Abstract
Solid malignancies contain subsets of multipotent cells that grow as spheres and efficiently propagate tumors in xenograft models, reflecting a stem-like, self-renewing and tumor-propagating phenotype. These cancer ‘stem cells (SCs)’ have been shown to maintain tumor growth, contribute to resistance and drive tumor recurrence. Cancer cell stemness is dynamically influenced by epigenetic mechanisms and differentially regulated coding and noncoding RNAs. How these mechanisms specifically contribute to the generation and/or maintenance of cancer SCs remains unclear. This study identifies a novel epigenetically regulated circuit that integrates microRNA, chromatin remodeling and the reprogramming transcription factor Sox2 to regulate glioblastoma (GBM)-propagating SCs. We show that miR-296-5p expression is repressed in a DNA methylation-dependent manner under conditions that promote GBM cell stemness and that miR-296-5p inhibits GBM cell stemness and their capacity to self-renew as spheres and propagate glioma xenografts in vivo. We show that the chromatin remodeling protein HMGA1 functions as a downstream effector of these biological responses to miR-296-5p and regulates Sox2 expression, a master driver of cell stemness, by modifying chromatin architecture at the Sox2 promoter. These results show for the first time that miR-296-5p inhibits transcriptional mechanisms that support GBM SCs and identify a miR-296-5p:HMGA1:Sox2 axis as a novel regulator of GBM SCs and candidate pathway for targeting therapies directed at depleting tumors of their tumor-propagating stem cell subsets.
Similar content being viewed by others
Introduction
Cancer results from multiple complementary defects in regulatory pathways that control cell proliferation and tissue homeostasis. It is well accepted that genetic modifications such as tumor suppressor inactivation and proto-oncogene hyperactivation can disrupt these regulatory pathways and thereby drive tumorigenesis.1 Non-mutational mechanisms of gene expression dysregulation such as DNA methylation, histone modification and expression of noncoding RNA are also critical regulators of the neoplastic phenotype.2, 3 These emerging epigenetic mechanisms present new opportunities for developing novel anticancer therapeutics.
Solid neoplasms are heterogeneous at the cellular level and contain cells that vary in their capacity to support tumor growth. Among these different cell sub-populations are multipotent stem-like cells that have particularly important roles in tumor growth, therapeutic resistance and recurrence following treatment.4, 5, 6, 7 Substantial evidence indicates that these stem-like tumor-propagating cells (also referred to as cancer stem cells or CSCs) are highly plastic and in response to epigenetic events dynamically transition between stem-like/tumor-propagating and more differentiated/non-tumor-propagating states. DNA and histone modifications (e.g. methylation, acetylation) regulate gene expression networks and the transition of stem cells from pluripotent to more differentiated phenotypes,8 and aberrant patterns of chromatin modification characterize many cancers.9 In glioblastoma (GBM), high expression of DNA methyltransferases (DNMTs) has been associated with tumor suppressor gene hypermethylation,10 and DNMT dysregulation has been reported to contribute to the GBM-propagating SC phenotype.11 Interestingly, the expression of a defined set of transcription factors involved in development is sufficient to recapitulate the neoplastic epigenetic landscape and reprogram non-tumorigenic cancer cells to display a tumor-propagating stem-like phenotype,12 highlighting the growing view that tumorigenesis results from both genetic and epigenetic changes.3 The specific epigenetic pathways involved in the acquisition and/or maintenance of the cancer SC phenotype remain unclear.
MicroRNAs regulate cell fate and oncogenesis by selectively inhibiting gene expression by either blocking mRNA translation or targeting mRNA for degradation.13, 14, 15, 16 MicroRNAs (miRNAs) are differentially expressed in GBM and re-expression of specific miRNAs can induce cell cycle arrest and the differentiation of tumor-derived SCs.17, 18, 19, 20, 21 The expression of miRNAs, as with coding genes, is regulated by the epigenetic landscape and there is cross-talk between epigenetic chromatin modification, miRNA networks and fate-determining transcription factors.22, 23 These observations support a role for specific epigenetic mechanisms for maintaining tumor-propagating stem-like cell subsets through miRNA regulation.
The focus of this study is to further understand how cross-talk between specific DNA methylation events, miRNA expression and SC-inducing transcription factors regulate GBM-propagating SCs. We present the novel findings that miR-296-5p, a miRNA not previously known to regulate cancer cell stemness, is repressed in a DNMT-dependent manner under conditions that promote GBM cell stemness. miR-296-5p is shown to inhibit efficiently the self-renewal capacity of GBM SCs in vitro and the growth of SC-derived glioma xenografts in vivo. We show for the first time that miR-296-5p inhibits the transcriptional mechanisms that support GBM SCs and identify a miR-296-5p:HMGA1:Sox2 axis as a novel regulator of GBM SCs and as a candidate pathway for targeting therapies directed at depleting tumors of their tumor-propagating stem cell subsets.
Results
Oct4/Sox2 coexpression inhibits miR-296-5p via promoter DNA methylation
It is becoming increasingly evident that cancer SC pools are maintained via dynamic processes dependent on the delicate interplay between multiple molecular regulators, including the expression of fate-determining transcription factors, changes in epigenetic landscape and expression of specific coding and noncoding RNAs.24 We recently demonstrated that reprogramming transcription factors Oct4 and Sox2 directly activate DNMT transcription, resulting in methylation events that induce the SC phenotype in GBM cells. Moreover, we identified a subset of miRNAs that are regulated by promoter DNA methylation and influence tumorigenesis.17 MiR-296-5p was among the miRNA found to be repressed by Oct4/Sox2 in a DNMT-dependent manner, suggesting that miR-296-5p might function as a stemness-inhibiting miRNA.
