Abstract
Ribosomes, which are important sites for the synthesis of proteins related to expression and transmission of genetic information in humans, have a complex structure and diverse functions. They consist of a variety of ribosomal proteins (RPs), ribosomal RNAs (rRNAs) and small nucleolar RNAs. Owing to the involvement of ribosomes in many important biological processes of cells, their major components, rRNAs and RPs, have an important role in human diseases, including the initiation and evolvement of malignancies. However, the main mechanisms underlying the involvement of ribosomes in cancer remain unclear. This review describes the crucial role of ribosomes in various common malignant tumors; in particular, it examines the effects of RPs, including S6, the receptor for activated C-kinase and RPS15A, on the development and progression of hepatocellular carcinoma.
Similar content being viewed by others
Introduction
The ribosome, a large macromolecular substance that has a complex structure and consists of >80 unique ribosomal proteins (RPs) of varying size, is the factory for protein biosynthesis; thus, it is essential for some important cellular processes.1, 2 This cellular organelle is primarily involved in the translation of genetic information. The key components of ribosomes, RPs, perform some extra-ribosomal functions related to proliferation, differentiation, DNA repair, apoptosis and other biological processes in cell.3 Ribosome biogenesis, which is fundamental to all life forms, involves highly coordinated processes, including the synthesis of RPs in the cytoplasm, the synthesis and modification of ribosomal RNAs (rRNAs; 5SRNA in the nucleoplasm; 5.8SRNA, 18SRNA and 28SRNA in the nucleolus), the importation of rRNAs into the nucleus, the assembly of RPs and rRNAs in the nucleoplasm and the transportation of the two mature subunits (40S and 60S) into the cytoplasm.4, 5, 6 Abnormalities in the synthesis or structure of ribosomes that affect protein synthesis are interrelated to a variety of human diseases, including hematologic disorders and cancer.7, 8, 9 As a result of its high recurrence rate and poor prognosis, cancer is an urgent and challenging research topic that has received considerable attention from researchers investigating its etiology, pathogenesis, early diagnosis and treatment. At present, scientific research on the pathology of tumor development has reached the micromolecular level and has begun to focus on the function and effect of ribosomes in cancer occurrence and progression.1, 10, 11 This review mainly elucidates the crucial role of ribosomes in various common malignant tumors; in particular, it concentrates on the role of three RPs (RPS6, receptor for activated C-kinase (RACK1) and RPS15A) in the occurrence and development of HCC with the aim of providing a reference for the discovery of new prognostic or diagnostic markers and therapeutic targets for tumors.12, 13, 14, 15, 16, 17, 18
Structure of ribosomes
Ribosomes, which are large macromolecule compounds that universally exist in cells, act as a molecular machine that regulates protein synthesis and accurately monitors the translation process in the cells.19 The ribosome’s complex structure is constructed in the nucleolus through a large number of temporally and spatially ordered steps, such as processing and modification.20 Both the structure of ribosomes and the rRNA/protein ratio vary among different parts of a cell, such as the mitochondria and cytoplasm.21 Mitochondria ribosomes (that is, ‘mitoribosomes’) have special structure, which are dedicated to the expression of genetic information encoded by mitochondria genomes.22 In addition, the ribosomes in prokaryotes (70S) and eukaryotes (80S) differ considerably in structure.19 Nevertheless, both of the 70S and 80S ribosomes assemble asymmetrically with two ribosomal subunits of different sizes. A prokaryotic 70S ribosome is built up of a 50S large subunit and a 30S small subunit with a molecular weight of 2.3 MDa. An eukaryotic 80S ribosome has a molecular weight of 4.3 MDa and consists of a 40S subunit containing approximately 33 proteins and one single RNA chain (18S, 1900 nucleotides) and a 60S subunit containing 28S, 5.8S and 5SRNAs, and approximately 47 proteins.4, 5, 19 Essentially, the ribosome is a kind of RNA-based, universally conserved macromolecule. In a mammalian system, a ribosome contains approximately 80 RPs, 4 rRNAs and 70 small nucleolar RNAs (snoRNAs), and each ribosomal component has an independent and special function. RPs have an important role in promoting the folding of rRNAs to form a functional three-dimensional structure during rRNA processing and in stabilizing the final spatial conformation of the ribosome. The main function of rRNA is to catalyze formation of peptide bond during the process of protein synthesis. Meanwhile, RPs collaborate with rRNA to catalyze the protein synthesis process. The role of snoRNAs is primarily to regulate chemical modifications of other RNAs.7 Exceptionally, compared with their prokaryotic counterparts, some ribosomes in multicellular eukaryotes have additional rRNA ingredients called ‘expansion segments (ESs)’, which modulate ribosome assembly and function, and some additional protein moieties that contribute to ribosome biogenesis.23, 24 Despite the abovementioned significant structural differences, prokaryotic and eukaryotic ribosomes share the same structural center for major functions, involving the decoding site, the peptidyl transferase center and the transfer ribonucleic acid (tRNA)-binding site.19 Although the composition of ribosomes has been preliminarily revealed, many unknown, complex areas need to be further explored.
