Abstract
Recent large-scale genomic studies have classified medulloblastoma into four subtypes: Wnt, Shh, Group 3 and Group 4. Each is characterized by specific mutations and distinct epigenetic states. Previously, we showed that a chromatin regulator SMARCA4/Brg1 is required for Gli-mediated transcription activation in Sonic hedgehog (Shh) signaling. We report here that Brg1 controls a transcriptional program that specifically regulates Shh-type medulloblastoma growth. Using a mouse model of Shh-type medulloblastoma, we deleted Brg1 in precancerous progenitors and primary or transplanted tumors. Brg1 deletion significantly inhibited tumor formation and progression. Genome-wide expression analyses and binding experiments indicate that Brg1 specifically coordinates with key transcription factors including Gli1, Atoh1 and REST to regulate the expression of both oncogenes and tumor suppressors that are required for medulloblastoma identity and proliferation. Shh-type medulloblastoma displays distinct H3K27me3 properties. We demonstrate that Brg1 modulates activities of H3K27me3 modifiers to regulate the expression of medulloblastoma genes. Brg1-regulated pathways are conserved in human Shh-type medulloblastoma, and Brg1 is important for the growth of a human medulloblastoma cell line. Thus, Brg1 coordinates a genetic and epigenetic network that regulates the transcriptional program underlying the Shh-type medulloblastoma development.
Similar content being viewed by others
Introduction
Medulloblastoma is the most common malignant childhood brain tumor. Although the survival rate of medulloblastoma patients is relatively high, current treatments have serious side effects.1, 2, 3 Several large-scale genomic studies have provided rich information about specific transcription profiles and mutations associated with medulloblastoma. On the basis of these analyses, medulloblastomas are classified into four subgroups: Wnt, Sonic hedgehog (Shh), Group 3 and Group 4.4, 5, 6, 7, 8 The subtypes are characterized by specific mutations, rely on distinct signaling pathways, and possibly have different cell origins. Interestingly, each also displays distinct epigenetic properties, which are likely important for generating and maintaining the transcription program characteristic of each subtype. The functional epigenome studies to identify the molecular mechanisms that regulate the growth of each tumor subtype are required to accelerate the pace of development of much-needed, subtype-specific treatment plans for medulloblastoma patients.
The Shh-type medulloblastoma accounts for 25% of all medulloblastomas. Mutations that result in constitutively active Shh signaling in cerebellum granule neuron precursors (CGNPs) are the essential genetic causes of Shh-type medulloblastoma.9, 10 During early postnatal development, Shh expressed from Purkinje neurons induces the rapid expansion of CGNPs in the external granule layer. CGNPs differentiate into granule neurons that migrate to and reside in the internal granule layer. Active Shh signaling is required for normal CGNP proliferation and for maintenance of CGNPs in an undifferentiated state;11, 12, 13, 14 however, elevated or constitutively active Shh signaling leads to CGNP over-proliferation and medulloblastoma.
Although various mutations cause Shh-type medulloblastoma and Shh-type medulloblastoma occurs in infants, children and adults, these tumors share certain subtype-specific gene expression patterns that are likely required for tumor proliferation and identity.4, 5, 6, 7, 8, 10 These include high levels of expression of Shh signaling target genes such as Gli1 and CGNP specification genes such as Atoh1, and relatively low levels of expression of neuronal differentiation genes. The Shh signaling mediated by patched (Ptch1) and smoothened (Smo) regulates development and cancer by modulating the activities of Gli family transcription factors.15, 16, 17, 18 The Shh activation releases inhibition of Smo by Ptch1 to activate Gli1/2 transcription activators (GliA) and downstream target genes. Shh-induced GliA activities provide a driving force for tumor proliferation by activating mitogenic genes such as Gli1, CcnD1 and N-myc.9, 18 The acquisition of CGNP identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma;19 therefore, Atoh1, an essential transcription factor in CGNP specification, has an important role in medulloblastoma development.20, 21 Atoh1 also regulates medulloblastoma proliferation by controlling Gli2 expression.21 Compared with Group 3 and Group 4 medulloblastoma, Shh subtype expresses low levels of neuronal genes. The master repressor NRSF/REST is expressed at high levels in Shh-type medulloblastoma and may function as an oncogene to repress expression of certain neuronal genes leading to inhibition of differentiation.22, 23, 24 Thus, GliA, Atoh1 and REST form a core transcriptional regulatory circuitry to control the identity and proliferation of medulloblastoma. The targetome of this transcription circuitry likely produces medulloblastoma phenotypes, and its coordinated regulation is critical to tumor development.
Previously, we and others have identified several GliA-interacting epigenetic regulators to activate target genes in response to Shh signaling.25, 26, 27 We have reported that a SWI2/SNF2-like chromatin remodeler SMARCA4/Brg1 functions as a GliA co-activator and is required for Shh-induced target gene activation.27 As a core subunit of the SWI/SNF-like chromatin remodeling BAF complexes, Brg1 contains an ATPase domain that provides the enzymatic activity to remodel chromatin structures to regulate transcription.28, 29 In addition, BAF complexes interact with many proteins, including other chromatin regulators and histone modifiers, that might mediate ATPase-independent activities of Brg1 and BAF complexes. BAF complexes have been shown to function as transcription activators or as repressors and have diverse roles during the development.29 In cancers, BAF complexes also have important but context-dependent roles.29, 30, 31 Recent genomic studies indicated that BAF complexes are mutated in ~20% of all cancer types and represent the most highly mutated chromatin regulator in human cancer.2, 30 Brg1 biallelic inactivating mutations have been identified in many cancers such as atypical teratoid/rhabdoid tumors and ovarian cancers.32, 33 Heterozygous missense mutations of Brg1 have been identified in the Wnt and Group 3 medulloblastomas,2, 30 suggesting a tumor suppressor function of Brg1 in these cancers. In other cancers, however, BAF complexes are required for cancer development and excluded from mutations. Brg1 has been shown to be required for leukemia and small cell lung cancer maintenance by regulating Myc expression or Myc activities.34, 35 The requirement of Brg1 for Shh/GliA activation suggests that Brg1 may be required for Shh-type medulloblastoma growth.
Recently, we discovered that under basal conditions Shh/Gli target genes poised for activation are marked by a bivalent chromatin domain containing H3K27me3/H3K4me3. The H3K27me3 demethylase Jmjd3/Kdm6b has a crucial role in removing H3K27me3, recruiting other co-activators, and activating Gli target genes in response to Shh signaling.36 It has been shown that Brg1 functions together with Jmjd3 to activate transcription of certain genes.37, 38 Notably, the non-enzymatic activities of both Brg1 and Jmjd3 are required for their co-activator functions in Shh signaling;27, 36 these two proteins may interact with each other or recruit other cofactors to activate Shh target genes coordinately. Consistent with their co-activator functions in Shh signaling, few mutations in Brg1 and Jmjd3 have been identified from pediatric Shh-type medulloblastomas despite a potential tumor suppressor function for both proteins in other medulloblastoma subtypes and other types of cancer.6, 30, 39
In this report, using a mouse model of Shh-type medulloblastoma that closely resembles the human tumor, we demonstrate that the chromatin remodeler Brg1 coordinates the core transcriptional circuitry that controls Shh-type medulloblastoma identity and proliferation. Deletion of Brg1 in CGNPs impaired normal cerebellum development, but considerably extended the survival time of tumor-bearing animals. Brg1 deletion after medulloblastoma formation significantly inhibited tumor proliferation and growth. Using genome-wide expression and binding experiments, we discovered that Brg1 coordinates with Gli1, Atoh1 and REST to regulate expression of both oncogenes and tumor suppressors specifically required for tumor growth. Brg1-regulated pathways are conserved in human Shh-type medulloblastomas, and Brg1 was also important for the growth of a human medulloblastoma cell line. In addition, we showed that Brg1 modulates activities of H3K27me3 modifiers in regulating medulloblastoma genes. Thus, Brg1 regulates genetic and epigenetic networks that control the transcription program underlying Shh-type medulloblastoma development.