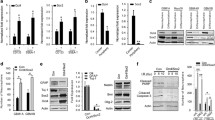
As a first step toward examining the role of miR-296-5p as a potential regulator of the glioma SC phenotype, we measured the effects of Oct4 and Sox2 on the expression of precursor (pre-) miR-296-5p in GBM-derived cells. Oct4 and Sox2 coexpression significantly decreased pre-miR-296-5p levels (50–90% reduction) in each of three culture models tested (Figure 1a). Treating cells with the pan-DNMT inhibitor 5-azacytidine abrogated the inhibition of pre-miR-296-5p levels in Oct4/Sox2-induced glioma SCs (A172-iGSC) (see Materials and Methods and Lopez-Bertoni et al.17) without affecting pre-miR-296-5p levels in control parental A172 cells, implicating a mechanism involving transcriptional repression in response to promoter DNA methylation (Figure 1b). This was confirmed using bisulfite sequencing that revealed an increase in miR-296-5p promoter CpG methylation from 50 to 83% in A172-iGSCs compared with parental A172 control cells (Figure 1c). Furthermore, treating A172-iGSCs with 5-azacytidine induced miR-296-5p expression and significantly decreased their capacity to self-renew as evidenced by a decrease in sphere formation (Figure 1d) and this coincided with a decrease in the expression of regulators and markers of cell stemness—Nanog, Prominin and Olig2 (Figure 1e).
miR-296-5p is regulated by promoter DNA methylation in GBM neurospheres. (a) Expression of precursor (pre-) miR-296-5p was measured using qRT–PCR in several GBM cell lines coexpressing Oct4 and Sox2. (b) A172 cells expressing transgenic Oct4/Sox2 (A172-Oct4/Sox2) or control cells were treated±DNA methylation inhibitors 5-azacytidine (5-Aza) (1 μm) for 72 h. Expression of pre-miR-296-5p was quantified by qRT–PCR. (c) Methylation status of the putative promoter region for miR-296-5p was determined by bisulfite sequencing in A172-iGSC and A172 control cells. Five clones were sequenced for each condition, and each row represents one clone. (d) A172-iGSCs or controls were treated with DNMT inhibitors 5-Aza (1 μm) and sphere formation capacity was measured 7 days after treatment. (e) DNMT inhibition results in reduced expression of Prominin, Nanog and Olig2 as quantified by qRT–PCR. (f) GBM neurosphere cells expressing stem cell markers CD133 or SSEA were isolated by flow cytometry. CD133+ and SSEA+ cells express lower levels of miR-296-5p compared with CD133− or SSEA− cells as determined by normalized qRT–PCR. (g) Forced differentiation induces pre-miR-296-5p expression in GBM neurospheres as determined by qRT–PCR. (h) Methylation status of the putative promoter region for miR-296-5p was determined by bisulfite sequencing following forced differentiation of GBM1A neurospheres. Five clones were sequenced for each condition, each row represents one clone. *P<0.05.
The above results reveal an inverse relationship between miR-296-5p expression and the SC phenotype in GBM neurospheres. This relationship was examined further by measuring miR-296-5p levels in GBM neurosphere cell fractions enriched for tumor-propagating SCs.6, 25 CD133+ and SSEA-1+ neurosphere cell subsets were found to express substantially lower levels of pre-miR-296-5p compared with their CD133− and SSEA-1− counterparts (Figure 1f). Conversely, forced differentiation of GBM neurosphere isolates induced a 4–9-fold increase in miR-296-5p expression (Figure 1g and Supplementary Figure S1) coincident with a decrease in DNA methylation from 67 to 33% in the putative miR-296-5p promoter (Figure 1h). Taken together, these results show that Oct4 and Sox2 repress miR-296-5p expression in a DNA methylation-dependent manner and suggest that endogenous miR-296-5p functions as an inhibitor of the SC phenotype in GBM.
miR-296-5p functions as a negative regulator of the CSC phenotype
To test the hypothesis that endogenous miR-296-5p functions as an inhibitor of the GBM SC phenotype, GBM-derived neurospheres were transduced with the lentivirus expressing either a scrambled RNA sequence or an antisense RNA oligo designed to target miR-296-5p (miR-296-5p sponge). The miR-296-5p sponge efficiently decreased the levels of pre-miR-296-5p in two independent GBM-derived neurosphere models by ~80%, as measured by real-time reverse transcription–PCR (qRT–PCR) (Figure 2a). Inhibition of endogenous miR-296-5p concurrently increased self-renewal as evidenced by significant increases in the number and size of neurospheres (Figure 2b) and consistently increased the expression of SC drivers Sox2, the pluripotency transcription factor Nanog and stem cell markers Nestin and Olig2 (Figure 2c).
miR-296-5p inhibition promotes the stem cell phenotype of GBM neurospheres. (a) GBM1A and Mayo39 neurospheres were transduced with a scrambled miRNA sequence (control sponge) or a miR-296-5p antisense sequence (miR-296-5p sponge). qRT–PCR shows decreased expression of pre-miR-296-5p 72 h after transduction in GBM1A and Mayo39 neurospheres compared with control neurospheres. (b) Equal numbers of GBM1A or Mayo39 neurospheres transduced with the control sponge or the miR-296-5p sponge were cultured in the neuropshere medium for 10 days and the number of spheres was quantified by computer-assisted image analysis. (c) Inhibition of miR-296-5p induces the expression of reprogramming transcription factors and stem cell markers in GBM1A and Mayo39 neurospheres. Gene expression was measured using qRT–PCR 72 h after transduction. *P<0.05.
A doxycycline (Dox)-inducible vector system was used to test directly the effects of miR-296-5p expression on the GBM SC phenotype. Pre-miR-296-5p expression peaked 3 days after adding Dox to the medium on day 0 and then returned to baseline by day 7 (Figure 3a). Induction of miR-296-5p robustly inhibited the sphere-forming capacity (Figure 3b) concurrent with the decreased expression of SC drivers and markers Sox2, Bmi-1, Klf-4, Nanog and Olig2 (Figure 3c). miR-296-5p expression significantly impaired the proliferation capacity of GBM neurosphere cells without affecting the cell viability (Figure 3d).
