DHX15 affected androgen binding to AR. Knockdown of DHX15 in LNCaP cells did not influence the affinity of [3H]-labeled dihydrotestosterone for endogenous AR (Supplementary Figure S4C), indicating that DHX15 did not affect androgen-AR interaction. Because knockdown of DHX15 decreased AR activity significantly even in the absence of androgens (Figures 2a and b), we argued that AR trafficking might not be the main mechanism involved in the regulation of DHX15 on AR signaling.
AR transcriptional activity is modulated by multiple posttranslational modifications including ubiquitination.22, 32, 33, 34, 35 As our previous work demonstrated that NESAR has a key role in AR ubiquitination,11 we examined whether DHX15 modulates AR activity by regulating its ubiquitination. First, we determined the ubiquitination of endogenous AR in LNCaP cells treated with siRNA targeting DHX15 and in C4-2 cells with shRNA. Consistent with a previous report,35 androgen treatment enhanced the ubiquitination of endogenous AR (Figures 3a and b). Notably, DHX15 knockdown led to a decrease in AR ubiquitination in both LNCaP and C4-2 cells (Figures 3a and b). Overexpression of DHX15 in C4-2 cells increased ubiquitination of transfected AR (Figure 3c). To confirm that DHX15 regulated AR ubiquitination independent of ATPase activity, we assessed whether ATPase-deficient mutants of DHX15 influenced AR ubiquitination. As shown in Figure 3d, DHX15-K166A increased AR ubiquitination similarly to that of wild-type DHX15.
DHX15 regulates AR ubiquitination and Siah2 expression. (a) LNCaP cells were transfected with siRNAs targeting DHX15 or control for 24 h, subsequently cultured in CS for 24 h, and then treated with 1 nm R1881 or vehicle for another 24 h. Cell lysates were immunoprecipitated with AR antibody, and subjected to western blotting with ubiquitin and AR antibodies. The whole cell lysates were immunoblotted with DHX15, AR and tubulin antibodies. (b) C4-2 cells stably transfected with control or DHX15 shRNA were cultured in CS for 48 h, then treated with 1 nm R1881 or vehicle for 24 h. Cell lysates were analyzed as described in a. (c) C4-2 cells were transfected with Flag-AR, Myc-DHX15 and HA-Ub for 24 h, and treated with 5 μm MG132 for 16 h. Cell lysates were immunoprecipitated with Flag antibody and subjected to western blotting with ubiquitin and Flag antibody. (d) C4-2 cells were transfected with HA-Ub, Myc-AR and Flag-DHX15-WT or Flag-DHX15-K166A, the analysis was performed as described for c. (e) LNCaP and C4-2 cells were transfected with two different DHX15 siRNAs for 72 h and subjected to western blotting and probed with DHX15, MDM2, Siah2 and RNF6 antibodies. (f) C4-2 cells were transfected with siRNAs targeting DHX15 or/and Siah2 as indicated for 72 h, the analysis was performed as described for a. (g) C4-2 cells were transfected with siRNAs targeting DHX15 or/and Siah2 as indicated for 72 h, then subjected to western blotting and probed with DHX15, Siah2, PSA and AR. (h) C4-2 cells were transfected with siRNAs targeting DHX15 or/and Siah2 as indicated for 24 h, then cultured in CS for 24 h and treated with or without 1 nm R1881 for another 24 h. The luciferase activity was analyzed. Data represent average of a minimum of three technical replicates representative of three different experiments. (*P<0.05, **P<0.01).
DHX15 modulation of AR ubiquitination is likely to require AR E3 ligase(s). Thus, we determined the expression of three important AR E3 ligases MDM2, RNF6 and Siah2 (reviewed in ref. 36) in PCa cells in response to DHX15 knockdown. Knockdown of DHX15 led to a dramatic decrease of Siah2, but not of MDM2 and RNF6 expression in both LNCaP and C4-2 cells (Figure 3e). To determine whether Siah2 mediated the modulation of AR ubiquitination by DHX15, Siah2 and DHX15 were both knocked down in C4-2 cells. Although single knockdown of either DHX15 or Siah2 resulted in a significant decrease of AR ubiquitination, double knockdown of DHX15 and Siah2 did not further reduce ubiquitination of AR compared with single knockdown of Siah2 (Figure 3f). These results indicated that DHX15 regulation of AR ubiquitination is Siah2 dependent. Siah2 targets a selective pool of NCOR1-bound, transcriptionally inactive AR for ubiquitin-dependent degradation, thus enhancing AR activity and contributing to CRPC progression.22 However, this study did not address if DHX15 also impacts AR bound to NCOR1 and this question will need to be addressed in future studies. Given that the regulation of AR ubiquitination by DHX15 was mediated by Siah2, we tested whether the inhibition of AR activity by DHX15 knockdown was abrogated by Siah2 downregulation. Both single knockdown of DHX15 and Siah2 downregulated PSA expression and luciferase activity in C4-2 cells, but knockdown of DHX15 in Siah2-knockdown cells did not further reduce PSA and luciferase activity (Figures 3g and h). Although Siah2 acts as the E3 ubiquitin ligase of AR, it does not affect the expression of endogenous AR, but does decreases exogenous AR expression.22 Interestingly, knockdown of DHX15 did not change the expression of endogenous AR (Figure 2c and Supplementary Figure S4D), but increased exogenous AR significantly (Supplementary Figures S4D and E), which is consistent with what Siah2 did. These data indicate that DHX15 regulation of AR activity is mediated, at least partly, through Siah2.