Results
Brg1 is required for cerebellum development and the proliferation of precancerous CGNPs
To determine whether the GliA co-activator Brg1 is required for Shh-dependent CGNP proliferation in vivo, we deleted Brg1 using the CGNP-specific Atoh1-Cre transgene.40 Atoh1-Cre-induced Brg1 deletion in CGNPs led to significantly smaller cerebellums at postnatal day 12 (P12) compared with those in wild-type mice despite an overall similar foliation pattern (Figures 1a, b and a′). Brg1 protein was deleted in most CGNPs in the external granule layer and subsequently in granule neurons in the internal granule layer (Figure 1c, c′). Brg1-deleted CGNPs also displayed abnormal morphologies such as larger nuclei (Figure 1c, 1c′). The external granule layer was thinner in Brg1-deleted cerebellum, due to impaired CGNP proliferation as indicated by reduced BrdU incorporation (Figure 1d, 1d′). This result indicates an essential function of Brg1 in Shh/Gli-dependent normal cerebellum development. Recently, another genetic study also demonstrated that Brg1 and SMARCB1/BAF47/INI1/SNF5 are required for cerebellum development.41
Brg1 is required for CGNP proliferation and cerebellum development. (a and a′) Conditional deletion of Brg1 in CGNPs using Atoh1-Cre led to reduced cerebellum size at postnatal day 12 (P12) compared with wild-type mice. (b and b′) H&E staining of sagittal sections of P12 wild-type and Brg1-mutant cerebellum. The Brg1-mutant cerebella are significantly smaller than the wild type. (c and c′) Immunostaining of P12 wild-type and Brg1-mutant cerebellum demonstrates the deletion of Brg1 from many CGNPs and granule neuron progenies in the mutant. The external granule layer (EGL) is thinner in Brg1-mutant cerebellum. The higher magnification images of the EGL regions in the boxes are shown at the bottom. (d and d′) BrdU labeling in P8 Brg1-mutant cerebellum EGL is decreased relative to that in wild-type EGL. BrdU was injected 2 h before they were killed. The quantification of BrdU-positive cells in EGL is plotted. Student’s t-test; **P<0.01. H&E, hematoxylin and eosin.
The expression of a Cre-inducible SmoM2 (an activating mutation in Smo) transgene in CGNPs results in constitutively active Shh signaling, over-proliferative CGNPs and medulloblastoma development.19, 42 In precancerous SmoM2 CGNPs, Brg1 was required for proliferation (Supplementary Figure 1A). Cultured CGNPs were obtained from P4 CAG-CreER SmoM2 pups with Brg1F/F or Brg1+/+. In both the genotypes, tamoxifen treatment induced expression of SmoM2, and, in the Brg1F/F cells, deletion of Brg1 was induced simultaneously, which significantly reduced CGNP proliferation as indicated by reduced BrdU incorporation (Supplementary Figure 1A). Consistently, Brg1 deletion in SmoM2 CGNPs significantly inhibited the expression of Shh target genes that are induced by the SmoM2 mutant protein (Supplementary Figures 1B and C).
Brg1 is required for SmoM2 medulloblastoma formation and progression
To determine how Brg1 functions in Shh-induced medulloblastoma formation, Brg1 was deleted in CGNPs in the SmoM2 mouse model of Shh-type medulloblastoma using Atoh1-Cre at the same time that SmoM2 was induced. Induction of SmoM2 expression led to abnormal cerebellum development and medulloblastoma growth. At P28, when normal cerebellum development is finished and CGNPs are all differentiated, the cerebellum in Atoh1-Cre SmoM2 mice had no foliation, indicating the requirement of proper Shh signaling for CGNP proliferation and differentiation. Multiple foci of Ki67-positive cells indicated the initiation of medulloblastoma growth (Figure 2a). In Atoh1-Cre SmoM2 Brg1F/F mice, the cerebellum is smaller and also lack of foliation. However, no Ki67-positive foci were observed (Figure 2a). In survival studies, all Atoh1-Cre SmoM2 Brg1+/+ mice developed medulloblastoma and died within 3 to 4 months of age (Figure 2b). Deletion of Brg1 in CGNPs significantly extended survival time of Atoh1-Cre SmoM2 mice (Figure 2b). Despite the abnormal cerebellum, the tumor-free mice looked normal. In those few Atoh1-Cre SmoM2 Brg1F/F mice that did suffer from medulloblastoma growth, the Brg1 allele was not completed deleted in the tumor cells (Supplementary Figure 2). Deletion of one Brg1 allele also significantly reduced the lethality rate compared with the mice with wild-type Brg1 (Figure 2b).
Brg1 is required for SmoM2 medulloblastoma formation and progression. (a) H&E and Ki67 staining of sagittal sections of cerebella from P28 Atoh1-Cre SmoM2 Brg1+/+or Atoh1-Cre SmoM2 Brg1F/F mice. Arrows indicate a few Ki67-positive tumor initiation areas. (b) Deletion of Brg1 significantly extended the survival time for mice harboring SmoM2 medulloblastoma. Shown are the survival curves of Atoh1-Cre SmoM2 mice with Brg1+/+, Brg1F/+ or Brg1F/F alleles (n=15, 19 and 13, respectively). Log-rank test was used to determine the significance. (c) The genetic strategy for deletion of Brg1 in SmoM2 medulloblastoma. LoxP sites flanking the stop signal and Brg1 exons are shown as empty triangles. Leaky CAG-CreER activity induces SmoM2 expression and medulloblastoma formation with an intact Brg1 floxed allele. Treatment with tamoxifen activates CreER to induce Brg1 deletion in tumors. Tamoxifen applied before tumor formation will induce the expression of SmoM2 and deletion of Brg1 simultaneously. (d) CAG-CreER SmoM2 Brg1F/F medulloblastomas grown in the absence of tamoxifen induction were transplanted subcutaneously into SCID-NOD mice. The mice were injected with tamoxifen (Tam) or oil control after the tumors became visible at 20 days after transplantation. Examples of tumors after oil or tamoxifen treatment are shown in the top panels. Genotyping results indicate the presence of the recombined Brg1Δ allele only in tamoxifen-treated samples. The remaining Brg1F allele results from partial recombination of Brg1 allele. The Brg1+ alleles are from the host tissues. Shown in the right is the quantifications of transplanted Brg1F/F tumor sizes after tamoxifen or oil treatment; n=6 for oil treatment and n=5 for tamoxifen treatment. (e) Tamoxifen-induced Brg1 deletion from transplanted CAG-CreER SmoM2 Brg1F/F medulloblastoma led to decreased tumor proliferation as indicated by decreased staining of proliferation markers H3P and BrdU (n=3). Student’s t-test; **P<0.01. H&E, hematoxylin and eosin.
To determine whether Brg1 is required for medulloblastoma maintenance and progression, we deleted Brg1 after tumor formation by taking advantage of the leaky CAG-CreER activities (Figure 2c). CAG-CreER SmoM2 medulloblastomas with Brg1+/+, Brg1F/+ or Brg1F/F alleles were induced by weak Cre activities without tamoxifen treatment. These tumors contain intact Brg1 alleles (Figures 2c and d). When transplanted into SCID-NOD immunodeficient mice subcutaneously, deletion of one or both Brg1 alleles induced by tamoxifen injection significantly inhibited the growth of tumor allografts (Figure 2d). Brg1 deletion significantly inhibited tumor cell proliferation as indicated by reduced staining of proliferation markers such as phosphorylated histone H3 (H3P) and by BrdU incorporation (Figure 2e). Deletion of Brg1 in primary medulloblastomas using the same strategy also significantly inhibited tumor proliferation (Supplementary Figure 3). Thus, Brg1 is required for both the formation and progression of SmoM2 medulloblastoma.
Brg1 is required for the expression of a transcription program specific to SmoM2 medulloblastoma
In medulloblastoma transplantation, as expected, deletion of Brg1 significantly inhibited the expression of Shh/Gli target genes Gli1, Ptch1, Ccnd1 and N-myc (Figures 3a and b), which is consistent with the function of Brg1 as a co-activator of Gli1/2. Interestingly, Brg1 deletion also inhibited other genes known to be important for medulloblastoma identity and proliferation such as Atoh1 and Gli2 (Figure 3b). We next performed RNA-seq to compare the transcription profiles from primary medulloblastomas with or without Brg1 deletion. The mice with CAG-CreER SmoM2 medulloblastomas with Brg1F/F or Brg1+/+ were injected with tamoxifen to induce Brg1 deletion or to serve as controls. Differential expression analyses showed that levels of expression from 1517 genes were significantly changed upon Brg1 deletion (Supplementary Table 1). These Brg1-regulated genes significantly overlapped with the genes differentially expressed in SmoM2 medulloblastoma versus normal cerebellum42 (47%, P=2.3e−161, Figure 3c). Of the 1517 Brg1-regulated genes, levels of expression of 440 genes were decreased upon Brg1 deletion (these are Brg1-activated genes), and levels of expression of 1077 genes were increased (these are Brg1-repressed genes). Ranked fold-change analyses showed an almost perfect correlation between Brg1-regulated genes and the SmoM2-specific gene set (Figure 3d) indicating that Brg1 activates genes specifically expressed in SmoM2 medulloblastoma and inhibits genes repressed in tumors. These include genes important for cell proliferation and differentiation as shown by the gene ontology analyses (Figure 3e). Thus, the inhibition of tumor growth that resulted from Brg1 deletion is due to the destruction of the specific transcription program controlling SmoM2 medulloblastoma.