miR-296-5p inhibits the stem cell phenotype of GBM neurospheres. Neurospheres were engineered to express pre-miR-296-5p under the control of a Dox-inducible promoter. (a) Cells were treated by adding Dox to medium once at day 0 and samples were collected at days 0 (untreated), 3 and 7. qRT–PCR shows expression of pre-miR-296-5p in Mayo39 and GBM1A neurospheres compared with uninduced neurospheres. (b) Equal numbers of cells were cultured in neuropshere medium for 10 days±Dox (1 μg/ml) and the number of spheres was quantified by computer-assisted image analysis. (c) Expression of pre-miR-296-5p inhibits expression of reprogramming transcription factors and stem cell markers and drivers in Mayo39 and GBM1A neurospheres. Gene expression was measured using qRT–PCR 72 h after Dox (1 μg/ml) treatment. (d) Equal numbers of cells were dissociated into single-cell suspensions and cultured in neuropshere medium±Dox (1 μg/ml). Cells were counted at the indicated intervals using Trypan blue exclusion. (e) Mayo39-Tet-miR-296-5p cells were cultured in the presence (or absence) of Dox (1 μg/ml) for 4 days, and then 3.0 × 106 viable cells were implanted into the flanks of nude mice (top panel). Mice were fed Dox-containing diet for the duration of the experiment. Representative image of tumors generated by Mayo39-Tet-miR-296-5p±Dox (bottom panel). (f) Tumor volume was monitored over time using caliper measurements (N=8 for each group). (g) The table showing the percentage of animals with detectable subcutaneous tumors from limiting dilution experiment (N=6 for each group). *P<0.05.
Depleting GBM neurosphere cultures of their most stem-like cells is expected to reduce tumor-propagating capacity.24 Pre-miR-296-5p was induced in GBM neurospheres for 4 days and equal number of viable neurosphere cells was implanted subcutaneously to the flanks of immune-compromised mice. Mice were then fed Dox-containing diet for the duration of the experiment. As predicted by our in vitro findings, forced expression of pre-miR-296-5p resulted in tumor growth inhibition and overall reduction of tumor burden (Figures 3e and f). Using a similar experimental paradigm, we asked if miR-296-5p expression inhibits the tumor-propagating capacity of GBM neurospheres. To evaluate the effect of miR-296-59 on the relative numbers of tumor-propagating cells, serially diluted single-cell suspensions of neurosphere cells (1 × 106, 1 × 105 or 1 × 104 viable cells) were implanted subcutaneously to the flanks of immune-compromised mice and mice were fed Dox-containing diet for the duration of the experiment. As before, we observed a significant reduction in the tumor burden in the presence of miR-296-5p (Supplementary Figure S2), and, most importantly, miR-296-5p expression significantly decreased the tumor-propagating capacity of GBM cells (Figure 3g and Supplementary Figure S2).
HMGA1, a miR-296-5p target gene, regulates the GBM stem cell phenotype
A bioinformatics approach (www.targetscan.org) identified 10 high-confidence candidate miR-296-5p target genes (Supplementary Table S1).26 Two criteria, (1) expression downregulation following forced neurosphere cell differentiation and (2) expression upregulation in CD133+ and SSEA-1+ SCs, were used to identify which of the 10 candidate target genes were likely to function downstream of miR-296-5p. Forced differentiation, shown above to induce pre-miR-296-5p expression (Figure 1g), consistently decreased the expression of HMGA1, Sox13, Wnt7b and KCTD15 in three neurosphere models (Figure 4a). Of these four candidate miR-296-5p targets, only HMGA1 and KCTD15 were enriched in CD133+ (13-fold and 6-fold, respectively) and SSEA+ (8-fold and 2-fold, respectively) SC subsets (Figure 4b). We observed no significant enrichment of either Sox13 or Wnt7b in either of these cell subsets. HMGA1 expression was found to be upregulated in a panel of 12 independently isolated primary human GBM-derived neurosphere isolates compared with neurospheres derived from normal human fetal brain (Figure 4c). Thus, HMGA1 expression was increased in GBM SCs and inversely correlated with miR-296-5p expression. We asked if HMGA1 expression levels influence neurosphere cell phenotypes and the expression patterns of SC drivers. GBM neurospheres were transduced with lentivirus-expressing HMGA1 and transgenic HMGA1 significantly enhanced GBM self-renewal (Figure 4d). HMGA1 expression also induced molecular drivers and markers of GBM SCs including Oct4, Sox2, Klf-4, Nanog, Prominin, Nestin, Bmi-1 and Olig2 (Figure 4e). Conversely, targeting HMGA1 expression using a Dox-inducible shRNA (short hairpin RNA) hairpin robustly inhibited GBM sphere-forming capacity and decreased the expression of Sox2, Bmi-1, Olig2, Prominin and Nestin (Figures 4f and g). These responses were replicated in an additional neurosphere isolate and using a second shRNA hairpin (Supplementary Figure S3). Thus, HMGA1 modulates GBM cell phenotypes and HMGA1 inhibition mimics the effects of forced miR-296-5p expression. Taken together, these results are consistent with HMGA1 being a target by which miR-296-5p regulates GBM SCs.
HMGA1 regulates the stem cell phenotype of GBM neurospheres. (a) qRT–PCR analysis showing gene expression of predicted miR-296-5p targets in GBM1A, Mayo39 and 609 neurospheres following forced differentiation. (b) GBM1A neurospheres expressing stem cell markers CD133 or SSEA were isolated by flow cytometry. CD133+ and SSEA+ cells express higher levels of HMGA1 and KCTD15 compared with CD133− or SSEA− cells as determined by normalized qRT–PCR. (c) qRT–PCR to measure HMGA1 gene expression in GBM neurospheres lines and primary GBM neurosphere isolates. (d) GBM1A neurospheres were transduced with lentivirus expressing either GFP or HMGA1. (Left panel) Western blot showing the expression of HMGA1. (Right panel) Equal numbers of neurospheres were cultured in the neuropshere medium for 7 days and the number of spheres was quantified by computer-assisted image analysis. (e) qRT–PCR analysis showing the expression of reprogramming transcription factors and stem cell markers in response to transgenic HMGA1. (f) GBM1A cells were engineered to express an shRNA hairpin targeting HMGA1 under the control of a Dox-inducible promoter. (Left panel) Immunoblot shows HMGA1 knockdown and decreased expression of stem cell markers (Sox2, Nestin, Bmi-1 and Olig2) 72 h after the addition of Dox. (Right panel) qRT–PCR analysis showing decrease in Prominin in response to HMGA1 knockdown. (g) Equal numbers of viable GBM1A-Tet-shHMGA1 neurospheres were cultured for 7 days with or without Dox (1 μg/ml) and the number of spheres was quantified by computer-assisted image analysis. *P<0.05.