DHX15 stabilizes Siah2 and enhances its E3 ligase activity
To elucidate the mechanisms of Siah2 regulation by DHX15, we first showed that DHX15 knockdown did not change Siah2 mRNA level (Figure 4a). We then co-transfected HA-Siah2 with different amount of Flag-DHX15 into 293 cells, and found Siah2 protein level was increased with increasing amount of DHX15 expression vectors (Figure 4b). As expected, the mutant DHX15-K166A showed the similar effect on Siah2 expression (Figure 4b). We also monitored Siah2 half-life in the presence of the protein synthesis inhibitor cycloheximide and found that DHX15 co-transfection prolonged Siah2 half-life from ~1 h to >3 h (Figure 4c). These findings suggested that DHX15 enhances Siah2 expression by inhibiting its degradation.
DHX15 regulates Siah2 protein stabilities and its E3 ligase activity. (a) C4-2 cells were transfected with two different siRNAs targeting DHX15 or control for 72 h. The mRNA expression of DHX15 and Siah2 were detected by qPCR. (b) 293 cells were transfected with HA-Siah2, GFP and different dose of Flag-DHX15-WT or Flag-DHX15-K166A for 48 h, cell lysates were subjected to western blotting and probed with anti-Flag and anti-HA antibodies. GFP was served as transfection efficiency control. (c) 293 cells were transfected with HA-Siah2 or/and Flag-DHX15 as indicated for 48 h and then treated with cycloheximide (CHX, 50 μg/ml) for 1, 3, 6 h and cell lysates were subjected to western blotting for detecting HA-Siah2 and Flag-DHX15 expression, GFP was served as control. The degradation curves of Siah2 by cycloheximide chase in the presence of DHX15 are shown on the right side. (d) 293 cells were transfected with Flag-DHX15, Myc-AR and HA-Siah2 as indicated, the analysis was performed as described for c and the degradation curve of Myc-AR was presented. (e) 293 cells were transfected with Flag-DHX15, Myc-Spry2 and HA-Siah2 as indicated, the analysis was performed as described for d. (f) C4-2 cells were transfected with siRNAs targeting DHX15 or/and Siah2 as indicated for 72 h and subjected to western blotting and probed with DHX15, Siah2 and Sprouty2 antibodies. (g) PC3 cells were infected with lentivirus encoding for shRNA to DHX15 or scramble, and cells lysates were subjected to western blotting and probed with DHX15, Siah2 and Spry2 antibody. Data represent average of a minimum of three technical replicates representative of three different experiments.
Like other RING finger E3 ubiquitin ligases, Siah2 limits its own expression by self-ubiquitination and autodegradation, which is a sign of its ubiquitin-ligase activity.37, 38 To test the effect of DHX15 stabilization on Siah2 on its E3 ligase activity, we monitored Siah2-mediated degradation of AR, a recently identified Siah2 substrate.22 Surprisingly, overexpression of Siah2 effectively reduced AR half-life from~6 h to ~3 h, and co-overexpression of DHX15 with Siah2 significantly upregulated Siah2 expression while further reducing AR half-life to 2 h (Figure 4d), which suggested that although Siah2 was stabilized by DHX15, its E3 ligase activity was enhanced rather than decreased. To further confirm this unexpected result, we monitored changes in Spry2, one of the classic substrates of Siah2 and a marker for Siah2 ligase activity.19, 39 Overexpression of Siah2 reduced Spry2 half-life from >4 h to 3 h, and co-expression of DHX15 with Siah2 further reduced Spry2 half-life to ~2 h (Figure 4e). In addition, as shown in Figure 4f, knockdown of either Siah2 or DHX15 in C4-2 cells increased endogenous Spry2 expression, whereas double knockdown of DHX15 and Siah2 did not result in a further decrease Spry2 expression, suggesting DHX15-modulated Spry2 expression through Siah2. To exclude the possibility that these effects were cell specific, we showed that knockdown of DHX15 by shRNA in PC3 cells also increased Spry2 expression (Figure 4g). Collectively, these data strongly suggest that DHX15 can stabilize Siah2 expression and enhance its E3 ligase activity.
