Abstract
The canonical WNT signaling pathway is crucial for intestinal stem cell renewal and aberrant WNT signaling is an early event in colorectal cancer (CRC) development. Here, we show for the first time that WNT2 is one of the most significantly induced genes in CRC stroma as compared to normal stroma. The impact of stromal WNT2 on carcinoma formation or progression was not addressed so far. Canonical WNT/β-catenin signaling was assessed using a 7TGP-reporter construct. Furthermore, effects of WNT2 on fibroblast migration and invasion were determined using siRNA-mediated gene silencing. Tumor cell invasion was studied using organotypic raft cultures and in vivo significance was assessed via a xenograft mouse model. We identified cancer-associated fibroblasts (CAFs) as the main source of WNT2. CAF-derived WNT2 activated canonical signaling in adenomatous polyposis coli/β-catenin wild-type colon cancer cells in a paracrine fashion, whereas no hyperactivation was detectable in cell lines harboring mutations in the adenomatous polyposis coli/β-catenin pathway. Furthermore, WNT2 activated autocrine canonical WNT signaling in primary fibroblasts, which was associated with a pro-migratory and pro-invasive phenotype. We identified FZD8 as the putative WNT2 receptor in CAFs. Three-dimensional organotypic co-culture assays revealed that WNT2-mediated fibroblast motility and extracellular matrix remodeling enhanced cancer cell invasion of cell lines even harboring mutations in the adenomatous polyposis coli/β-catenin pathway. Thus, suggesting a tumor-promoting influence on a broad range of CRC. In line, WNT2 also promotes tumor growth, invasion and metastasis in vivo. Moreover, high WNT2 expression is associated with poor prognosis in human CRC. The identification of the pro-malignant function of stromal derived WNT2 in CRC classifies WNT2 and its receptor as promising stromal targets to confine cancer progression in combination with conventional or targeted therapies.
Similar content being viewed by others
Introduction
Active WNT signaling has been shown to be crucial for normal intestinal development and homeostasis. Constitutive activation of the canonical WNT pathway caused by mutations in the adenomatous polyposis coli gene, in β-catenin (CTNNB1) or in AXIN is an early event in more than 90% of colorectal carcinomas. These mutations lead to expression of WNT target genes in a ligand independent manner. However, also a number of upstream WNT signaling components are deregulated in some cancer types,1 which suggests additional modes of action during tumorigenesis. WNT2 mRNA overexpression has been shown in fibroadenomas,2 breast-3 and colorectal carcinomas (CRC).4 WNT2 displayed transforming activity in breast epithelial cells5 and downregulation of WNT2 by siRNA suppressed growth of malignant glioma cells.6 Interestingly, injection of breast epithelial cells ectopically expressing WNT2 into mice resulted in hyperplastic tumors with significant fibrosis7 and WNT2 was also elevated in a mouse model for cystic fibrosis.8 WNT2 knockout mice display poorly differentiated lung mesenchyme (airway smooth muscle cells) and lack proper lung development.9 These data and activation of WNT2 in mesenchymal stem cells10 suggest a role of WNT2 as a regulator of mesenchymal cell fate, and might influence the activation of mesenchymal cells in the tumor stroma. However, the cellular origins of tumor-specific WNT2 expression remain obscure and the impact of its expression on cancer growth or invasion was scarcely addressed.
We evaluated an expression-profiling study with laser capture microdissected (LCM) CRC patient material11 and found WNT2 as one of the most significantly induced genes in fibroblast activation protein-positive (FAP+) tumor stroma. In this study, knockdown experiments and overexpression studies were performed together with three-dimensional culture models to functionally analyze the role of stromal derived WNT2 in CRC. Moreover, xenograft experiments and human patient data demonstrated the in vivo significance of our findings.
Results
WNT2 is overexpressed in the tumor stroma of colorectal carcinomas
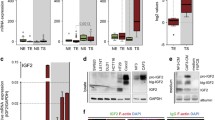
In previous studies WNT2 mRNA was shown to be elevated in CRC as compared to normal colon.4, 12, 13 We confirmed these results using an independent data set (Figure 1a) and data from Oncomine14 (Supplementary Table S1). WNT2 mRNA was also significantly elevated in other cancers of the digestive tract (esophagus, larynx, pancreas, stomach, Supplementary Figure S1A and Supplementary Table S2), whereas no induction was seen in breast, lung, prostate, bladder and cervix carcinomas (Supplementary Figure S1B and Supplementary Table S2). These data were derived from non-dissected tumors and did not reveal the cellular origin of WNT2 expression. Interestingly, comparison of WNT2 expression and ESTIMATE stromal score of 218 CRC patients revealed a positive correlation suggesting a stromal origin of WNT2 mRNA (Figure 1b). A colon cancer mRNA expression data set dissected by LCM obtained from Gene Expression Omnibus (GEO15) confirmed significantly upregulation of WNT2 in the tumor stroma (Nishida data set,16 Figure 1c and Supplementary Table S4). Using immunohistochemically guided LCM (iLCM)17 activated FAP+ tumor stroma (FAP+ TS) and FAP- normal stroma (FAP− NS) was dissected from epithelial cells of CRC (tumor epithelium, TE) and their normal counterparts (normal epithelium, NE), respectively.11 Here, we demonstrate that WNT2 mRNA was significantly upregulated in the tumor stroma compared to normal stroma, normal epithelium and tumor cells (Rupp data set,11 Figure 1d and Supplementary Table S3). These results were further validated by FAP+ stromal cells sorted by flow cytometry, which were compared to epithelial cell adhesion molecule-positive cancer cells from CRC (Calon data set,18 Figure 1e and Supplementary Table S5). Furthermore, species-specific RNA-seq profiles of patient-derived xenografts from CRC (Isella data set)19 substantiated our finding. The patient-derived xenografts transcriptome is comprised of a mixture of human (tumor cell derived) and mouse (stromal cell derived) mRNA. WNT2 expression was predominantly assigned to the mouse reads demonstrating its stromal specific expression (Supplementary Figure S1C). Hence, we provide strong evidence that WNT2 expression in vivo is exclusively induced in the tumor stroma of CRC.