HMGA1 is a bonafide target of miR-296-5p in GBM SCs
miRNAs negatively regulate gene expression primarily by binding to complementary sites in the 3′-untranslated regions (UTRs) of their target mRNAs and inducing degradation.27 In silico analysis of the 3′-UTR of HMGA1 shows a conserved binding site for miR-296-5p (Figure 5a). To characterize rigorously the direct functional interactions between miR-296-5p and the HMGA1 transcript in GBM SCs, we transfected a luciferase reporter cloned with the 3′-UTR region of HMGA1 containing the predicted miR-296-5p binding site into 293T cells and GBM-derived neurospheres. Coexpressing mir-296-5p significantly reduced luciferase activity in 293T cells transfected with the luciferase reporter containing the wild-type miR-296-5p binding site and had no effect in cells expressing a negative control luciferase construct lacking the miR-296-5p binding site (Figure 5b). Conversely, inhibiting endogenous miR-296-5p activated expression of the same luciferase construct ~16-fold with no effect on control luciferase reporter expression (Figure 5c). To test if miR-296-5p regulates HMGA1 expression in GBM SCs under physiological conditions, GBM neurospheres were transfected with the luciferase reporter constructs and forced to differentiate. Luciferase levels were substantially inhibited in response to differentiation conditions that induce miR-296-5p expression (Figure 5d). Finally, directly inhibiting miR-296-5p expression by transducing GBM neurosphere cells with miR-296-5p sponge increased HMGA1 mRNA levels ~4–5-fold (Figure 5e). HMGA1 belongs to the HMGA family of chromatin remodeling proteins, which consists of HMGA1 and HMGA2.28 Forced expression of miR-296-5p significantly reduced gene expression levels of HMGA1 (>70%) but did not affect levels of HMGA2 (Figure 5f), which lacks miR-296-5p binding sites (as determined by TargetScan analysis; Supplementary Figure S4). These results confirm that miR-296-5p specifically and efficiently targets the HMGA1 3′-UTR in response to differentiating signals in GBM cells.
HMGA1 is a bonafide target of miR-296-5p in GBM neurospheres. (a) Sequence alignment showing that the miR-296-5p binding site located in the HMGA1 3′-UTR is conserved among several species. (b) The 293T cells were co-transfected with luciferase reporter construct spanning the HMGA1 3′-UTR containing the miR-296-5p binding site or a control region and miR-296-5p. miR-296-5p expression decreases luciferase activity of the construct containing the HMGA1 3′-UTR but not the control region. (c) The 293T cells were co-transfected with luciferase reporter construct spanning the HMGA1 3′-UTR containing the miR-296-5p binding site or a control region and miR-296-5p inhibitor. miR-296-5p inhibition increases luciferase activity of the construct containing the HMGA1 3′-UTR but not the control region. (d) GBM1A and 609 neurospheres were transfected with luciferase reporter construct covering the HMGA1 3′-UTR containing the miR-296-5p binding site and forced to differentiate. Luciferase activity was measured 3 days after differentiation. (e) qRT–PCR showing increase in HMGA1 gene expression in response to miR-296-5p inhibition in GBM1A and Mayo39 neurospheres. (f) qRT–PCR shows decreased gene expression of HMGA1 but not HMGA2 72 h after miR-296-5p induction in GBM1A and Mayo39 neurospheres. (g) Mayo39-Tet-miR-296-5p neurospheres were transduced with lentivirus expressing HMGA1 lacking the endogenous 3′-UTR. (Left panel) Western blot showing the expression of HMGA1, Sox2, Bmi-1 and Nestin 72 h after miR-296-5p induction. Equal numbers of viable Mayo39-Tet-miR-296-5p neurospheres with or without transgenic HMGA1 were cultured for 10 days±Dox (1 μg/ml) and the number of spheres was quantified by computer-assisted image analysis. *P<0.05.
miRNAs typically have multiple targets and forced expression may have off-target effects. A lentiviral construct containing the full-length HMGA1 coding sequence without the 3′-UTR regulatory region was used to determine if the neurosphere responses to forced miR-296-5p expression are specifically dependent on inhibiting endogenous HMGA1 expression. Forced expression of miR-296-5p downregulated endogenous HMGA1 as well as the SC drivers Sox2 and Bmi-1, effects that were abolished by transgenic 3′-UTR-deficient HMGA1 (Figure 5g, left panel). Similarly, transgenic 3′-UTR-deficient HMGA1 rescued the inhibition of self-renewal and sphere-forming capacity observed in response to forced miR-296-5p expression (Figure 5g right panel). Taken together, these results identify HMGA1 as the main functional downstream target by which miR-296-5p regulates the GBM SC phenotype.
HMGA1 modulates chromatin architecture at the Sox2 promoter
HMGA1 has been shown to activate the expression of critical pluripotency genes in human pluripotent cells.29 We therefore hypothesized that HMGA1 regulates GBM SCs, at least in part, by maintaining basal levels of dedifferentiation transcription factors. Inhibiting endogenous HMGA1 expression with an shRNA construct by ~90% significantly inhibited the expression of reprogramming transcription factors Oct4, Sox2, KlF4 and Nanog (Figure 6a, left panel). However, inhibiting HMGA1 expression more modestly (~50%) with an alternate shRNA only inhibited Sox2 expression (Figure 6a, right panel), indicating that Sox2 is most sensitively and specifically activated by HMGA1. The Sox2 promoter was found to contain two regions with putative HMGA1 binding sites (http://alggen.lsi.upc.es/) (Figure 6b, top panel). Anti-HMGA1 chromatin immunoprecipitation-PCR-enriched promoter region 2 by ~17-fold in GBM1A neurospheres and promoter regions 1 and 2 by ~5- and ~7-fold, respectively, in a primary neurosphere isolate (Figure 6b). HMGA proteins are architectural transcription factors that regulate gene expression by binding to DNA and displacing histone H1 from these locations.30 This predicts H1 displacement if HMGA1 regulates Sox2 expression through promoter binding (Figure 6c). To test this hypothesis, we expressed HMGA1 in 293T cells and measured HMGA1 and H1 binding to the Sox2 promoter using chromatin immunoprecipitation-PCR. Quantitative chromatin immunoprecipitation showed increased HMGA1 binding and decreased histone H1 binding to the Sox2 promoter in response to forced HMGA1 expression (Figure 6d). The same pattern of H1 and HMGA1 binding was observed in GBM neurospheres coexpressing transgenic Oct4/Sox2 that inhibits miR-296-5p expression, increases HMGA1 expression and induces GBM SCs (Figure 6e). Conversely, H1 binding to the Sox2 promoter increased in response to forced miR-296-5p expression that directly targets HMGA1 expression and thereby inhibits Sox2 expression (Figures 5g and 6f). Taken together, these results demonstrate that miR-296-5p controls reciprocal chromatin binding of HMGA1 and histone H1 to affect Sox2 expression.
