Abstract
To date, the mutational status of EGFR and PTEN has been shown as relevant for favoring pro- or anti-tumor functions of STAT3 in human glioblastoma multiforme (GBM). We have screened genomic data from 154 patients and have identified a strong positive correlation between STAT3 and HDAC7 expression. In the current work we show the existence of a subpopulation of patients overexpressing HDAC7 and STAT3 that has particularly poor clinical outcome. Surprisingly, the somatic mutation rate of both STAT3 and HDAC7 was insignificant in GBM comparing with EGFR, PTEN or TP53. Depletion of HDAC7 in a range of GBM cells induced the expression of tyrosine kinase JAK1 and the tumor suppressor AKAP12. Both proteins synergistically sustained the activity of STAT3 by inducing its phosphorylation (JAK1) and protein expression (AKAP12). In absence of HDAC7, activated STAT3 was responsible for significant imbalance of secreted pro-/anti-angiogenic factors. This inhibited the migration and sprouting of endothelial cells in paracrine fashion in vitro as well as angiogenesis in vivo. In a murine model of GBM, induced HDAC7-silencing decreased the tumor burden by threefold. The current data show for the first time that silencing HDAC7 can reset the tumor suppressor activity of STAT3, independently of the EGFR/PTEN/TP53 background of the GBM. This effect could be exploited to overcome tumor heterogeneity and provide a new rationale behind the development of specific HDAC7 inhibitors for clinical use.
Similar content being viewed by others
Introduction
Glioblastoma multiforme (GBM) is classified as the most aggressive type of glial tumors. The clinical outcome of GBM patients is daunting, especially because there are no effective therapies that can slow down the progression (median survival of 15 months post diagnosis). Molecular profiling studies have surfaced several driver-genes (for example, EGFR, KDR and PDGFR), overexpressed in GBM, that decisively impact the ability of the cancer cell to survive, proliferate and invade.1, 2, 3, 4 Unfortunately, none of these genes could be successfully used as targets in clinical practice.5 This suggests that GBM is not a homogenous disease and that it consists of various subtypes.6, 7, 8 One strategy in developing novel and effective drugs against GBM is to understand and individually target the possible tumor subtypes.9 Recent findings on GBM heterogeneity demonstrate that such personalised therapy might be very difficult because many mutations originally thought to be mutually exclusive do occur in the same tumor.10, 11 This calls for broader and more robust therapies that target proteins involved in more basic processes shared by most if not all cancer cells. Promising categories of such targets are proteins implicated in embryonic development that are frequently hijacked and reactivated by cancer.12, 13 In this regard, a key step in the process of activating embryonic programs is the re-expression of specific transcription factors that control many embryonic genes. One such factor is signal transducer and activator of transcription 3 (STAT3), which particularly in the developing brain directs the differentiation of neural stem cells into astrocytes.14 In GBM, STAT3 has been long considered as a tumor promoter especially in conjunction with the v3 isoform of EGFR.15, 16 The tumor suppressive role has been shown dependent on the functional PTEN.17 Unfortunately, PTEN is a developmentally important gene frequently mutated and deactivated in cancer.18, 19 In glioblastoma, only 40% of GBM are considered as PTEN wild type, 50% as overexpressed EGFR and 11% are positive for EGFRv3 isoform.20 These circumstances, specific for GBM, leave little room for STAT3 to develop its tumor-supressing potential.
HDAC7 is a class IIa histone deacetylase that has essential role in the development of normal vasculature during embryogenesis as well as angiogenesis in adult tissues.21 Silencing HDAC7 inhibits angiogenesis in endothelial cells both in vivo and in vitro.22, 23, 24 Although angiogenesis is one of the key hallmarks of GBM, to our knowledge, there are no studies addressing the potential role of HDAC7 in GBM biology. Most of them describe the effects of broad HDAC inhibitors on GBM. In the clinics, broad HDAC inhibitors have shown only modest results when used as single agents in GBM patients.25 As HDAC function through protein-deacetylation and affect many cellular processes along the way, their concomitant targeting may not be the best solution for achieving a potent anti-tumor effect. HDAC form complexes with histone acetyltransferases to modulate the acetylation of many protein targets beyond histones or nuclear proteins.26 STAT3 activity is directly dependent on K685 acetylation that enables the dimerization of the protein into a functional unit. HDAC3/p300 (HDAC/histone acetyltransferases) complex has been shown responsible for modulating this acetylation on STAT3 establishing a direct link between HDAC and STAT3.27 Xiao et al.28 have demonstrated that HDAC7 can specifically interact with Tip60 to suppress the activity of STAT3.
We have previously shown that HDAC7-silencing induces activation of STAT3 in endothelial cells, resulting in stalled endothelial cell migration and differentiation.24 The results suggested that HDAC7 and STAT3 might assume very basic and broad functions in angiogenesis. High-grade gliomas are particularly prone to angiogenesis and this feature is used in their pathological classifications. Consequently, anti-angiogenic therapy has been strongly advocated in the past for high-grade gliomas treatment. However, the results of clinical trials with VEGF-targeting biologics proved disappointing. The reason behind this may be partly due to a rescue mechanism mediated by other equally potent angiogenic molecules. On the basis of previous findings, we hypothesized that HDAC7 targeting in cancer cells may activate STAT3 and provoke a very broad anti-angiogenic effect that would be difficult to overcome by cancer cells.
Results
High STAT3 and HDAC7 expression delineates a subset of human GBM with poor outcome
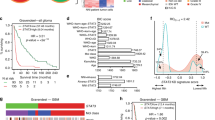
We have conducted a meta-analysis of publicly deposited gene-expression data (cBioPortal and the cancer genome atlas (TCGA) data set;20 N=154) to assess the relationship between STAT3 and HDAC7 expression in GBM. Correlation analysis (Pearson correlation coefficient) revealed that nine HDAC positively correlated with STAT3 expression, while HDAC7 displayed the strongest degree of correlation with STAT3 (Figure 1a). The data showed that approximately one third of GBM had a modulation of STAT3 and HDAC7 genes, half of which displayed downregulation and the other half showed upregulation (Figure 1a). Patient alignment according to the HDAC7/STAT3 expression in the GBM highlighted five distinct groups: (1) high expression of HDAC7 and STAT3 (HDAC7high/STAT3high), (2) high expression of STAT3 (STAT3high), (3) low expression of STAT3 and HDAC7 (STAT3low/HDAC7low), (4) high expression of HDAC7 (HDAC7high) and (5) low expression of HDAC7. Having identified these clusters, we next asked the question if the clinical evolution of the patients would be different in those subgroups. We have therefore examined the overall survival and the progression-free survival of the patients found in each individual group (Figure 1a). Patients expressing low levels of HDAC7 (group 5) were not further considered as their low numbers were not sufficient for statistical analysis. As shown in the Figure 1a, GBM patients who have upregulated either HDAC7 or STAT3 or both have significantly lower survival rate (overall survival and progression-free survival) than patients who downregulate these two genes. Next, we looked for enrichment of any typical gene mutations or/and amplifications/deletions in each GBM cluster (Figure 1a). Surprisingly, the analysis highlighted that HDAC7high/STAT3high patients were delineated by high frequency of EGFR amplification (over 90% of cases) and by absence of TP53 mutations or deletions. As far as PTEN is concerned, no mutations were detectable (in contrast to other subgroups) and deletions were present in 17% of cases. This frequency was not different from other groups of GBM patients. Given the large number of different mutations in GBM we sought to investigate how frequently are STAT3 and HDAC7 mutated in GBM. The analysis of the TCGA patient cohort (N=154) revealed that both genes are mutated in <1% of cases. This was in evident contrast with the mutation frequency of EGFR, PTEN and TP53, the three being mutated in >50, 30 and 20% of cases respectively (Supplementary Figure S1, Supplementary Data).