WNT2 induction in CRC is due to high WNT2 expression in CAFs. (a) GeneLogic expression database analysis of WNT2 mRNA in human colon adenocarcinomas (CRC, 209 cases) as compared to normal colon (NC, 92 cases). (b) Correlation of WNT2 mRNA expression and ESTIMATE stromal score of 218 human CRC samples derived from TCGA. Gray dots represent individual samples; black line illustrates linear regression; dashed lines represent 95% confidence interval. (c–e) Human CRC samples, laser capture microdissected into normal epithelium (NE), normal stroma (NS), tumor epithelium (TE) and tumor stroma (TS) were re-evaluated (c) Nishida data set,16 NE, four cases; TE, 13 cases; NS, four cases; TS, 13 cases; (d) Rupp data set,11 NE, six cases; TE, 26 cases; NS, six cases; TS, 26 cases; (e) FAC-sorted EpCAM+ epithelial cells and FAP+ fibroblasts from human CRC were also re-evaluated (Calon data set,18 EpCAM+, six cases; FAP+, six cases). Statistical analysis is provided in Supplementary Tables S3-5. (f) WNT2 mRNA expression in skin fibroblasts (BJ1), colon myofibroblasts (CCD18Co) and primary CAFs (CAF1, CAF2). Results are from three independent experiments. Center line, median; Box, interquartile range (IQR); Whiskers, 1.5 times the IQR; Dots, outliers. For statistics see Supplementary Table S6. (g) WNT2 mRNA expression in NCF and CAFs of three different patients (NCF and CAF3-5). RT-qPCR analysis was carried out in three different passages (n=3). Data were normalized to GAPDH, expression in NCF3 was set to 1. Scale bars, means; error bars, s.e.m. EpCAM+, epithelial cell adhesion molecule-positive; FACS, fluorescence-activated cell sorting; NCF, normal colon fibroblasts; RT-qPCR, quantitative PCR with reverse transcription.
CAFs from colorectal carcinomas express WNT2
To substantiate these findings primary human cancer-associated fibroblast (CAFs)20 from CRC were analyzed. They showed high levels of WNT2 mRNA compared to intermediate WNT2 expressing normal colon myofibroblasts (CCD18Co), whereas dermal fibroblasts (BJ1) did not express significant levels of WNT2 (Dolznig data set,20 Figure 1f and Supplementary Table S6). Furthermore, three matched pairs of normal colon fibroblasts (NCF) and CAFs displayed high WNT2 expression in CAFs, whereas NCFs were virtually negative as demonstrated by quantitative PCR (Figure 1g). Moreover, transcript analysis showed minimal expression of WNT2 mRNA in different tumor cell lines compared to CAF3 (Supplementary Figure S1DE). Thus we conclude that CAFs, and not tumor cells, are a major source of WNT2 in CRC.
Expression of WNT signaling components in CRC cells and stromal fibroblasts
Expression profiling of cancer cells and CAFs (Dolznig data set,20 Supplementary Figure S2), iLCM of tumor cells and FAP+ tumor stroma (Rupp data set,11 Supplementary Figure S3) and sorted epithelial cell adhesion molecule-positive tumor cells and FAP+ CAFs (Calon data set,18 Supplementary Figure S4) revealed that essential WNT/β-catenin signaling pathway components (Supplementary Figure S5) display similar expression patterns in the three data sets (Figure 2). WNT2, WNT2B and WNT5A were consistently upregulated in the tumor stroma compared to tumor cells. Frizzled receptors, LRPs and intracellular signaling components were expressed in tumor cells and fibroblasts. This ubiquitous expression of the WNT/β-catenin signaling compounds implicates potentially active Wnt signaling in epithelial cells and CAFs.
Profiling of WNT/β-catenin signaling pathway components in CRC. Data from three expression-profiling screens11, 18, 20 were re-evaluated. In HG-U133 chips, black squares: high expression (>200 arbitrary expression units), gray squares: intermediate expression (>50, <200), white squares: absent or low expression (<50). In HT HG-U133+ PM chips, black squares: high expression (>27.5), gray squares: intermediate expression (>12.5, <27.5), white squares: absent or low expression (<12.5). Expression patterns are indicated by arrows. TE, tumor epithelium; TS, tumor stroma.
Stromal WNT2 induces canonical signaling in CRC cells wild-type for the WNT/β-catenin pathway
First, we determined whether WNT2 was able to hyperactivate canonical WNT-signaling in CRC lines stably expressing a 7TGP reporter construct.21 7TGP reporter cells express GFP in response to canonical WNT signaling (Supplementary Figure S5). As LS174T, HT29, HCT116 and DLD1 cells already harbor mutations constitutively activating the β-catenin pathway, all cells were GFP-positive after selection (Supplementary Figure S6A, no conditioned medium, CM). In order to measure canonical WNT signaling activation 7TGP reporter tumor cells were co-cultivated with either naïve mouse fibroblasts (L-parental) or mouse fibroblasts overexpressing WNT2 (L-WNT2), WNT3A (L-WNT3A) or WNT5A (L-WNT5A). GFP expression did not change during direct contact with L-WNT2 cells (Figure 3a and Supplementary Figure S6B). L-parental and WNT5A expressing cells (L-WNT5A), which do not activate canonical WNT signaling, served as negative control. L-WNT3A cells were used as positive control. Next, RKO colon cancer cells that do not harbor mutations in the WNT/β-catenin pathway22 were analyzed. RKO-7TGP reporter cells were GFP negative in normal growth medium (Supplementary Figure S6A, no CM). However, direct cell–cell contact with L-WNT3A and L-WNT2 induced GFP expression (Figure 3a and Supplementary Figure S6B). These direct co-culture experiments did not reveal the exact percentage of responsive tumor cells since a mixture of WNT-producing L-cells and 7TGP reporter cells were analyzed via flow cytometry. Thus, we used transwell inserts populated on one side of the membrane with producer L-cells and on the other side with RKO-7TGP reporter cells to exclude cell intermingling in the presence of direct cell–cell contact. Reporter cells were selectively harvested and subjected to flow cytometric analysis (Supplementary Figure S6C). In this setup, WNT2 from L-cells induced canonical WNT signaling in 50% of RKO-7TGP reporter cells and WNT3A in 30% of the cells (Supplementary Figures S6D and E). Incubation with WNT2 containing medium or rhWNT2 did not induce reporter activity in RKO-7TGP cells, only L-WNT3A CM induced reporter activation (Figure 3b and Supplementary Figures S6A, F and G). In other CRC cell lines, WNT signaling could not be hyperactivated with any CM (Supplementary Figure S6A). Addition of the WNT signaling antagonist DKK1 resulted in effective inhibition of canonical WNT signaling in RKO-7TGP co-cultured with L-WNT3A and L-WNT2 (Figure 3c and Supplementary Figure S6H). Since expression profiles indicated that CAFs from the tumor stroma express WNT2B, albeit at significantly lower levels than WNT2 (see Figure 2 and Supplementary Figures S2), we tested our NCF/CAF pairs derived from three different patients and found varying expression of WNT2B by quantitative PCR with reverse transcription (Figure 3d). CAF3 expressed significantly more WNT2B mRNA compared to NCF3, whereas NCF4 and CAF4 expressed comparable amounts. CAF5 expressed less WNT2B mRNA than NCF5. Thus, there was no consistent WNT2B CAF specific expression detectable as it was found for WNT2. However, CAF3, CAF4 and CAF5 but neither NCF3, NCF4 nor NCF5 could activate the 7TGP reporter in RKO cells (Figure 3e and Supplementary Figure S6I). These data demonstrate that WNT2, but not WNT2B, activates canonical WNT signaling in CRC cells harboring wild-type adenomatous polyposis coli and β-catenin.
