Abstract
Signal transducer and activator of transcription 3 (STAT3) is constitutively activated in malignant tumors and has important roles in multiple aspects of cancer aggressiveness. Thus targeting STAT3 promises to be an attractive strategy for treatment of advanced metastatic tumors. Although many STAT3 inhibitors targeting the SH2 domain have been reported, few have moved into clinical trials. Targeting the DNA-binding domain (DBD) of STAT3, however, has been avoided due to its ‘undruggable‘ nature and potentially limited selectivity. In a previous study, we reported an improved in silico approach targeting the DBD of STAT3 that resulted in a small-molecule STAT3 inhibitor (inS3-54). Further studies, however, showed that inS3-54 has off-target effect although it is selective to STAT3 over STAT1. In this study, we describe an extensive structure and activity-guided hit optimization and mechanistic characterization effort, which led to identification of an improved lead compound (inS3-54A18) with increased specificity and pharmacological properties. InS3-54A18 not only binds directly to the DBD and inhibits the DNA-binding activity of STAT3 both in vitro and in situ but also effectively inhibits the constitutive and interleukin-6-stimulated expression of STAT3 downstream target genes. InS3-54A18 is completely soluble in an oral formulation and effectively inhibits lung xenograft tumor growth and metastasis with little adverse effect on animals. Thus inS3-54A18 may serve as a potential candidate for further development as anticancer therapeutics targeting the DBD of human STAT3 and DBD of transcription factors may not be ‘undruggable‘ as previously thought.
Similar content being viewed by others
Introduction
Upon stimulation by cytokines and growth factors, signal transducer and activator of transcription 3 (STAT3) is phosphorylated at tyrosine residue 705 (Tyr705), leading to dimerization, nuclear entry, binding to specific elements in genomic DNA and activation of gene transcription.1 In contrast to the transient and strictly controlled activation of STAT3 in normal cells, overexpression and/or constitutive activation of STAT3 is frequently observed in many human cancers.2 Aberrant STAT3 signaling contributes to oncogenic transformation by upregulating its downstream target genes important for proliferation, survival, migration, invasion, angiogenesis and immune evasion.3, 4, 5, 6 Analysis of clinical specimens revealed that persistent STAT3 signaling correlates with enhanced tumor progression.7, 8, 9 Subcutaneous injection of cells harboring constitutively activated STAT3 resulted in tumor formation10 and inhibition of STAT3 expression using antisense oligonucleotides significantly impaired the growth of human and mouse nucleophosmin-anaplastic lymphoma kinase tumors in xenograft models.11 Therefore, STAT3 represents an attractive target for the development of novel antineoplastic drugs.
Indeed, various STAT3 inhibitors have been identified over the past 15 years,2, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 with some inhibiting xenograft tumor growth in animal models. However, few have moved into clinical trials and none is approved for clinical use. The reasons for the failures are currently unknown. It is noteworthy that the common feature of many of these inhibitors is that they are designed to inhibit the binding of SH2 domain to pTyr705 residue for dimerization or to inhibit phosphorylation of Tyr705 for activation. This approach is problematic because unphosphorylated STAT3 can bind to DNA and may still be functional.26, 27, 28 Thus disrupting the interactions between SH2 domain and pTyr705 of STAT3 or STAT3 activation alone may not completely inhibit STAT3.
Although targeting the DNA-binding domain (DBD) of STAT3 may be more effective to eliminate STAT3 signaling, it has not been in the main stream of research perhaps due to the belief that targeting DBD of transcription factors has potentially limited selectivity.29, 30 Hence, DBDs are considered ‘undruggable‘. In a previous study, we reported an improved in silico approach targeting the DBD of STAT3 and identified a small-molecule inhibitor (inS3-54), which is selective to STAT3 over STAT1.31 However, further study showed that it has off-target effects. In the present study, we performed an extensive structure and activity-guided hit optimization and mechanistic study and identified an improved lead compound (inS3-54A18) with increased specificity and pharmacological properties. We also showed that inS3-54A18 indeed binds directly to the DBD of STAT3. Thus inS3-54A18 may serve as a potential lead candidate for further development of anticancer therapeutics by targeting the DBD of STAT3 to eliminate STAT3 signaling.
Results
Identification of active analogs of inS3-54 targeting the DBD of STAT3
To further investigate the feasibility of targeting DBD of STAT3 and identify inhibitors with improved potency, selectivity and pharmacological properties, we first searched the virtual Chemdiv database for inS3-54 analogs with a criterion of 80% structural similarity using the Chemfinder module in Chemoffice 8.0. In total, 79 commercially available analogs (see Supplementary Table S1) were acquired and tested for their activity in suppressing STAT3-dependent luciferase expression in comparison to inS3-54 and dimethyl sulfoxide (DMSO) vehicle controls. Figure 1a shows that 15 analogs (A1, A13, A18, A20, A26, A27, A36, A41, A45, A51, A68, A69, A72, A75 and A79) significantly inhibited STAT3-dependent luciferase expression compared with DMSO vehicle control. Among these 15 analogs, 3 (A18, A26 and A69) appear to be more potent than the original compound inS3-54.