Expression of HDAC7 and STAT3 in human GBM patients. (a) Gene expression analysis of the TCGA GBM data set (154 patients) conducted with cBioPortal (www.cbioportal.org). From left to right: Pearson correlation analysis of STAT3 and HDAC expression; clusters of patients are discernable with different expression levels of HDAC7 and STAT3 (*denotes a cohort of limited size insufficient for statistical analysis; the colors indicate: upregulation (red), downregulation (blue), no change (grey)); overall (OS) and progression-free survival (PFS) among the four major GBM subgroups; mutation, amplification and deletion of EGFR, PTEN and TP53 genes in different patient cohorts. (b) Western blot (WB) analysis of STAT3, HDAC7, EGFR and PTEN expression in a selection of GBM human tissues (N=22). HSC70 was used as a loading control. From left to right: WB analysis; Hierarchical clustering based on Pearson correlation and protein quantities from WB analysis (color scheme: upregulation (red), downregulation (green), no change (black)); OS in two major patient clusters. (a and b) Results are expressed as a mean, error bars indicate s.e.m., *P<0.05 (N=154 for a and N=22 for b). (c) Immunohistochemical analysis of HDAC7 and STAT3 expression in normal human brain and human GBM samples (matched patients specimen used in WB analysis). Of six patients analyzed two representative cases are shown. Immunohistochemistry (IHC) images of normal brain are obtained from publicly available data (source: Protein Atlas; www.proteinatlas.org).
Taking into account that gene expression does not always translate to protein expression, we next examined the expression of STAT3, HDAC7, EGFR and PTEN proteins in a cohort of 22 GBM patients (Figure 1b). The clustering analysis revealed high similarity to gene-expression data (Figure 1a). Namely, most STAT3 overexpressing patients were also highly expressing HDAC7 and EGFR. No particular clustering was observable for PTEN. In contrast to gene-expression data, the patients mainly segregated into two groups: HDAC7high/STAT3high and HDAC7low/STAT3low. The survival analysis in this patient cohort confirmed the trend observed with TCGA data, underlying the bad clinical prognosis for HDAC7high/STAT3high patients. Given the fact that GBM are composed of both tumoral and stromal cells (for example, microglia and endothelial cells), we next examined which cell types expressed HDAC7 and STAT3 in GBM. For this purpose, we performed immunohistochemistry (IHC) analysis on the GBM from the first six patients analyzed in western blot (HDAC7high/STAT3high subtype). We have also examined publicly deposited IHC data from Human Protein Atlas project. The results showed that both proteins are barely expressed or absent in normal brain tissue and that, in fact, cancerous cells account for the majority of HDAC7 and STAT3 protein expression in GBM (Figure 1c).
To further assess the biological relationship between STAT3 and HDAC7, we have proceeded with silencing experiments in a set of human GBM cell lines (both established and primary). The cell lines were chosen to mimic several genetic backgrounds, previously described relevant for STAT3 function in GBM. We have aimed to cover several EGFR statuses (WT, amplified and expression of variant V3) and we have selected only PTEN deficient/mutated GBM cell lines. Dysfunctional PTEN was showed to be important for the pro-tumorigenic activity of STAT3 (ref. 17; Figure 2a). In addition, and based on our findings with the patient data (Figure 1), we have also selected two primary GBM cell lines that had particularly high-HDAC7/STAT3 expression (Supplementary Figure S2, Supplementary Data). In general and similarly to the patient findings, we observed a positive correlation trend between STAT3 and HDAC7 protein levels in the different GBM cell lines. Having these different models, we next investigated the interplay between HDAC7 and STAT3 as well as their biological relevance in GBM.
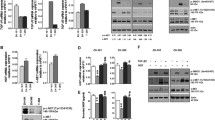
HDAC7-silencing activates STAT3 in PTEN-deficient GBM cells. (a) Overview of mutation status of different cell lines used. The information on mutations was obtained from the public DNA-sequencing repository (COSMIC, http://cancer.sanger.ac.uk/cell_lines). HDAC7-silencing increases STAT3 phosphorylation on Y705 and expression in PTEN-deficient GBM cells lines. (b) Same as in a with primary GBM cultures. (c) HDAC7 depletion triggers acetylation on K685. (a–c) Irrelevant siRNA was used as a negative control. (d) Western blot (WB) analysis of cytoplasmic/nuclear fractionation of U87-MG transfected with control or siRNA HDAC7. CYTO, cytoplasmic fraction; NUC, nuclear fraction. MEK2 and SP1 were used as a cytoplasmic and nuclear fraction markers, respectively. All panels: WB images are representative of five biological replicates and HSC70 was used as a loading control; values under WB images indicate fold changes based on densitometry evaluations of the representative image (control is set to 1.0).
Silencing HDAC7 activates STAT3 through JAK1 pathway
Silencing HDAC7 induced both STAT3 phosphorylation on the tyrosine 705 and augmented STAT3 expression in all GBM cell lines tested (Figures 2a and b). Moreover, depletion of HDAC7 triggered the acetylation of STAT3 on lysine 685, which is essential for stability of the STAT3 homodimer complex (Figure 2c). As a result of this activation, increased levels of phosphorylated STAT3 were observable in the nucleus of GBM cells (Figure 2d). To further clarify which kinase is responsible for STAT3 phosphorylation we sought to test the modulation of several proteins that could directly (JAK1, JAK2 and SRC) or indirectly (PKCα) activate STAT3. As indicated in the Figure 3a, HDAC7 depletion in U87-MG cells did not affect the protein levels of most protein kinases, except JAK1. HDAC7-silencing induced the upregulation of JAK1, whereas only the silencing of JAK1 was able to diminish the phosphorylation of STAT3 in U87-MG cells. Depletion of JAK2 and SRC paradoxically increased the levels of total- and phospho-STAT3 (Figure 3b). On the basis of the findings outlined in Figures 3a and b we hypothesized that HDAC7-silencing induces STAT3 phosphorylation through JAK1. To verify this we have co-transfected HDAC7 and JAK1 or HDAC7 and JAK2 in U87-MG cells (Figure 3c). Indeed, only the co-silencing of JAK1 with HDAC7 was able to reverse the induction of STAT3 phosphorylation. However, the silencing of JAK1 did not affect the total levels of STAT3 protein (Figure 3c) or its messenger RNA (mRNA) expression (Figure 3d). Induction of JAK1 post HDAC7 silencing was also observable using immunofluorescence (Figure 3e). Immunostaining of HDAC7-depleted U87-MG cells evidenced strong perinuclear staining of JAK1 and nuclear staining of phospho-STAT3.
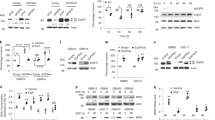
JAK1 phosphorylates STAT3 following HDAC7 silencing. (a) Western blot (WB) analysis of selected protein kinase expression after HDAC7 silencing. (b) Modulation of STAT3 phosphorylation and total protein expression following targeted silencing of selected protein kinases. (c) WB evaluation of JAK1 and JAK2 contribution to phosphorylation of STAT3 following the induction by HDAC7 silencing. (d) Reverse transcription PCR quantification of STAT3 mRNA levels following HDAC7 and JAK1 silencing. Results are shown as means where error bars indicate s.e.m. (N=3 biological replicates; **P<0.01 & NS=non significant). (e) Immunofluorescence (IF) analysis of pSTAT3 and JAK1 protein expression following HDAC7 depletion in GBM cells. Magnification used: × 400. (a–c and e) Shown are representative pictures of three biological replicates; WB: HSC70 was used as a loading control; All experiments were performed using U87-MG cells.