Canonical WNT signaling is activated in wild-type WNT/β-catenin CRC cells. (a–c and e) Measurement of reporter gene activation via FACS analysis. (a) DLD1-7TGP, HCT116-7TGP, HT29-7TGP LS174T-7TGP and RKO-7TGP tumor cells co-cultivated (CC) with WNT expressing fibroblasts (L-WNT5A, L-WNT3A, L-WNT2) and control fibroblasts (L-par) (n=3). (b) RKO-7TGP treated with 0.2 μg/ml rhWNT2 or CC with WNT2 expressing L-cells (n=3). (c) WNT pathway inhibition with 0.5 μg/ml rhDKK1 in RKO-7TGP cells upon CC with different WNT expressing L-cells (n=3). (d) WNT2B mRNA expression of three pairs of NCF and CAFs. RT-qPCR analysis of three different passages (n=3); normalized to GAPDH. (e) Reporter activation of RKO-7TGP CC with NCF and CAFs of three different patients (n=4). Bar charts, mean; error bars, s.e.m. FACS, fluorescence-activated cell sorting.
WNT2 induces canonical signaling in colon fibroblasts
Next we analyzed whether WNT2 could activate canonical WNT-signaling in fibroblasts. Direct co-culture of CCD18Co-7TGP cells with L-WNT3A and L-WNT2 cells induced WNT/β-catenin signaling (Figure 4a and Supplementary Figure S7A). Co-culture experiments using transwell inserts in order to determine exact percentages of reporter activation resulted in 96% of GFP-positive CCD18Co-7TGP cells by WNT3A, whereas WNT2 induced signaling in about 50% of the fibroblasts (Supplementary Figure S7BC). Consistent with 7TGP activation, nuclear β-catenin increased upon co-culture of CCD18Co with WNT2 and WNT3A producers, in contrast to untreated and WNT5A treated controls (Figure 4b). Quantification of nuclear β-catenin revealed an almost fourfold induction by WNT2 and a ninefold induction by WNT3A. Importantly, colon CAFs (CAF3) endogenously expressing high levels of WNT2 (see Figure 1d) induced canonical WNT-signaling in the CCD18Co-7TGP fibroblasts, whereas NCFs (NCF3) from the same patient with undetectable WNT2 expression did not (Figure 4c and Supplementary Figure S7). In contrast to WNT3A activation of BJ1-7TGP skin fibroblast reporters, co-cultivation with WNT2 producers had no effect (Figure 4d and Supplementary Figure S7E). L-WNT2 CM did not induce canonical signaling in BJ1-7TGP or CCD18Co-7TGP reporter cells (Supplementary Figures S7F and G).
Canonical WNT signaling is activated in colonic fibroblasts by WNT2 in co-cultures. (a) FACS analysis of CCD18Co-7TGP upon co-culture (CC) with different L-cells (n=3) (b) β-catenin (top) and DAPI (bottom) staining of CCD18Co CC with L-WNT5A, L-WNT3A and L-WNT2 cells. CCD18Co nuclei are indicated by dashed white ellipses (CCD18Co nuclear mask). β-catenin expression in the nucleus (middle) was visualized by subtracting cytoplasmic expression using the CCD18Co nuclear mask. Scale bar, 100 μm. β-catenin nuclear translocation was measured by subtracting cytoplasmic intensity from nuclear intensity (no co-culture, n=54 cells; +L-WNT5A n=56; +L-WNT3A n=59; +L-WNT2 n=70). (c) Reporter activation of CCD18Co-7TGP CC with NCF3 and CAF3. (d) FACS analysis of BJ1-7TGP CC with WNT expressing L-cells (n=3). (e) Expression data from Dolznig data set20 in BJ1 and CCD18Co. High expression: black squares (>200), intermediate expression: gray squares (>50, <200), low or absent expression: white squares (<50). (f) Reporter activation of BJ1-7TGP cells ectopically expressing FZD4, FZD6 or FZD8, CC with L-par or L-WNT2 cells (n=3). Bar graph, means; error bars, s.e.m. FACS, fluorescence-activated cell sorting.
To elucidate the different responsiveness of BJ1 and CCD18Co cells to WNT2 treatment, expression profiles of the Dolznig data set20 were re-evaluated. Only Frizzled-4 (FZD4) and FZD8 were differentially expressed in dermal (BJ1) (non responders, absent FZD4 and FZD8) versus colonic (CCD18Co) fibroblasts (responders, FZD4 and FZD8 present, Figure 4e). We therefore ectopically expressed these receptors individually in BJ1-7TGP and re-tested WNT2 responsiveness. BJ1-7TGP-FZD8 cells displayed GFP expression upon co-culture with L-WNT2, whereas FZD4 transfected cells did not (Figure 4f). This strongly indicated that FZD8 is a functional WNT2 receptor in fibroblasts. Ectopic expression of FZD6 was included as control, being already endogenously expressed at high levels in both cell types and had no influence on signaling. Taken together, these data demonstrate that WNT2 can activate canonical signaling specifically in colon fibroblasts involving FZD8. To enable signaling, direct contact of WNT2 producing cells with the responding fibroblasts was necessary.
WNT2 induces migration and invasion in colon fibroblasts
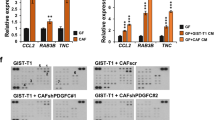
Co-culture with WNT2 expressing L-cells or WNT2-KD in CAF3 (Supplementary Figure S8A) did not alter proliferation (Supplementary Figures S8B and C) or apoptosis (Supplementary Figures S8D and E). Furthermore, CAF3-NTC or CAF3-WNT2-KD were subjected to transwell-migration assays. A significant decrease of migrated CAF3-WNT2-KD cells was detectable compared to CAF3-NTC (Figure 5a). Quantitation of invading cells from spheroids revealed that length and number of invasive structures were significantly reduced in CAF3-WNT2-KD as compared to controls (Figure 5b). To demonstrate the functionality of the FZD8-WNT2 cascade CAF3-FZD8-KD cells (Supplementary Figure S8F) were subjected to transwell-migration and spheroid invasion assays. Indeed, CAF3-FZD8-KD displayed reduced migratory potential compared to CAF3-NTC (Supplementary Figure S8G) and decreased number and length of invasive sprouts (Supplementary Figure S8H). In line, BJ-1 cells engineered to respond to WNT2 by stable FZD8 expression (Supplementary Figure S8I) showed a higher tendency to invade in this assay as compared to controls (Supplementary Figure S8J). Taken together these results demonstrate that autocrine WNT2 signals via FZD8 receptor, thereby leading to a pro-migratory and pro-invasive phenotype of CAFs.