RP synthesis and ribosome biogenesis
In a living cell, a ribosome acts as a cellular translational machinery that is primarily in charge of translating messenger RNA (mRNA), which carries the genetic information as a template for protein synthesis, into amino-acid chains in the cytoplasm during protein synthesis. During the translation process, they have different roles and function independently, but they coordinate closely with one another, including their capability to contact at initiation, revolve during elongation and separate after protein release.25, 26 The small subunit, which is the initial site of translation and is responsible for decoding genetic information, recruits not only translation factors, tRNAs, and mRNA, but also binds with the large subunit to begin protein production.27 It contains three main functional sites, including the mRNA path that leads mRNA to translate, the decoding center where mRNA is decoded and three tRNA-binding sites (entry site, A; peptidyl-tRNA site, P; and exit site, E). Site A, which is the entrance for the first procedure of nascent peptide chain extension, binds to the aminoacyl-tRNA selected by the mRNA sequence entering the ribosome. The combination of mRNA with aminoacyl-tRNA to the ribosome lead to a translation initiation complex formation. During the process of translation, tRNA translocates from site A to site P and then to site E. Site P holds peptidyl-tRNA, which carries the nascent polypeptide chain. Site E (exit site) is where tRNA separates from the ribosome after completion of the translation process. The large subunit of the ribosome participates in catalyzing the peptide bond formation. This subunit also has three functional centers including A, P and E sites, a peptidyl transferase center (PTC), and a peptide exit passage where the nascent polypeptide chain extends from the middle of the large subunit. The PTC and the A, P and E sites all located on the interface between the two different ribosomal subunits. The PTC, which is situated at the entrance to the peptide passage in a conserved area mainly consisting of rRNA, is the core catalyzing site contributing to peptide bond formation. As a result of forming peptide bond in the PTC, the nascent polypeptide chain is transferred from the P site to the AA-tRNA in A site, thus extending from one amino acid to a peptide chain. At the end, the growing peptide chain emerges at the solvent side from the peptide exit passage before completion of the translation process.19
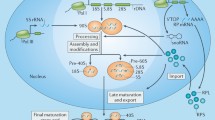
As a fundamental factor involved in all life activities, synthesis of protein is crucial for cell survival, and its rate determines the speed of cell growth and proliferation. Similarly, the rapid proliferation of cancer cells requires an increased rate of protein synthesis, which depends on the number of ribosomes. To achieve the increasing demand for protein in the process of growth and proliferation, cells have to enhance their ability to synthesize proteins by upregulating ribosomal biosynthesis.28 In eukaryotic cells, ribosome biosynthesis is an essential and complex process involving a large amount of temporally and spatially linked steps (Figure 1), including the regulation of RP binding and rRNA remodeling in the nucleoli, nucleus and cytoplasm.29 The assembly of a mature 80S eukaryotic ribosome with two mature subunits requires 3 RNA polymerases, 75 snoRNAs and approximately 200 non-ribosomal factors, including helicases, isomerases, methyltransferases, and exo- and endonucleases that modify the nascent rRNA, all of which are involved in transcription, translation, and the import and export of functional factors. First, the rRNA gene (rDNA) is transcribed to 47S pre-rRNA. This process requires the involvement of RNA polymerase I (RNA pol I) and a lot of transcription factors (TFs), such as selectivity factor 1, upstream binding factor and so forth. Subsequently, through highly specific chemical modifications (pseudouridylation and methylation) and processing by hundreds of non-RPs and snoRNAs, and 47S pre-rRNA is carved to produce 18S RNA, which is demanded for the synthesis of the pre-40S ribosomal small subunit, and the 5.8S and 28S RNAs, which are required for the synthesis of the pre-60S ribosomal large subunit. Post-transcription modification of 47S pre-rRNA requires the involvement of small nucleolar ribonucleoproteins and extra protein-processing factors. Moreover, 47S pre-rRNA synthesis is precisely regulated by RNA pol I, and the process is mediated by small nucleolar ribonucleoproteins, whereas 5S rRNA synthesis in the nucleus is monitored by RNA pol III. The pre-40S ribosomes and pre-60S ribosomes are assembled from a large number of RPs before their export. Finally, the 40S and 60S ribosome subunits are transported to cytoplasm, respectively, where they combined with mRNA to build up a functional ribosome.20, 28 RPs are the key component in the basic assembly of ribosomes. Recent studies have found that RPs not just have a role in the cell translation and protein synthesis. They also have extra-ribosomal functions that are involved in cell proliferation,30, 31, 32, 33differentiation,34, 35 apoptosis,36, 37 DNA repair,38, 39 the modulation of cell migration and invasion40 and other cellular processes.
A brief schematic of eukaryotic ribosome biogenesis. In eukaryotic cells, ribosome biosynthesis is a complex cellular process involving a great number of temporally and spatially steps with three RNA polymerases. First, the rRNA gene (rDNA) will transcribe to 47S pre-rRNA, which is synthesized by the RNA polymerase I (RNA pol I), then the 47S pre-rRNA is processed and modified by small nucleolar ribonucleoproteins (snoRNPs) in the nucleolus. After that the 47S pre-rRNA is subsequently cleaved to form the 18S, 5.8 S and 28SRNAs that are required for the synthesis of the pre-40S ribosomal small subunit and the pre-60S ribosomal large subunit. Meanwhile, 5S rRNA is synthesized by RNA polymerase III in the nucleus, which incorporates into pre-60S subunit. Finally, pre-40S and pre-60S subunits are formed and continue to mature to the final 80S complex, then exported to the cytoplasm. RPs (RPSs and RPLs) are the key component of ribosomes and participate in the basic assembly of ribosomes.
Ribosomes and diseases
Syntheses of ribosome and protein are indispensable to cell survival. The meticulous interaction and coordination between protein and ribosomes synthesis is intricately associated with the processes of cell growth, proliferation, differentiation and biological development. The disorders associated to ribosomal dysfunction or deficits during nascent ribosome biogenesis are collectively known as ribosomopathies. Ribosomopathies are caused by mutations of genes involved in the synthesis of integral ribosomal ingredients such as RPs and rRNA, or defected in biogenesis factors participating in the assembly, modification and processing of nascent ribosomes.8, 41, 42 At the end of the twentieth century, researchers reported the discovery of recurrent mutations in a RP S19 gene in Diamond–Blackfan anemia, a chronic congenital aregenerative anemia syndrome characterized by absent or reduced erythroid precursors.43 Later, ribosomopathies were found to be closely related to a number of other congenital diseases, such as Schwachman–Diamond syndrome, X-linked dyskeratosis congenita,2 etc. Moreover, various tumor suppressors inhibit tumor growth and proliferation through the interference of ribosome biosynthesis and inhibition of protein synthesis in vivo. For example, the tumor-suppressor proteins retinoblastoma (RB) and p130, the two retinoblastoma protein (Rb) family members, reduce rRNA production by inhibiting upstream binding factor that RNA polymerase I (RNA pol I) required, thereby slowing down the growth of cell.44 Tumor-suppressor p53 also inhibits cell growth by suppressing the interaction between selectivity factor 1 and upstream binding factor that influence the function of RNA polymerases.45 RB and p53 are closely related to RNA pol III, which is another key polymerase in ribosomal synthesis and gene transcription for protein synthesis. The two can intervene in the synthesis of ribosomes and proteins by blocking the interaction of RNA pol III with initiation factors and others. Conversely, carcinogenic genes, such as oncogenic Myc, which regulates the transcriptional programs,46, 47 can accelerate the synthesis of ribosomes and proteins to promote cell growth and proliferation. All the above findings demonstrate an inseparable relationship between tumor progression and ribosomes.
Ribosomes and HCC
Hepatocellular carcinoma (HCC), which is accounting for the largest proportion of liver cancer and the fifth leading cause of cancer-related death all over the world, is characterized by high rates of relapse, metastasis and mortality.48 HCC is treatment resistant and involves a multifaceted molecular pathogenesis.49 Various factors are responsible for the occurrence of HCC, and infection of hepatitis virus is a major risk factor, particularly in China. Research has found that the interaction between viruses and ribosomes has an important role in HCC occurrence and progression.50, 51 For example, hepatitis virus expression can activate rRNA transcription, thus interfering with cellular functions that have a certain impact on cell differentiation and cell growth.52
rRNA transcription in HCC
Dysregulation of rRNA transcription has been implicated in cancers.53 rRNA synthesis is upregulated in a number of cancers and transformed cells and is most likely involved in the initiation stage of tumorigenesis.45 This reveals that rRNA transcription might have a crucial role in hepatocarcinogenesis.
rRNA transcription, a critical step in ribosomal biosynthesis mediated by RNA pol I, finally determines the amount of ribosomes, thereby affecting growth, proliferation and the response to environmental stimuli as well as the physiological state of cells.54, 55, 56 The HBx oncoprotein of hepatitis B virus, which is involved in the evolution of hepatitis virus-related liver cancer, is reported to motivate RNA pol I-dependent promoters by activating Ras and TATA-binding protein,57 thereby promoting rRNA transcription and upregulating rRNA synthesis and ribosome biogenesis.50 In addition, it can enhance nucleolar phosphoprotein (nucleophosmin) acetylation and affect nucleosome occupancy to promote rDNA transcription and cell proliferation.58 Furthermore, hepatitis C viruses can activate rRNA transcription through the phosphorylation of upstream binding factor 1 (Ser484) induced by activation of cyclin D1, thereby promoting the formation of liver cancer.52 These findings suggest that rRNA transcription is markedly linked to HCC, particularly hepatitis virus-associated HCC, and rRNA transcription may emerge as a novel target for anticancer therapy.