HMGA1 regulates basal levels of Sox2. (a) qRT–PCR to measure the expression of reprogramming transcription factors and Nanog following HMGA1 knockdown using two different shRNA hairpins. (b, left panel) DNA purified from chromatin immunoprecipitation was analyzed by qRT–PCR using primer pairs designed to amplify fragments containing HMG binding sites on the Sox2 promoter. (Right panel) Agarose gel electrophoresis shows relative efficiencies of HMGA1 binding to Sox2 promoters. (c) Schematic showing effects of changes in HMGA1 levels on chromatin. High levels of HMGA1 result in open chromatin and low levels of HMGA1 result in closed chromatin configuration (adapted from Catez et al.42). (d) Binding of histone H1 and HMGA1 to the portion of the Sox2 promoter containing HMG binding sites was measured following chromatin immunoprecipitation (Ch-IP) using qRT–PCR in 293T cells. (Left panel) Western blot analysis showing HMGA1 expression and (right panel) agarose gel electrophoresis shows relative efficiencies of histone H1 and HMGA1 binding to Sox2 promoters. (e) Ch-IP-PCR to measure binding of histone H1 and HMGA1 to the portion of the Sox2 promoter containing HMG binding sites in GBM1A neurospheres coexpressing Oct4/Sox2. (Left panel) Western blot analysis showing HMGA1 expression. (f) Binding of histone H1 and HMGA1 to the portion of the Sox2 promoter containing HMG binding sites following induction of miR-296-5p in Mayo39 cells. *P<0.05.
Discussion
Solid malignancies consist of phenotypically plastic cells that differ in tumor-propagating capacity. Tumor cells dynamically convert between these diverse cell phenotypes via mechanisms involving epigenetic responses to autocrine/paracrine signaling events within the context of an aberrant neoplastic genome. How these components function together to induce and/or maintain the tumor-propagating cancer SC phenotype remains only partially understood. We recently demonstrated that Oct4 and Sox2 drive glioma cell dedifferentiation to a tumor-propagating SCs, in part, by directly inducing DNMT expression, resulting in methylation-dependent repression of miRNA networks. We now demonstrate that one of these methylation-regulated miRNAs, miR-296-5p, functions as an inhibitor of the GBM SC phenotype. We also identify HMGA1, a remodeler of chromatin architecture, as a functional target of miR-296-5p and an intermediary by which miR-296-5p regulates Sox2 expression (Figure 7). Taken together, our findings identify the miR-296-5p:HMGA1:Sox2 transcriptional axis as a previously unrecognized regulator of GBM SCs.
Model summarizing how the Oct4/Sox2:miR-296-5p:HMGA1 axis coordinates the stem cell phenotype of GBM neurospheres. As cells transition between stem- and non stem-like states, Oct4 and Sox2 induce promoter methylation events that inactivate expression of miR-296-5p. miR-296-5p targets HMGA1 in GBM neurospheres and the miR-296-5p:HMGA1 axis regulates self-renewal and tumor growth capacity of GBM stem-like cells, in part, by controlling the relative levels of histone H1 binding to the promoter region of genes that regulate cell phenotype (i.e. Sox2).
Our findings add significantly to the small amount of current knowledge regarding miR-296-5p in the contexts of malignancy and cancer cell phenotype regulation. It is interesting within the context of methylation-dependent miR-296-5p silencing that the miR-296-5p sequence is found in a genomic region regulated by imprinting in both mouse and human.31 Low levels of miR-296-5p had been associated with resistance to chemotherapy and poor clinical outcomes in patients with colon cancer and esophageal squamous cell carcinoma.32, 33 Similarly, miR-296 expression levels were found to correlate inversely with more advanced tumor grade and stage in patients with prostate cancer and ectopic miR-296 expression inhibited prostate cancer cell growth and migration in vitro.34, 35 Our work provides a mechanistic basis for these previous observations by identifying a function pathway linking miR-296-5p to cancer SC regulation and demonstrating that miR-296-5p inhibits the propagation and growth of GBM tumors in vivo.
Our findings that upregulating Oct4 and Sox2 represses miR-296-5p expression and that forced expression of miR-296-5p downregulates Sox2 expression indicate that miR-296-5p resides within a positive feedback loop that augments the expression of Sox2, a critical driver of the cancer SC phenotype. We further identify HMGA1 as a miR-296-5p target and regulator of this feedback loop. HMGA proteins modulate gene expression by altering chromatin structure and are among the most abundant non-histone chromatin-binding proteins found in cancer.28, 36, 37 HMGA1 was first discovered in rapidly growing cervical carcinoma cells and HMGA1 overexpression induces the formation of multiple types of tumors in mice.37, 38 HMGA proteins bind to AT-rich DNA regions, relax chromatin and thereby allow transcription complexes to access DNA and modulate gene expression. Interestingly, HMGA1 is a component of an embryonic stem cell-like gene signature compiled from poorly differentiated human tumors and murine leukemias.39, 40 HMGA1 has recently been shown to activate reprogramming transcriptional networks in embryonic SCs and is thought to induce stem-like properties in colon cancer cells.29, 41 Despite the growing spectrum of biological and oncogenic functions attributed to HMGA proteins, little has been reported about their mechanisms of action and expression regulation. Emerging evidence points to chromosomal rearrangement as a determinant of HMGA protein dysregulation, with HMGA1 and HMGA2 gene rearrangement a common occurrence in several tumors.42 The predominant view regarding chromosomal translocation focuses on rearrangements of coding sequences, but often fails to take into consideration loss of regulatory signals found in the 3′-UTR. miRNAs are negative regulators of gene expression that function primarily by binding to complementary sites in the 3′-UTRs of the target mRNAs to induce degradation.27 We show that loss of miR-296-5p expression induces stemness concurrent with a 4–6-fold increase in HMGA1 mRNA levels. Similarly, expressing mutant HMGA1 lacking its 3′-UTR regulatory region enhances cell stemness and activates reprogramming signals. Taken together, these findings establish an important role for HMGA1 targeting by miR-296-5p in cancer SC regulation and highlight the potential oncogenic consequence of losing 3′-UTR regulatory regions during HMGA1 chromosomal translocation. We also show that HMGA1 expression induces reprogramming signals in GBM and is required for maintaining the GBM SC phenotype. Our findings build upon the current knowledge of HMGA1 biology by describing its role in regulating Sox2 promoter function through changes in the chromatin architecture. We show that HMGA1 associates with and displaces histone H1 from the Sox2 promoter and in doing so regulates Sox2 expression. These observations provide a detailed mechanistic explanation that integrates the cross-talk between miR-296-5p, HMGA1 and regulation of the CSC phenotype.