DHX15, Siah2 and AR form a complex
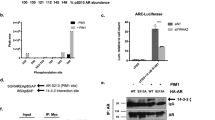
To better understand the potential mechanism of DHX15 regulating Siah2 and AR, we tested whether these three proteins form a complex. First, co-immunoprecipitation assays demonstrated that transfected AR and DHX15 interacted in COS-1 cells (Figure 5a), and endogenous AR could co-precipitate with endogenous DHX15 in LNCaP cells in the absence or presence of R1881 (Figure 5b). The endogenous AR-DHX15 interaction was enhanced in the presence of R1881 (Figure 5b), possibly owing to an increase in AR level in response to R1881 treatment.40 Considering that DHX15 is an RNA-binding protein,41 we tested whether RNA was required for DHX15 association with AR by including RNase in cell lysates to degrade RNA. In the presence of RNase, AR protein was still co-precipitated by DHX15 antibody (Supplementary Figure S5A), indicating that DHX15 binds to AR in the absence of RNA and suggesting DHX15 associates with AR independent of its RNA helicase function. To identify the regions of AR that interact with DHX15, flag-DHX15 was co-transfected with GFP-fusion proteins containing various domains of AR (Supplementary Figure S5B) separately in COS-1 cells. The GFP-constructs containing NESAR, but not those without NESAR, were co-precipitated by anti-flag antibody, suggesting that the NESAR region mediated the interaction of AR and DHX15 (Figure 5c). Co-immunoprecipitation of 293 cells co-transfected with DHX15 and Siah2 also revealed an interaction between these two proteins (Figure 5d). Siah2 consists of three different domains: N-terminal domain, central RING domain/zinc finger domain and C-terminal SBD.15 To map the Siah2 domains required for DHX15 interaction, we generated truncation mutants of Siah2 (Supplementary Figure S5C) and co-transfected them individually with DHX15 into 293 cells. The co-immunoprecipitation assay revealed that the SBD domain interacted with DHX15 (Supplementary Figure S5D). Given that AR is a substrate of Siah2 and AR binds to DHX15 as well, we investigated whether AR, DHX15 and Siah2 could form a protein complex. GFP-AR, Flag-DHX15 and HA-Siah2 were co-transfected into 293 cells, and the cell lysates from nuclear fractions were immunoprecipitated with anti-HA antibody, and immunoblotted with anti-GFP or anti-Flag antibody. As expected, these three proteins co-precipitated (Figure 5e). We also showed GFP-AR, Flag-DHX15-K166A and HA-Siah2 co-precipitated (Figure 5f), further suggesting that DHX15 form a complex with AR and Siah2 independent of its RNA helicase function.
DHX15, Siah2 and AR form a complex. (a) COS-1 cells were transiently transfected with Flag-vector, Myc-vector, Flag-DHX15 and/or Myc-AR as indicated for 48 h. Cell lysates were immunoprecipitated with antibodies against Flag or Myc. The associated proteins were immunoblotted to detect existence of Flag-DHX15 and Myc-AR in the precipitate. (b) LNCaP cells were treated with vehicle or 1 nm R1881 for 24 h, Cell lysates were immunoprecipitated with antibodies against AR or DHX15, and the associated proteins were probed with DHX15 and AR antibodies. (c) COS-1 cells were transiently co-transfected with Flag-DHX15 and different AR domains as indicated. Cell lysates were immunoprecipitated with antibodies against Flag. The associated proteins were immunoblotted with GFP antibody. (d) 293 cells were transfected with Flag-DHX15 and/or HA-Siah2 for 48 h. Cell lysates were immunoprecipitated with Flag antibody and subjected to western blotting. (e) 293 cells were transfected with Flag-DHX15, GFP-AR and/or HA-Siah2 for 48 h, cell lysates were immunoprecipitated with HA antibody and subjected to western blotting and probed with anti-Flag, GFP and HA antibodies. (f) 293 cells were transfected with Flag-DHX15-K166A, GFP-AR and/or HA-Siah2 for 48 h, nuclear fractions were extracted and were immunoprecipitated with HA antibody and subjected to western blotting and probed with anti-Flag, GFP and HA antibodies. Data are representative of three different experiments.
DHX15 is required for PCa cell growth in vitro and in vivo
As AR plays a key role in regulating PCa cell growth and motility, and cell growth was inhibited by mutation of Prp43 in the yeast mutants (see Figure 1b), we next evaluated the effect of DHX15 on PCa cells growth and motility. Knockdown of DHX15 reduced proliferation of AR-positive PCa cells LNCaP and C4-2, but not of AR-negative cells PC3 and DU145 (Figure 6a and Supplementary Figure S6A). Furthermore, single knockdown of DHX15 or AR caused similar inhibition of proliferation, but double knockdown of DHX15 and AR did not further reduce cell proliferation in either LNCaP or C4-2 cells (Figures 6b and c). These findings suggested the DHX15 effect on cell proliferation was AR-dependent and vice versa. Because Siah2 mediates DHX15 regulation of AR activity, we then evaluated whether Siah2 could mediate DHX15 effect on proliferation. As expected, knockdown of DHX15 or Siah2 individually reduced cells proliferation and double knockdown of these two genes did not further inhibit proliferation (Figure 6d). These findings suggest that Siah2 can mediate the DHX15 regulation of AR-positive cell growth and are consistent with the report that Siah2 effect on cell proliferation is AR-dependent.22 Similarly, knockdown of DHX15 reduced colony formation in C4-2 cells both in the presence or absence of 1 nm dihydrotestosterone, but no apparent effects were observed in PC3 cells (Figure 6e and Supplementary Figures S6B and C). Further, knockdown of DHX15 inhibited C4-2 cells transwell migration, whereas, knockdown of DHX15 in PC3 cells showed no effect on cell migration (Figure 6f and Supplementary Figure S6D). Collectively, these data suggest that DHX15 plays an important role in regulating AR-dependent growth and motility in PCa cells, and that this regulation is mediated through Siah2.