RPS6 in HCC
The mammalian target of rapamycin (mTOR) is a 289-kDa serine/threonine (Ser/Thr) kinase that regulates a wide spectrum of cellular processes, such as proliferation and metabolism.59, 60 mTOR complex 1 (mTORC1) controls several steps positively during the period of ribosome biogenesis, including transcription of rRNA and the synthesis of RPs and other ingredients necessary for ribosome assembly.61 Normally, both the AKT/mTOR and Ras/mitogen-activated protein kinase (MAPK) cascades are activated simultaneously in human HCC. The AKT-mTORC1-RPS6 signaling pathway was proven to be a key pathway in the initiation and evolvement of HCC by accelerating cell growth and proliferation, angiogenesis62 and de novo lipogenesis,13 which are believed to be involved in oncogenesis (Figure 2a). Moreover, RPS6 and eIF4E act as mTORC1 effectors in AKT/Ras-induced liver cancer.12 As the result of activation of mTORC1, the p70S6K/RPS6 axis triggers cell proliferation via boosting lipid and protein synthesis, cell metabolism, and DNA transcription.63 For instance, it is confirmed that the AKT-mTORC1-RPS6 pathway stimulate lipogenesis through inhibition of fatty acid synthase ubiquitination by ubiquitin-specific protease 2a de-ubiquitinase and disruption of sterol regulatory element-binding protein 1/2 (SREBP1/2) degradation complexes. Meanwhile, Calvisi et al.13 found that suppression of the gene SREBP1 reduced AKT-dependent cell proliferation and survival of HCC cell lines. It revealed that the AKT-mTORC1-RPS6 pathway and lipogenesis have pathogenic and prognostic significance for HCC. Moreover, suppression of lipogenic signaling, including those hinder the AKT-related pathway, will probably be effective treatment for HCC. However, despite extensive studies, the functional significance of RPS6 phosphorylation remains to be further unveiled.
RPs (RPS6, RACK1 and RPS15A) involved signaling pathways. (a) the AKT-mTORC1-RPS6 signaling pathway involves in proliferation, angiogenesis and de novo lipogenesis. (b) P70S6K can be inhibited by deleted in liver cancer 2 (DLC2) gene that encodes a Rho GTPase-activating protein (RhoGAP) through inhibiting the activity of Raf-1–ERK1/2–p70S6K by its RhoGAP function to suppress the proliferation. (c) RACK1 can interact with MKK7, thereby activating MKK7 and JNK, which ultimately enhance cell proliferation and resistance to TRAIL-mediated apoptosis or Fas-mediated apoptosis; overexpression of RACK1 increased the phosphorylation level of IRE1 and enhanced XBP1 mRNA splicing activity to protect the HCC cells from sorafenib-induced apoptosis. (d) RPS15A is associated with Wnt/beta-catenin-fibroblast growth factor (FGF) signaling pathway that promotes angiogenesis of HCC.
The RP S6 kinases (S6K1/2), which are named for their function to phosphorylate RPS6, and the protein initiation factor 4E binding proteins (4EBP1/2/3) are two key downstream effectors of mTORC1.64 The mTOR pathway was proven to be an attractive promotional target for cancer therapeutics as rapamycin inhibits tumor growth by blocking mTOR phosphorylation of S6K.65 RPS6K expression is generally negative or weakly positive in the normal liver, but it is significantly increased in most HCC patients,65 indicating that S6K might have a critical role in hepatic malignancy. By analyzing p-AKT and p70S6K expression associated with clinicopathological features, Li et al. found that overexpression of p-AKT and p70S6K in the sinusoidal endothelial cells in HCC was markedly involved in venous and capsular invasion.62 Using cell cycle analysis and the western blotting technique, Baba et al.66 revealed that rapamycin interferes with cell cycle arrest and apoptosis to enhance proliferation through impeding the phosphorylation of p70S6k kinase. As the upstream of mTOR, AKT has been shown to promote de novo lipogenesis by hampering proteasomal degradation of SREBP1/2 (Figure 2a).13 Similarly, Li et al. discovered that activation of p-p70S6K upregulates angiogenesis in HCC.62 P70S6K can be also inhibited via the deleted in liver cancer 2 gene, which encodes a Rho GTPase-activating protein (RhoGAP). Using flow cytometry and western blotting, it was found that the deleted in liver cancer 2 gene obstructs the proliferation and metastasis of liver malignant cells in vitro by hindering the progression of the G0/G1 phase in the cell cycle and inhibiting the Raf-1–ERK1/2–p70S6K activity by its RhoGAP function (Figure 2b).67, 68
RACK1 in HCC
Mass spectrometry and electron cryo-microscopy (cryo-EM) have identified the RACK1 as a component located at the ‘ occiput’ region in the 40S subunit of eukaryotic ribosomes.69, 70 RACK1 belongs to the Trp-Asp (WD) repeat protein family and acts as a scaffold protein for a variety of kinases and receptors. Thus, RACK1 has a pivotal impact on far-ranging biological responses, such as the immune response, signal transduction, and cell growth, differentiation, as well as migration.71 Activated PKC and eIF6 could be recruited by RACK1 and both of them will stimulate translation and recruit some signaling molecules such as Src and the integrin β subunit, which have a part in cancer progression.70, 72 Research has found that RACK1 is frequently overexpressed in HCC and is closely related to its progression. In the study by Ruan et al.,15 the RACK1 level was highly associated with the Ki67 expression and the serum α-fetoprotein level, suggesting that RACK1 might contribute to the tumorigenesis and prognosis of HCC. c-Jun N-terminal protein kinase (JNK) is a member of the MAPK superfamily. Guo et al.14 found that RACK1 directly interacts with MKK7 (the JNK-specific MAPK kinase), thereby activating MKK7 and JNK and ultimately promoting HCC growth by enhancing proliferation and resisting tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis or Fas-mediated apoptosis (Figure 2c).73 Another study found that RACK1 can interact with and regulate carbonyl reductase 1 (CBR1) to suppress the generation of reactive oxygen species, thus protecting HCC cells from tumor necrosis factor-α-induced cell death.74 In addition, RACK1 can affect sorafenib-induced apoptosis through regulating the activity of IRE1. XBP1s is a potent TF that regulates many target genes as a key regulators of the unfolded protein response, including endoplasmic reticulum (ER) chaperones, ER-associated degradation components and other TFs. The upregulation of RACK1 can promote the phosphorylation of IRE1, which enhances XBP1 mRNA splicing activity and thus inhibits sorafenib-induced apoptosis. On the contrary, the inhibition of RACK1 substantially enhances the lethality of sorafenib to HCC cells.75 This evidence indicates that targeting RACK1 probably become a potential therapeutic strategy for HCC, particularly when combined with sorafenib treatment.