Invasive capacity of fibroblasts is increased by WNT2. (a) Migration assessed using transwell chambers (8 μm pore size) shown as scatter dot plots (n=10). Representative images of migrated CAF3-NTC and WNT2-KD stained with crystal violet are depicted; scale bars, 250 μm. (b) Collagen gel spheroid invasion assay. CAF3-WNT2-KD or CAF3-NTC spheroids were embedded into collagen gels; pictures were taken at 0 and 17 h; length and number of invasive structures were evaluated (NTC n=10, WNT2-KD n=7). Representative images are shown; scale bars, 250 μm. (c) CAF3-NTC and CAF3-WNT2-KD were used in raft culture assays with DLD1 tumor cells seeded on top of the gels in duplicates. After 21 days gels were subjected to FFPE and sections were H&E stained. Number of invasive structures was determined of five sections per gel (n=10). Details of representative images are shown, scale bars, 125 μm. (d) Effect of rhDKK1 (0.5 ug/ml) in the same assay as in c and e. Invasion of DLD-1 in response to WNT2 overexpressing BJ1 was assessed in the same assays as in c, d, f and g Sirius. Red staining of collagen fibers and three-dimensional surface plots of red staining density of CAF3-NTC and CAF3-WNT2-KD (e) or BJ1-GFP and BJ1-WNT2 (f) co-cultivated with DLD1 tumor cells. Scale bars, 500 μm. Scatter dot plots: center line, means; whiskers, s.e.m.; circles/squares, individual values. H&E, hematoxylin and eosin.
WNT2 expressed in CAFs stimulates invasion of CRC cells in organotypic 3D co-cultures
Fibroblasts display a leading role for collective tumor cell invasion.23 Hence, we addressed if WNT2 has an indirect effect on CRC cell invasion by modulating the invasive capacity of stromal fibroblasts. Therefore, we set up organotypic raft culture assays (OTAs) of DLD1 CRC cells with CAF3-NTC or CAF3-WNT2-KD to analyze tumor cell invasion. Indeed, despite having no direct effect on WNT signaling in DLD1 cells (Figure 3a), the presence of WNT2 in fibroblasts resulted in a threefold increase of invasive DLD1 structures as compared to CAFs with WNT2-KD (Figure 5c). Similar results were obtained with LS174T (Supplementary Figure S8K). In line, treatment of DLD1/CAF3 OTAs with the WNT inhibitor DKK1 resulted in the same inhibition of invasion (Figure 5d). Ectopic expression of WNT2 in dermal fibroblasts, which were unresponsive to WNT2 (Figure 4d), expectedly failed to promote DLD1 cell invasion (Figure 5e). Sirius Red staining revealed higher matrix remodeling by increased collagen fibers in CAF-NTC compared to CAF3-WNT2-KD conditions (Figure 5f and Supplementary Figure S8L); in BJ1-GFP and BJ1-WNT2 no differences were visible (Figure 5g). This was further substantiated by decreased collagen gel contractility of CAF3-WNT2-KD (Supplementary Figure S8M). These data suggest that CAF-derived WNT2 led to enhanced fibroblast invasiveness and extracellular matrix remodeling. We conclude that direct interaction at the tumor/stroma border and WNT2-induced signaling in CAFs promotes CRC invasion.
WNT2 expression increases tumor growth, invasion and metastasis in CRC xenografts and is a predictor for poor prognosis in CRC patients
To validate our findings in vivo CAF3-WNT2-KD or CAF3-NTC were co-implanted together with HT29 in NOG mice, but no difference in tumor growth was detected (Supplementary Figure S9A). Since human CAFs were completely lost (Supplementary Figure S9B), which is in concordance with previous reports on early disappearance of stromal cells during xenograft growth,24 WNT2 overexpressing HT29 and HCT116 CRC cells were used for constant WNT2 supply (Supplementary Figure S9C and D). Overexpression effects of WNT2 in the tumor cells would exert its function on mice stroma, rather than the WNT2 expressing tumor cells, as there was no hyperactivation of β-catenin signaling in tumor cells (see Figure 3a and Supplementary S5A and B) and WNT2 expression in HT29 and HCT116 showed no changes in cell cycle distribution in vitro compared to GFP controls (Figure 6a and Supplementary Figure S9E). HT29 (Figure 6BC) and HCT116 (Supplementary Figures S9F and G) displayed increased tumor growth in the presence of WNT2 in xenografts and resulted in doubled tumor weight. WNT2 overexpressing tumors showed augmented Ki67 staining (Figure 6d) indicating increased proliferation but displayed the same amount of necrotic areas (Figure 6e and Supplementary Figure S9H). Mouse stroma was present at the outer rim of xenografted tumors at comparable amounts in WNT2 expressing and control tumors as shown by IHC for PDGFRB and alpha-SMA (Supplementary Figure S9I).
WNT2 has a tumor-promoting effect in vivo. (a) Cell cycle histogram of HT29-GFP (gray) and HT29-WNT2 (black) cells. One representative experiment is shown. (b) 1 × 105 cells subcutaneously injected in SCID mice were assessed for tumor growth (n=5). After sacrifice, tumors were weighed. Growth curves and bar charts, means; error bars, s.e.m. (c) Macroscopic appearance of HT29 tumors; scale bar, 0.5 cm. (d) Ki67 quantification using Tissue Studio. Mean percentages of KI67+ cells in sections of whole tumors of five different animals are shown. Scale bar, 50 μm; error bars, s.e.m. (e) Determination of viable versus necrotic tissue using Tissue Studio; scale bar, 100 μm. SCID, severe combined immunodeficient.
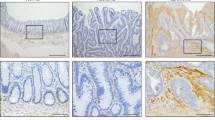
Underscoring these results, HT29-WNT2 cells orthotopically inoculated into the cecum showed increased tumor growth (IVI, Figure 7a), tumor budding and invasiveness (Figures 7b and c). Quantification of immunohistochemically stained cancer cells with a specific antibody against human Cytokeratin 18 revealed an 2.2-fold increase in muscle layer invasion and a 5.5-fold elevated mucosa infiltration rate of HT29-WNT2 compared to HT29-GFP (Figure 7d). Moreover liver metastasis (IVI, Figures 7e and f) was elevated compared the HT29-GFP controls.
WNT2 induces invasion and liver metastasis in orthotopic mouse tumor xenograft models. 1 × 105 HT29-GFP or HT29-WNT2 cells were injected into the cecum of NOG mice. After 29 days mice were injected with a human-specific HLA-ABC antibody, sacrificed and IVI performed on different organs. (a) Mean photon efflux in the colon (horizontal line, error bars: SD; HT29-GFP n=8, HT29-WNT2 n=6). Representative images are shown. (b, h and e) stain of colon sections. Arrowheads indicate tumor budding (top, middle), normal mucosa infiltration (top, right), mucosa displacement (bottom, middle) and strand like infiltration into the muscularis (bottom, right). (c) Human-specific Cytokeratin 18 staining (brown) of tumor cells, nuclei were counterstained with hematoxylin (blue). (d) Relative values of muscle layer infiltrating areas (HT29-GFP n=6; HT29-WNT2 n= 8) as well as mucosa invasion (HT29-GFP n=8; HT29-WNT2 n=9) based on the area of invasive areas/total tissue. (e) Mean photon efflux in the livers (horizontal line, error bars: s.d.). (f, h and e) stain of liver sections. Metastatic lesions are encircled. The number of metastatic nodules was determined per section; mean values are shown (n=6).