Brg1 deletion inhibits the transcription program specifically expressed in SmoM2 medulloblastoma. (a and b) Tamoxifen (Tam) treatment of SCID-NOD recipient mice with CAG-CreER SmoM2 Brg1F/F medulloblastoma transplantation led to (a) decreased Gli1 expression as indicated by western blot and (b) decreased expression of Shh target genes and other known medulloblastoma oncogenes as shown by RT–qPCR. The controls were treated with vehicle (Oil). (c) Significant overlap between Brg1-regulated genes and genes differentially expressed in SmoM2 tumors versus normal cerebellum tissues. Brg1-regulated genes were identified by comparing gene profiles between SmoM2 medulloblastoma with or without Brg1 deletion. (d) Brg1 activates and represses gene sets specifically activated or repressed in SmoM2 tumors. The genes regulated by Brg1 and SmoM2 were placed in a 20 × 20 matrix with ranked fold changes on both axes. The color key indicates the number of genes falling into each unit. (e) Gene ontology analysis of the Brg1-regulated genes indicates the main categories of genes activated or repressed by Brg1 in SmoM2 medulloblastoma. Significance was determined by Student’s t-test. **P<0.01.
Brg1 regulates key transcriptional regulatory circuits in SmoM2 medulloblastoma
To identify direct Brg1 target genes, we performed Brg1 chromatin immunoprecipitation (ChIP)-seq in SmoM2 medulloblastoma. Approximately 37 million reads were obtained, and more than 20 million reads were uniquely mapped to the genome. We analyzed the data using the SICER program that is specifically designed for the analysis of ChIP-seq data of chromatin regulators.43 We identified 5727 significant Brg1-binding regions with an average size of 2.4 kb; this number of binding regions is similar to those obtained in Brg1 ChIP-seq studies performed in other tissues.44, 45 Most Brg1-binding regions (70%) are in gene bodies or within 5 kb upstream or downstream; 30% are in the intergenic regions (Figure 4a). The analysis of the distribution of Brg1-binding regions in gene units showed enrichment close to the transcription start sites (Figure 4b). From the 5727 Brg1-binding regions, we identified 3841 genes with a Brg1-binding region overlapping with the gene body or 5-kb surrounding regions (Supplementary Table 2). The intersection of the gene set bound by Brg1 with the Brg1-regulated genes identified from primary medulloblastoma yielded 399 genes that are likely direct targets of Brg1 (Figure 4c, Supplementary Table 3).
Brg1 is required for the specific transcriptional regulatory circuitry controlling SmoM2 medulloblastoma. (a) Genomic distribution of Brg1 ChIP-seq binding regions identified in SmoM2 medulloblastoma using SICER program. (b) Distribution of Brg1-binding regions in a gene unit. The center of the Brg1-binding regions was used for the analyses. (c) Venn diagram indicating the overlap between Brg1-binding genes and Brg1-regulated genes in medulloblastoma. Also indicated are genes containing both Brg1 and Gli1 binding regions. (d) Significant overlap between Brg1-binding genes and Atoh1-binding genes. The number in parentheses indicates the number of genes regulated by Brg1 in medulloblastoma. (e) Comparison of the percentages of target genes of Gli1, Atoh1 and Gli1/Atoh1 that are bound by Brg1. A significant enrichment of Brg1 occupancy was observed in the genes with Gli1 and/or Atoh1-binding sites. (f) A significant number of potential REST target genes are bound and repressed by Brg1 in medulloblastoma. (g) Snapshots of Brg1 occupancy on representative Gli1 and Atoh1 target oncogenes. The genomic structures of the genes are shown below graphs with 5′ on the left. Brg1-binding regions identified using SICER are shown as black bars under the ChIP-seq signals. The Gli1 and Atoh1-binding sites close to the Brg1-binding regions are shown as red and green bars, respectively. (h) Confirmation of the differential expression of Brg1 target genes by RT–qPCR in SmoM2 medulloblastoma with or without Brg1 deletion. The classification of the selected genes is listed below the gene names. TS, tumor suppressors. Significance was determined by Student’s t-test. *P<0.05; **P<0.01.
The analyses of Brg1 targetome provide insights into the mechanisms underlying the essential function of Brg1 in Shh-type medulloblastoma. We found that Brg1 regulates important transcriptional regulatory pathways in medulloblastoma including the targetomes of Gli1, Atoh1 and REST. Gli binding sites have been identified in another mouse model of Shh-type medulloblastoma.46 Despite using the different tumor models, we observed a high overlap between Gli1 binding genes and Brg1-binding genes. Within 1169 Gli binding genes, 557 contain Brg1-binding regions (48%, P=1.2e−206, Figure 4c). In many target genes, putative Brg1 and Gli1 binding regions are in close proximity (Figure 4g), confirming our previous findings27 that Gli1 recruits the Brg1 co-activator complex to the Gli regulatory regions. Consistently, we observed an interaction between endogenous Gli1 and Brg1 in medulloblastoma cells (Supplementary Figure 4A). These data indicate that the essential co-activator function of Brg1 is not limited to a few known Gli1 target genes, but can be applied to the Gli1 targetome in medulloblastoma.
Of the 557 genes co-occupied by Brg1 and Gli1, 82 genes were regulated by Brg1. Brg1 deletion led to decreases in expression of Gli1-activated known oncogenic genes Gli1, N-myc and Ccnd1 (Figure 3b, Supplementary Table 1) and potential oncogenic genes such as Akna, Foxo6 and Sox18 (ref. 46; Figure 4h, Supplementary Table 1). In addition, the expression levels of Notch and Hippo pathway genes such as Hes5, Hey1 (also Gli1 targets), Jag1 and Tead1 as well as Wnt pathway inhibitors Wif1 and SFRP1 (also Gli1 targets) decreased after Brg1 deletion (Figure 4h). It has been shown that active Notch and Hippo signaling pathways positively regulate Shh-type medulloblastoma,47, 48, 49 whereas the Wnt pathway inhibits it.50 Therefore, these data suggest that Brg1 coordinates the crosstalk between signaling pathways to facilitate Shh-activated medulloblastoma development.
In CGNPs and Shh-type medulloblastoma, Atoh1 is necessary for cell-type specification and cell identity maintenance.20, 21 It also regulates CGNP and medulloblastoma proliferation, mainly by activating Gli2 expression.21 The Atoh1 targetome has been identified in early postnatal CGNPs, where Shh signaling is active.51 Many Atoh1 target genes in CGNPs likely also have important roles in maintaining medulloblastoma cell identity and proliferation. Within the 582 high stringency Atoh1 targets, 251 genes (43%) were also bound by Brg1 (Figure 4d). Of these, 48 were regulated by Brg1 including potent medulloblastoma oncogenes Gli2, Atoh1, CXCR4, Tgif1 and Aurkb (Figures 3b and 4d, g, h),21, 52, 53 which are likely direct targets of Brg1. The recruitment of Brg1 to Atoh1 target genes is likely mediated through the interaction between Atoh1 and Brg1. We observed that endogenous Atoh1 co-immunoprecipitated with Brg1 in SmoM2 medulloblastoma (Supplementary Figure 4A). We next analyzed the specificity of Brg1 in regulating target genes and non-target genes of Gli1 and Atoh1. Of the genes containing neither Gli1 nor Atoh1-binding sites (Gli1−/Atoh1−), less than 15% were occupied by Brg1. In contrast, 38% of the Gli1−/Atoh1+ genes, 46% of the Gli1+/Atoh1− genes and 66% of the Gli1+/Atoh1+ (73/110) genes were co-occupied by Brg1 (Figure 4e). The highly enriched Brg1 occupancy in the target genes of Gli1 and/or Atoh1 indicates that Brg1 specifically controls the transcriptional circuitry involved in medulloblastoma identity and proliferation.