RPS15A in HCC
RPS15A is a highly conservative protein that belongs to the 40S ribosomal subunit. The RPS15A gene responds to transforming growth factor beta and facilitates cell proliferation in the A549 cell line of human lung cancer.76 In addition, RPS15A advances the combination of capped mRNA with the 40S ribosomal subunit during the early period of translation by interacting with the cap-binding subunit of eukaryotic initiation factor 4 F,77, 78 which is indispensable for cell survival and proliferation. Chen et al.79 revealed that RPS15A may promote the malignant transformation of colorectal cancer (CRC) through the p53 signaling pathway. Recent studies showed that in addition to lung adenocarcinoma and CRC,76, 79 malignant tumors in the liver also aberrantly express RPS15A.17 As previously described, hepatitis virus infection is the primary risk factor for HCC, and the expression of S15A is upregulated by HBx antigen (HBxAg). Lian et al.16 believed that S15A might be an innate target of HBxAg in vivo because the elevated expression of S15A is very common in nontumor hepatitis B virus-infected livers. Based on these findings, we conjecture that RPS15A participates in the development of HCC, particularly hepatitis virus-associated HCC. In order to further explore the role of RPS15A in HCC, our team investigated the effect of RPS15A knockdown by short hairpin RNA in hepatoma cells in vitro and found that the downregulation of RPS15A can hamper the growth of hepatoma cells. There are two possible mechanisms (the HBxAg-dependent manner and the cell cycle-mediated manner) underlying the role of RPS15A in promoting hepatocellular cell growth.17
In addition, because HCC is a hypervascular tumor, it is of great significance to verify whether RPS15A has a crucial role in HCC angiogenesis. Our research group conducted further experiments and verified that RPS15A enhances fibroblast growth factor expression via the Wnt/beta-catenin signaling pathway and thereby promotes angiogenesis and the evolvement of HCC by activating the FAK and AKT pathways of vascular endothelial cells after fibroblast growth factor ligand-receptor binding (Figure 2d).
Other RPs in HCC
In addition to RPS6, RACK1 and RPS15a, other RPs may participate in the evolution of liver cancer, such as RPL36 and RPS2. Kim et al.31 detected that the expression of RPL36A mRNA was rose in 85% of HCC cases (34 of 40, P<0.001) and in all of eight HCC cell lines, suggesting a possible role of RPL36A overexpression in hepatocarcinogenesis and tumor cell proliferation. In a study evaluating the RPL36 expression in tumor tissue and normal tissues on a tissue microarray, Song et al.80 further confirmed that the overexpression of RPL36 in HCC is related to the maintenance of the liver’s synthetic function and that RPL36 might be a potential biomarker for predicting the prognosis of HCC. The study by Kowalczyk et al.81 found that RPS2 was overexpressed in mouse HCC samples and could potentially affect the fidelity of mRNA translation associated with aminoacyl-tRNA binding to ribosome, thereby facilitating cell proliferation.
Ribosomes and other malignancies
Colorectal cancer
A number of RP genes, involving S3, S6, S8, S12 and L5, are overexpressed in CRC.82 S8, S12 and another 10 RPs (RPSa, S11, S18, S24, L7, L13a, L18, L28, L32 and L35a) are also found expressed differentially in human colon and neoplastic colorectal tissues.83 The overexpression of L13 mRNA was found in 41% of CRC tissue samples compared with normal tissue samples.84 In the study by Kobayashi et al.,84 silencing the L13 gene using small interference RNA inhibits cell growth, indicating that tumor cell growth rate is positively correlated with the expression level of RPL13 in vitro. Huang et al.85 found that fecal RPL19 expression is related to an advanced tumor stage and can be a prognostic predictor for CRC patients. Later, the same research group also discovered that an increased S27L level contribute to a better prognosis for CRC patients.86 A recent study reported that RP15a can promote malignant cell transformation via misregulation of the p53 signaling pathway, causing a poor prognosis for colon cancer.79
Gastric cancer
Gastric cancer (GC) exhibits differential expression of RPs. Academics have reported the upregulation of RPS1387 and RPL688, 89 in multidrug-resistant GC cells, respectively, which prevented GC cells from drug-induced apoptosis and thus produce drug tolerance and facilitated cell survival. In in vitro experiments, upregulation of RPL6 and RPS13 stimulated the cell growth and strengthened the colony-forming ability of GC cells yet downregulation of RPL6 displayed the adverse effect. Furthermore, low expression of RPL6 and RPS13 inhibits the G1 to S transformation in the cell cycle, leading to cell growth arrest.87, 88 Meanwhile, using the Kaplan–Meier method, the researchers found that, patients with RPL6-positive expression had a shorter survival time than those with RPL6 negative expression.87, 88 Zhang et al.90 stated that RPL23 can stabilize wild-type (wt) p53 through inhibiting the interaction of p53–MDM2, resulting in G1-S cell cycle arrest and/or apoptosis of human GC cells carrying wt p53 gene. In addition, similar to RPS13, RPL23 can protect GC cells from drug-induced apoptosis, resulting in drug resistance.91
Pancreatic cancer
KRAS is one of the important oncogenes contributing to pancreatic cancer tumorigenesis, progression and maintenance; therefore, it is a potential target for antitumor therapy. Li et al.92 found that, the expression of RPL26 and RPL29 notably increased in response to the knockdown of KRAS by short hairpin RNA in pancreatic cancer PANC-1 cells. In addition, silencing of RPL26 or RPL29 gene by small interference RNA triggered cell arrest at G0/G1 period, markedly repressed cell proliferation, and enhanced cell apoptosis. These findings suggested that, the inhibitors against RPL26 and RPL29 have potential therapeutic value for pancreatic cancer. More importantly, a recent study by Muro et al.93 indicated the role of anti-60S RPL29 antibody (anti-RPL29) in human sera being a new prognostic marker for unresectable pancreatic cancer. The researchers found the anti-RPL29 antibody could retard the pancreatic cancer cells proliferation in vitro and reduce the serum anti-RPL29 level corresponded to the degree of spontaneous immune response to autologous cancer cells, suggesting the serum anti-RPL29 level is a potential candidate prognostic marker for pancreatic cancer. RPS6KA2 is also involved in KRAS-mediated pathways. RPS6KA2, a downstream factor of the EGFR/RAS/MAPK kinase (MEK)/extracellular-signal regulated kinase (ERK) signaling pathway, can be activated independently by EGF at the presence of KRAS mutations.94 Milosevic et al.94 found that cell apoptosis increased after knockdown of RPS6KA2 by small interference RNA in erlotinib treatment. This indicates that RPS6KA2 can be a potential treatment target because its inhibition can synergize with erlotinib to enhance the treatment efficacy. Recent study found that RPL34 was also overexpressed in human pancreatic cancer tissues and cells. Moreover, through restraining the expression of RPL34 by lentivirus-delivered small interference RNA, Wei et al.95 discovered that knockdown of RPL34 are accompanied by G2 period cell cycle arrest and induction of cell apoptosis, resulting in distinct suppression of pancreatic cancer cells proliferation, migration and drug resistance.