HDAC7 depletion inhibits angiogenesis in vitro regardless of the EGFR status in GBM cells
To test the functional significance of the activated STAT3, we have next verified the expression of several known/potential STAT3 target genes (Figure 4a). The PCR analysis confirmed that HDAC7 silencing leads to induction of STAT3 expression and to a modulation of expression of angiogenesis-relevant genes. We observed a significant increase in expression of the anti-angiogenic factor IGFBP3 and downregulation of pro-angiogenic factors IL8, TGFα and FGF1. Looking for further biological impact of HDAC7 silencing on cancer cells, we have examined for possible alteration in proliferation (Supplementary Figure S3, Supplementary Data) and migration capacity (not shown). None of the two assays showed any significant modulation, raising the question of possible paracrine effects. As the pro-angiogenic factors tested are secreted in nature, we have next sought to verify if the modulation of mRNA expression of TGFα translates in a drop of protein quantity secreted in the medium of cancer cells. TGFα was selected as it was univocally downregulated in all the four cell lines tested. Enzyme-linked immunosorbent assay analysis has confirmed the decrease of TGFα protein levels (Figure 4b), justifying further investigation of possible effects on endothelial cells. Along this line, we have collected conditioned media from U87-MG and U251-MG cancer cells silenced for HDAC7 and have added this media to human umbilical vein endothelial cells (HUVEC), testing their ability to form sprouts and to migrate (Figures 4c and d). The analysis revealed a strong inhibition of both migration and sprouting capacity of HUVEC when exposed to media from GBM cancer cells previously silenced for HDAC7. These in vitro findings clearly indicated that HDAC7 silencing in GBM cells results in a switch toward an anti-angiogenic phenotype, posing the question if such effects could be translated in an animal model.
HDAC7 depletion in PTEN-deficient GBM cell lines impairs angiogenesis in vitro. (a) Relative gene-expression analysis of anti- (IGFBP3) and pro- (IL8, TGFα and FGF1) angiogenic factors following HDAC7 silencing in several GBM cell lines. (b) Enzyme-linked immunosorbent assay analysis of TGF alpha levels in conditioned media of U87-MG depleted for HDAC7. (c) HUVEC-sprouting assay prepared from endothelial cells stimulated with conditioned media collected from control or HDAC7-depleted glioblastoma cells. Formation of radially growing cell sprouts was monitored during 24 h and they were counted on individual pictures. Control cell sprouts were set to 100%. (d) Wound-healing assay conducted on HUVEC monolayers treated with conditioned media from control or HDAC7-depleted glioblastoma cells. Pictures were taken directly and 24 h after wounding. Quantitative analysis was performed using ImageJ software. All panels: results are expressed as a mean, error bars indicate s.e.m. (N=3 biological replicates; ***P<0.001, **P<0.01 and *P<0.05).
STAT3 activation following HDAC7-silencing inhibits angiogenesis in vivo
To examine the observations made in vitro as well as to gain mechanistic insight we have decided to employ the chorioallantoic membrane assay (CAM) model that enables rapid analysis of tumor-induced angiogenesis. Indeed, and as shown in the Figure 5a, silencing HDAC7 in U87 GBM cells fully supressed tumor development on CAM. Moreover, the HDAC7-silenced tumors appeared as small white nodules compared with the control, suggesting a deficient angiogenesis. To verify this we have excised the tumors from the CAM and stained these with Sambucus nigra agglutinin-lectin to highlight the tumor vasculature. As outlined in the Figure 5a and in sharp contrast to the controls, HDAC7-silenced tumors displayed no vasculature network (green signal) despite the presence of tumoral cells in the examined nodules (red signal). We next hypothesized that the anti-tumor effect caused by HDAC7 silencing depends on the activation/phosphorylation of STAT3. To test this hypothesis we have employed a chemical inhibitor STAT3-i6 that blocks the phosphorylation of STAT3 on Y705 residue (Figure 5b). As demonstrated in the Figure 5c, STAT3-i6 efficiently reversed the anti-sprouting and anti-migratory effect induced by HDAC7 silencing on HUVEC. Consequently, we showed that anti-tumor effect caused by HDAC7 silencing on CAM was partially rescued by adding STAT3-i6 to tumor cells and thus blocking STAT3 activation (Figure 5d).
STAT3 activation mediated through HDAC7-silencing inhibits angiogenesis in vivo. (a) CAM-based angiogenesis/tumor-growth assay following HDAC7 silencing in U87-MG cells. (b) Chemical inhibition of STAT3 (S3I-201/STAT3-i6) reverses HDAC7-induced STAT3 phosphorylation on Y705 in U87-MG cells. (c) HUVEC sprouting and wound-healing assay prepared from endothelial cells stimulated with conditioned media collected from glioblastoma cells. (d) Representative pictures of macroscopic/microscopic tumor appearance (CAM) on STAT3-i6 treatment of U87-MG cells. Dimethyl sulfoxide (DMSO) is used as a vehicle control. (c and d) Results are expressed as a mean, error bars indicate s.e.m. (N=5 biological replicates; **P<0.01 and *P<0.05). Quantitative evaluation of tumor volume showed that inhibition of STAT3 leads to partial tumor regrowth in absence of HDAC7. (a and d) The deficiency in tumor-associated vasculature was visualized using FITC-lectin staining and following tumor collection on day 7 of tumor development. For IF the magnification used was: × 200 and × 400. Results are expressed as mean of the tumor size. Error bars indicate s.e.m. (N=10; **P<0.01).
HDAC7 depletion induces the upregulation of tumor suppressor AKAP12
Inspired by our previous data in endothelial cells, showing that tumor suppressor protein AKAP12 is epigenetically regulated by HDAC7 and that AKAP12 can regulate STAT3,24 we hypothesized that in GBM a similar regulatory loop could take place. To test this hypothesis we have verified if AKAP12 protein levels change following HDAC7 silencing. The results shown in the Figure 6a confirmed the strong upregulation of AKAP12 following HDAC7 silencing in GBM cells. The co-silencing of HDAC7 and AKAP12 markedly diminished STAT3 expression levels. Indeed, AKAP12 silencing alone was able to significantly decrease the STAT3 protein and mRNA levels. This demonstrated the strong regulatory role of AKAP12 in controlling STAT3 expression and thus inevitably STAT3 activity (Figure 6a). In line with this observation is the CAM experiment, which critically showed that the co-silencing of HDAC7 and AKAP12 rescued the tumor growth on CAM (Figure 6b). This rescue was specifically related to the reestablishment of angiogenesis in HDAC7 deficient tumors. Indeed, histological data displayed in Figure 6c evidence vasculature reappearance in CAM following the HDAC7/AKAP12 co-silencing and in comparison with HDAC7-silencing condition alone.
AKAP12 overexpression mediated by HDAC7-silencing induces STAT3 activation and angiogenesis inhibition in vivo. (a) (Upper panel) western blot analysis of STAT3 expression and phosphorylation levels following HDAC7 and AKAP12 co-silencing in U87-MG cells. Concomitant depletion of HDAC7 and AKAP12 diminishes STAT3 protein expression. (Lower panel) AKAP12-silencing reduces STAT3 gene expression. (b) Representative macroscopic pictures of U87-MG CAM tumors on HDAC7 and/or AKAP12 silencing. Quantitative evaluation of tumor volume showed a significant decrease in HDAC7-depleted tumors, which is abolished in absence of AKAP12. Results are expressed as means of the tumor sizes. Error bars indicate s.e.m. (N=10; ***P<0.001 and *P<0.05). (c) Histological analysis of U87-MG-collected tumors displayed a decreased vascular density in HDAC7-depleted tumors. From left to right: Hematoxylin–eosin (H&E) staining of paraffin embedded tumors; Sambucus nigra agglutinin (SNA)-lectin staining of tumor-endothelial cells; overlay of SNA-staining with nuclei of U87 cells (red). For H&E and IF the magnification used was: × 200 and × 400 for high-power view only.