DHX15 knockdown inhibits PCa cell growth in vitro and in vivo. (a) LNCaP, C4-2, PC3 and DU145 cells were transfected with DHX15 siRNA for 72 h, then treated with 10 μm BrdU for 5 h (LNCaP) or 2 h (C4-2, PC3 and DU145). BrdU assay was performed as described in materials and methods. (b, c) LNCaP and C4-2 cells were transfected with siRNAs targeting DHX15 or/and AR as indicated for 72 h, BrdU assay was performed as described for a. (d) C4-2 cells were transfected with siRNAs targeting DHX15 or/and Siah2 as indicated for 72 h, BrdU assay was performed as described above. (e) C4-2 cells stably transfected with control or DHX15 shRNA were cultured in CS with or without 1 nm DHT in six-well plate for 2 weeks. Cells were then stained with 0.1% crystal violet for 30 min, and the number of colonies was quantified. (f) C4-2 or PC3 cells were transfected with DHX15 siRNA or shRNA as indicated and cultured in chambers as described in materials and methods. After 24 h (PC3) or 36 h (C4-2), cells were stained with crystal violet and the number of cells per field was quantified. (g) Tumor growth curve. (h) The mice survival curve. The number of animals is 10 in shSCR group and 9 in shDHX15 group, no animals were excluded from study. (*P<0.05, **P<0.01).
To evaluate the role of DHX15 in vivo, we established C4-2 tumor xenografts with DHX15 stable knockdown (C4-2/shDHX15) and control cells (C4-2/shScramble) in severe combined immunodeficiency mice. Knockdown efficiency of DHX15 in C4-2/shDHX15 cells was confirmed with western blotting and real-time PCR before injection (Supplementary Figures S7A and B). C4-2/shScramble cells established tumors much earlier and more frequently (8/10 animals had tumors at day 21), whereas 2/9 (22.2%) animals had tumors at day 21 in C4-2/shDHX15 cells. Consistently, tumors derived from DHX15 knockdown cells grew slower than those from the control group (Figure 6g). The mice in shDHX15 group had a significantly shorter mean survival time compared with those in shScramble group (Figure 6h). Unfortunately, DHX15 protein levels eventually rebounded in several of the shDHX15 xenograft tumors as determined by western blotting (Supplementary Figure S7C), which might alleviate part of the tumor repression in the shDHX15 group. Nevertheless, the significant decrease in the establishment and the lower growth rate of C4-2 xenografts in the shDHX15 group suggests that DHX15 is important for prostate xenograft tumor establishment and growth.
DHX15 expression is upregulated in human PCa and is correlated to PSA recurrence
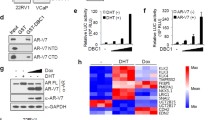
To evaluate the expression of DHX15 in clinical prostate tumor specimens, we performed immunostaining analysis of a tissue microarray (TMA) of PCa specimens from the Prostate Cancer Biorepository Network (PCBN). The TMA contained 199 different Gleason grade tumors, 11 benign prostate hyperplasia and 24 normal prostate samples. DHX15 showed a nuclear expression pattern both in tumors and in benign tissues (Figure 7a). Significantly, the expression of DHX15 was higher in PCa tissues than in benign prostate hyperplasia and normal tissues (P=0.0004) (Figure 7b). DHX15 exhibited a much higher expression in the tumor cells compared with the adjacent normal epithelial cells in PCa tissue cores (Figure 7a), and DHX15 expression was higher in the cases of Gleason score ⩾7 compared with those of Gleason score <7 (P=0.0056) (Figure 7c). Notably, cases with strong and moderate DHX15 expression had PSA recurrence at a rate of 23.4% and 19.8%, respectively, whereas only 5.1% of the cases with weak DHX15 staining developed PSA recurrence (P=0.0258) (Figure 7d), indicating that high expression of DHX15 in PCa tissues was associated with biochemical recurrence. In a separate TMA obtained from the University of Pittsburgh Prostate Tumor Bank consisting of 89 prostate tumor specimens, DHX15 immunostaining was found to correlate with Siah2 immunostaining (Figures 7e and f), which was consistent with the results in vitro that DHX15 stabilized Siah2 expression.
DHX15 expression in PCa samples. (a) Representative images of DHX15 IHC staining on the PCa TMA. (b) Quantification of DHX15 IHC staining on the BPH, PCa or tumor adjacent normal samples. P=0.0004. (c) Quantification of DHX15 IHC staining on PCa samples of different Gleason scores. P=0.0056. (d) Quantification of recurrence in different DHX15 expression groups. P=0.0258. (e) Representative images of DHX15 and SIAH2 IHC staining on the HSTB TMA. (f) Scatter plot of DHX15 and SIAH2 IHC staining in HSTB TMA. Statistical Pearson correlation was −0.651 (P = 0.003) and Spearman Coefficient was 0.21 (P < 0.05). All the above data were analyzed with Kruskal−Wallis test.