Breast cancer
Breast cancer is among the foremost causes of cancer-caused deaths globally. In some breast and prostate cancers, cell proliferation is highly reliant on steroid hormone levels (especially androgen and estrogen) in the body. In other words, hormone receptor (HR) signaling is the major stimulus for cell growth. Activation of rRNA gene transcription (the initial stage of ribosome biogenesis) in breast cancer cells is estrogen receptor dependent and can be suppressed by hormone antagonists. The enhanced initiation rather than extension of transcription is the primary mechanism underlying the HR-dependent activation of rRNA synthesis. This has laid an important foundation for the therapy of hormone-dependent breast cancer, as well as the discovery of novel targeted therapeutic agents.96 In addition, p90 RPS6 kinase 4 (RSK4) expression is significantly upregulated in breast cancer of MMTV-c-Myc transgenic mice, which increases cumulation of G0-G1 phase cells and decreases cell proliferation. Moreover, expression levels of claudin-2 and CXCR4, both of which effect on invasion and chemotaxis, are, respectively, upregulated and downregulated in RSK4-overexpressing cells, indicating that RSK4 overexpression can lead to an inhibited cell proliferation, decreased colony-forming ability and suppressed invasive ability of tumor cells.97 In additional, 75% of breast cancers showed a downregulation of L41 mRNA expression, which is related to malignant transformation.98
Prostate cancer
RPL19 is an element of the 60S ribosomal subunit in eukaryotes belonging to the L19E superfamily of proteins. RPL19 gene is highly overexpressed in prostate cancer cell lines. Using in situ hybridization data from the prostatic tissue samples and a Kaplan–Meier analysis, RPL19 was proven to be a powerful and independent prognostic marker.99 Prostate cancer has a considerably high RPL19 expression level, which functionally involved in maintaining the malignant phenotype; therefore, RPL19 can be a potential target for therapeutic intervention.100 RPS2, a 33 kDa RP, was discovered to be upregulated in malignant prostate cancer cell lines and archived tumor specimens, and its overexpression is closely links to formation of prostate tumor and is a key factor for tumor cell survival.101 Moreover, Shen et al.102 revealed a close relationship of proliferation, migration and invasion of cell and increased RACK1 expression in prostate cancer in vitro, suggesting RACK1 has an of importance part in the progress of prostate cancer. However, despite of the significant progresses in related research, the majority of the existing studies was descriptive and did not clarify the main mechanism of the abovementioned proteins in carcinogenesis.
Lung cancer
Most RPs show an increased expression in malignancies. For example, phospho-RPS6 is overexpressed in lung adenocarcinoma, which enhances the metastasis ability of tumor.103 Yang et al.104 observed that RPL22 expression is downregulated in non-small cell lung cancer. RPL22 is a micromolecular protein that consists of 128 amino acids, and it is a component of 60S ribosomal subunits. RPL22 is not necessary for protein synthesis, but increased expression of RPL22 inhibits gene transcription, and dysfunction of RPL22 increases tumor susceptibility.104, 105, 106 Protein kinase CK2, a conservative Ser/Thr protein kinase, regulates a lot of significant signaling pathways, such as PI3K/Akt and WNT. Elevated protein kinase CK2 expression can promote the proliferation of both tumor and normal cells, and maladjusted CK2 expression may affect cell proliferation and apoptosis.107, 108 CK2α is one subunit of protein kinase CK2 tetramer complex. RPL22 can bind to CK2α and affect the process of phosphorylation of CK2α, resulting in the formation and progression of lung cancer.109
Others
Ribosome has a role in various other tumors as well. Researchers found that hypermethylation of the 18S and 28S ribosomal DNAs can serve as a prognosis predictor for the progression-free survival of ovarian cancer patients.110 In addition, absence of RPS4X inhibits the cell growth and enhances the cisplatin tolerance in ovarian cancer cell lines.111 RPS6 expression is downregulated to reduce the ability of esophageal cancer metastasis and invasion,112 whereas it is upregulated in diffuse large B cell lymphomas and acts as a critical regulator of the translation of 5’ terminal oligopyrimidine tract mRNA.113
The findings regarding the roles of RPs in other malignancies are summarized in Table 1.
Conclusions
As an in vivo factory for protein synthesis, ribosomes are involved in a great number of cellular processes. A ribosome has a complex structure and possesses diverse functions; as a result, pathological changes in ribosomes are associated with the initiation and progression of many human diseases, such as Diamond–Blackfan anemia, Schwachman–Diamond syndrome and cancer. As described above, ribosome dysregulation occurs in various common malignant tumors, such as breast, prostate and lung cancers, and in gastrointestinal tumors, particularly HCC. Research has found that RPS6, RACK1 and RPS15A have a crucial part in liver cancer and that they affect the development of various aspects of HCC, including tumor cell proliferation, angiogenesis and drug resistance. These discoveries indicated that RPs might be potential promising prognostic markers and therapeutic targets for HCC. However, ribosome disorders in malignancies are complicated and remain to be completely explored.
References
Ruggero D, Pandolfi PP . Does the ribosome translate cancer? Nat Rev Cancer 2003; 3: 179–192.
Shenoy N, Kessel R, Bhagat TD, Bhattacharyya S, Yu Y, McMahon C et al. Alterations in the ribosomal machinery in cancer and hematologic disorders. J Hematol Oncol 2012; 5: 32.
Wang W, Nag S, Zhang X, Wang MH, Wang H, Zhou J et al. Ribosomal proteins and human diseases: pathogenesis, molecular mechanisms, and therapeutic implications. Med Res Rev 2015; 35: 225–285.
Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M . The structure of the eukaryotic ribosome at 3.0 A resolution. Science 2011; 334: 1524–1529.
Khatter H, Myasnikov AG, Natchiar SK, Klaholz BP . Structure of the human 80 S ribosome. Nature 2015; 520: 640–645.
Brar GA, Weissman JS . Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol 2015; 16: 651–664.
Nakhoul H, Ke J, Zhou X, Liao W, Zeng SX, Lu H . Ribosomopathies: mechanisms of disease. Clin Med Insights Blood Disorders 2014; 7: 7–16.
Danilova N, Gazda HT . Ribosomopathies: how a common root can cause a tree of pathologies. Dis Models Mech 2015; 8: 1013–1026.
Teng T, Thomas G, Mercer CA . Growth control and ribosomopathies. Curr Opin Genet Dev 2013; 23: 63–71.
Stumpf CR, Ruggero D . The cancerous translation apparatus. Curr Opin Genet Dev 2011; 21: 474–483.
Bruno PM, Liu Y, Park GY, Murai J, Koch CE, Eisen TJ et al. A subset of platinum-containing chemotherapeutic agents kills cells by inducing ribosome biogenesis stress. Nat Med 2017; 23: 461–471.
Wang C, Cigliano A, Jiang L, Li X, Fan B, Pilo MG et al. 4EBP1/eIF4E and p70S6K/RPS6 axes play critical and distinct roles in hepatocarcinogenesis driven by AKT and N-Ras proto-oncogenes in mice. Hepatology (Baltimore, MD) 2015; 61: 200–213.
Calvisi DF, Wang C, Ho C, Ladu S, Lee SA, Mattu S et al. Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology 2011; 140: 1071–1083.