Induced HDAC7 silencing in vivo significantly diminishes GBM growth in mouse brain
Considering the in vitro and in vivo CAM data, we next sought to determine the therapeutic value of targeting HDAC7 for inhibiting the GBM growth. To this end we have decided to employ an orthotopic model of GBM in nonobese diabetic-severe combined immunodeficiency (NOD-SCID) mice, with U87-MG cells which were transformed with the inducible short hairpin RNA (shRNA) against HDAC7. Following the orthotopic grafting of GBM cells, the tumors were allowed to develop without treatment for 2 weeks. After this period, the mice were randomized in two groups, one treated with placebo and the second with isopropyl β-d-1-thiogalactopyranoside (IPTG; inducer of shRNA) for 2 weeks. As demonstrated in the Figures 7a and b both by bioluminescence and magnetic resonance imaging, induction of HDAC7 silencing in growing GBM reduced the tumor volume by threefold. Expectedly, the GBM with lowered HDAC7 levels showed a marked decrease in vascular density (CD31 staining, Figure 7c), which was associated with decreased proliferation of glioma cells (Ki67 staining, Figure 7c).
HDAC7-silencing inhibits GBM growth in vivo. (a) Bioluminescence signal of xenografted mice depleted (+IPTG) or not (−IPTG) of HDAC7 accrued at 2 weeks post-treatment using Xenogen-IVIS system. Quantification of bioluminescent signal shows that HDAC7-induced silencing decreases tumor growth in vivo. Western blot analysis shows efficient silencing (−60%) of HDAC7 in U87-shRNA-HDAC7i cells on IPTG treatment. (b) Magnetic resonance imaging of GBM (in situ), coronal view and 3D reconstruction of tumor volume of U87-MG xenografts. The computed tumor volume confirms that HDAC7 silencing significantly decreases GBM growth in vivo. (a and b) Results are expressed as mean of the tumor size. Error bars indicate s.e.m. (N=6; **P<0.01 and *P<0.05). (c) Immunohistochemistry (IHC)-based evaluation of CD31 (tumor-associated vasculature/angiogenesis) and Ki67 expression (proliferation) in GBM from U87-shRNA-HDAC7i cells. Inhibition of HDAC7 (+IPTG) provokes a significant reduction of tumor angiogenesis and proliferation in vivo. Results of IHC quantification are expressed as means and error bars indicate s.e.m. (N=3; **P<0.01 and *P<0.05).
Discussion
STAT3 is one of the most potent transcription factors modulating well over 300 different genes involved in paramount biological processes.29 Intriguingly, STAT3 appears to exhibit both tumor suppressor and promoter functions in several cancers including glioma.16, 17, 30, 31 It has been proposed that this dual activity might be explained, at least in part, by the intervention of specific proteins able to interact and activate JAK-STAT pathway and/or induce post-translational modifications of STAT3.32 In glioma, the expression of STAT3 along with EGFR isoform v3 is considered pro-oncogenic. The two proteins associate in a complex that translocates to the nucleus and promotes tumor growth in vivo.17 The nuclear translocation of STAT3-EGFRv3 complex is further enhanced by additional phosphorylation of EGFRv3 by EGFR.16 This pro-tumorigenic role of STAT3 observed in a majority of gliomas has qualified this transcription factor as a potential therapeutic target.33 However, tumors are heterogeneous entities composed of cells with different molecular background.34 For example, STAT3 has a tumor suppressive role in glioma cells with wild-type PTEN.17 Inhibiting STAT3 in such tumors might trigger rapid resistance. In the current work, we unveiled a mechanism able to reprogram STAT3 into a tumor suppressor regardless of EGFR, TP53 or PTEN status. To our knowledge, this is the first example of reprogramming a known tumor promoter to a tumor suppressor in the genetic environment that actually favors its tumor promoting function. Key to this step is HDAC7, whose depletion in glioma triggers STAT3 upregulation, post-translational modification (acetylation and phosphorylation) and nuclear localization. Interestingly, in sharp contrast to EGFR, TP53 or PTEN, both STAT3 and HDAC7 have very few to no somatic mutations in glioblastoma. We know today only a handful of genes that are not mutated and yet assume a very important function in cancer. An example is NFkB transcription factor family, which is almost never mutated in solid tumors.35 The existence of such genes, that are somewhat protected from mutations in cancer, is indeed surprising. The present data supports the notion that HDAC7 and STAT3 may assume very fundamental functions in the GBM biology and that their mutation is potentially detrimental to the cancer cell. If true, such mutation events are not evolutionary selected, which would explain their absence in tumors. We show here that silencing HDAC7 activates STAT3 that in turn rapidly induces anti-angiogenic genes while suppressing the pro-angiogenic ones. This imbalance of angiogenic factors inhibits angiogenesis and tumor growth in vivo. Mechanistically, HDAC7 induces the expression of JAK1 that in turn phosphorylates STAT3. HDAC7 depletion also leads to STAT3 acetylation. This is not surprising because previous studies have shown that HDAC7, together with acetyl-transferase Tip60, is physically interacting with STAT3.28 The authors hypothesized about how this complex may lead to a modulation of STAT3 acetylation levels, but failed to provide final evidence. We demonstrated previously in endothelial cells that HDAC7 silencing results in STAT3 acetylation.24 Here we show for the first time that HDAC7 depletion causes STAT3 acetylation on a specific lysine residue (K685) in human glioma cells. This modification is known to be critical for the dimerization of this transcription factor and hence its function.27
A further important aspect of the observed anti-tumor phenotype is the activation of tumor suppressor AKAP12. AKAP12 is a key gene ensuring proper angiogenesis during embryonic development of the brain.36 AKAP12 protein functions mainly by scaffolding PKC, PKA and SRC kinases to distinct sub-cellular locations, thus fine-tuning their activity.37, 38 In high-grade glioma, AKAP12 is usually silenced by hyper-methylation of the promoter.39 We found in the current work that this silencing can be reversed very efficiently on HDAC7 depletion. Overexpression of AKAP12 is essential for sustained STAT3 expression both at mRNA and protein level. At present, we are unable to explain how AKAP12 regulates STAT3 mRNA expression and future studies should clarify this link. Thus, silencing of AKAP12 following HDAC7 depletion leads to decrease of STAT3 levels and full regrowth of the tumor in vivo. We were able to prove that inhibiting STAT3 activation (phosphorylation) results in partial rescue of tumor growth. These results suggest that the tumor suppressive action of AKAP12 depends at least in part on activated STAT3 (Figure 8). In addition, STAT3-independent and AKAP12-mediated, mechanism of tumor suppression cannot be excluded and should be examined in further studies. Such investigations are warranted by our findings, showing that the depletion of AKAP12 alone resulted in a major decrease of STAT3 levels while having only a minor effect on the tumor volume (−20%). This rather muted inhibition of tumor growth could result from compensatory tumor proliferation promoted by the removal of the oncogenic suppressor AKAP12.
Schematic overview of the STAT3/HDAC7 reprogramming and function in glioma. In normal astrocytes, when PTEN is functional, STAT3 assumes anti-tumor functions. In GBM PTEN is frequently mutated and inactivated while EGFR is active and particularly oncogenic. In these circumstances STAT3 is pro-tumorigenic. Low activity of JAK1 and tumor suppressor AKAP12 as well as high expression of HDAC7 sustains the pro-tumor role of STAT3. HDAC7 takes a central role in this process by suppressing the expression of JAK1 and AKAP12. It also deacetylates STAT3. Thus, HDAC7 silencing can mediate efficient reprogramming of STAT3 to tumor suppressor. On HDAC7 depletion JAK1 and AKAP12 are overexpressed and STAT3 is acetylated. High levels of JAK1 are responsible for phosphorylation of STAT3 on Y705 residue while AKAP12 upregulates STAT3 mRNA and protein levels. In addition to its role in reprogramming STAT3, AKAP12 is an omnipotent tumor suppressor that probably exerts additional anti-tumor pressure.