In conclusion, we describe a novel transcriptional circuit involving DNA methylation, miRNA expression regulation and HMGA1-dependent chromatin remodeling at the level of the Sox2 promoter to regulate the GBM SC phenotype. We show that miR-296-5p is a novel inhibitor of GBM cell stemness and tumor-propagating potential and identify HMGA1 as the miR-296-5p target responsible for these effects on the GBM phenotype. Our findings offer new insights into the dynamic regulation of cancer cell stemness and identify miR-296-5p as a potential therapeutic tool for silencing oncogenic HMGA1 functions and inhibiting stem cell populations in GBM.
Materials and methods
GBM neurosphere, neurosphere formation and differentiation assay
GBM-derived neurosphere lines (GBM1A and GBM1B) were originally derived and characterized by Vescovi and colleagues.5 The GBM-KK neurosphere line was derived from a single GBM patient and kindly provided by Dr Jaroslaw Maciaczyk (University of Freiburg, Freiburg im Breisgau, Germany). Low-passage primary neurospheres were derived directly from human GBM clinical specimens and from patient-derived xenografts obtained from pathological GBM specimens obtained during clinically indicated surgeries at Johns Hopkins Hospital using established methods.5 The human GBM xenograft line, Mayo39, was originally obtained from the Mayo Clinic (Rochester, MN, USA).43 Neurospheres were cultured in serum-free medium containing DMEM/F-12 (Invitrogen, Carlsbad, CA, USA), supplemented with 1% bovine serum albumin, 20 ng/ml epidermal growth factor and 10 ng/ml fibroblast growth factor. Human glioma cell lines A172 was originally obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in Dulbecco's modified Eagle's medium (high glucose) supplemented with 10% penicillin and streptomycin. GBM1A, GBM1B and GBM-KK lines have all been tested for mycoplasma contamination.
For neurosphere formation, cells were dissociated into single cell and cultured in ultra-low attachment flasks (2.5 × 104 cells per ml). After 10–12 days, GBM neurospheres were embedded in 1% agarose and stained with 0.1% Wright stain solution for 1–2 h at 37 °C. Cells were washed 4 × with phosphate-buffered saline and incubated at 4 °C overnight (in phosphate-buffered saline) before quantification. Spheres larger than 50 μm were quantified using computer-assisted image analysis.44 To induce differentiation, GBM neurospheres were dissociated to single-cell suspensions and cultured on Matrigel-coated plates in neurosphere medium lacking epidermal growth factor for 2 days and then grown in medium containing 2.5% fetal bovine serum without epidermal growth factor/fibroblast growth factor for 5–7 days.
Generation of luciferase reporter constructs
BAC clones spanning the HMGA1 (RP11-957A23) genomic region were purchased from Life Technologies Corporation (Grand Island, NY, USA) and used as a template to amplify the 3′-UTR region containing the miR-296-5p binding site. DNA fragments were cloned into the XhoI and BglII sites of the pGL4.2 vector Promega Biosciences, LLC (San Luis Obispo, CA, USA). Luciferase activity was measured 48 h after transfection using a Luciferase Assay Kit (Promega Biosciences, LLC).
Immunoblotting
Western blot was performed using quantitative Western Blot System (LI-COR Bioscience, Lincoln, NE, USA) following the manufacturer’s instruction. Membranes were blotted with Sox2 (cat. no. 4900), and Bmi-1 (cat. no. 6964), PTEN (9559), Dnmt3a (2160) and Dnmt1 (5032) were purchased from Cell Signaling Technology (Boston, MA, USA); nestin (cat. no. sc-23927), olig2 (cat. no. sc-48817) and histone H1 (cat. no. sc-34464) were obtained from Santa Cruz Biotechnology, Inc (Dallas, TX, USA); actin (cat. no. A1978) was bought from Sigma, GFAP (cat. no. Z0334) was purchased from Dako North America, Inc (Carpinteria, CA, USA) and Tuj1 (cat. no. 05-661) was obtained from Millipore (Billerica, MA, USA); and HMGA1 (cat. no. AB4078) antibody was obtained from Abcam (Cambridge, MA, USA). IR dyes IRDye 800CW and IRDye 680CW secondary antibodies were purchased from LI-COR Biosciences.
Bisulfite sequencing
Genomic DNA was isolated using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA) and DNA was subjected to bisulfite treatment using EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA). The bisulfite-converted DNA was amplified using primers described in Supplementary Table 2. The PCR products were cloned into pCR II TA vectors using the TOPO-TA Cloning Kit (Invitrogen, Grand Island, NY, USA) and sequenced using Sanger sequencing. The sequencing data was analyzed using the BISMA (http://services.ibc.uni-stuttgart.de/BDPC/BISMA/).