STAT3-dependent luciferase reporter assay of inS3-54 analogs. (a) Effects of inS3-54 analogs on STAT3-dependent luciferase activity. MDA-MB-231 cells with stable STAT3-dependent luciferase expression were treated with DMSO control, 20 μM inS3-54 or its analogs (A1–A79), followed by measurement of luciferase activity. (*P<0.05; **P<0.01; ***P<0.001, n=3). (b) Core structure of inS3-54 analogs and representative active and inactive R1 and R2 side groups.
Structural and activity analysis of these analogs revealed some potential active and inactive side R1 and R2 groups with a 5-phenyl-1H-pyrrol-2(3H)-ketone core structure (Figure 2b). For example, it appears that compounds with R1 being p-hydroxybenzene or p-carboxybenzene have high STAT3-inhibitory activity while compounds with R1 being m-hydroxybenzene or m-carboxybenzene have low or no activity. When the R2 group is nitrobenzene, p-chlorobenzene or benzenamine, the compounds appear to have higher STAT3-inhibitory activity than the compounds with R2 being p-methoxylbenzene. Clearly, both R1 and R2 groups have important roles in inhibiting STAT3 activity.
STAT3 selectivity and specificity of active analogs A18, A26 and A69. (a, b) Effects of A18, A26 and A69 on colony formation of hematopoietic progenitor cells. Hematopoietic progenitor cells isolated from STAT3+/+ (a) and STAT3−/− (b) mice were subjected to colony-formation assay in the presence of DMSO control or 20 μM inS3-54, A18, A26 or A69 (*P<0.001 compared with DMSO group of each cell type). CFU-GEMM, colony-forming unit-granulocyte erthyroid macrophage megakaryocyte (multipotential progenitor cell); CFU-GM, colony-forming unit-granulocyte macrophage (granulocyte macrophage progenitor cell); BFU-E, burst-forming unit-erthyroid (erthyroid progenitor cell). (c) Effects of A18, A26 and A69 on in vitro DNA-binding activity of STAT3 and STAT1. Whole cell lysate of H1299 cells with transient expression of FLAG-STAT3 or STAT1 were pretreated without or with excess cold probe, antibody or different concentrations of active analogs A18, A26 and A69 followed by incubation with [32P]-labeled probe and analysis on non-denaturing PAGE. (d) Relative DNA-binding activity of STAT3 and STAT1 in the presence of different concentrations of A18, A26 and A69 derived from three experiments.
Validation of active analogs
Next we validated A18, A26 and A69 using resynthesized compounds in the STAT3-dependent luciferase reporter assay. As shown in Supplementary Figures S1A and B, the resynthesized A18, A26 and A69 all suppressed STAT3-dependent luciferase expression in dose- and time-dependent manners with IC50 of 8.8–12.6 μM and t1/2 of 12.7–49.9 h. It appears that A18 and A26 have significant lower IC50 and thus may be slightly more potent than inS3-54. Although the IC50 of A69 did not change from that of inS3-54, its t1/2 is significantly lower than that of inS3-54. Similar to inS3-54, none of these three analogs inhibited the reporter expression driven by a p27 promoter containing no STAT3-binding site (Supplementary Figure S1C), suggesting that the inhibition of reporter expression by these compounds is unlikely due to non-specific effect on the reporter gene or due to cell death induced by the compounds.
Selectivity and specificity of active inS3-54 analogs
To determine the specificity of A18, A26 and A69, we performed colony-formation assay of hematopoietic progenitor cells isolated from bone marrows of wild-type (STAT3+/+) and bone marrow-specific STAT3 knockout (STAT3−/−) mice.32 Recently, it has been shown that STAT3 knockout in bone marrow reduces 50–70% colony-formation activity of granulocyte macrophage, erythroid and multi-potential hematopoietic progenitor cells.33 We reasoned that a STAT3-specific inhibitor should not further reduce the proliferative activity of STAT3−/− hematopoietic progenitor cells due to lack of STAT3 in these cells. As shown in Figure 2a, inS3-54 and the three analogs reduced ~40–80% colony-formation efficiency of hematopoietic progenitor cells from STAT3+/+ mice, consistent with the effects of STAT3 knockout.33 However, inS3-54 further reduced the colony formation of the cells from STAT3−/− mice, consistent with the finding that inS3-54 inhibits colony formation of wild-type cells more than its analogs (Figure 2b). This finding suggests that inS3-54 may not be specific to STAT3. Interestingly, none of the active analogs had any significant effect on the colony formation of hematopoietic progenitor cells from STAT3−/− mice (Figure 2b), suggesting that these analogs unlikely inhibit other targets important for proliferation of hematopoietic progenitor cells. These findings indicate that the analogs A18, A26 and 69 are more specific to STAT3 than the parent inS3-54.