Overall, our study highlights HDAC7 as a promising target in a subset of glioblastoma that overexpress this protein along with STAT3. Animal studies conducted with orthotopically xenografted tumors show that even after initial tumor development, the induced inhibition of HDAC7 can rapidly (within 2 weeks) shrink the tumor volume by threefold. Glioblastoma are addicted to angiogenesis. Suppression of HDAC7 inhibits angiogenesis, cutting off cancer cells from oxygen and nutrients. Unfortunately, to date, no specific inhibitors exist for HDAC7, making its targeting extremely difficult.40 Successful translation of these results would therefore depend on the future development of novel targeted therapies aiming at gene silencing or functionally inhibiting intracellular proteins. Some examples of such technologies include targeted delivery of shRNA by viral vectors or use of zinc-finger nucleases for genome editing. Further studies are needed to examine the feasibility of such targeting in the in vivo model of human glioma.
Materials and methods
Patients
In the current study, the Ethics Committee of University Hospital Liege has approved the use of human samples, which were obtained from the institutional biobank (Protocol No.: 2009/69). Patients received the information that the residual material could be used for research purpose and that the consent is presumed as long as the patient does not oppose (opting-out). All 22 patients were characterized histologically as astrocytoma grade IV according to the WHO (World Health Organization) standards. A clinical 10-year follow-up was available for all cases. In addition to the institutional cases, the current study also uses publicly deposited data from 154 GBM cases collected by the TCGA network and made available for analysis through the cBioPortal.20, 41 No power analysis was used to determine the size of the patient cohort. The patients were selected based on material availability and clinical follow-up.
Cell culture
U251-MG and U87-MG human glioma cell lines were obtained from American Type Cell Collection (ATCC) and cultured in Eagle's minimum essential medium (EMEM) with 10% heat-inactivated fetal bovine serum (Life Technologies, Carlsbad, CA, USA). The cells were authenticated through DNA profiling of eight different and highly polymorphic short-tandem repeat loci (DSMZ, Braunschweig, Germany) and were regularly tested for mycoplasma contamination. U87-wtEGFR cells that overexpress wild-type EGFR as well as the U87-ΔEGFR cells that express the mutated and constitutively active EGFRv3 (deletion of exons 2–7 of the EGFR) were both a kind gift from Dr Frank Furnari (Ludwig Institute for Cancer Research, UCSD, San Diego, USA) and were generated as described before.8 The cells were maintained in EMEM with 10% fetal bovine serum. The primary GBM cell cultures (0627 and 0913) were derived from tumor biopsies and were a kind gift from Dr Angelo Vescovi (Università degli Studi Bicocca-Millan, Italy). The full description of the primary GBM is detailed elsewhere.42 The original GBM cells were cultured in Neurocult NS-A medium (Stem Cell, Vancouver, BC, Canada) in the presence of EGF (20 ng/ml) and b-FGF (10 ng/ml) and maintained at 37 °C in a humidified incubator with an atmosphere of 5% CO2. For the purpose of present experiments, the primary GBM cells were differentiated in EMEM with 15% of fetal bovine serum for 2 weeks. Following this, the primary GBM cells were maintained for maximum five passages. HUVEC were isolated from human umbilical cords as described previously.43 The cells were used at early passages (passages I–IV) and grown on a plastic surface coated with porcine gelatin (Sigma-Aldrich, St. Louis, MO, USA), in M199 medium (Life Technologies) supplemented with 20% fetal calf serum (Life Technologies), EC growth factor (100 μg/ml; Sigma-Aldrich) and porcine heparin (100 μg/ml; Sigma-Aldrich).
Antibodies and reagents
Anti-STAT3 (#9139), anti-phospho-STAT3 (Tyr-705; #9138), anti-acetyl-STAT3 (lys-685; #2523), anti-SRC (#2108), anti-JAK1 (#3344) and anti-JAK2 (#3230) antibodies were purchased from Cell Signaling (Carlsbad, CA, USA). Anti-PTEN (#04–035) and anti-P53 (#05–224) antibodies were purchased from Millipore (Bedford, MA, USA). Anti-HSC70 (sc-7298), anti-HDAC7 (sc-11421) and anti-EGFR (sc-03) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-BrdU (B 8434) was purchased from Sigma-Aldrich. Anti-AKAP12 (#A301–401 A) antibody was purchased from Bethyl (Montgomery, TX, USA). Bromodeoxyuridine (BrdU) and anti-PKCα (610107) was purchased from BD Bioscience (Erembodegem, Belgium). S3I-201, a selective STAT3 inhibitor (hereafter referred as inhibitor 6 (STAT3-i6), was purchased from Santa Cruz Biotechnology (sc-204304).
SiRNA transfection
Calcium phosphate-mediated transfections were performed using following small interfering RNA (siRNA) synthesized by Eurogentec (Liège, Belgium): HDAC7 ((#1) 5′-CCU-GAA-GCU-GCG-CUA-UAA-G-3′, (#2) 5′-GGA-CAA-GAG-CAA-GCG-AAG-U-3′ and (#3) 5′-GGC-UGG-AAA-CAG-AAA-CCC-A-3′) and AKAP12 (5′-UCU-GCA-GAA-UCU-CCG-ACU-AUU-3′). The siRNA for JAK1 (SMART pool L-003145-00-0005), JAK2 (SMART pool L-003146-00-0005), SRC (SMART pool L-003175-00-0005) and PKCα (SMART pool L-003523-00-0005) were purchased from GE Healthcare (Buckinghamshire, England, UK). All siRNA were used at a concentration of 40 nm and for 48 h before cell lysis. GL3 luciferase siRNA (5′-CUU-ACG-CUG-AGU-ACU-UCG-A-3′) was used as a negative control (siRNA Irr).
Construction of U87-MG with inducible shRNA HDAC7
U87-MG cells were first transformed using pLenti6-Luc plasmid that was generated by cloning the luciferase gene into the pLenti6/V5-D-Topo vector (K4955-10, Life Technologies). Following selection of the positive clones, the U87-MG-luc+ cells were further transformed using pLKO-shHDAC7i plasmid that was generated by cloning the shRNA sequence in the pLKO-puro-IPTG 3xLacO vector. The shRNA vector (sequence: 5′-CCGGGCTGATCTATGACTCG-GTCATCTCGAGATGACCGAGTCATAGATCAGCTTTTT-3′) was purchased from Sigma (TRCN0000004847). The transformations were conducted using Lentiviral vectors. The vectors were generated by co-transfecting the Lenti-X 293 T cells (Clontech, Mountain View, CA, USA) with pLenti6-Luc or pLKO-shHDAC7i, pSPAX2 (a gift from Didier Trono (Addgene plasmid # 12260)) and VSV-G-encoding plasmids.44 Lentiviral supernatants were collected 48, 72 and 96 h post transfection, filtered and concentrated 100 × by ultracentrifugation. The lentiviral vectors were then titrated with qPCR Lentivirus Titration (Titer) Kit (LV900; Applied Biological Materials, Richmond, BC, Canada) and used to transduce U87-MG cells. Stably transduced U87-MG cells expressing luciferase were isolated and maintained in medium supplemented with 10 μg/ml of Blasticidin (ant-bl-1; Invivogen, San Diego, CA, USA,). Following this, U87-MG-luc+ were transduced with shRNA-HDAC7i lentiviral vectors, selected and maintained in medium supplemented with 10 μg/ml of Puromycin (ant-pr-1; Invivogen) and 10 μg/ml of Blasticidin.