Inhibition of several known tumor suppressors significantly enhances the formation and growth of mouse models of Shh-type medulloblastoma.54, 55, 56 Notably, Brg1 deletion led to an increase in expression of specific tumor suppressor genes and genes known to be involved in neuronal function and synaptogenesis (Figure 3e). In medulloblastoma, the tumor cells maintain a progenitor state and inhibit neuronal gene expression. It is thought that REST functions as an oncogene by repressing expression of neuronal genes and inhibiting differentiation.22, 23, 24 Many REST target genes (for example, Nrxn2, Nrxn3 and NeuroD2) were derepressed upon Brg1 deletion (Figures 4f and h). Although the target genes of REST in CGNPs and medulloblastomas are not known, the REST targetome identified in embryonic stem cells, where most of the neuronal genes are repressed, may encompass many REST target genes in the cerebellum.57 A comparison between Brg1-repressed genes (1077) with REST binding genes (1775) in embryonic stem cells yielded 198 (P=1.8e−32) commonly repressed genes. Of these, 62 are also bound by Brg1 (P=4.9e−18; Figure 4f, Supplementary Table 3). This indicates that Brg1 positively regulates the repression function of REST to inhibit neuronal gene expression and to allow medulloblastoma to grow. We observed the interaction between endogenous Brg1 and REST in SmoM2 medulloblastoma (Supplementary Figure 4A). Deletion of Brg1 led to reduced REST binding to one target gene NeuroD2 (Supplementary Figure 4B). These results are consistent with a previous report that REST recruits Brg1 to target genes, whereas Brg1 further facilitates REST binding.58 In addition, Brg1 repressed potential tumor suppressors with mutations identified in Shh-type medulloblastoma such as LRP1B, Syne1 and Pde4D.10 In contrast, potential tumor suppressor genes such as DDX3x, TCF4 and NcoR found in multiple types of medulloblastomas4, 5, 6, 7, 8 were not significantly changed upon Brg1 deletion (Figure 4h). Therefore, by a coordinated regulation of specific oncogenic and tumor-suppressing transcription circuits, Brg1 supports SmoM2 medulloblastoma development.
Brg1 regulates the activities of H3K27me3 modifiers in medulloblastoma
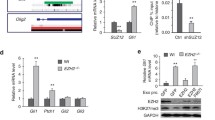
The human Shh-type medulloblastoma is characterized by low global H3K27me3 levels,8 which may allow the expression of many H3K27me3-regulated oncogenic genes. Similarly, we observed low global H3K27me3 levels in CGNPs and in SmoM2 medulloblastoma (Figure 5a).36 Interestingly, human medulloblastoma and SmoM2 tumors contain relatively high levels of SUZ12, a component of the H3K27 methyltransferase complex PRC2 (Figure 5a).8 This paradox indicates that high H3K27me3 demethylase activities such as those of Jmjd3 must be present in Shh-type medulloblastoma to remove H3K27me3 and to keep H3K27me3 levels low. Interestingly, we observed a global increase of H3K27me3 levels but decreased PRC2 levels (as indicated by decreased levels of an essential subunit Suz12) when tumor growth was inhibited by Brg1 deletion (Figure 5a). Quantitative PCR with reverse transcription (RT–qPCR) analyses showed that Brg1 deletion in SmoM2 tumors led to reduced PRC2 subunits mRNA levels, but not the Jmjd3 levels (Figure 5b). Thus, Brg1 may facilitate Jmjd3-mediated removal of H3K27me3 at specific loci to activate oncogenic genes, whereas Brg1 may also help maintain the H3K27me3 levels at the repressed gene regulatory regions by positively support PRC2 expression in Shh-type medulloblastoma. In support, target gene-specific functions of Brg1 in H3K27me3 regulation, ChIP–qPCR showed increased H3K27me3 levels at Brg1-activated genes such as Gli1 and Atoh1 and decreased H3K27me3 levels at Brg1-repressed genes such as NeuroD2 upon Brg1 deletion in SmoM2 medulloblastoma (Figures 5c–e). We have previously shown that Jmjd3 directly binds to Gli1 regulatory regions in Shh-activated CGNPs and is required for maintaining the low local H3K27me3 level and activating Gli1 expression.36 Our data indicate that Brg1 is required for Jmjd3 binding to Gli1 regulatory regions as Jmjd3 binding in Shh-activated CGNPs was significantly impaired when Brg1 was deleted (Figure 5f). This reduction of Jmjd3 binding was not due to a decrease of Jmjd3 expression level, as Jmjd3 mRNA level was even slightly increased in Brg1-deleted medulloblastoma (Figure 5b). By regulating Jmjd3 activities and PRC2 expression levels, Brg1 may control expression of certain key target genes through modulation of H3K27me3 levels.
Brg1 regulates the activities of H3K27me3 modifiers in medulloblastoma. (a) Tamoxifen-induced Brg1 deletion in primary CAG-CreER SmoM2 medulloblastoma led to increased global H3K27me3 and decreased levels of PRC2 subunits as shown by immunostaining. The quantifications of immunohistochemistry densities normalized to Brg1+/+ tumors are shown on the right. (b) RT–qPCR analyses of expression levels of PRC2 subunits and H3K27me3 demethylases in SmoM2 medulloblastoma with or without Brg1 deletion. (c–e) ChIP–qPCR experiments indicate increased or decreased H3K27me3 levels at the regulatory regions of Brg1 target genes in SmoM2 medulloblastoma upon Brg1 deletion. (f) Decreased Jmjd3 binding to the Gli1 gene regulatory region in Shh-treated, Brg1-deleted CGNPs as shown by ChIP–qPCR. A region upstream of Gli1 gene was used as a negative control for Jmjd3 binding. Significance was determined by Student’s t-test. *P<0.05, **P<0.01.
Conserved functions of Brg1 in human Shh-type medulloblastoma
To determine whether Brg1-regulated transcriptional pathways in SmoM2 tumors are also specifically expressed in human Shh-type medulloblastoma, we compared the expression levels of human homologs of Brg1 targets in different medulloblastoma subtypes. Using a publicly available microarray data set of 76 pediatric medulloblastoma samples (GSE37418),8 we ranked the average expression levels of the human homologs of each Brg1-activated Gli1 or Atoh1 targets as well as Brg1-repressed REST targets in four types of human medulloblastoma (Supplementary Table 4). Both Brg1-activated Gli1 and Atoh1 targets have significantly more #1 ranked genes in Shh-type medulloblastoma than other subgroups, whereas significantly more Brg1-repressed REST targets have lowest expression in Shh-type tumors compared with the other subgroups (Figures 6a and b). Brg1 was expressed at relatively high levels in all the four subgroups (Figure 6b). These results indicate that in Shh-type medulloblastoma, Brg1-regulated Gli1 and Atoh1 target genes are specifically activated and many REST targets are specifically repressed. Thus, the gene programs coordinately regulated by Brg1 and these transcription factors in SmoM2 tumors are also specifically associated with human Shh-type medulloblastoma and are likely to be important for human tumor growth.
Brg1 function is conserved in human Shh-type medulloblastoma. (a) The transcription pathways regulated by Brg1 in SmoM2 mouse medulloblastoma are also specific to human Shh-type medulloblastoma. Using a microarray data set of 76 human pediatric medulloblastoma samples with known subtypes (GSE37418), the average expression levels of human homologs of Brg1-activated Gli1 or Atoh1 targets as well as Brg1-repressed REST targets in each of the four types of medulloblastoma were compared. The subgroup with the highest expression level for each Brg1-activated genes and the subgroup with the lowest expression for each Brg1-repressed genes were identified and counted. The distributions of #1 or #4 ranked genes in each subgroup for the three Brg1-regulated gene lists are shown. The dash line indicates the expected 25% for each subgroup if there are no specificities. (b) The average expression levels of example genes in each medulloblastoma subtype in the human microarray data set. SMARCA4/Brg1 is highly expressed in all the tumor samples. The expression levels of human GLI1 and ATOH1 were significantly higher, whereas that of a REST target CACAN1G was significantly lower, in Shh-type medulloblastoma than in the other subgroups. The numbers in parentheses indicate the number of tumor samples in each subgroup. (c and d) RNAi-mediated Brg1 inhibition in Gli1-activated human medulloblastoma cell line DaoY led to (c) reduced Gli1/Atoh1 target gene expression as shown by RT–qPCR and (d) decreased cell growth as analyzed by ATP cell viability assays. (e) RNAi-mediated Brg1 inhibition did not affect the growth of D283 human medulloblastoma cell line. (f) RT–qPCR analyses of mRNA levels of Brg1 in DaoY and D283 cells with or without Brg1 RNAi inhibition. (g) Western blot assays show the endogenous Brg1 and exogenous HA-Gli1 protein levels in DaoY cells with or without inhibition of Brg1 expression. (h) Western blot assays show global H3K27me3 levels in DaoY and D283 cells with or without inhibition of Brg1 expression. Significance was determined using t-test or analysis of variance with post hoc t-test. **P<0.01 and *P<0.05.