Discussion
Understanding the mechanisms AR activation in prostate carcinogenesis is fundamentally important and clinically relevant. Here, based on a yeast genetic screen, we identified the RNA helicase DHX15 as a novel AR co-activator and revealed that DHX15 regulates AR activity by modulating E3 ligase Siah2-mediated ubiquitination independent of its ATPase activity in PCa cells. Our studies showed AR, DHX15 and Siah2 form a complex, most likely through the scaffold role of NESAR (see Figure 5). DHX15 stabilizes Siah2 and enhances its E3 ubiquitin-ligase activity, resulting in activation of AR. We demonstrated DHX15 is required for the growth of C4-2 tumor xenografts (see Figure 6). Importantly, we further found DHX15 was upregulated in PCa and its expression was highly correlated with Gleason scores and PSA recurrence (see Figure 7). Furthermore, expression of DHX15 was positively correlated with Siah2 expression in prostate tumor specimens. Collectively, our findings suggest that DHX15 functions as an important co-activator of AR via Siah2 and its upregulation may contribute to the PCa progression.
A growing body of evidence shows that abnormal expression of splicing factors correlates with cancer development and progression.42 Several splicing factors including p54nrb, p68, Sam68 and PSF have been identified as AR co-activators or co-repressors.40, 43, 44 In the present study, we showed for the first time that DHX15 was upregulated in human PCa samples and promoted AR activity. DHX15 stimulation of AR activity should be mediated by its binding to AR through NESAR. Although DHX15 knockdown appeared to slow down androgen-induced AR nuclear localization, most of the AR was translocated into the nuclei in DHX15 knockdown cells when treated with androgens for a longer time. This suggests that DHX15 can regulate the dynamics of AR intracellular trafficking but not the final state of AR distribution between nuclei and cytoplasm following androgen induction. Further studies will be needed to test whether regulation of AR trafficking dynamics could be an important mechanism for DHX15 to modulate AR activity.
AR activity is modulated by posttranslational modifications including ubiquitination. We have previously reported that NESAR has an important role in the ubiquitin-dependent degradation of AR, however, it was unclear how NESAR mediates AR ubiquitination. In this study, we found knockdown of DHX15 resulted in a decreased ubiquitination of AR both in androgen-sensitive LNCaP and in CRPC C4-2 cells. Given that DHX15 binds to AR through NESAR, we hypothesized that NESAR plays the key role in DHX15 stimulation of AR ubiquitination. However, protein sequence analysis did not identify any recognizable ubiquitination signal within NESAR.11 Also, mutation of the only two lysine residues within NESAR did not block the ubiquitination of NESAR fusion protein (unpublished observations). One possibility is that NESAR itself is not the target of ubiquitination, but functions as a scaffold for the recruitment of an ubiquitin ligase. As DHX15 is not predicted as an E3 ligase based on its a.a. sequence, DHX15 modulation of NESAR-mediated ubiquitination is indirect and involves another protein with E3 ligase activity. Several E3 ubiquitin ligases are capable of controlling AR stability and activity, including CHIP, MDM2, RNF6 and Siah2, among which RNF6 and Siah2 are considered contributing to the progression of CRPC.22, 35 As knockdown of DHX15 leads to a significant decreased expression of Siah2, but not of MDM2 and RNF6, Siah2 is likely to be a key E3 ligase mediating DHX15 regulation of AR ubiquitination. It is noteworthy that both of the two major Siah2-binding sites on AR locate in the NESAR region,22 which means Siah2 should bind to NESAR. Our co-IP assay demonstrated that through NESAR, AR binds to DHX15 and Siah2 to form a complex, supporting a model that DHX15 enhances Siah2 level and the latter promotes the ubiquitination of AR via NESAR in LBD. This model is also consistent with the finding that DHX15 knockdown or overexpression did not affect the transactivation of PSA-luciferase reporter by GFP-NAR (AR lacking LBD), which does not bind to DHX15 or Siah2, in C4-2 cells. Interestingly, we found that knockdown of DHX15 increases the expression of transfected AR, but not of endogenous AR (see Figures 2c–e), which was consistent with the results from Qi et al.22, 45 that knockdown of Siah2 increases the expression of transfected AR, but not of endogenous AR, and the mechanisms still need to be elucidated in the future.
Siah2 is very unstable owing to its self-ubiquitination and autodegradation.37, 38, 45 Like other Ring finger E3 ubiquitin ligases, the ligase activity of Siah2 is reflected by its protein stability. Several factors including deubiquitinating enzyme USP13, Ski and vitamin K3 have been reported to stabilize Siah2 expression, simultaneously inhibit its E3 ligase activity.39, 46, 47 In contrast, the E3 ligase activity of Siah2 was enhanced, when its expression was stabilized by DHX15. This unexpected result was based on the reduction of two different substrates of Siah2, AR and Spry2, when Siah2 is stabilized by DHX15. In addition, knockdown of DHX15 led to a significant decrease of endogenous Siah2 and an increase of Spry2 both in C4-2 and PC3 cells. Overall, these findings point to a positive correlation between Siah2 expression and activity, which is contrary to the previous reports. Based on our findings that DHX15 binds to Siah2 through the SBD domain, but not the Ring finger domain which is critical for the ligase activity, one of the possibilities is the binding spatially blocks the self-ubiquitination sites on SBD, whereas the ubiquitin-ligase activity of Siah2 and its ability of binding to the other substrates are not inhibited.