Western blot analysis
Crushed snap-frozen tissue powder (22 patients with GBM) or cells from culture were lysed using SDS buffer (SDS 1%, Tris-HCl 40 mm (pH 7.5), containing protease and phosphatase inhibitor mixture (Roche, Basel, Switzerland)). Equal amounts of proteins were resolved by SDS-polyacrylamide gel electrophoresis. Membranes were probed with primary antibodies (see antibodies and reagents), followed by horseradish peroxidase-conjugated secondary antibodies, and developed by chemiluminescence detection. When necessary, relative density of the bands was calculated using ImageJ software V1.48 (National Institutes of Health (NIH), Bethesda, MA, USA).
Cytoplasmic and nuclear protein extraction
Cells were washed twice with phosphate-buffered saline (PBS), scraped and centrifuged. The pellets were resuspended in 500 μl of cold hypotonic buffer (Hepes-KOH 10 mm (pH 7.4), MgCl2 2 mm, EDTA 0.1 mm, KCl 10 mm, NP40 0.2% and protease inhibitor mixture Complete (Roche)), incubated on ice for 30 s. The lysates were vortexed and centrifuged for 5 min, 4000 g at 4 °C. The supernatants containing cytoplasmic proteins were stored at −80 °C. Next, the pellets were resuspended in 100 μl of cold hypertonic buffer (Hepes-KOH 20 mm (pH 7.4), MgCl2 1.5 mm, EDTA 0.2 mm, NaCl 630 mm, glycerol 25%, dithiothreitol 5 mm and protease inhibitor mixture Complete (Roche), incubated on ice for 45 min and centrifuged for 30 min at 20,000 g at 4 °C.
Quantitative real-time PCR
Total mRNA was isolated from cells using NucleoSpin RNA (Macherey-Nagel, Hoerdt, France) according to the manufacturer’s instructions. First-strand complementary DNA was synthesized using 2 μg of total RNA in 20 μl of reverse transcription (RT) reaction mixture containing 0.2 μg of pd(N)6 random hexamers (GE Healthcare), 2 mm of each deoxynucleotide triphosphate (Eurogentec), 1x first-strand buffer (Tris-HCl 50 mm (pH 8.3), KCl 75 mm, MgCl2 3 mm, dithiothreitol 10 mm and SuperScriptTM II RNase H RT (Life Technologies; 100 units). The reverse transcriptase reaction was performed at 42 °C for 50 min followed by a 15 min inactivation step at 70 °C. Quantitative real-time PCR was performed in triplicate using the ABI Prism 7700 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. HDAC7 primers (Frw: 5′-CTGCATTGGAGGAATGAAGCT-3′; Rev: 5′-CTGGCACAGCGGATGTTTG-3′) and probe (5′-TGTCAGTGTCCACCCCAACCCCA-3′) were purchased from Eurogentec. STAT3 (Frw: 5′-CCCTTGGATTGAGAGTCAAGA-3′; Rev: 5′-AAGCGGCTATACTGCTGGTC-3′; UPL #14), IGFBP3 (Frw: 5′-AACGCTAGTGCCGTCAGC-3′; Rev: 5′-CGGTCTTCCTCCGACTCAC-3′; UPL #1), IL8 (Frw: 5′-GAGCACTCCATAAGGCACAAA-3′; Rev: 5′-ATGGTTCCTTCCGGTGGT-3′; UPL #72), TGFA (Frw: 5′-TTGCTGCCACTCAGAAACAG-3′; Rev: 5′-ATCTGCCACAGTCCACCTG-3′; UPL #63) and FGF1 (Frw: 5′-AGGAGCGACCAGCACAAC-3′; Rev 5′-TGCATGCTTCTTGGATATATAGGT-3′; UPL #19) human specific primers were purchased from Integrated DNA Technology (Coralville, IA, USA) and UPL probes were purchased from Roche. Complementary DNA samples (100 ng each) were mixed with 1 × gene kit and TaqMan Universal PCR Master Mix (Roche) in a total volume of 20 μl. Real-time PCR was conducted using the following parameters: 94 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Acquired data were analyzed by Sequence Detector software version 1.9 (Applied Biosystems).
Enzyme-linked immunosorbent assay
After 48 h of transfection, media of U87-MG were collected, centrifuged at 1000 g to remove any cell material, and the supernatant was stored at −20 °C. The conditioned media were analyzed for human TGFA using commercially available enzyme-linked immunosorbent assay kit (R&D Systems, Minneapolis, MN, USA) according to the manufacturer’s instructions.
EC sprouting and wound repair assay
The conditioned media were obtained from the transfected/treated with STAT3-i6 (80 μm) or wild-type glioblastoma cell lines by starving them (0% serum) for 72 h. The conditioned media were collected, filtered (0.45 μm) and used in the subsequent experiments. For sprouting assay, HUVEC cell spheroid aggregates were prepared in 20% methylcellulose medium, embedded in fibrin gel and stimulated with 50% of conditioned media from transfected glioblastoma cells in the presence of 5% fetal calf serum. Formation of radially growing cell sprouts was observed during the next 24 h, photographed at a × 200 magnification using an Axiovert 200 M microscope and counted. For wound closure test, confluent HUVEC cells were scraped with a 200-μl tip to obtain a 2-mm-thick denuded area and cultured in the presence of 100% conditioned media from transfected or wild-type glioblastoma cells in the presence of 3.5% fetal calf serum. After 24 h, wounded monolayers were photographed and the width of the wounds was measured in three-independent sites per group and the percentage of repair was quantified with ImageJ software.
Immunofluorescence
U87-MG cells were grown on sterilized 12 mm glass coverslips (CB00120RA1, Menzer Gläser, Braunschweig, Germany). The cells were fixed and permeabilized in the ice-cold methanol/acetone mix. The samples were then incubated overnight at 4 °C with primary antibodies against STAT3 phospho-Y705 and JAK1 (same as used for western blot). After incubation with the primary antibodies, the samples were incubated for 1 h with Alexa Fluor 488 and 546 conjugated secondary antibodies (A11029 & A11010, Life Technologies). The counterstaining was performed using the nuclear stain ToPRO (T3605, Life Technologies). Coverslips were mounted onto microscope slides with Fluorescent Mounting Medium (S0323, Dako, Glostrup, Denmark). Images were obtained with a Nikon A1R (Nikon Instruments, Melville, NY, USA) laser scanning confocal microscope. False color attribution and contrast adjustments were done using ImageJ software.
Animal experiments
All the experimental procedures and protocols involving CAM and mouse models were reviewed and approved by the Institutional Animal Care and Ethics Committee of the University of Liège (Belgium; protocol no.: 1590 (CAM) and 1182 (mice)). The animal experimentation adhered to the ‘Guide for the Care and Use of Laboratory Animals’ prepared by the Institute of Laboratory Animal Resources, National Research Council, and published by National Academy Press, as well as to European and local legislation. ARRIVE ethical guidelines were followed.45 NOD-SCID mice were purchased from Janvier Labs (Saint Berthevin Cedex, France) and housed in the animal facility of the University of Liege under standard conditions (12 h light/dark cycle, lights on at 7:00 a.m.). They were acclimated to the room 1 week before the beginning of the experiment. Food and water were provided ad libitum. Five animals for mice and 10 animals for CAM per replicate and condition were estimated as sufficient to account for the biological variability. However, considering the lethality during the experiments, 10 animals for mice and 15 animals for CAM were used (per replicate and condition). For the treatment, no specific inclusion/exclusion criteria were used and the animals were randomized based on simple randomization technique.46 The investigator was not blinded during the study.