To further determine the function of Brg1 in human medulloblastoma growth, we performed RNAi-mediated Brg1 inhibition in human medulloblastoma cell lines and examined the effects on target gene expression and cell growth. The DaoY human cell line resembles Shh-type medulloblastoma cells.59 Although DaoY cells express low levels of Gli1 and Atoh1, exogenous Gli1 significantly activated Gli1 and Atoh1 targets and accelerated growth of these cells (Figures 6c and d). RNAi-mediated inhibition of Brg1 expression in DaoY cells significantly decreased the expression of these oncogenes and impaired the cell growth in the absence or presence of exogenous Gli1 (Figures 6c–g). In contrast, the reduction of Brg1 levels in a Group 3/4-like medulloblastoma cell line D283 (ref. 60) did not affect the cell growth (Figures 6e and f). Notably, similar to in SmoM2 tumors, inhibition of Brg1 in DaoY cells but not in D283 cells also increased global H3K27me3 levels (Figure 6h), suggesting that Brg1 may also regulate human medulloblastoma through modulating H3K27me3 modifier activities. These results indicate that Brg1 has conserved and specific role in coordinating the genetic and epigenetic regulatory programs in Shh-type medulloblastoma.
Discussion
In this report, we demonstrated that Brg1 is essential for both normal cerebellum development and Shh-type medulloblastoma growth. In both mouse and human Shh-type medulloblastoma, Brg1 regulates expression of GliA and Atoh1 target genes to maintain tumor identity and proliferation. Brg1 also inhibits expression of genes with potential tumor suppressor functions as well as REST-repressed neuronal genes. Furthermore, Brg1 may regulate the activities of H3K27me3 modifiers to maintain the distinct chromatin environment specific to Shh-type medulloblastoma. Therefore, Brg1 is part of a genetic and epigenetic network that controls the specific transcriptional program underlying Shh-type medulloblastoma (Figure 7). Our study demonstrates that a novel epigenetic mechanism controls subtype-specific medulloblastoma development.
Chromatin regulator Brg1 controls the transcriptional regulatory circuits underlying Shh-type medulloblastoma growth. A model representing specific functions of Brg1 in regulating the transcriptional regulatory circuits that control Shh-type medulloblastoma. Brg1 activates target genes of two key regulators of Shh-type medulloblastoma, Gli1 and Atoh1. Brg1 represses the expression of Shh-type medulloblastoma-specific tumor suppressors and REST-targeted neuronal genes. Brg1 deletion led to the inhibition of Shh-type medulloblastoma growth by deregulation of the essential transcription program.
BAF complexes are known to target many developmentally important loci, mainly by interacting with transcription factors through its diverse subunits.28, 29 Our studies provide genetic and genomic evidence indicating an essential function of Brg1 in regulating Shh/Gli signaling and Atoh1 pathway in cancer. The interactions between Brg1 and Gli1 or Atoh1 and the significant enrichment of Brg1 occupancy in genes targeted by Gli1 and/or Atoh1 relative to non-target genes suggests that in medulloblastoma, Brg1 could be recruited by Gli1 or Atoh1 and functions specifically to co-regulate Gli1 and Atoh1 target genes. Brg1 also represses a large number of REST target genes in medulloblastoma and prevents tumor cell differentiation. Brg1 interacts with REST and is also required for maximum binding of REST to certain target genes. This is consistent with a previous report that REST recruits BAF complexes whereas BAF complexes further facilitate REST binding.58 Gli1 and Atoh1 as well as their target genes are overrepresented in human Shh-type medulloblastoma than the other subtypes, whereas REST target genes are underrepresented, indicating that Brg1-regulated transcription network is also conserved in human cancers. The mechanisms identified in mouse models are likely applicable to human cancers. A direct test in human cancer xenograft model would be very useful to directly test Brg1 function in human medulloblastoma.
The activities of BAF complexes in tumor development appear to be tumor-type dependent, consistent with its diverse subunit compositions and functions in different tissues.61 Using the ATPase-dependent chromatin remodeling activities or ATPase-independent functions by interacting with other epigenetic cofactors, BAF complexes could activate or repress specific target genes in a context-dependent manner.29 Therefore, depending on the specific roles of BAF subunits in regulating different oncogenes or tumor suppressors that are associated with each type of tumors, BAF subunits may function either to suppress or to support tumor growth. The tumor suppressor functions of BAF subunits have been extensively studied in atypical teratoid/rhabdoid tumors, where biallelic mutations of SMARCB1/BAF47 as well as SMARCA4/Brg1 cause cancer development.32, 62 In atypical teratoid/rhabdoid tumors with Brg1 or BAF47 mutated, increased Shh target gene expression was observed, which may contribute to the atypical teratoid/rhabdoid tumors growth.63 This notion is consistent with our previous finding that Brg1/BAF complex has a dual role in Shh signaling by repressing the basal expression and activating signaling-induced target gene expression.27 In cancers that require active Shh signaling such as Shh-type medulloblastoma, Brg1 is then required for tumor growth. Therefore depending on the Shh signaling activities in different cell types, BAF deletion may cause different effects on Shh target genes and cancer growth.29 In medulloblastoma, the potential tumor suppressor functions of Brg1 in Wnt and Group 3 subtypes remain unknown.
The transcription profiles classify medulloblastomas into four groups. Interestingly, mutations in a significant number of epigenetic regulators have been identified. The mutations in H3K4me3 methyltransferase MLL2/3 occur in multiple medulloblastoma subgroups,6, 64 whereas mutations in other epigenetic regulators occur with strong subgroup bias. For example, the mutations in H3K27me3 demethylase UTX/Kdm6A are highly enriched in Group 4 tumors, but are largely absent from Shh-type medulloblastoma.4, 8, 64 SMARCA4/Brg1 mutations have been identified in Wnt and Group 3 subgroups but are rare in Shh-type medulloblastoma, likely because of the essential functions that Brg1 has in the regulation of the Shh-type medulloblastoma transcriptional program. Although mutations in other BAF subunits are rare in pediatric Shh-type medulloblastoma, they are present in adult Shh-type medulloblastoma.10 It is not clear how these mutations regulate adult tumor progression; they may contribute to the tumor heterogeneity between pediatric and adult medulloblastoma. In Shh-type medulloblastoma with BAF subunit mutations, the function of Brg1 remains to be explored. Interestingly, it has been shown that Brg1 is required for the oncogenesis caused by loss of the SMARCB1/SNF5 tumor suppressor.65 Therefore, the aberrant BAF complexes formed with mutated BAF subunits may misregulate oncogenic genes, and further deleting Brg1 would inhibit tumor growth.
The extensive and subgroup specific mutations in epigenetic regulators in medulloblastoma implicate the importance of epigenetic regulation in medulloblastoma development and epigenetic therapy holds great promise for the future treatment of medulloblastoma. In our study, we also showed an important function of Brg1 in coordinating the activities of H3K27me3 regulators. Brg1 deletion caused increased H3K27me3 at the regulatory regions of Gli1 and Atoh1 and possibly other oncogenes, which may lead to decreased expression of these genes. Brg1 deletion also led to a significant decrease of PRC2 levels. PRC2 has been shown to repress tumor suppressors and is oncogenic in other cancers. In addition, PRC2 can function as a co-repressor for REST.66 It is possible that the decreased PRC2 levels contribute to the de-repression of certain tumor suppressor genes and REST target genes such as NeuroD2 in Brg1-deleted tumors. Thus, Brg1 regulates local H3K27me3 levels in a target gene-specific manner. The coordinated regulation of H3K27me3 by Brg1 is likely one of the key mechanisms underlying Brg1 function in medulloblastoma transcription regulation. The availability of the chemical inhibitors to Jmjd3 and PRC2 may provide new treatment options for medulloblastoma. In addition to regulating H3K27me3 levels, other ATPase-dependent and/or independent functions of Brg1 in regulating medulloblastoma genes remain to be elucidated.