To confirm the STAT3 specificity of these analogs, we performed electrophoretic mobility shift assay (EMSA) using [32P]-labeled sis-inducible element (SIE) probe and H1299 cells transiently transfected with FLAG-STAT3 or STAT1. As shown in Figure 2c, the specific binding of DNA probe to STAT3 was demonstrated using super-shift and competition analyses. A18, A26 and A69 all inhibited the DNA-binding activity of STAT3 in a dose-dependent manner although A18 and A69 appear to be more effective than A26 (Figures 2c and d). On the other hand, the specific binding of DNA probe to STAT1 as shown by interference of binding using cold probe and STAT1 antibody that is known to interfere DNA-binding activity of STAT1(refs 28, 29) was not affected by any of these compounds at concentrations up to 100 μM. Thus A18, A26 and A69 all selectively inhibit the DNA-binding activity of STAT3 over STAT1.
InS3-54 analogs bind to STAT3
To determine whether the analogs (A18, A26 and A69) bind to STAT3, we took advantage of A26, which contains a secondary amine group, and conjugated it to CNBr-activated Sepharose 4B. Successful conjugation of A26 to CNBr-activated Sepharose was confirmed by the change in color of the beads due to the intrinsic yellow color of A26. The A26-conjugated beads were then used to pull down STAT3 from the lysates of Flag-STAT3-transfected H1299 cells followed by western blotting analysis as described previously.31 Figure 3a shows the expression of Flag-STAT3. Figure 3b shows that the A26-conjugated beads successfully pull down STAT3, whereas the vehicle control beads or the beads conjugated with an irrelevant compound do not. Furthermore, pretreatment of the cell lysate using excess free A26 but not the irrelevant compound inhibited the pull down of STAT3 by A26-conjugated beads (Figure 3c). To determine whether A18 and A69 also bind to STAT3, we performed a similar competition analysis due to lack of usable group for immobilization of A18 and A26. As shown in Figure 3c, excess amount of both A18 and A69 completely inhibited STAT3 binding to A26-conjugated beads. Thus, we conclude that A18, A26 and A69 all bind to STAT3.
Binding of inS3-54 analogs to STAT3. (a) Western blotting analysis of STAT3 from total lysate of H1299 cells transfected with vector control (VC) or FLAG-STAT3 (F-STAT3). (b) Pull-down assay of F-STAT3 from total lysate of H1299 cells using Sepharose 4B-immobilized vehicle control, A26 or irrelevant negative control compound (IC). (c, d) Competition of STAT3 binding to immobilized A26 by excess free A26, A18, A69, DMSO vehicle control or an irrelevant negative control compound (IC). (d) Schematic domain structures of STAT3 and of recombinant proteins. (e) Pull-down assay of purified recombinant STAT3 proteins with different deletions by immobilized A26. The doublet of ΔSH2-C protein may be due to degradation.
A26 binds to the DBD of STAT3
Although the parent compound inS3-54 was identified from in silico screening by targeting the DBD of STAT3(ref. 31) and its active analogs also bind to STAT3 as shown above using pull-down assay, it is unknown whether inS3-54 and its active analogs bind directly to DBD of STAT3. To show the direct binding and to map the binding domain in STAT3, we performed pull-down assays of purified recombinant STAT3 protein with different domains (Figure 3d) using A26-conjugated beads. As shown in Figure 3e, purified STAT3 proteins lacking carboxyl terminal domains, including transactivation domain (ΔTAD) and both TAD and SH2 domains (ΔSH2), were successfully pulled down by A26-conjugated beads. However, further deletion from carboxyl terminus including DBD eliminated the pull down by A26-conjugated beads. These findings suggest that A26 can directly bind to STAT3 and its binding site in STAT3 is likely located in DBD as anticipated.
InS3-54 analogs inhibit cancer cell survival by inducing apoptosis
To determine the effectiveness of the analogs A18, A26 and A69 in suppressing cancer cell survival and to determine the potential therapeutic window, we performed sulforhodamine B assay of these compounds using cancer cells of lung (A549 and H1299) and breast (MDA-MB-231 and MDA-MB-468) as well as a non-cancerous mammary epithelial cell line (MCF10A1) and lung fibroblast cell (IMR90) in comparison with the parent compound inS3-54. Figure 4a shows that the IC50 of these compounds range from 1.8 to 5.6 μM for cancer cells and form 4.0 to 12.0 μM for non-cancerous cells. The in vitro therapeutic window for inS3-54, A18 and A69 has a range of ~2–5 fold (Supplementary Figure S2). However, A26 appears to be toxic to non-cancerous lung fibroblast with an IC50 of 4.0 μM and a small in vitro therapeutic window for lung cancer cells.