We showed significant upregulation of DHX15 in PCa samples compared to benign prostate hyperplasia samples or tumor adjacent normal tissues. In addition, DHX15 expression showed a positive correlation to Gleason scores and PSA recurrence, suggesting that DHX15 has an important role in PCa progression. Furthermore, DHX15 expression correlated positively to Siah2, although the correlation was not strong. This probably reflects differential regulation of the two genes in prostatic cells. The reason for a weak correlation may be in part due to the stabilization of Siah2 by DHX15 in PCa cells. One limitation of our study was the lack of CRPC specimens, and it will be worthwhile to investigate the expression of DHX15 in CRPC specimens.
In summary, our study provided insights into an important role for DHX15 in PCa progression, and also revealed a novel mechanism that DHX15 stimulates AR activity through Siah2-mediated ubiquitination independent of its ATPase activity. DHX15 overexpression is associated with PCa recurrence and provides a potential new molecular target for the treatment of advanced PCa.
Materials and methods
Cell culture
The human PCa cell lines LNCaP, PC3 and DU145, and HEK 293 cells and COS-1 cells were obtained from American Type Culture Collection (Manassas, VA, USA), and C4-2 cells were a generous gift from Dr Leland WK Chung. Cells were maintained in the appropriate medium (Roswell Park Memorial Institute medium-1640 for LNCaP, PC3, DU145 and C4-2; Dulbecco's Modified Eagle's medium for HEK 293 and COS-1) supplied with 10% fetal bovine serum, 5% antibiotics and 1% l-glutamine at 37 °C with 5% CO2. Cells were verified as mycoplasma free by PCR. Cells were cultured in phenol red-free medium supplied with 5% dextran-coated charcoal-stripped fetal bovine serum for 24 h before treatment with methyltrienolone (R1881, PerkinElmer, Waltham, CA, USA) or dihydrotestosterone (Sigma-Aldrich, St Louis, MO, USA). In some experiments, cells were treated with protein synthesis inhibitor cycloheximide (Sigma-Aldrich) at 50 μg/ml and/or proteasome inhibitor MG132 (Sigma-Aldrich) at 5 μm for various times as described in the figure legends.
Immunohistochemistry and TMA
The TMAs were obtained from the PCBN and the surgical pathology archives of the University of Pittsburgh Prostate Tumor Bank from de-identified tumor specimens consented for research at time of treatment. Use of these prostate tissues was approved by the PCBN and the University of Pittsburgh Institutional Review Board. Immunohistochemical staining was performed on the paraffin embedded TMA using DHX15 (sc-271686, (E-6), Santa Cruz Biotechnologies, Santa Cruz, CA, USA) and Siah2 antibodies (NB110-88113, (24E6H3), Novus Biologicals, Littleton, CO, USA). TMAs were scored on a standard intensity (negative, weak, moderate, strong) proportion (0–100%) scale by two investigators in a blinded fashion (FMD, LEP). Staining intensity was correlated with clinicopathologic variables including disease, Gleason Score and recurrence.
Animals and xenograft tumors
Male severe Combined immunodeficiency mice at 8 weeks of age were randomized into two groups, the knockdown group (n=10) and the control group (n=9), for subcutaneous injection of 300 μl of C4-2/shDHX15 or C4-2/shScramble cells (1 × 106) mixed 1:1 with Matrigel (Invitrogen) in the flank of each mouse. Tumors were measured with calipers every other day. Tumor volume was calculated using the formula length × width2 × 0.52. Tumor take rate was calculated 10 weeks after injection. Mice were castrated once the tumor volume reached 0.8 ml. All animal studies were conducted in accordance with the University of Pittsburgh Institutional Animal Care and Use Committee guidelines.
Statistical analysis
The data are presented as mean±s.e.m. or mean±s.d. Statistical analyses were performed with Student’s t-test, one-way analysis of variance or Kruskal–Wallis test. P<0.05 was considered statistically significant.
References
Siegel RL, Miller KD, Jemal A . Cancer statistics, 2017. CA Cancer J Clin 2017; 67: 7–30.
Hellerstedt BA, Pienta KJP . The cturrent state of hormonal therapy for prostate cancer. CA Cancer J Clin 2002; 52: 154–179.
Chan SC, Dehm SM . Constitutive activity of the androgen receptor. Adv Pharmacol 2014; 70: 327–366.
Kobayashi T, Inoue T, Kamba T, Ogawa O . Experimental evidence of persistent androgen-receptor-dependency in castration-resistant prostate cancer. Int J Mol Sci 2013; 14: 15615–15635.
Gelmann EP . Molecular biology of the androgen receptor. J Clin Oncol 2002; 20: 3001–3015.
Lonergan PE, Tindall DJ . Androgen receptor signaling in prostate cancer development and progression. J Carcinog 2011; 10: 20.
Picard D, Yamamoto KR . Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J 1987; 6: 3333–3340.
Poukka H, Karvonen U, Yoshikawa N, Tanaka H, Palvimo JJ, Janne OA . The RING finger protein SNURF modulates nuclear trafficking of the androgen receptor. J Cell Sci 2000; 113: 2991–3001.