In human CRC high WNT2 expression was associated with decreased disease-free survival and higher numbers at risk (Figures 8a and b) and improved the predictive power (Supplementary Figure S9J) of a bona fide stromal gene (FAP) associated with bad prognosis.25
Survival analysis of 160 CRC patients using GSE24551 data set.51 Data were bifurcated for high and low WNT2 expression at the median and a Kaplan–Meier survival plot was generated with the SurvExpress online tool.52 (a) Red, high; green, low expression of WNT2. Numbers at risk and statistic parameters are indicated. (b) Expression levels of Wnt high (red) versus low (green) in five probe sets.
Discussion
Our finding that CRC express high levels of WNT2 mRNA supports earlier reports that WNT2 was induced in colon cancers.4, 13, 26, 27, 28, 29 In addition, WNT2 is consistently induced in malignant lesions of the digestive tract as reported in esophageal and gastric carcinomas,4, 13, 27, 28, 29, 30 whereas the normal epithelium of the digestive system did not show WNT2 expression.4 This notion is manifested by our data mining approach demonstrating that WNT2 was not upregulated in other cancer types compared to normal tissue.
We show that WNT2 is selectively expressed in stromal fibroblasts of CRC. Another CRC study proposed macrophages as the source of WNT2 by in situ hybridization analysis.27 The stromal source of WNT2 in tumors of the digestive tract is indirectly supported by lack of WNT2 mRNA in different gastric cancer cell lines26, 30 and CRC cells as demonstrated in our study. Recently, WNT2 expression was found in CAFs isolated from esophageal carcinomas,29 further substantiating our hypothesis that induction of WNT2 in the tumor stroma might be common in carcinomas of the digestive tract.
The tumor stroma can influence the development of malignant tumors.31, 32 However, the functional role of stromal WNT2 in gastrointestinal cancer progression was barely addressed so far. We demonstrate here that paracrine WNT2 signaling from CAFs could not hyperactivate canonical WNT signaling in CRC cell lines harboring mutations in the WNT/β-catenin pathway, which applies to the majority (>70%) of CRC cases.33 As expected, non-treated CRC cell lines already displayed activated canonical signaling. Nonetheless, we cannot rule out that paracrine effects of WNT2 on CRC cells with altered WNT/β-catenin signaling were too subtle to be detected. WNT2 treatment induced canonical WNT pathway activation in CRC cells bearing wild-type adenomatous polyposis coli and CTNNB1 genes (RKO cells). This is in agreement with stromal derived HGF, which induced β-catenin activation in cancer cells from primary patient material.34 Therefore, a promising testable hypothesis is, whether WNT2 is besides HGF another stromal factor that mediates the canonical response in primary CRC cells, which can display a mixed WNT/β-catenin mutation status. This is supported by results from esophageal squameous cell carcinoma, in which epithelial cells were responsive to WNT2 thereby promoting tumor progression.29 In this case, the lack of mutations in adenomatous polyposis coli35 or CTNNB136 are potentially enabling activation of β-catenin upon WNT2 stimulation.
WNT2 induced canonical signaling in the colonic myofibroblasts CCD18Co upon co-cultivation with WNT2 producer cells. In line, WNT2 activates canonical WNT signaling in rat lung fibroblasts37 and in NIH-3T3.38 However, WNT2 did not activate canonical signaling in skin fibroblasts, which could be explained by differential receptor expression. In non-small cell lung cancer cells, WNT2 signals through FZD8.39 Indeed, FZD8 mRNA was present in colonic fibroblasts, whereas BJ1 lacked its expression. Ectopic expression of FZD8 restored WNT2 responsiveness in BJ1 indicating that FZD8 is the receptor for WNT2 in colonic fibroblasts.
Canonical signaling response to WNT2 in colonic fibroblasts was only seen in co-culture with WNT2 producing cells. The lack of reporter activation using WNT2 CM is not surprising, as WNT140 and WNT25 can be tightly bound to heparan sulfate proteoglycans on the cell surface. In contrast, the activity of WNT3A in CM could be explained by association of WNT3A with lipoprotein particles.41 A recent report strongly supports the predominant insolubility of WNTs in cell culture supernatants as they convincingly demonstrate that WNTs are passed to responder cells via filopodia.42
WNT2 induced proliferation and invasion of esophageal squameous cell carcinoma cells29 and promoted metastasis in pancreatic cancer.43 We are the first to demonstrate that WNT2 changes the phenotype of fibroblasts thereby affecting CRC progression. First, WNT2 depletion in CAFs decreased migration, indicating that WNT2 has a pro-migratory effect on colonic fibroblasts. Furthermore, CAF3-WNT2-KD resulted in reduced invasion. Second, WNT2-KD in CAFs reduced the number of invasive structures of DLD1 cells in co-cultures. Convincingly, WNT2 expression in BJ1 had no effect on DLD1 tumor cell invasion, as these fibroblasts were non-responsive to canonical signaling. In organotypic co-cultures of squameous cell carcinoma cells and fibroblasts, invading tumor cells were following leading fibroblasts, providing tracks within the remodeled extracellular matrix,23 which are sufficient to increase collective invasion providing a possible explanation for our results. Hence, the increased extracellular matrix remodeling capacity of WNT2 expressing CAFs might provide more tracks for tumor cells to invade. Third, xenografted tumors expressing WNT2 growing to double the size of controls strongly support the in vivo relevance of our findings. In the presence of WNT2 there was an increase in proliferation of HT29 cells detectable. As these cells do not respond to WNT2 with further increased canonical signaling and lack altered cell cycle distribution in monocultures in vitro, we speculate that WNT2 expression in the CRC cells could have effects in the mouse stroma leading to increased proliferative capacity in the tumor cells as secondary effect. The mechanisms of this cancer promoting effect remain to be identified in further studies. Finally, high WNT2 levels in CRC were associated with decreased patient survival consistent with poor survival in esophageal squamous cell carcinomas. In those tumors WNT2 was also highly expressed in CAFs.29
In conclusion, the identification of the pro-malignant function of WNT2 (for a summary scheme see Supplementary Figure S10) might represent an encouraging stromal target, for example, by blocking FZD8 function to efficiently inhibit tumor progression in combination with conventional and targeted anti-cancer therapies.