Guo Y, Wang W, Wang J, Feng J, Wang Q, Jin J et al. Receptor for activated C kinase 1 promotes hepatocellular carcinoma growth by enhancing mitogen-activated protein kinase kinase 7 activity. Hepatology (Baltimore, MD) 2013; 57: 140–151.
Ruan Y, Sun L, Hao Y, Wang L, Xu J, Zhang W et al. Ribosomal RACK1 promotes chemoresistance and growth in human hepatocellular carcinoma. J Clin Invest 2012; 122: 2554–2566.
Lian Z, Liu J, Li L, Li X, Tufan NL, Wu MC et al. Human S15a expression is upregulated by hepatitis B virus X protein. Mol Carcinog 2004; 40: 34–46.
Xu M, Wang Y, Chen L, Pan B, Chen F, Fang Y et al. Down-regulation of ribosomal protein S15A mRNA with a short hairpin RNA inhibits human hepatic cancer cell growth in vitro. Gene 2014; 536: 84–89.
Zeng M, Zheng M, Lu D, Wang J, Jiang W, Sha O . Anti-tumor activities and apoptotic mechanism of ribosome-inactivating proteins. Chin J Cancer 2015; 34: 325–334.
Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M . One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol 2012; 19: 560–567.
Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI . The multifunctional nucleolus. Nat Rev Mol Cell Biol 2007; 8: 574–585.
Gruschke S, Ott M . The polypeptide tunnel exit of the mitochondrial ribosome is tailored to meet the specific requirements of the organelle. BioEssays 2010; 32: 1050–1057.
Desai N, Brown A, Amunts A, Ramakrishnan V . The structure of the yeast mitochondrial ribosome. Science 2017; 355: 528–531.
Ramesh M, Woolford JL Jr . Eukaryote-specific rRNA expansion segments function in ribosome biogenesis. RNA 2016; 22: 1153–1162.
Robledo S, Idol RA, Crimmins DL, Ladenson JH, Mason PJ, Bessler M . The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA 2008; 14: 1918–1929.
Orelle C, Carlson ED, Szal T, Florin T, Jewett MC, Mankin AS . Protein synthesis by ribosomes with tethered subunits. Nature 2015; 524: 119–124.
Zeng F, Chen Y, Remis J, Shekhar M, Phillips JC, Tajkhorshid E et al. Structural basis of co-translational quality control by ArfA and RF2 bound to ribosome. Nature 2017; 541: 554–557.
Hinnebusch AG, Lorsch JR . The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harbor Perspect Biol 2012; 4: pii: a011544.
Orsolic I, Jurada D, Pullen N, Oren M, Eliopoulos AG, Volarevic S . The relationship between the nucleolus and cancer: current evidence and emerging paradigms. Semin Cancer Biol 2016; 37-38: 36–50.
Ruggero D . Revisiting the nucleolus: from marker to dynamic integrator of cancer signaling. Sci Signal 2012; 5: pe38.
Volarevic S, Stewart MJ, Ledermann B, Zilberman F, Terracciano L, Montini E et al. Proliferation, but not growth, blocked by conditional deletion of 40 S ribosomal protein S6. Science 2000; 288: 2045–2047.
Kim JH, You KR, Kim IH, Cho BH, Kim CY, Kim DG . Over-expression of the ribosomal protein L36a gene is associated with cellular proliferation in hepatocellular carcinoma. Hepatology (Baltimore, MD) 2004; 39: 129–138.
Donati G, Montanaro L, Derenzini M . Ribosome biogenesis and control of cell proliferation: p53 is not alone. Cancer Res 2012; 72: 1602–1607.
Wang H, Zhao LN, Li KZ, Ling R, Li XJ, Wang L . Overexpression of ribosomal protein L15 is associated with cell proliferation in gastric cancer. BMC Cancer 2006; 6: 91.
Zhan Y, Melian NY, Pantoja M, Haines N, Ruohola-Baker H, Bourque CW et al. Dystroglycan and mitochondrial ribosomal protein L34 regulate differentiation in the Drosophila eye. PLoS One 2010; 5: e10488.
Da Costa L, Narla G, Willig TN, Peters LL, Parra M, Fixler J et al. Ribosomal protein S19 expression during erythroid differentiation. Blood 2003; 101: 318–324.
He H, Sun Y . Ribosomal protein S27L is a direct p53 target that regulates apoptosis. Oncogene 2007; 26: 2707–2716.
Jang CY, Lee JY, Kim J . RpS3, a DNA repair endonuclease and ribosomal protein, is involved in apoptosis. FEBS Lett 2004; 560: 81–85.
Hegde V, Wang M, Deutsch WA . Human ribosomal protein S3 interacts with DNA base excision repair proteins hAPE/Ref-1 and hOGG1. Biochemistry 2004; 43: 14211–14217.
Kim J, Chubatsu LS, Admon A, Stahl J, Fellous R, Linn S . Implication of mammalian ribosomal protein S3 in the processing of DNA damage. J Biol Chem 1995; 270: 13620–13629.
Yang ZY, Jiang H, Qu Y, Wei M, Yan M, Zhu ZG et al. Metallopanstimulin-1 regulates invasion and migration of gastric cancer cells partially through integrin beta4. Carcinogenesis 2013; 34: 2851–2860.
Narla A, Ebert BL . Ribosomopathies: human disorders of ribosome dysfunction. Blood 2010; 115: 3196–3205.
Fumagalli S, Thomas G . The role of p53 in ribosomopathies. Semin Hematol 2011; 48: 97–105.
Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet 1999; 21: 169–175.
Sutcliffe JE, Brown TR, Allison SJ, Scott PH, White RJ . Retinoblastoma protein disrupts interactions required for RNA polymerase III transcription. Mol Cell Biol 2000; 20: 9192–9202.
Donati G, Bertoni S, Brighenti E, Vici M, Trere D, Volarevic S et al. The balance between rRNA and ribosomal protein synthesis up- and downregulates the tumour suppressor p53 in mammalian cells. Oncogene 2011; 30: 3274–3288.
Zhou X, Liao WJ, Liao JM, Liao P, Lu H . Ribosomal proteins: functions beyond the ribosome. J Mol Cell Biol 2015; 7: 92–104.
van Riggelen J, Yetil A, Felsher DW . MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 2010; 10: 301–309.
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A . Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108.
Whittaker S, Marais R, Zhu AX . The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene 2010; 29: 4989–5005.
Shukla SK, Kumar V . Hepatitis B virus X protein and c-Myc cooperate in the upregulation of ribosome biogenesis and in cellular transformation. FEBS J 2012; 279: 3859–3871.
Fatima G, Mathan G, Kumar V . The HBx protein of hepatitis B virus regulates the expression, intracellular distribution and functions of ribosomal protein S27a. J Gen Virol 2012; 93: 706–715.
Raychaudhuri S, Fontanes V, Barat B, Dasgupta A . Activation of ribosomal RNA transcription by hepatitis C virus involves upstream binding factor phosphorylation via induction of cyclin D1. Cancer Res 2009; 69: 2057–2064.
Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA . Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol 2005; 7: 295–302.
Sollner-Webb B, Tower J . Transcription of cloned eukaryotic ribosomal RNA genes. Annu Rev Biochem 1986; 55: 801–830.