Chorioallantoic membrane assay
At day 3 post-fertilization, 8 ml of albumin was removed from the egg and a window was cut in the eggshell exposing the CAM. Eggs were then sealed with Durapore tape (3 M, Diagem, Belgium) and kept closed at 37 °C (80% humidity) until implantation of the cells. U87 cells were transfected with 90 nm siRNA (HDAC7 siRNA#1 was used), subjected to medium change at 16 h post transfection and were implanted on CAM 4 h later. All implantations were conducted at day 11 post-fertilization. For this purpose, 5 × 106 human U87-MG cells were suspended in 20 μl of EMEM. STAT3-i6 (dissolved in pure dimethyl sulfoxide) was diluted at a final concentration of 80 μm in the cell suspension. For the control condition an equivalent volume of dimethyl sulfoxide was used. This cells suspension was deposited within a plastic ring placed on the CAM, after prior gentle laceration of the membrane. Tumors were allowed to develop for an additional 7 days. At day 18, the tumors were collected and tumor volume was estimated using calipers. Tumor volume was calculated assuming an ellipsoid shape and using the formula V=4/3.π.((l.w.h))/8), where l, w, h are the length, width and height of the tumor. Following volume measurement, the tumors were collected and subjected to histology analysis as described below.
Orthotopic model of GBM in mouse
40,000 cells/μl suspended in PBS were injected in the right striatum of 6-week-old male NOD-SCID mice. Two weeks following the tumor engraftment, silencing of HDAC7 was induced by IPTG (Cat. no. I6758-5; Sigma-Aldrich) that was diluted in drinking water at a concentration of 10 mm. In addition, mice were treated with a daily intraperitoneal injection of 200 μl 20 mm IPTG solution. Tumor growth was monitored using bioluminescence on an IVIS 200 imaging system (Xenogen-Caliper, Hopkinton, MA, USA) after luciferine injection (Cat. no. E1605; Promega, Fitchburg, WI, USA); Intraperitoneal injection of 50 μl of 0.1 mm luciferine solution in PBS, signal acquisition at 12 min post injection. Magnetic resonance imaging was performed on a 9.4T Biospec (Bruker, Ettlingen, Germany) using a volume coil (23 mm diameter). Animals were kept anesthetized with 1–1.5% isoflurane delivered in oxygen (LA no. 1500589, protocol no. CMMI-2011–07). Their respiratory rate was monitored during the entire imaging session. T2-weighted TurboRARE (TR: 2500 ms, TE: 33 ms) and T1-weighted RARE (TR: 467.8 ms, TE: 8.5 ms) pre and 3 min post-contrast agent injection were the sequences used to visualize the brain on 20 slices (0.8 mm thickness). The contrast agent (Dotarem from Guerbet, Aulnay-sous-Bois, France) was injected intravenously (0.1 mmol/kg) to improve tumor conspicuity on T1-weighted acquisitions. Tumors were manually contoured and the volumes were calculated with the ROI tool of the Osirix software (version 5.6, Pixmeo, Bernex, Switzerland). Four weeks post-xenografting, mice were killed and the brains containing GBM were prepared for histology analysis as described below.
Histology analysis
Mice orthotopic model
euthanized mice were perfused first with saline (0.9% NaCl) and then with 4% paraformaldehyde solution. Brains were removed and further fixed in paraformaldehyde 4% for 2 h and then embedded in paraffin. Paraffin tissue blocks were cut with the microtome in 5-μm-thick sections and then subjected to xylene bath and rehydratation steps in a series of graded methanol dilutions. The sections were washed in PBS with 0.25% triton X-100 and boiled in citrate buffer (0.342 g citric acid and 2.378 g sodium citrate in 1 l distilled water, pH 6) using 95 °C water bath for 40 min. The sections were blocked for 30 min in PBS-normal goat serum solution (150 μl normal goat serum and 20 μl Tween 20 (Sigma-Aldrich) in 10 ml PBS) and incubated with the primary antibodies at 4 °C overnight. Following antibodies were used: CD31 (clone JC70A; Dako) and Ki67 (clone MIB-1; Dako). Following this, the slides were washed once with the blocking solution and the sections were incubated in biotinylated anti-rabbit IgG (dilution 1:500) for 30 min. Subsequently, the sections were washed in PBS and incubated in avidin-biotin complex kit (Vector Labs, Peterborough, UK) for 30 min. Finally, the tissue sections were stained with 3,3′-diaminobenzidine (2 mg 3,3′-diaminobenzidine and 5 μl H2O2 in 5 mL PBS) and counter-stained in hematoxylin. Pictures of representative fields were taken under a light microscope Leica DMRB (Leica, Diegem, Belgium). The microscopy pictures of the stained sections (× 100 magnification) were automatically evaluated for staining extension (CD31) and percent-positive nuclei (Ki67) using ImageJ software.
CAM model
The collected tumors were fixed in a 4% paraformaldehyde for 30 min and then embedded in paraffin. Paraffin tissue blocks were first cut in 5-μm-thick sections and then subjected to xylene bath and rehydratation steps in a series of graded methanol dilutions (100–95-70- and 50%). Following this the samples were further incubated for 30 min in the dark with 0.05% Triton X-100 in PBS containing Sambucus nigra agglutinin (4 μg/ml). The sections were washed with 0.05% Triton X-100 in PBS, counter-stained with ToPRO (Molecular Probes) and visualized with fluorescence microscope (EVOS, Thermo Fisher Scientific, Grand Island, NY, USA) or confocal microscope (only high-power view; A1R, Nikon Instruments).
Statistical analysis
Unless otherwise indicated, statistical analysis was performed using a two-sided, unpaired Student’s t-test, assuming equal variances using GraphPadPrism (GraphPad Software, Inc., La Jolla, CA, USA; version 5.01). The t-test was used because data followed a normal distribution (Shapiro-Wilk test, threshold 0.05). All results were reported as means with s.e.m. The number of biological replicates is described in the figure legends. Pearson correlation clustering of protein expression patterns in GBM patients was conducted using the Multi Experiment Viewer software (version 4.8.1, Dana-Farber Cancer Institute, Boston, MA, USA).47 For this purpose western blot results for individual patients and proteins were quantified and normalized against the HSC70.
References
Lund-Johansen M, Bjerkvig R, Humphrey PA, Bigner SH, Bigner DD, Laerum OD . Effect of epidermal growth factor on glioma cell growth, migration, and invasion in vitro. Cancer Res 1990; 50: 6039–6044.
Lokker NA, Sullivan CM, Hollenbach SJ, Israel MA, Giese NA . Platelet-derived growth factor (PDGF) autocrine signaling regulates survival and mitogenic pathways in glioblastoma cells: evidence that the novel PDGF-C and PDGF-D ligands may play a role in the development of brain tumors. Cancer Res 2002; 62: 3729–3735.
Talasila KM, Soentgerath A, Euskirchen P, Rosland GV, Wang J, Huszthy PC et al. EGFR wild-type amplification and activation promote invasion and development of glioblastoma independent of angiogenesis. Acta Neuropathol 2013; 125: 683–698.
Frattini V, Trifonov V, Chan JM, Castano A, Lia M, Abate F et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet 2013; 45: 1141–1149.
De Witt Hamer PC . Small molecule kinase inhibitors in glioblastoma: a systematic review of clinical studies. Neuro Oncol 2010; 12: 304–316.
Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci USA 2012; 109: 3041–3046.
Inda MM, Bonavia R, Seoane J . Glioblastoma multiforme: a look inside its heterogeneous nature. Cancers 2014; 6: 226–239.
Inda MM, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev 2010; 24: 1731–1745.
Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci USA 2005; 102: 5814–5819.
Little SE, Popov S, Jury A, Bax DA, Doey L, Al-Sarraj S et al. Receptor tyrosine kinase genes amplified in glioblastoma exhibit a mutual exclusivity in variable proportions reflective of individual tumor heterogeneity. Cancer Res 2012; 72: 1614–1620.
Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell 2011; 20: 810–817.
Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP . Pten is essential for embryonic development and tumour suppression. Nat Genet 1998; 19: 348–355.
Monk M, Holding C . Human embryonic genes re-expressed in cancer cells. Oncogene 2001; 20: 8085–8091.
Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I et al. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science 1997; 278: 477–483.
de la Iglesia N, Konopka G, Lim KL, Nutt CL, Bromberg JF, Frank DA et al. Deregulation of a STAT3-interleukin 8 signaling pathway promotes human glioblastoma cell proliferation and invasiveness. J Neurosci 2008; 28: 5870–5878.
Fan QW, Cheng CK, Gustafson WC, Charron E, Zipper P, Wong RA et al. EGFR phosphorylates tumor-derived EGFRvIII driving STAT3/5 and progression in glioblastoma. Cancer Cell 2013; 24: 438–449.
de la Iglesia N, Konopka G, Puram SV, Chan JA, Bachoo RM, You MJ et al. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev 2008; 22: 449–462.
Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 1997; 275: 1943–1947.
Hollander MC, Blumenthal GM, Dennis PA . PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer 2011; 11: 289–301.
Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR et al. The somatic genomic landscape of glioblastoma. Cell 2013; 155: 462–477.
Turtoi A, Peixoto P, Castronovo V, Bellahcène A . Histone deacetylases and cancer-associated angiogenesis: current understanding of the biology and clinical perspectives. Crit Rev Oncog 2015; 20: 119–137.
Chang S, Young BD, Li S, Qi X, Richardson JA, Olson EN . Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell 2006; 126: 321–334.
Mottet D, Bellahcène A, Pirotte S, Waltregny D, Deroanne C, Lamour V et al. Histone deacetylase 7 silencing alters endothelial cell migration, a key step in angiogenesis. Circ Res 2007; 101: 1237–1246.
Turtoi A, Mottet D, Matheus N, Dumont B, Peixoto P, Hennequiere V et al. The angiogenesis suppressor gene AKAP12 is under the epigenetic control of HDAC7 in endothelial cells. Angiogenesis 2012; 15: 543–554.
Bezecny P . Histone deacetylase inhibitors in glioblastoma: pre-clinical and clinical experience. Med Oncol 2014; 31: 985.
Xu WS, Parmigiani RB, Marks PA . Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene 2007; 26: 5541–5552.
Yuan ZL, Guan YJ, Chatterjee D, Chin YE . Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science 2005; 307: 269–273.
Xiao H, Chung J, Kao HY, Yang YC . Tip60 is a co-repressor for STAT3. J Biol Chem 2003; 278: 11197–11204.
Dauer DJ, Ferraro B, Song L, Yu B, Mora L, Buettner R et al. Stat3 regulates genes common to both wound healing and cancer. Oncogene 2005; 24: 3397–3408.
Wheeler SE, Suzuki S, Thomas SM, Sen M, Leeman-Neill RJ, Chiosea SI et al. Epidermal growth factor receptor variant III mediates head and neck cancer cell invasion via STAT3 activation. Oncogene 2010; 29: 5135–5145.
Schneller D, Machat G, Sousek A, Proell V, van Zijl F, Zulehner G et al. p19(ARF) /p14(ARF) controls oncogenic functions of signal transducer and activator of transcription 3 in hepatocellular carcinoma. Hepatology 2011; 54: 164–172.
Yu H, Lee H, Herrmann A, Buettner R, Jove R . Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer 2014; 14: 736–746.
Atkinson GP, Nozell SE, Benveniste ET . NF-kappa Band STAT3 signaling in glioma: targets for future therapies. Expert Rev Neurother 2010; 10: 575–586.
Turtoi A, Blomme A, Castronovo V . Intratumoral heterogeneity and consequences for targeted therapies. Bull Cancer 2015; 102: 17–23.
DiDonato JA, Mercurio F, Karin M . NF-κB and the link between inflammation and cancer. Immunol Rev 2012; 246: 379–400.
Lee SW, Kim WJ, Choi YK, Song HS, Son MJ, Gelman IH et al. SSeCKS regulates angiogenesis and tight junction formation in blood-brain barrier. Nat Med 2003; 9: 900–906.
Gelman IH . Suppression of tumor and metastasis progression through the scaffolding functions of SSeCKS/Gravin/AKAP12. Cancer Metastasis Rev 2012; 31: 493–500.
Su B, Gao L, Meng F, Guo LW, Rothschild J, Gelman IH . Adhesion-mediated cytoskeletal remodeling is controlled by the direct scaffolding of Src from FAK complexes to lipid rafts by SSeCKS/AKAP12. Oncogene 2013; 32: 2016–2026.
Goeppert B, Schmidt CR, Geiselhart L, Dutruel C, Capper D, Renner M et al. Differential expression of the tumor suppressor A-kinase anchor protein 12 in human diffuse and pilocytic astrocytomas is regulated by promoter methylation. J Neuropathol Exp Neurol 2013; 72: 933–941.
West AC, Johnstone RW . New and emerging HDAC inhibitors for cancer treatment. J Clin Invest 2014; 124: 30–39.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404.
Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 2004; 64: 7011–7021.
Jaffe EA, Nachman RL, Becker CG, Minick CR . Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J Clin Invest 1973; 52: 2745–2756.
Emi N, Friedmann T, Yee JK . Pseudotype formation of murine leukemia virus with the G protein of vesicular stomatitis virus. J Virol 1991; 65: 1202–1207.
Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG . Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol 2010; 8: e1000412.
Suresh K . An overview of randomization techniques: an unbiased assessment of outcome in clinical research. J Hum Reprod Sci 2011; 4: 8–11.
Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA et al. TM4 microarray software suite. Methods Enzymol 2006; 411: 134–193.
Acknowledgements
The authors acknowledge the experimental support of Dr Chantal Humblet and Ms Alice Marquet (GIGA-histology platform, ULg), Dr Sandra Ormenese (GIGA-imaging platform, ULg) as well as of Evgenia Turtoi (Metastasis Research Laboratory) for the immunohistochemistry analysis and of Naima Maloujahmoum (Metastasis Research Laboratory) for the cell culture. The authors are also thankful to Dr Stephanie Gofflot from the institutional Biobank of the University Hospital Liege for providing GBM fresh tissues and to Dr Gilles Doumont (group of Prof Serge Goldman, Université libre de Bruxelles) for help with animal experiments. The authors are grateful to Ms. Ana Turtoi for proofreading the manuscript. The results shown in this work are in part based on data generated by the TCGA Research Network: http://cancergenome.nih.gov/. This work was supported with grants from the University of Liège (Concerted Research Action Program (IDEA project)) and from the National Fund for Scientific Research (FNRS/TELEVIE). Andrei Turtoi is a post-doctoral research fellow (FNRS/TELEVIE) and Akeila Bellahcène is a senior research associate (FNRS). Arnaud Blomme is a post-doctoral research fellow (FNRS/TELEVIE). The Center for Microscopy and Molecular Imaging (CMMI) is supported by the European Regional Development Fund and Wallonia. No funding bodies had any role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Peixoto, P., Blomme, A., Costanza, B. et al. HDAC7 inhibition resets STAT3 tumorigenic activity in human glioblastoma independently of EGFR and PTEN: new opportunities for selected targeted therapies. Oncogene 35, 4481–4494 (2016). https://doi.org/10.1038/onc.2015.506
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.506
- Springer Nature Limited
This article is cited by
-
Bioinformatic identification and experiment validation reveal 6 hub genes, promising diagnostic and therapeutic targets for Alzheimer’s disease
BMC Medical Genomics (2024)
-
AKAP12 promotes cancer stem cell-like phenotypes and activates STAT3 in colorectal cancer
Clinical and Translational Oncology (2023)
-
MiR-489 inhibited the development of gastric cancer via regulating HDAC7 and PI3K/AKT pathway
World Journal of Surgical Oncology (2020)
-
HDAC7 promotes the oncogenicity of nasopharyngeal carcinoma cells by miR-4465-EphA2 signaling axis
Cell Death & Disease (2020)
-
ZNF326 promotes malignant phenotype of glioma by up-regulating HDAC7 expression and activating Wnt pathway
Journal of Experimental & Clinical Cancer Research (2019)