Material and methods
Cells and cell lines
STR-profiled and authenticated (PowerPlex-16-HS-System, Promega, Madison, WI, USA) LS174T, DLD1, HCT116, HT29, RKO, BJ1, CCD18Co, L-WNT3A, L-WNT5A and parental L-cells were obtained from the master-cell-bank at Boehringer Ingelheim RCV and used within 6 months. Primary CAF cultures were used as described.20 For establishment of WNT2 overexpressing cells pLKO.1-CMV-WNT2 lentiviral particles were used. For further details please see Supplementary Methods. A list of all cell lines used in this study is given in Supplementary Table S7.
Correlation of WNT2 mRNA expression and ESTIMATE stromal score
ESTIMATE stromal scores (http://bioinformatics.mdanderson.org/estimate/)44 were calculated for 218 CRC patients (TCGA-COAD and TCGA-READ data sets, Agilent G4502A chip, Agilent Technologies, Santa Clara, CA, USA) and corresponding mRNA expression data generated by the TCGA Research Network (http://cancergenome.nih.gov/) was downloaded from UCSC Cancer Genomics Browser (COADREAD gene expression AgilentG4502A_07_3; https://genome-cancer.ucsc.edu). WNT2 mRNA expression and stromal score were plotted, linear regression was performed (black line) and 95% confidence interval is shown (dashed line).
Expression-profiling data
Data were re-evaluated from Affymetrix expression-profiling screens of dissected human CRC specimens performed earlier (Rupp,11 Nishida, GSE3560216 and Calon, GSE3939718). Expression-profiling data of three-dimensional-cultures of LS174T, CAF1, CAF2, BJ1 and CCD18Co were re-evaluated (Dolznig et al.)20
7TGP reporter assays
A 7TGP lentiviral plasmid21 (Addgene, Cambridge, MA, USA, #24305) was used. BJ1-7TGP, CCD18Co-7TGP or the tumor cell-7TGP reporters were treated with conditioned medium of WNT overexpressing L-cells (L-WNT5A, L-WNT3A, L-WNT2), control L-cells (L-par);45 co-cultivated with the different L-cells, NCF3, CAF3; treated with 0.2 μg/ml rhWNT2 (Novus Biologicals, Littleton, CO, USA) or 0.5 μg/ml rhDKK1 (R&D Systems, Minneapolis, MN, USA). Pathway activation was determined by GFP+ cells using flow cytometry. To quantify actual percentages of GFP+ cells, cell culture inserts (0.4 μm pore size, Merck Millipore, Billerica, MA, USA) were used. The membrane undersurface was covered with WNT2-expressing or control cells and reporter-7TGP cells were seeded on the top surface. Reporter cells were harvested and percentages of GFP+ cells were determined. For further details refer to Supplementary Methods.
Ectopic FZD expression
FZD4 (#42256), FZD6 (#42258), FZD8 (#42260)46 and GFP expression vectors were obtained from Addgene. BJ1-7TGP cells were transfected using ViaFect according to manufacturers’ protocol (Promega). Transfected BJ1-7TGP cells were co-cultivated with L-par or L-WNT2 cells. GFP expression was measured by flow cytometry. Transfection efficiency was determined in parallel using a GFP expression plasmid. Percentages of BJ1-7TGP GFP+ cells were calculated using the transfection efficiency from the control approach. Stable FZD8 expression was achieved with lentiviral transduction and selection (Genecopeia, Rockville, MD, USA).
RT-qPCR
mRNA was isolated using the ReliaPrep Kit (Promega). First-strand cDNA synthesis was carried out using GoScript Reverse Transcriptase and Oligo(dT)15 primer (Promega). WNT2 mRNA levels were assessed using qPCR-Master Mix (Promega) in a Bio-Rad CFX96 Real-Time PCR System. Relative expression levels were calculated using the ΔΔCq method.47 WNT2-fw-5′-CCAGCCTTTTGGCAGGGTC-3′; WNT2-rev-5′-GCATGTCCTGAGAGTCCATG-3′; WNT2B-fw-5′-AAGATGGTGCCAACTTCACCG-3′; WNT2B-rev-5′-CTGCCTTCTTGGGGGCTTTGC-3′; FZD8-fw-5′-GCGTTTCCAGAACCCGAGAT-3′; FZD8-rev-5′-CCAAACCGTGCAATCCCATC-3′; GAPDH-fw-5′-AACAGCGACACCCACTCCTC-3′; GAPDH-rev-5′-CATACCAGGAAATGAGCTTGACAA-3′. For further details see Supplementary Methods.
Nuclear β-catenin levels
CCD18Co cells were co-cultivated with WNT expressing and control L-cells and stained according to Cell Signaling’s Immunofluorescence Protocol with methanol permeabilization. Antibodies: Vimentin (V9, 1:300, #MA5-11883, Thermo Scientific, Waltham, MA, USA); β-catenin (1:100, #8480, Cell Signaling Technologies, CST, Danvers, MA, USA), Alexa Fluor-594 and -488 (1:500, Thermo Scientific). Nuclei were counterstained with 1 μg/ml DAPI (Sigma-Aldrich, St Louis, MO, USA). β-catenin nuclear intensity in fluorescence images (AxioObserver, Zeiss, Wetzlar, Germany) was measured (ImageJ64, NIH, Bethesda, MD, USA) and calculated.48 Refer to Supplementary Methods.
siRNA-mediated knockdown
ON-TARGETplus-SMARTpool siRNAs (GE Dharmacon, Lafayette, CO, USA) were used for WNT2 and FZD8 knockdown (KD) experiments according to an established protocol.49 See Supplementary Methods.
Migration
2.5 × 104 CAF3 were seeded in serum-free medium in the upper chamber of transwell-migration inserts (8 μm, Sigma-Aldrich), lower chambers contained complete medium. After 22 h, non-migrated cells were removed and migrated cells were fixed (Roti-Histofix 4%). CAF3 cells were stained with 0.006% (w/v) Crystal violet-solution (Sigma-Aldrich) for 30 min. Five random images were taken and membrane coverage was determined (ImageJ64, NIH).
Invasion
1.5x103 CAF3-NTC, CAF3-WNT2-KD or CAF3-FZD8-KD cells were seeded per well of a v-shaped 96-well plate in medium containing 0.3% (v/v) methylcellulose (Sigma-Aldrich). After 6 h, spheroids were collected and embedded in Collagen I gel as described.20 After 16 h pictures were taken, length and number of invaded structures was measured (ImageJ64, NIH).
Organotypic assays
Organotypic raft cultures were used as described50 with minor changes. Fibroblasts were embedded in collagen gels. DLD1 or LS174T tumor cells were added on top, gels were transferred onto metal grids and incubated at the air-liquid interphase. After 21 days gels were fixed (Roti-Histofix 4%), paraffin embedded and perpendicular 5 μm sections were cut, every fifth section was hematoxylin and eosin and Sirius Red stained for assessment of collagen fiber architecture. Numbers and areas of invasive structures were measured and three-dimensional surface plots were generated (ImageJ64, NIH). For details see Supplementary Methods.