In summary, our study reveals an epigenetic mechanism that controls specific transcription programs essential for Shh-type medulloblastoma development. Future functional epigenome studies will elucidate the specific epigenetic regulations of each tumor subtypes and will provide targets for the development of the much-needed subtype-specific treatments for medulloblastoma patients.
Materials and Methods
Mice
The SmoM2 mice42 and CAG-CreER67 mice were purchased from Jackson Laboratory. Owing to the CreER leakage, the CAG-CreER SmoM2 mice have a high rate of spontaneous medulloblastoma development before 2 months of age (~40%) even without tamoxifen induction. These mice were crossed to Brg1+/+, Brg1F/+ or Brg1F/F mice27, 68 and are maintained on a mixed genetic background at University of Texas Southwestern Medical Center Animal Facility. All the procedures were performed in accordance with the IACUC-approved protocols. The Atoh1-Cre mice were generated by knock-in of the Cre gene at the Atoh1 locus40 and were provided by Dr Lin Gan (Rochester University) and Dr Jane Johnson (University of Texas Southwestern). The SCID-NOD mice were purchased from the University of Texas Southwestern Mouse Breeding Core Facility.
Cell cultures and lentiviral infection
The primary CGNP cultures were performed as previously described.27 Briefly, CGNPs were derived from dissociated P4–7 mouse cerebella and cultured in DMEM/F12 media containing 25 mm KCl, N2 supplement and 10% fetal bovine serum. For Shh stimulation, the Shh-conditioned medium produced from Shh-CM 293T cells69 was added at a 1:20 dilution to the CGNP cultures. The CGNP cells were treated with Shh in high serum media for 2–3 days. The induction of the Brg1 conditional deletion was performed by treating the cultures with 1 μm 4-hydroxytamoxifen for 3 days. The medulloblastoma cell lines DaoY and D283 were purchased from ATCC and cultured as suggested by the supplier. The lentiviral constructs pGIPZ, shBrg1 and shCtrl70 were generously provided by Dr Chin-ping Chang (Indiana University) and packaged as described in ref. 27. Polyjet (Signagen, Gaithersburg, MD, USA) was used for plasmid transfection in the cultured cells. The DaoY and D283 cells were infected with viruses at a multiplicity of infection of 5 for 24 h in media with 8 μg/ml polybrene. The CellTiter-Glo Luminescent Cell Viability Assay (ATP viability assay) kit (Promega, Fitchburg, WI, USA) was used to measure the tumor cell survival signals.
Tumor transplantation
Medulloblastoma grown in CAG-CreER SmoM2 mice with Brg1+/+, Brg1F/+ or Brg1F/F alleles were dissected and dissociated. The tumor cells (106) were mixed with Matrigel (BD Biosciences, San Jose, CA, USA) and injected subcutaneously in the flank region of SCID-NOD immunodeficient mice. The recipient mice were monitored daily for tumor growth. Tamoxifen (75 mg/kg) or oil solvent were injected intraperitoneally when tumor was visible at ~20 days post transplantation. Tamoxifen was injected every other day during a 20-day period before killing the recipient mice for tumor analyses.
Immunoblotting
For immunoblotting, the cells or ground tissues were lysed in RIPA buffer (50 mm Tris, pH 8, 250 mm NaCl, 0.05% SDS, 0.5% DOC, 1% NP-40). The histone fractions were prepared with standard acid extraction (0.2 n HCl). The cell lysates or histone fractions were separated on SDS–PAGE gels. The antibodies used were against Gli1 (Cell Signaling, Danvers, MA, USA), GAPDH (Sigma-Aldrich, St Louis, MO, USA), H3K27me3 (Active Motif, Carlsbad, CA, USA), histone H3 (ab1791, Abcam, Cambridge, UK), Brg1 (G7, Santa Cruz, Dallas, TX, USA), beta-tub3 (Tuj1, Covance, San Diego, CA, USA), Nestin (Rat-401, BD Pharmingen, San Diego, CA, USA), GFAP (BD Pharmingen), HSP90 (Pierce, Rockford, IL, USA), HA tag (ab9110, Abcam), beta-catenin (Santa Cruz), Atoh1 (Gift from Dr Jane Johnson) and REST (07-579, Millipore, Billerica, MA, USA). HRP-conjugated secondary antibodies were purchased from Jackson ImmunoResearch (West Grove, PA, USA).
Immunohistology
Hematoxylin and eosin staining and immunostaining were performed on paraffin sections of tumor or brain tissues. Antibodies used were against H3K27me3 (Millipore), H3P (Millipore), Ki67 (eBioscience), BrdU (BD Pharmingen), SUZ12 (Cell Signaling) and Brg1 (G7, Santa Cruz). The images were visualized using an Olympus BX50 microscope (Tokyo, Japan). H3P-, Ki67- or BrdU-positive cells were counted from four sections (>2000 nuclei counted for each stain) and the data are given as percentage of total cells (n=3). The semi-quantitative immunohistochemistry densities of H3K27me3 staining were measured using ImageJ program (NIH).
Co-immunoprecipitation experiments
The SmoM2 medulloblastoma samples were lysed with Buffer A (25 mm Tris, pH 7.5, 25 mm KCl, 5 mm MgCl2, 10% glycerol, 0.1% NP-40, with protease inhibitor freshly added). The nuclear extracts were prepared in RIPA buffer (150 mm NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS and 50 mm Tris, pH 8.0) with rotation at 4 °C for 1 h. After centrifugation, the rabbit polyclonal antibodies against Brg1/Brm (J1)71, 72 were added to pre-cleared nuclear extracts and incubated at 4 °C overnight. The samples were incubated with protein A beads (GE Healthcare, Chalfont, UK) for 1 h; the beads were washed with RIPA buffer four times. The precipitated proteins were eluted by boiling in 2 × sample buffer before SDS–PAGE and western blot analysis.
RT–PCR and qPCR
The RNA from cells or ground tissues was extracted with TRIZOL (Invitrogen, Carlsbad, CA, USA). The complementary DNAs were synthesized by reverse transcription using Iscript (Bio-Rad, Hercules, CA, USA), followed by PCR or quantitative PCR analysis. A Bio-Rad real-time PCR system (C1000 Thermal Cycler) was used for quantitative PCR. The levels of GAPDH mRNA were used to normalize input RNA. The graphics shown are representative of experiments performed in triplicate. The experiments were repeated at least three times. The standard errors were calculated according to a previously described method.27
RNA-seq analyses
The CAG-CreER SmoM2 mice harboring spontaneous medulloblastoma with Brg1F/F or Brg1+/+ alleles were injected with tamoxifen. The Brg1+/+ and Brg1-deleted (Brg1iko) SmoM2 medulloblastomas were used for RNA-seq analyses. Total RNAs were extracted, and RNA-seq libraries were prepared using the Illumina RNA-Seq Preparation Kit and were sequenced on a HiSeq 2000 sequencer at the University of Texas Southwestern Sequencing Core Facility. The RNA-seq reads were mapped using TopHat with default settings (http://tophat.cbcb.umd.edu). The mapped reads with the Phred quality score <20 were filtered out, whereas the duplicates were marked but not removed using SAMTOOLS73 and PICARD (http://picard.sourceforge.net). Transcript assembly and transcript abundance quantification were carried out using CUFFLINKS, and then differential expression analysis between Brg1+/+ and Brg1iko SmoM2 medulloblastomas was performed using CUFFDIFF.74 The differentially expressed genes with fold change larger than 2 and P<0.05 were selected as Brg1-regulated genes (Supplementary Table S1). Gene ontology analysis was performed using DAVID tools (http://david.abcc.ncifcrf.gov/).
ChIP and ChIP-seq analyses
The ChIP experiments were performed as described previously.27 Dounced tissue or dissociated cells were crosslinked with PFA or double crosslinked with DSG (Pierce), and sonicated to fragments (200–500 bp). The antibodies used were against H3K27me3 (Millipore), Jmjd3 (Abcam) and Brg1/Brm (J1).71 J1 antibody has been used previously for Brg1 ChIP-seq analyses.44, 45 The precipitated DNA was purified and subjected to either real-time PCR or next-generation sequencing. NEBNext ChIP-Seq Sample Prep Master Mix Set 1 was used for library generation and a Hiseq 2500 sequencer was used for sequencing at the University of Texas Southwestern Medical Center Sequencing Core Facility.