Yu F, Shen X, Fan L, Yu Z . Analysis of histone modifications at human ribosomal DNA in liver cancer cell. Sci Rep 2015; 5: 18100.
Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V . A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell Mol Life Sci 2007; 64: 29–49.
Wang HD, Trivedi A, Johnson DL . Regulation of RNA polymerase I-dependent promoters by the hepatitis B virus X protein via activated Ras and TATA-binding protein. Mol Cell Biol 1998; 18: 7086–7094.
Ahuja R, Kapoor NR, Kumar V . The HBx oncoprotein of hepatitis B virus engages nucleophosmin to promote rDNA transcription and cellular proliferation. Biochim Biophys Acta 2015; 1853: 1783–1795.
Gentilella A, Kozma SC, Thomas G . A liaison between mTOR signaling, ribosome biogenesis and cancer. Biochim Biophys Acta 2015; 1849: 812–820.
Zinzalla V, Stracka D, Oppliger W, Hall MN . Activation of mTORC2 by association with the ribosome. Cell 2011; 144: 757–768.
Iadevaia V, Liu R, Proud CG . mTORC1 signaling controls multiple steps in ribosome biogenesis. Semin Cell Dev Biol 2014; 36: 113–120.
Li W, Tan D, Zhang Z, Liang JJ, Brown RE . Activation of Akt-mTOR-p70S6K pathway in angiogenesis in hepatocellular carcinoma. Oncol Rep 2008; 20: 713–719.
Magnuson B, Ekim B, Fingar DC . Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J 2012; 441: 1–21.
Plas DR, Thomas G . Tubers and tumors: rapamycin therapy for benign and malignant tumors. Curr Opin Cell Biol 2009; 21: 230–236.
Sahin F, Kannangai R, Adegbola O, Wang J, Su G, Torbenson M . mTOR and P70 S6 kinase expression in primary liver neoplasms. Clin Cancer Res 2004; 10: 8421–8425.
Baba HA, Wohlschlaeger J, Cicinnati VR, Hilgard P, Lang H, Sotiropoulos GC et al. Phosphorylation of p70S6 kinase predicts overall survival in patients with clear margin-resected hepatocellular carcinoma. Liver Int 2009; 29: 399–405.
Ching YP, Wong CM, Chan SF, Leung TH, Ng DC, Jin DY et al. Deleted in liver cancer (DLC) 2 encodes a RhoGAP protein with growth suppressor function and is underexpressed in hepatocellular carcinoma. J Biol Chem 2003; 278: 10824–10830.
Leung TH, Yam JW, Chan LK, Ching YP, Ng IO . Deleted in liver cancer 2 suppresses cell growth via the regulation of the Raf-1-ERK1/2-p70S6K signalling pathway. Liver Int 2010; 30: 1315–1323.
Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR et al. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol 1999; 17: 676–682.
Sengupta J, Nilsson J, Gursky R, Spahn CM, Nissen P, Frank J . Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat Struct Mol Biol 2004; 11: 957–962.
McCahill A, Warwicker J, Bolger GB, Houslay MD, Yarwood SJ . The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol Pharmacol 2002; 62: 1261–1273.
Wu J, Meng J, Du Y, Huang Y, Jin Y, Zhang J et al. RACK1 promotes the proliferation, migration and invasion capacity of mouse hepatocellular carcinoma cell line in vitro probably by PI3K/Rac1 signaling pathway. Biomed Pharmacother 2013; 67: 313–319.
Wang WD, Wen Z, Ji W, Ma Y . RACK1 expression contributes to JNK activity, but JNK activity does not enhance RACK1 expression in hepatocellular carcinoma SMMC-7721 cells. Oncol Lett 2015; 9: 2767–2770.
Zhou S, Cao H, Zhao Y, Li X, Zhang J, Hou C et al. RACK1 promotes hepatocellular carcinoma cell survival via CBR1 by suppressing TNF-alpha-induced ROS generation. Oncol Lett 2016; 12: 5303–5308.
Zhou T, Lv X, Guo X, Ruan B, Liu D, Ding R et al. RACK1 modulates apoptosis induced by sorafenib in HCC cells by interfering with the IRE1/XBP1 axis. Oncol Rep 2015; 33: 3006–3014.
Akiyama N, Matsuo Y, Sai H, Noda M, Kizaka-Kondoh S . Identification of a series of transforming growth factor beta-responsive genes by retrovirus-mediated gene trap screening. Mol Cell Biol 2000; 20: 3266–3273.
Lavoie C, Tam R, Clark M, Lee H, Sonenberg N, Lasko P . Suppression of a temperature-sensitive cdc33 mutation of yeast by a multicopy plasmid expressing a Drosophila ribosomal protein. J Biol Chem 1994; 269: 14625–14630.
Jimenez L, Becerra A, Landa A . Cloning, expression and partial characterization of a gene encoding the S15a ribosomal protein of Taenia solium. Parasitol Res 2004; 92: 414–420.
Chen J, Wei Y, Feng Q, Ren L, He G, Chang W et al. Ribosomal protein S15A promotes malignant transformation and predicts poor outcome in colorectal cancer through misregulation of p53 signaling pathway. Int J Ocol 2016; 48: 1628–1638.
Song MJ, Jung CK, Park CH, Hur W, Choi JE, Bae SH et al. RPL36 as a prognostic marker in hepatocellular carcinoma. Pathol Int 2011; 61: 638–644.
Kowalczyk P, Woszczynski M, Ostrowski J . Increased expression of ribosomal protein S2 in liver tumors, posthepactomized livers, and proliferating hepatocytes in vitro. Acta Biochim Pol 2002; 49: 615–624.
Pogue-Geile K, Geiser JR, Shu M, Miller C, Wool IG, Meisler AI et al. Ribosomal protein genes are overexpressed in colorectal cancer: isolation of a cDNA clone encoding the human S3 ribosomal protein. Mol Cell Biol 1991; 11: 3842–3849.
Kasai H, Nadano D, Hidaka E, Higuchi K, Kawakubo M, Sato TA et al. Differential expression of ribosomal proteins in human normal and neoplastic colorectum. J Histochem Cytochem 2003; 51: 567–574.
Kobayashi T, Sasaki Y, Oshima Y, Yamamoto H, Mita H, Suzuki H et al. Activation of the ribosomal protein L13 gene in human gastrointestinal cancer. Int J Mol Med 2006; 18: 161–170.
Huang CJ, Chien CC, Yang SH, Chang CC, Sun HL, Cheng YC et al. Faecal ribosomal protein L19 is a genetic prognostic factor for survival in colorectal cancer. J Cell Mol Med 2008; 12: 1936–1943.
Huang CJ, Yang SH, Lee CL, Cheng YC, Tai SY, Chien CC . Ribosomal protein S27-like in colorectal cancer: a candidate for predicting prognoses. PLoS One 2013; 8: e67043.
Guo X, Shi Y, Gou Y, Li J, Han S, Zhang Y et al. Human ribosomal protein S13 promotes gastric cancer growth through down-regulating p27(Kip1). J Cell Mol Med 2011; 15: 296–306.
Wu Q, Gou Y, Wang Q, Jin H, Cui L, Zhang Y et al. Downregulation of RPL6 by siRNA inhibits proliferation and cell cycle progression of human gastric cancer cell lines. PLoS ONE 2011; 6: e26401.