Xenograft tumor model
1 × 106 HCT116 and HT29 tumor cells ectopically expressing WNT2 or GFP were subcutaneously injected into severe combined immunodeficient mice. After killing, tumor weight was assessed and pictures were taken. Sections were immunohistochemically stained with Ki67 (Life Technologies), quantitative image analysis was performed with the Tissue Studio software (Definiens, Munich, Germany). Before quantification, necrotic areas were subtracted and only viable tumor parts were used for further analysis. For orthotopic implantation 1 × 105 HT29-GFP or HT29-WNT2 cells were injected in 50 μl of collagen I/Matrigel mixture into the cecum of NOG mice. In vivo imaging (IVI) on live animals and on organs after resection was performed 29 days post injection with the Image Station in vivo FX (Kodak, Rochester, NY, USA) using hHLA-ABC-Alexa750 antibody (0.02 mg, BeckmannCoulter, Indianapolis, IN, USA). Non-tumor bearing mice were used as control. Organs were subjected to FFPE and sections were stained with hematoxylin and eosin or a human-specific Cytokeratin 18 antibody (clone DC10, #M7010, DAKO, Glostrup, Denmark).
Patient data
Survival analysis was performed using the GSE24551 data set51 with the SurvExpress online tool.52
Statistical analysis
Graphs were generated using GraphPad Prism5 (GraphPad Software, Inc., La Jolla, CA, USA). Significance (P-values<0.05) was calculated using the unpaired t-test. Data were normal distributed.
References
Anastas JN, Moon RT . WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer 2013; 13: 11–26.
Huguet EL, McMahon JA, McMahon AP, Bicknell R, Harris AL . Differential expression of human Wnt genes 2, 3, 4, and 7B in human breast cell lines and normal and disease states of human breast tissue. Cancer Res 1994; 54: 2615–2621.
Dale TC, Weber-Hall SJ, Smith K, Huguet EL, Jayatilake H, Gusterson BA et al. Compartment switching of WNT-2 expression in human breast tumors. Cancer Res 1996; 56: 4320–4323.
Vider BZ, Zimber A, Chastre E, Prevot S, Gespach C, Estlein D et al. Evidence for the involvement of the Wnt 2 gene in human colorectal cancer. Oncogene 1996; 12: 153–158.
Blasband A, Schryver B, Papkoff J . The biochemical properties and transforming potential of human Wnt-2 are similar to Wnt-1. Oncogene 1992; 7: 153–161.
Pu P, Zhang Z, Kang C, Jiang R, Jia Z, Wang G et al. Downregulation of Wnt2 and beta-catenin by siRNA suppresses malignant glioma cell growth. Cancer Gene Ther 2009; 16: 351–361.
Humphreys RC, Rosen JM . Stably transfected HC11 cells provide an in vitro and in vivo model system for studying Wnt gene function. Cell Growth Differ 1997; 8: 839–849.
Bayle J, Fitch J, Jacobsen K, Kumar R, Lafyatis R, Lemaire R . Increased expression of Wnt2 and SFRP4 in Tsk mouse skin: role of Wnt signaling in altered dermal fibrillin deposition and systemic sclerosis. J Invest Dermatol 2008; 128: 871–881.
Goss AM, Tian Y, Tsukiyama T, Cohen ED, Zhou D, Lu MM et al. Wnt2/2b and beta-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Dev Cell 2009; 17: 290–298.
Matushansky I, Hernando E, Socci ND, Mills JE, Matos TA, Edgar MA et al. Derivation of sarcomas from mesenchymal stem cells via inactivation of the Wnt pathway. J Clin Invest 2007; 117: 3248–3257.
Rupp C, Scherzer M, Rudisch A, Unger C, Haslinger C, Schweifer N et al. IGFBP7, a novel tumor stroma marker, with growth-promoting effects in colon cancer through a paracrine tumor-stroma interaction. Oncogene 2015; 34: 815–825.
Ma XR, Edmund Sim UH, Pauline B, Patricia L, Rahman J . Overexpression of WNT2 and TSG101 genes in colorectal carcinoma. Trop Biomed 2008; 25: 46–57.
Holcombe RF, Marsh JL, Waterman ML, Lin F, Milovanovic T, Truong T . Expression of Wnt ligands and Frizzled receptors in colonic mucosa and in colon carcinoma. Mol Pathol 2002; 55: 220–226.
Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007; 9: 166–180.
Edgar R, Domrachev M, Lash AE . Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002; 30: 207–210.
Nishida N, Nagahara M, Sato T, Mimori K, Sudo T, Tanaka F et al. Microarray analysis of colorectal cancer stromal tissue reveals upregulation of two oncogenic miRNA clusters. Clin Cancer Res 2012; 18: 3054–3070.
Rupp C, Dolznig H, Puri C, Schweifer N, Sommergruber W, Kraut N et al. Laser capture microdissection of epithelial cancers guided by antibodies against fibroblast activation protein and endosialin. Diagn Mol Pathol 2006; 15: 35–42.
Calon A, Espinet E, Palomo-Ponce S, Tauriello DV, Iglesias M, Cespedes MV et al. Dependency of colorectal cancer on a TGF-beta-driven program in stromal cells for metastasis initiation. Cancer Cell 2012; 22: 571–584.
Isella C, Terrasi A, Bellomo SE, Petti C, Galatola G, Muratore A et al. Stromal contribution to the colorectal cancer transcriptome. Nat Genet 2015; 47: 312–319.
Dolznig H, Rupp C, Puri C, Haslinger C, Schweifer N, Wieser E et al. Modeling colon adenocarcinomas in vitro a 3D co-culture system induces cancer-relevant pathways upon tumor cell and stromal fibroblast interaction. Am J Pathol 2011; 179: 487–501.
Fuerer C, Nusse R . Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS One 2010; 5: e9370.
da Costa LT, He TC, Yu J, Sparks AB, Morin PJ, Polyak K et al. CDX2 is mutated in a colorectal cancer with normal APC/beta-catenin signaling. Oncogene 1999; 18: 5010–5014.
Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol 2007; 9: 1392–1400.
Hylander BL, Punt N, Tang H, Hillman J, Vaughan M, Bshara W et al. Origin of the vasculature supporting growth of primary patient tumor xenografts. J Transl Med 2013; 11: 110.
Wikberg ML, Edin S, Lundberg IV, Van Guelpen B, Dahlin AM, Rutegard J et al. High intratumoral expression of fibroblast activation protein (FAP) in colon cancer is associated with poorer patient prognosis. Tumour Biol 2013; 34: 1013–1020.
Katoh M . Frequent up-regulation of WNT2 in primary gastric cancer and colorectal cancer. Int J Oncol 2001; 19: 1003–1007.