Saporita AJ, Zhang Q, Navai N, Dincer Z, Hahn J, Cai X et al. Identification and characterization of a ligand-regulated nuclear export signal in androgen receptor. J Biol Chem 2003; 278: 41998–42005.
Zhou ZX, Sar M, Simental JA, Lane MV, Wilson EM . A ligand-dependent bipartite nuclear targeting signal in the human androgen receptor. Requirement for the DNA-binding domain and modulation by NH2-terminal and carboxyl-terminal sequences. J Biol Chem 1994; 269: 13115–13123.
Gong Y, Wang D, Dar JA, Singh P, Graham L, Liu W et al. Nuclear export signal of androgen receptor (NESAR) regulation of androgen receptor level in human prostate cell lines via ubiquitination and proteasome-dependent degradation. Endocrinology 2012; 153: 5716–5725.
Nguyen MM, Harmon RM, Wang Z . Characterization of karyopherins in androgen receptor intracellular trafficking in the yeast model. Int J Clin Exp Pathol 2014; 7: 2768–2779.
Ferraldeschi R, Welti J, Luo J, Attard G, de Bono JS . Targeting the androgen receptor pathway in castration-resistant prostate cancer: progresses and prospects. Oncogene 2015; 34: 1745–1757.
Shafi AA, Yen AE, Weigel NL . Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol Ther 2013; 140: 223–238.
Reed JC, Ely KR . Degrading liaisons: Siah structure revealed. Nat Struct Biol 2002; 9: 8–10.
Fukuba H, Takahashi T, Jin HG, Kohriyama T, Matsumoto M . Abundance of aspargynyl-hydroxylase FIH is regulated by Siah-1 under normoxic conditions. Neurosci Lett 2008; 433: 209–214.
Matsuzawa SI, Reed JC . Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol Cell 2001; 7: 915–926.
Nadeau RJ, Toher JL, Yang X, Kovalenko D, Friesel R . Regulation of Sprouty2 stability by mammalian Seven-in-Absentia homolog 2. J Cell Biochem 2007; 100: 151–160.
Nakayama K, Frew IJ, Hagensen M, Skals M, Habelhah H, Bhoumik A et al. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell 2004; 117: 941–952.
Zhang J, Guenther MG, Carthew RW, Lazar MA . Proteasomal regulation of nuclear receptor corepressor-mediated repression. Genes Development 1998; 12: 1775–1780.
Nakayama K, Qi J, Ronai Z . The ubiquitin ligase Siah2 and the hypoxia response. Mol Cancer Res 2009; 7: 443–451.
Qi J, Tripathi M, Mishra R, Sahgal N, Fazli L, Ettinger S et al. The E3 ubiquitin ligase Siah2 contributes to castration-resistant prostate cancer by regulation of androgen receptor transcriptional activity. Cancer Cell 2013; 23: 332–346.
Freeman MR . The ubiquitin ligase Siah2 is revealed as an accomplice of the androgen receptor in castration resistant prostate cancer. Asian J Androl 2013; 15: 447–448.
Arenas JE, Abelson JN . Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc Nat Acad Sci USA 1997; 94: 11798–11802.
Combs DJ, Nagel RJ, Ares M Jr, Stevens SW . Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol Cell Biol 2006; 26: 523–534.
Tanaka N, Aronova A, Schwer B . Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev 2007; 21: 2312–2325.
Fourmann JB, Schmitzova J, Christian H, Urlaub H, Ficner R, Boon KL et al. Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev 2013; 27: 413–428.
Koodathingal P, Novak T, Piccirilli JA, Staley JP . The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5' splice site cleavage during pre-mRNA splicing. Mol Cell 2010; 39: 385–395.
Walbott H, Mouffok S, Capeyrou R, Lebaron S, Humbert O, van Tilbeurgh H et al. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J 2010; 29: 2194–2204.
Brown JD, Beggs JD . Roles of PRP8 protein in the assembly of splicing complexes. EMBO J 1992; 11: 3721–3729.
Mosallanejad K, Sekine Y, Ishikura-Kinoshita S, Kumagai K, Nagano T, Matsuzawa A et al. The DEAH-box RNA helicase DHX15 activates NF-kappaB and MAPK signaling downstream of MAVS during antiviral responses. Sci Signal 2014; 7: ra40.
Coffey K, Robson CN . Regulation of the androgen receptor by post-translational modifications. J Endocrinol 2012; 215: 221–237.
Chymkowitch P, Le May N, Charneau P, Compe E, Egly JM . The phosphorylation of the androgen receptor by TFIIH directs the ubiquitin/proteasome process. EMBO J 2011; 30: 468–479.
Gaughan L, Logan IR, Neal DE, Robson CN . Regulation of androgen receptor and histone deacetylase 1 by Mdm2-mediated ubiquitylation. Nucleic Acids Res 2005; 33: 13–26.
Xu K, Shimelis H, Linn DE, Jiang R, Yang X, Sun F et al. Regulation of androgen receptor transcriptional activity and specificity by RNF6-induced ubiquitination. Cancer Cell 2009; 15: 270–282.
Li B, Lu W, Chen Z . Regulation of androgen receptor by E3 ubiquitin ligases: for more or less. Receptors Clin Investig 2014; 1; doi:10.14800%2Frci.122.