Du J, Shi Y, Pan Y, Jin X, Liu C, Liu N et al. Regulation of multidrug resistance by ribosomal protein l6 in gastric cancer cells. Cancer Biol Ther 2005; 4: 242–247.
Zhang Y, Shi Y, Li X, Du W, Luo G, Gou Y et al. Inhibition of the p53-MDM2 interaction by adenovirus delivery of ribosomal protein L23 stabilizes p53 and induces cell cycle arrest and apoptosis in gastric cancer. J Gene Med 2010; 12: 147–156.
Shi Y, Zhai H, Wang X, Han Z, Liu C, Lan M et al. Ribosomal proteins S13 and L23 promote multidrug resistance in gastric cancer cells by suppressing drug-induced apoptosis. Exp Cell Res 2004; 296: 337–346.
Li C, Ge M, Yin Y, Luo M, Chen D . Silencing expression of ribosomal protein L26 and L29 by RNA interfering inhibits proliferation of human pancreatic cancer PANC-1 cells. Mol Cell Biochem 2012; 370: 127–139.
Muro S, Miyake Y, Kato H, Tsutsumi K, Yamamoto K . Serum anti-60 S ribosomal protein L29 antibody as a novel prognostic marker for unresectable pancreatic cancer. Digestion 2015; 91: 164–173.
Milosevic N, Kühnemuth B, Mühlberg L, Ripka S, Griesmann H, Lölkes C et al. Synthetic lethality screen identifies RPS6KA2 as modifier of epidermal growth factor receptor activity in pancreatic cancer. Neoplasia 2013; 15: 1354–1362.
Wei F, Ding L, Wei Z, Zhang Y, Li Y, Qinghua L et al. Ribosomal protein L34 promotes the proliferation, invasion and metastasis of pancreatic cancer cells. Oncotarget 2016; 7: 85259–85272.
Ray S, Johnston R, Campbell DC, Nugent S, McDade SS, Waugh D et al. Androgens and estrogens stimulate ribosome biogenesis in prostate and breast cancer cells in receptor dependent manner. Gene 2013; 526: 46–53.
Thakur A, Sun Y, Bollig A, Wu J, Biliran H, Banerjee S et al. Anti-invasive and antimetastatic activities of ribosomal protein S6 kinase 4 in breast cancer cells. Clin Cancer Res 2008; 14: 4427–4436.
Wang S, Huang J, He J, Wang A, Xu S, Huang S-F et al. RPL41, a small ribosomal peptide deregulated in tumors, is essential for mitosis and centrosome integrity. Neoplasia 2010; 12: 284–IN288.
Bee A, Ke Y, Forootan S, Lin K, Beesley C, Forrest SE et al. Ribosomal protein l19 is a prognostic marker for human prostate cancer. Clin Cancer Res 2006; 12: 2061–2065.
Bee A, Brewer D, Beesley C, Dodson A, Forootan S, Dickinson T et al. siRNA knockdown of ribosomal protein gene RPL19 abrogates the aggressive phenotype of human prostate cancer. PLoS One 2011; 6: e22672.
Wang M, Hu Y, Stearns ME . RPS2: a novel therapeutic target in prostate cancer. J Exp Clin Cancer Res 2009; 28: 6.
Shen F, Yan C, Liu M, Feng Y, Chen Y . RACK1 promotes prostate cancer cell proliferation, invasion and metastasis. Mol Med Rep 2013; 8: 999–1004.
McDonald JM, Pelloski CE, Ledoux A, Sun M, Raso G, Komaki R et al. Elevated phospho-S6 expression is associated with metastasis in adenocarcinoma of the lung. Clin Cancer Res 2008; 14: 7832–7837.
Yang M, Sun H, Wang H, Zhang S, Yu X, Zhang L . Down-regulation of ribosomal protein L22 in non-small cell lung cancer. Med Oncol 2013; 30: 646.
Rao S, Lee SY, Gutierrez A, Perrigoue J, Thapa RJ, Tu Z et al. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood 2012; 120: 3764–3773.
Ni JQ, Liu LP, Hess D, Rietdorf J, Sun FL . Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev 2006; 20: 1959–1973.
Lin KY, Tai C, Hsu JC, Li CF, Fang CL, Lai HC et al. Overexpression of nuclear protein kinase CK2 alpha catalytic subunit (CK2alpha) as a poor prognosticator in human colorectal cancer. PLoS One 2011; 6: e17193.
Siddiqui-Jain A, Drygin D, Streiner N, Chua P, Pierre F, O'Brien SE et al. CX-4945, an orally bioavailable selective inhibitor of protein kinase CK2, inhibits prosurvival and angiogenic signaling and exhibits antitumor efficacy. Cancer Res 2010; 70: 10288–10298.
Yang M, Sun H, He J, Wang H, Yu X, Ma L et al. Interaction of ribosomal protein L22 with casein kinase 2alpha: a novel mechanism for understanding the biology of non-small cell lung cancer. Oncol Rep 2014; 32: 139–144.
Chan MW, Wei SH, Wen P, Wang Z, Matei DE, Liu JC et al. Hypermethylation of 18 S and 28 S ribosomal DNAs predicts progression-free survival in patients with ovarian cancer. Clin Cancer Res 2005; 11: 7376–7383.
Tsofack SP, Meunier L, Sanchez L, Madore J, Provencher D, Mes-Masson AM et al. Low expression of the X-linked ribosomal protein S4 in human serous epithelial ovarian cancer is associated with a poor prognosis. BMC Cancer 2013; 13: 303.
Kim SH, Jang YH, Chau GC, Pyo S, Um SH . Prognostic significance and function of phosphorylated ribosomal protein S6 in esophageal squamous cell carcinoma. Mod Pathol 2013; 26: 327–335.
Hagner PR, Mazan-Mamczarz K, Dai B, Balzer EM, Corl S, Martin SS et al. Ribosomal protein S6 is highly expressed in non-Hodgkin lymphoma and associates with mRNA containing a 5' terminal oligopyrimidine tract. Oncogene 2011; 30: 1531–1541.
Acknowledgements
This work was supported by the grants from the Natural Science Foundation of Zhejiang Province (LY17H160047), the National Natural Science Foundation of China(81201953, 81772628, 81703310) and the Research Found for the Doctoral Program of High Education of China from the Ministry of Education (20113321120003).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Xie, X., Guo, P., Yu, H. et al. Ribosomal proteins: insight into molecular roles and functions in hepatocellular carcinoma. Oncogene 37, 277–285 (2018). https://doi.org/10.1038/onc.2017.343
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.343
- Springer Nature Limited
This article is cited by
-
CircPRKD3/miR-6783-3p responds to mechanical force to facilitate the osteogenesis of stretched periodontal ligament stem cells
Journal of Orthopaedic Surgery and Research (2024)
-
Ribosomal protein L34 is a potential prognostic biomarker and therapeutic target in hilar cholangiocarcinoma
Cell & Bioscience (2020)
-
DNA methylation signature of smoking in lung cancer is enriched for exposure signatures in newborn and adult blood
Scientific Reports (2019)