Smith K, Bui TD, Poulsom R, Kaklamanis L, Williams G, Harris AL . Up-regulation of macrophage wnt gene expression in adenoma-carcinoma progression of human colorectal cancer. Br J Cancer 1999; 81: 496–502.
Park JK, Song JH, He TC, Nam SW, Lee JY, Park WS . Overexpression of Wnt-2 in colorectal cancers. Neoplasma 2009; 56: 119–123.
Fu L, Zhang C, Zhang LY, Dong SS, Lu LH, Chen J et al. Wnt2 secreted by tumour fibroblasts promotes tumour progression in oesophageal cancer by activation of the Wnt/beta-catenin signalling pathway. Gut 2011; 60: 1635–1643.
Katoh M . WNT2 and human gastrointestinal cancer (review). Int J Mol Med 2003; 12: 811–816.
Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR . Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res 1999; 59: 5002–5011.
Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 2005; 121: 335–348.
Suraweera N, Robinson J, Volikos E, Guenther T, Talbot I, Tomlinson I et al. Mutations within Wnt pathway genes in sporadic colorectal cancers and cell lines. Int J Cancer 2006; 119: 1837–1842.
Vermeulen L, De Sousa EMF, van der Heijden M, Cameron K, de Jong JH, Borovski T et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol 2010; 12: 468–476.
Ogasawara S, Maesawa C, Tamura G, Satodate R . Lack of mutations of the adenomatous polyposis coli gene in oesophageal and gastric carcinomas. Virchows Arch 1994; 424: 607–611.
de Castro J, Gamallo C, Palacios J, Moreno-Bueno G, Rodriguez N, Feliu J et al. beta-catenin expression pattern in primary oesophageal squamous cell carcinoma. Relationship with clinicopathologic features and clinical outcome. Virchows Arch 2000; 437: 599–604.
Miller MF, Cohen ED, Baggs JE, Lu MM, Hogenesch JB, Morrisey EE . Wnt ligands signal in a cooperative manner to promote foregut organogenesis. Proc Natl Acad Sci U S A 2012; 109: 15348–15353.
Najdi R, Proffitt K, Sprowl S, Kaur S, Yu J, Covey TM et al. A uniform human Wnt expression library reveals a shared secretory pathway and unique signaling activities. Differentiation 2012; 84: 203–213.
Bravo DT, Yang YL, Kuchenbecker K, Hung MS, Xu Z, Jablons DM et al. Frizzled-8 receptor is activated by the Wnt-2 ligand in non-small cell lung cancer. BMC Cancer 2013; 13: 316.
Bradley RS, Brown AM . The proto-oncogene int-1 encodes a secreted protein associated with the extracellular matrix. EMBO J 1990; 9: 1569–1575.
Neumann S, Coudreuse DY, van der Westhuyzen DR, Eckhardt ER, Korswagen HC, Schmitz G et al. Mammalian Wnt3a is released on lipoprotein particles. Traffic 2009; 10: 334–343.
Stanganello E, Hagemann AI, Mattes B, Sinner C, Meyen D, Weber S et al. Filopodia-based Wnt transport during vertebrate tissue patterning. Nat Commun 2015; 6: 5846.
Jiang H, Li Q, He C, Li F, Sheng H, Shen X et al. Activation of the Wnt pathway through Wnt2 promotes metastasis in pancreatic cancer. Am J Cancer Res 2014; 4: 537–544.
Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013; 4: 2612.
Chen W, ten Berge D, Brown J, Ahn S, Hu LA, Miller WE et al. Dishevelled 2 recruits beta-arrestin 2 to mediate Wnt5A-stimulated endocytosis of Frizzled 4. Science 2003; 301: 1391–1394.
Yu H, Ye X, Guo N, Nathans J . Frizzled 2 and frizzled 7 function redundantly in convergent extension and closure of the ventricular septum and palate: evidence for a network of interacting genes. Development 2012; 139: 4383–4394.
Livak KJ, Schmittgen TD . Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25: 402–408.
Burgess A, Vigneron S, Brioudes E, Labbe JC, Lorca T, Castro A . Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc Natl Acad Sci USA 2010; 107: 12564–12569.
Rosner M, Siegel N, Fuchs C, Slabina N, Dolznig H, Hengstschlager M . Efficient siRNA-mediated prolonged gene silencing in human amniotic fluid stem cells. Nat Protoc 2010; 5: 1081–1095.
Timpson P, McGhee EJ, Erami Z, Nobis M, Quinn JA, Edward M et al. Organotypic collagen I assay: a malleable platform to assess cell behaviour in a 3-dimensional context. J Vis Exp 2011; 56: e3089.
Sveen A, Agesen TH, Nesbakken A, Rognum TO, Lothe RA, Skotheim RI . Transcriptome instability in colorectal cancer identified by exon microarray analyses: Associations with splicing factor expression levels and patient survival. Genome Med 2011; 3: 32.
Aguirre-Gamboa R, Gomez-Rueda H, Martinez-Ledesma E, Martinez-Torteya A, Chacolla-Huaringa R, Rodriguez-Barrientos A et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One 2013; 8: e74250.
Acknowledgements
HD was supported by the Herzfelder Family Foundation and the Niederösterreichische Forschungs-und Bildungsges.m.b.H. AW was a recipient of a DOC Fellowship (24032) of the Austrian Academy of Sciences. RM was supported by the Austrian Science Fund (FWF, SFB F28 & F47). RE was supported by FWF (P25925-B20, P26908-B20, DK Inflammation and Immunity) and the Comprehensive Cancer Center (CCC) Vienna Research Grant.
Author contributions
NK and HD conceived and designed the research. NK, JS, CU, HN, AR, MS, DU, AW, MA, IC, TS, BP and JuSch performed the research. NK and HD analyzed the data and wrote the manuscript. MH, RE, XH, RM and WS were involved in data analysis and manuscript preparation. All authors reviewed and approved the manuscript for publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Kramer, N., Schmöllerl, J., Unger, C. et al. Autocrine WNT2 signaling in fibroblasts promotes colorectal cancer progression. Oncogene 36, 5460–5472 (2017). https://doi.org/10.1038/onc.2017.144
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.144
- Springer Nature Limited
This article is cited by
-
Colorectal cancer cells secreting DKK4 transform fibroblasts to promote tumour metastasis
Oncogene (2024)
-
FEV-mediated WNT2 transcription is involved in the progression of colorectal cancer via the Wnt signaling
Cytotechnology (2024)
-
Investigating the effect of Fusobacterium nucleatum on the aggressive behavior of cancer-associated fibroblasts in colorectal cancer
Discover Oncology (2024)
-
Canonical WNT/β-catenin signaling upregulates aerobic glycolysis in diverse cancer types
Molecular Biology Reports (2024)
-
Signaling pathways involved in the biological functions of dendritic cells and their implications for disease treatment
Molecular Biomedicine (2023)