Short reads were mapped to UCSC reference mouse genome with bowtie75 and then SICER was used to detect the Brg1-binding regions.43 SICER uses clustering approach to pool together the enrichment signals of the ChIP-seq reads from neighboring nucleosomes to increase the power for detecting broad binding domains. Thus it is suitable for analyzing the ChIP-seq data of histone modifications and chromatin regulators, which are usually noisy and contain broad domains. SICER has been successfully used for detecting Brg1-binding regions in the hematopoietic cell lineages.76 Default parameter settings with three 200-bp windows were used to calculate the enrichment of Brg1-binding regions. The corresponding input sample was used as control. The duplicate reads were removed before peak calling by SICER. Statistically significant peaks (FDR<0.05) enriched in the Brg1-ChIP sample relative to its corresponding input sample were annotated for genomic location.
To calculate the distribution of binding regions in reference to nearby genes, we used the mouse gene annotation obtained from iGenomes (http://support.illumina.com/sequencing/sequencing_software/igenome.ilmn). The midpoint of each Brg1-binding region was compared with the location of genes to identify either the overlapping gene or the closest non-overlapped gene. The distance from the midpoint of each Brg1-binding region to the 5′ end of the overlapped or the nearest gene was calculated. The promoter region was defined as 5 kb upstream of the gene, and the downstream was defined as 5 kb downstream of the gene.
Bioinformatics analyses
Ranked fold-change analysis was performed to determine the association between Brg1-regulated genes and genes differentially expressed in SmoM2 MB. The differentially expressed genes detected from either Brg1 RNA-seq or SmoM2 expression array42 were ranked based on fold changes. Twenty bins with equal range of fold change were used to count the number of differentially expressed genes shared in both gene sets. To identify genes regulated by multiple proteins, including Brg1, Gli1 and Atoh1, we used the UCSC mouse genome build mm10 and mouse gene annotation from iGenomes as references to associate locations of binding regions of each protein with nearby genes. All the reported peak locations were converted to mm10 coordinates with UCSC liftOver tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver). We consider that a gene is bound by a protein if there is a binding region or peak overlapping its gene body or within the 5-kb flanking regions. Few previously reported target genes of these proteins were missed due to coordinate conversion of binding regions. Gli1 binding sites were identified using a FLAG-tagged Gli1 transgene in another similar mouse model of Shh-type medulloblastoma arising from Ptch1+/- mice.46 From 1059 Gli1 binding regions, we identified 1169 potential Gli1 binding genes. Atoh1-binding gene sets in P5 CGNPs were determined in a previous analysis.51 REST target genes in embryonic stem cells were determined by ChIP-PET and directly obtained from a previous report.57 To test the significance of the overlapping between gene sets bound by two proteins, we use Fisher’s exact test to calculate the P-value. The microarray data set GSE37418 with 76 subtype-characterized pediatric medulloblastoma samples8 was analyzed for expression levels of human homologs of Brg1-regulated medulloblastoma genes in different medulloblastoma subtypes. All the human homologs of Brg1-activated Gli1 targets (42 total) and Atoh1 targets (29 total) and Brg1-repressed REST targets (177 total) were analyzed for their average expression in each subgroup. The #1 ranked subgroup for each Brg1-activated gene or the #4 ranked subgroup for each Brg1-repressed genes were identified, counted and compared (Supplementary Table 4, Figure 6a). The binominal test was used for calculating the P-value. The RNA-seq and ChIP-seq data generated in this study have been deposited in NCBI GEO repository with accession number GSE69674 (GSE69672 for RNA-seq and GSE69673 for ChIP-seq).
Statistical analysis
The data are expressed as means±s.d. (n=3, unless specifically indicated). Statistical analysis was performed by either analysis of variance with analysis of variance post hoc t-test for multiple comparisons or a two-tailed unpaired Student’s t-test. A P-value <0.05 was considered significant.
References
Gajjar A, Chintagumpala M, Ashley D, Kellie S, Kun LE, Merchant TE et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multicentre trial. Lancet Oncol 2006; 7: 813–820.
Northcott PA, Jones DT, Kool M, Robinson GW, Gilbertson RJ, Cho YJ et al. Medulloblastomics: the end of the beginning. Nat Rev Cancer 2012; 12: 818–834.
Rutkowski S, von Hoff K, Emser A, Zwiener I, Pietsch T, Figarella-Branger D et al. Survival and prognostic factors of early childhood medulloblastoma: an international meta-analysis. J Clin Oncol 2010; 28: 4961–4968.
Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M et al. Dissecting the genomic complexity underlying medulloblastoma. Nature 2012; 488: 100–105.
Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 2012; 488: 49–56.
Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC et al. The genetic landscape of the childhood cancer medulloblastoma. Science 2011; 331: 435–439.
Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 2012; 488: 106–110.
Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L et al. Novel mutations target distinct subgroups of medulloblastoma. Nature 2012; 488: 43–48.
Barakat MT, Humke EW, Scott MP . Learning from Jekyll to control Hyde: Hedgehog signaling in development and cancer. Trends Mol Med 2010; 16: 337–348.
Kool M, Jones DT, Jager N, Northcott PA, Pugh TJ, Hovestadt V et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 2014; 25: 393–405.
Corrales JD, Rocco GL, Blaess S, Guo Q, Joyner AL . Spatial pattern of sonic hedgehog signaling through Gli genes during cerebellum development. Development 2004; 131: 5581–5590.
Dahmane N, Ruiz i Altaba A . Sonic hedgehog regulates the growth and patterning of the cerebellum. Development 1999; 126: 3089–3100.
Wallace VA . Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr Biol 1999; 9: 445–448.
Wang VY, Zoghbi HY . Genetic regulation of cerebellar development. Nat Rev Neurosci 2001; 2: 484–491.
Briscoe J, Therond PP . The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol 2013; 14: 416–429.
Fuccillo M, Joyner AL, Fishell G . Morphogen to mitogen: the multiple roles of hedgehog signalling in vertebrate neural development. Nat Rev Neurosci 2006; 7: 772–783.
Ingham PW, McMahon AP . Hedgehog signaling in animal development: paradigms and principles. Genes Dev 2001; 15: 3059–3087.
Jiang J, Hui CC . Hedgehog signaling in development and cancer. Dev Cell 2008; 15: 801–812.
Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell 2008; 14: 123–134.
Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q et al. Math1 is essential for genesis of cerebellar granule neurons. Nature 1997; 390: 169–172.
Flora A, Klisch TJ, Schuster G, Zoghbi HY . Deletion of Atoh1 disrupts Sonic Hedgehog signaling in the developing cerebellum and prevents medulloblastoma. Science 2009; 326: 1424–1427.
Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC et al. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 1995; 80: 949–957.
Negrini S, Prada I, D'Alessandro R, Meldolesi J . REST: an oncogene or a tumor suppressor? Trends Cell Biol 2013; 23: 289–295.
Schoenherr CJ, Anderson DJ . The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 1995; 267: 1360–1363.
Canettieri G, Di Marcotullio L, Greco A, Coni S, Antonucci L, Infante P et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat Cell Biol 2010; 12: 132–142.
Malatesta M, Steinhauer C, Mohammad F, Pandey DP, Squartrito M, Helin K . Histone acetyltransferase PCAF is required for Hedgehog-Gli-dependent transcription and cancer cell proliferation. Cancer Res 2013; 73: 6323–6333.
Zhan X, Shi X, Zhang Z, Chen Y, Wu JI . Dual role of Brg chromatin remodeling factor in Sonic hedgehog signaling during neural development. Proc Natl Acad Sci USA 2011; 108: 12758–12763.
Hargreaves DC, Crabtree GR . ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res 2011; 21: 396–420.
Wu JI . Diverse functions of ATP-dependent chromatin remodeling complexes in development and cancer. Acta Biochim Biophys Sin (Shanghai) 2012; 44: 54–69.
Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 2013; 45: 592–601.
Wilson BG, Roberts CW . SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 2011; 11: 481–492.
Hasselblatt M, Nagel I, Oyen F, Bartelheim K, Russell RB, Schuller U et al. SMARCA4-mutated atypical teratoid/rhabdoid tumors are associated with inherited germline alterations and poor prognosis. Acta Neuropathol 2014; 128: 453–456.
Witkowski L, Carrot-Zhang J, Albrecht S, Fahiminiya S, Hamel N, Tomiak E et al. Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat Genet 2014; 46: 438–443.