Hu G, Fearon ER . Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol Cell Biol 1999; 19: 724–732.
Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM . RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA 1999; 96: 11364–11369.
Scortegagna M, Subtil T, Qi J, Kim H, Zhao W, Gu W et al. USP13 enzyme regulates Siah2 ligase stability and activity via noncatalytic ubiquitin-binding domains. J Biol Chem 2011; 286: 27333–27341.
Clark EL, Coulson A, Dalgliesh C, Rajan P, Nicol SM, Fleming S et al. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res 2008; 68: 7938–7946.
Fouraux MA, Kolkman MJ, Van der Heijden A, De Jong AS, Van Venrooij WJ, Pruijn GJ . The human La (SS-B) autoantigen interacts with DDX15/hPrp43, a putative DEAH-box RNA helicase. RNA 2002; 8: 1428–1443.
Grosso AR, Martins S, Carmo-Fonseca M . The emerging role of splicing factors in cancer. EMBO Rep 2008; 9: 1087–1093.
Dong X, Sweet J, Challis JR, Brown T, Lye SJ . Transcriptional activity of androgen receptor is modulated by two RNA splicing factors, PSF and p54nrb. Mol Cell Biol 2007; 27: 4863–4875.
Rajan P, Gaughan L, Dalgliesh C, El-Sherif A, Robson CN, Leung HY et al. The RNA-binding and adaptor protein Sam68 modulates signal-dependent splicing and transcriptional activity of the androgen receptor. J Pathol 2008; 215: 67–77.
Sequoia Ecosystem and Recreation Preserve Act of 1999. 106th Congress ed, US House of Representatives1999.
Haider AS, Kaye G, Thomson A . Autoimmune hepatitis in a demographically isolated area of Australia. Int MedJ 2010; 40: 281–285.
Zhao HL, Ueki N, Hayman MJ . The Ski protein negatively regulates Siah2-mediated HDAC3 degradation. Biochem Biophys Res Commun 2010; 399: 623–628.
Acknowledgements
We thank Aiyuan Zhang for technical support, Richard Gaber (Northwestern University) for advice on yeast screen, the PCBN for TMA with PCa specimens, Megan Lambert, Krystal Roskov and Robin Frederick for animal husbandry, and members of the Wang Lab for discussion. This work was supported in part by NIH R01 CA186780 (ZW), NIH R01 CA108675 (ZW), ACS #PF-05-229-01-CSM (MMN), the Department of Defense Prostate Cancer Research Program Award No W81XWH-14-2-0182, W81XWH-14-2-0183, W81XWH-14-2-0185, W81XWH-14-2-0186, and W81XWH-15-2-0062 PCBN, National Nature Science Foundation of China #81402091 (YJ), and National Natural Science Foundation of China Key Project 81130046 (JZ) and 2013GXNSFEA053004 (JZ). This work was also supported by a post-doctoral fellowship from the Urology Care Foundation of the American Urological Association (DW), the Mellam Family Fellowship (DW and KZM), the Tippens Scholarship (LEP) and NIH R50 CA211242-01 (LEP). In addition, this research was supported by the UPCI Animal Facility, Vector Core and Tissue and Research Pathology Services (TARPS) funded through NCI CCSG P30CA047904.
Author contributions
Yifeng Jing conceived and performed the majority of experiments and wrote manuscript. Minh M Nguyen performed the yeast screening. Dan Wang constructed the plasmids of flag-DHX15 and myc-DHX15, performed radioactive ligand-binding assay and ChIP and assisted with technical details and experimental design. Laura E Pascal performed animal experiments and analysis of the animal data, and helped with manuscript. Wenhuan Guo constructed the truncated Siah2 plasmids, performed parts of BrdU assay and several extractions of proteins and RNAs. Yadong Xu generated data for Supplementary Figures S1 and S3. Junkui Ai constructed the plasmid of Flag-AR and C4-2-AR, assisted with technical details. Fang-ming Deng performed IHC. Khalid Masoodi constructed and standardized the primers of DHX15. Xinpei Yu performed some of the ubiquitination IP assays. Shujie Xia, Jian Zhang, Joel B Nelson conceived study and helped with manuscript. Zhou Wang conceived and directed study, and helped with manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Jing, Y., Nguyen, M., Wang, D. et al. DHX15 promotes prostate cancer progression by stimulating Siah2-mediated ubiquitination of androgen receptor. Oncogene 37, 638–650 (2018). https://doi.org/10.1038/onc.2017.371
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.371
- Springer Nature Limited
This article is cited by
-
Fusobacterium nucleatum promotes tumor progression in KRAS p.G12D-mutant colorectal cancer by binding to DHX15
Nature Communications (2024)
-
The DNA/RNA helicase DHX9 contributes to the transcriptional program of the androgen receptor in prostate cancer
Journal of Experimental & Clinical Cancer Research (2022)
-
The RNA helicase DHX15 is a critical regulator of natural killer-cell homeostasis and functions
Cellular & Molecular Immunology (2022)
-
Functional roles of E3 ubiquitin ligases in prostate cancer
Journal of Molecular Medicine (2022)
-
DDX52 knockdown inhibits the growth of prostate cancer cells by regulating c-Myc signaling
Cancer Cell International (2021)