QRT–PCR and miRNA expression
Total RNA was extracted from cells using RNeasy Mini Kit (Qiagen). cDNA was made by reverse-transcribing 1–3 μg of total RNA using MuLV Reverse Transcriptase and Oligo (dT) primers Applied Biosystems (ThermoFisher, Grand Island, NY, USA). qRT–PCR was performed with an iQ5 detection System (Bio-Rad, Hercules, CA, USA) and expression of target genes was detected using Power SYBR green PCR kit (Applied Biosystems, ThermoFisher). Samples were amplified in triplicate and relative gene expression was analyzed using the Bio-Rad iQ5 software and normalized to 18S RNA. Primer sequences are listed in Supplementary Table S4.
For miRNA analysis, total RNA including small RNA was extracted using miRNeasy Kit from Qiagen and 1–3 μg of total RNA was used as a template to generate cDNA using miScript II RT Kit (Qiagen). To measure levels of pre-miRNAs, total RNA was extracted using miRNeasy and cDNA was synthesized from total RNA (1–2 μg) using gene-specific primers and High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, ThermoFisher). The gene- specific primers included a mixture of 10 μm each of the antisense primers to the selected miRNA and U6 RNA. Following an 80 °C denaturation step and 60 °C annealing, the cDNA was reverse transcribed for 45 min at 60 °C. cDNA was diluted 1:20 before detection using Power SYBR Green PCR Kit from Applied Biosystems (ThermoFisher). Primer sequences are listed in Supplementary Table 3.
Lentiviral transduction
The miR-296-5p precursor (pre-miRNA) sequence was amplified from genomic DNA obtained from GBM1A cells and cloned into the pTRIPZ vector using XhoI and MluI sites. HMGA1a (NM_145903) was amplified from cDNA obtained from GBM1A cells and cloned into the SpeI and XhoI sites of the pLEX vector (Thermo Scientific, ThermoFisher). MiRNA inhibitors were purchased from Genecopoeia (Rockville, MD, USA). Lentiviral particles were packaged using the Trans-lentiviral packaging system from Invitrogen Technologies (ThermoFisher). Upon infection, stable cells were selected using puromycin (1 μg/ml). miRNA expression was induced using Dox (1 μg/ml). Primer sequences are listed in Supplementary Table S5.
Chromatin immunoprecipitation
HMGA1 binding sites 2 kb upstream of the Sox2 translation start site were identified using the PROMO45 algorithm using search term ‘HMGI(Y)’ (Transcription factor identifier: T02368), which is specific for HMGA1.28 Chromatin immunoprecipitation was performed using the MAGnify Chromatin Immunoprecipitation System (Life Technologies Corporation). Briefly, DNA from GBM1A neurosphere coexpressing Oct4 and Sox2 was crosslinked using formaldehyde and chromatin was isolated and fragmented by sonication. DNA fragments (~300 bp) were incubated with anti-HMGA1, anti-H1 or IgG antibody overnight at 4 °C. Precipitation of DNA fragments complexed with HMGA1 or H1 at the Sox2 promoter was quantified using qRT–PCR and PCR products were visualized on agarose gels. Primers targeting promoter regions lacking HMGA1 binding sites were used as a negative control.
Tumor formation in vivo
Female athymic nude NCR Nu/Nu mice (8 weeks old) were injected subcutaneously with Mayo39 neurospheres and assigned into different treatment groups in a non-blinded, non-randomized manner.46 The control cohort received regular diet and the experimental group received Dox-containing diet (Harlan Laboratories, Frederick, MD, USA; cat. no. TD.01306) for the duration of the experiments. Number of animals used for each experiment is indicated in the corresponding figure legend. Tumor growth was monitored by serial caliper measurement at the indicated intervals and animals were killed for isolation of tumor tissue when the tumor reached 1000 mm3. Tumor volume was calculated as (a × b2)/2, where a and b were the large and small diameters, respectively. Data for all in vivo experiments are shown as the mean tumor volume±s.d. of all animals used in the study. All animal procedures were approved by the Johns Hopkins Institutional Animal Care and Use Committee (protocol no. MO14M307), and were in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Statistical analysis
All experiments were performed in triplicates and repeated at least two times in each cell model. Two group comparisons were analyzed for variation and significance using a two-tailed, type 1 t-test and P-values <0.05 were considered significant and symbolized by an asterisk in the graphs. All data shown are mean±s.d.
References
Hanahan D, Weinberg RA . The hallmarks of cancer. Cell 2000; 100: 57–70.
Pardal R, Molofsky AV, He S, Morrison SJ . Stem cell self-renewal and cancer cell proliferation are regulated by common networks that balance the activation of proto-oncogenes and tumor suppressors. Cold Spring Harb Symp Quant Biol 2005; 70: 177–185.
Choi JD, Lee JS . Interplay between epigenetics and genetics in cancer. Genome Inform 2013; 11: 164–173.
Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006; 444: 756–760.
Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 2004; 64: 7011–7021.
Uchida N, Buck DW, He D, Reitsma MJ, Masek M, Phan TV et al. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci USA 2000; 97: 14720–14725.
Nguyen LV, Vanner R, Dirks P, Eaves CJ . Cancer stem cells: an evolving concept. Nat Rev Cancer 2012; 12: 133–143.
Singh RP, Shiue K, Schomberg D, Zhou FC . Cellular epigenetic modifications of neural stem cell differentiation. Cell Transplant 2009; 18: 1197–1211.
Wolffe AP . Chromatin remodeling: why it is important in cancer. Oncogene 2001; 20: 2988–2990.
Rajendran G, Shanmuganandam K, Bendre A, Muzumdar D, Goel A, Shiras A . Epigenetic regulation of DNA methyltransferases: DNMT1 and DNMT3B in gliomas. J Neuro-Oncol 2011; 104: 483–494.
Fanelli M, Caprodossi S, Ricci-Vitiani L, Porcellini A, Tomassoni-Ardori F, Amatori S et al. Loss of pericentromeric DNA methylation pattern in human glioblastoma is associated with altered DNA methyltransferases expression and involves the stem cell compartment. Oncogene 2008; 27: 358–365.
Suva ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 2014; 157: 580–594.
Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 2008; 134: 521–533.
Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 2011; 8: 633–638.
Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 2011; 8: 376–388.
Lin SL, Chang DC, Lin CH, Ying SY, Leu D, Wu DT . Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res 2011; 39: 1054–1065.
Lopez-Bertoni H, Lal B, Li A, Caplan M, Guerrero-Cázares H, Eberhart CG et al. DNMT-dependent suppression of microRNA regulates the induction of GBM tumor propagating phenotype by Oct4 and Sox2. Oncogene 2014; 34: 3994–4004.
Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res 2008; 68: 9125–9130.
Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res 2009; 69: 7569–7576.
Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med 2008; 6: 14.
Xia H, Cheung WK, Ng SS, Jiang X, Jiang S, Sze J et al. Loss of brain-enriched miR-124 microRNA enhances stem-like traits and invasiveness of glioma cells. J Biol Chem 2012; 287: 9962–9971.
Sato F, Tsuchiya S, Meltzer SJ, Shimizu K . MicroRNAs and epigenetics. FEBS J 2011; 278: 1598–1609.
Baylin SB, Ohm JE . Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 2006; 6: 107–116.
Li Y, Laterra J . Cancer stem cells: distinct entities or dynamically regulated phenotypes? Cancer Res 2012; 72: 576–580.
Son MJ, Woolard K, Nam DH, Lee J, Fine HA . SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell 2009; 4: 440–452.
Lewis BP, Burge CB, Bartel DP . Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005; 120: 15–20.
Esquela-Kerscher A, Slack FJ . Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 2006; 6: 259–269.
Reeves R, Beckerbauer L . HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim Biophys Acta 2001; 1519: 13–29.
Shah SN, Kerr C, Cope L, Zambidis E, Liu C, Hillion J et al. HMGA1 reprograms somatic cells into pluripotent stem cells by inducing stem cell transcriptional networks. PLoS One 2012; 7: e48533.
Cleynen I, Van de Ven WJ . The HMGA proteins: a myriad of functions (Review). Int J Oncol 2008; 32: 289–305.
Robson JE, Eaton SA, Underhill P, Williams D, Peters J . MicroRNAs 296 and 298 are imprinted and part of the GNAS/Gnas cluster and miR-296 targets IKBKE and Tmed9. RNA 2012; 18: 135–144.
Shivapurkar N, Mikhail S, Navarro R, Bai W, Marshall J, Hwang J et al. Decrease in blood miR-296 predicts chemotherapy resistance and poor clinical outcome in patients receiving systemic chemotherapy for metastatic colon cancer. Int J Colorectal Dis 2013; 28: 887.
Hong L, Han Y, Zhang H, Li M, Gong T, Sun L et al. The prognostic and chemotherapeutic value of miR-296 in esophageal squamous cell carcinoma. Ann Surg 2010; 251: 1056–1063.
Lee KH, Lin FC, Hsu TI, Lin JT, Guo JH, Tsai CH et al. MicroRNA-296-5p (miR-296-5p) functions as a tumor suppressor in prostate cancer by directly targeting Pin1. Biochim Biophys Acta 2014; 1843: 2055–2066.
Wei JJ, Wu X, Peng Y, Shi G, Basturk O, Yang X et al. Regulation of HMGA1 expression by microRNA-296 affects prostate cancer growth and invasion. Clin Cancer Res 2011; 17: 1297–1305.
Fusco A, Fedele M . Roles of HMGA proteins in cancer. Nat Rev Cancer 2007; 7: 899–910.
Lund T, Holtlund J, Fredriksen M, Laland SG . On the presence of two new high mobility group-like proteins in HeLa S3 cells. FEBS Lett 1983; 152: 163–167.
Xu Y, Sumter TF, Bhattacharya R, Tesfaye A, Fuchs EJ, Wood LJ et al. The HMG-I oncogene causes highly penetrant, aggressive lymphoid malignancy in transgenic mice and is overexpressed in human leukemia. Cancer Res 2004; 64: 3371–3375.
Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 2008; 40: 499–507.
Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell 2009; 4: 129–140.
Belton A, Gabrovsky A, Bae YK, Reeves R, Iacobuzio-Donahue C, Huso DL et al. HMGA1 induces intestinal polyposis in transgenic mice and drives tumor progression and stem cell properties in colon cancer cells. PLoS One 2012; 7: e30034.
Catez F, Ueda T, Bustin M . Determinants of histone H1 mobility and chromatin binding in living cells. Nat Struct Mol Biol. 2006; 13: 305–310.
Pandita A, Aldape KD, Zadeh G, Guha A, James CD . Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes Chromosomes Cancer 2004; 39: 29–36.
Glas M, Rath BH, Simon M, Reinartz R, Schramme A, Trageser D et al. Residual tumor cells are unique cellular targets in glioblastoma. Ann Neurol 2010; 68: 264–269.
Messeguer X, Escudero R, Farre D, Núñez O, Martínez J, Albà MM . PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 2002; 18: 333–334.
Li Y, Lal B, Kwon S, Fan X, Saldanha U, Reznik TE et al. The scatter factor/hepatocyte growth factor: c-met pathway in human embryonal central nervous system tumor malignancy. Cancer Res 2005; 65: 9355–9362.
Acknowledgements
This work was supported by grants from the American Brain Tumor Association (to HL-B), and the United States NIH Grants R01NS073611 (to JL) and R01NS070024 (to AQ-H).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Dr Laterra’s and Dr Quinones-Hinojosa’s work has been funded by the NIH.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Lopez-Bertoni, H., Lal, B., Michelson, N. et al. Epigenetic modulation of a miR-296-5p:HMGA1 axis regulates Sox2 expression and glioblastoma stem cells. Oncogene 35, 4903–4913 (2016). https://doi.org/10.1038/onc.2016.22
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.22
- Springer Nature Limited
This article is cited by
-
Nucleic acid drug vectors for diagnosis and treatment of brain diseases
Signal Transduction and Targeted Therapy (2023)
-
Systems Medicine for Precise Targeting of Glioblastoma
Molecular Biotechnology (2023)
-
HMGA1 stimulates MYH9-dependent ubiquitination of GSK-3β via PI3K/Akt/c-Jun signaling to promote malignant progression and chemoresistance in gliomas
Cell Death & Disease (2021)
-
MicroRNA-296-5p inhibits cell metastasis and invasion in nasopharyngeal carcinoma by reversing transforming growth factor-β-induced epithelial–mesenchymal transition
Cellular & Molecular Biology Letters (2020)
-
Functional characterization of SOX2 as an anticancer target
Signal Transduction and Targeted Therapy (2020)