Romero OA, Torres-Diz M, Pros E, Savola S, Gomez A, Moran S et al. MAX inactivation in small cell lung cancer disrupts MYC-SWI/SNF programs and is synthetic lethal with BRG1. Cancer Discov 2014; 4: 292–303.
Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe JS et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev 2013; 27: 2648–2662.
Shi X, Zhang Z, Zhan X, Cao M, Satoh T, Akira S et al. An epigenetic switch induced by Shh signalling regulates gene activation during development and medulloblastoma growth. Nat Commun 2014; 5: 5425.
Li Q, Wang HY, Chepelev I, Zhu Q, Wei G, Zhao K et al. Stage-dependent and locus-specific role of histone demethylase Jumonji D3 (JMJD3) in the embryonic stages of lung development. PLoS Genet 2014; 10: e1004524.
Miller SA, Mohn SE, Weinmann AS . Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell 2010; 40: 594–605.
Popov N, Gil J . Epigenetic regulation of the INK4b-ARF-INK4a locus: in sickness and in health. Epigenetics 2010; 5: 685–690.
Yang H, Xie X, Deng M, Chen X, Gan L . Generation and characterization of Atoh1-Cre knock-in mouse line. Genesis 2010; 48: 407–413.
Moreno N, Schmidt C, Ahlfeld J, Poschl J, Dittmar S, Pfister SM et al. Loss of Smarc proteins impairs cerebellar development. J Neurosci 2014; 34: 13486–13491.
Mao J, Ligon KL, Rakhlin EY, Thayer SP, Bronson RT, Rowitch D et al. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res 2006; 66: 10171–10178.
Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W . A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 2009; 25: 1952–1958.
Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR . An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci USA 2009; 106: 5187–5191.
Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell 2013; 152: 248–261.
Lee EY, Ji H, Ouyang Z, Zhou B, Ma W, Vokes SA et al. Hedgehog pathway-regulated gene networks in cerebellum development and tumorigenesis. Proc Natl Acad Sci USA 2010; 107: 9736–9741.
Dakubo GD, Mazerolle CJ, Wallace VA . Expression of Notch and Wnt pathway components and activation of Notch signaling in medulloblastomas from heterozygous patched mice. J Neurooncol 2006; 79: 221–227.
Fernandez LA, Northcott PA, Dalton J, Fraga C, Ellison D, Angers S et al. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev 2009; 23: 2729–2741.
Natarajan S, Li Y, Miller EE, Shih DJ, Taylor MD, Stearns TM et al. Notch1-induced brain tumor models the sonic hedgehog subgroup of human medulloblastoma. Cancer Res 2013; 73: 5381–5390.
Anne SL, Govek EE, Ayrault O, Kim JH, Zhu X, Murphy DA et al. WNT3 inhibits cerebellar granule neuron progenitor proliferation and medulloblastoma formation via MAPK activation. PLoS One 2013; 8: e81769.
Klisch TJ, Xi Y, Flora A, Wang L, Li W, Zoghbi HY . In vivo Atoh1 targetome reveals how a proneural transcription factor regulates cerebellar development. Proc Natl Acad Sci USA 2011; 108: 3288–3293.
Markant SL, Esparza LA, Sun J, Barton KL, McCoig LM, Grant GA et al. Targeting sonic hedgehog-associated medulloblastoma through inhibition of Aurora and Polo-like kinases. Cancer Res 2013; 73: 6310–6322.
Sengupta R, Dubuc A, Ward S, Yang L, Northcott P, Woerner BM et al. CXCR4 activation defines a new subgroup of Sonic hedgehog-driven medulloblastoma. Cancer Res 2012; 72: 122–132.
Smits M, van Rijn S, Hulleman E, Biesmans D, van Vuurden DG, Kool M et al. EZH2-regulated DAB2IP is a medulloblastoma tumor suppressor and a positive marker for survival. Clin Cancer Res 2012; 18: 4048–4058.
Uziel T, Zindy F, Sherr CJ, Roussel MF . The CDK inhibitor p18Ink4c is a tumor suppressor in medulloblastoma. Cell Cycle 2006; 5: 363–365.
Wetmore C, Eberhart DE, Curran T . Loss of p53 but not ARF accelerates medulloblastoma in mice heterozygous for patched. Cancer Res 2001; 61: 513–51669.
Johnson R, Teh CH, Kunarso G, Wong KY, Srinivasan G, Cooper ML et al. REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol 2008; 6: e256.
Ooi L, Belyaev ND, Miyake K, Wood IC, Buckley NJ . BRG1 chromatin remodeling activity is required for efficient chromatin binding by repressor element 1-silencing transcription factor (REST) and facilitates REST-mediated repression. J Biol Chem 2006; 281: 38974–38980.
Triscott J, Lee C, Foster C, Manoranjan B, Pambid MR, Berns R et al. Personalizing the treatment of pediatric medulloblastoma: polo-like kinase 1 as a molecular target in high-risk children. Cancer Res 2013; 73: 6734–6744.
Sengupta S, Weeraratne SD, Sun H, Phallen J, Rallapalli SK, Teider N et al. alpha5-GABAA receptors negatively regulate MYC-amplified medulloblastoma growth. Acta Neuropathol 2014; 127: 593–603.
Wu JI, Lessard J, Crabtree GR . Understanding the words of chromatin regulation. Cell 2009; 136: 200–206.
Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 1998; 394: 203–206.
Jagani Z, Mora-Blanco EL, Sansam CG, McKenna ES, Wilson B, Chen D et al. Loss of the tumor suppressor Snf5 leads to aberrant activation of the Hedgehog-Gli pathway. Nat Med 2010; 16: 1429–1433.
Dubuc AM, Remke M, Korshunov A, Northcott PA, Zhan SH, Mendez-Lago M et al. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol 2013; 125: 373–384.
Wang X, Sansam CG, Thom CS, Metzger D, Evans JA, Nguyen PT et al. Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex. Cancer Res 2009; 69: 8094–8101.
Dietrich N, Lerdrup M, Landt E, Agrawal-Singh S, Bak M, Tommerup N et al. REST-mediated recruitment of polycomb repressor complexes in mammalian cells. PLoS Genet 2012; 8: e1002494.
Hayashi S, McMahon AP . Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol 2002; 244: 305–318.
Sumi-Ichinose C, Ichinose H, Metzger D, Chambon P . SNF2beta-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol Cell Biol 1997; 17: 5976–5986.
Chen JK, Taipale J, Young KE, Maiti T, Beachy PA . Small molecule modulation of Smoothened activity. Proc Natl Acad Sci USA 2002; 99: 14071–14076.
Stankunas K, Hang CT, Tsun ZY, Chen H, Lee NV, Wu JI et al. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev Cell 2008; 14: 298–311.
Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR . BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 1993; 366: 170–174.
Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron 2007; 55: 201–215.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25: 2078–2079.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010; 28: 511–515.
Langmead B, Trapnell C, Pop M, Salzberg SL . Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10: R25.
Hu G, Schones DE, Cui K, Ybarra R, Northrup D, Tang Q et al. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res 2011; 21: 1650–1658.
Acknowledgements
We thank Drs Brent Orr (St. Jude Hospital), Wenzhe Niu and Zilai Zhang, and Mou Cao for technical support and Dr Chao Xing and the University of Texas Southwestern Sequencing Facility for performing the next-generation sequencing and RNA-seq analyses. We thank Drs Lin Gan and Jane Johnson for providing the Atoh1-Cre mice and Dr Ching-Ping Chang for providing human Brg1 shRNA constructs. This work was supported by grants from March of Dimes Foundation (JW), American Cancer Society (JW), NIMH (JW) and Department of Defense Visionary postdoc fellowship (XS).
Author contributions
JW and XS designed the experiments. XS, QW and JW performed the experiments and collected the data. XS and JW analyzed the results. JG and ZX performed the main bioinformatics analyses. JW wrote the manuscript with help from all the authors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Shi, X., Wang, Q., Gu, J. et al. SMARCA4/Brg1 coordinates genetic and epigenetic networks underlying Shh-type medulloblastoma development. Oncogene 35, 5746–5758 (2016). https://doi.org/10.1038/onc.2016.108
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.108
- Springer Nature Limited
This article is cited by
-
Structural disruption of BAF chromatin remodeller impairs neuroblastoma metastasis by reverting an invasiveness epigenomic program
Molecular Cancer (2022)
-
Modeling medulloblastoma in vivo and with human cerebellar organoids
Nature Communications (2020)