Effect of inS3-54 and its active analogs on cancer cell survival. (a) IC50 of inS3-54 and its active analogs (A18, A26 and A69) in different cell lines derived from dose–response survival curves. (b) Effect of A18 and A69 on apoptosis of exponentially growing A549 and MDA-MB-231 cells. (*P<0.05; **P<0.01; ***P<0.001, n=3–4).
To determine whether apoptosis contributes to analog-induced loss of cancer cell viability, we performed enzyme-linked immunosorbent assay (ELISA) analysis to determine and quantify cytoplasmic histone-associated DNA fragments that are released from apoptotic cells following treatment with A18 or A69 for 72 h. As shown in Figure 4b, remarkable induction of apoptosis was observed in both lung and breast cancer cells following treatment by A18 or A69.
Selection of A18 as a potential lead for further investigation
Based on above studies, it is clear that the parent compound inS3-54 is less specific to STAT3 than its analogs A18, A26 and A69. A26 is less effective than A18 and A69 in EMSA assay and exerted a high level of cytotoxicity to non-cancerous lung fibroblasts. It was thus eliminated from further study. In acute in vivo toxicity studies, A69 performed poorly and caused death of mice at 0.5 mg/kg while inS3-54 could not be absorbed. Furthermore, inS3-54, A26 and A69 all have poor solubility. On the other hand, A18 was completely soluble in a commercial oral formulation (Pharmatek Laboratories, San Diego, CA, USA) for in vivo studies. It has little cytotoxicity to non-cancerous cells and can be tolerated by mice up to 200 mg/kg with multiple dosing. Therefore, A18 was selected as a potential lead for further investigation.
A18 inhibits cancer cell migration and invasion
To further characterize A18, we assessed A18 effects on cancer cell migration using wound-healing assay and invasion using Matrigel invasion assay. As shown in Figures 5a and b, at 24 h, 71% and 99% of wounds were healed in control-treated A549 and MDA-MB-231 cells, respectively. Wound healing was reduced to 64% and 76% by 5 μM A18 for A549 and MDA-MB-231 cells, respectively. A18 at 10 μM further reduced the healing to 47% and 39%, respectively.
A18 inhibits cancer cell migration and invasion. (a, b) Effect of A18 on migration. Panel (b) shows quantification analysis of wound-healing assay from triplicate measurements of three independent experiments shown in panel (a). (c, d) Effect of A18 on cell invasion. Panel (d) shows quantification of invasion from measurement of 10 random views each of three independent experiments shown in panel (c). (*P<0.05; **P<0.01; ***P<0.001, n=3).
Figures 5c and d show that treatment with 5 μM A18 for 6 h reduced invasion to 66% and 51% for A549 and MDA-MB-231 cells, respectively. Treatment with 10 μM A18 for 6 h further reduced invasion to 35% and 13%, respectively. Treatment for 24 h did not further reduce cell invasion. Although 100% confluent cells were used and the inhibition of invasion was observed at 6 h of treatment, the potential of contribution to reduced invasion by inhibition of proliferation and induction of apoptosis cannot be eliminated. To test if this is the case, we analyzed cell proliferation and apoptosis under the same condition as wound-healing and Matrigel invasion assays following A18 treatment for 6 and 24 h. Supplementary Figure S3 shows that treatment with 5 and 10 μM A18 for 6 or 24 h has no significant effect on proliferation and apoptosis of confluent A549 cells. Although A18 treatment for 24 h reduced <20% proliferation of MDA-MB-231 cells, it did not induce any apoptosis. Thus, A18 inhibition of cancer cell migration and invasion may not be due to its effect on apoptosis and cell proliferation.
A18 inhibits the expression of STAT3 downstream target genes and the binding of STAT3 to its endogenous target sequences
To investigate the potential effect of A18 on the expression of STAT3 downstream target genes and thereby validate its inhibitory effect on STAT3 in cells, we performed western blotting analysis of cyclin D1, survivin, matrix metalloproteinase-9 and vascular endothelial growth factor (VEGF) following A18 treatment of A549 and MDA-MB-231 cells. As shown in Figure 6a, the expression of all these genes was decreased in both cell lines following A18 treatment. A18, however, had no effect on the level of total and Tyr705-phosphorylated STAT3, indicating that it does not inhibit STAT3 activation.
A18 inhibits the expression of STAT3 downstream target genes and STAT3 binding to chromatin. (a) Effect of A18 on constitutive expression of STAT3 downstream target genes in A549 and MDA-MB-231 cells. (b) A18 inhibition of IL-6 (25 ng/ml)-stimulated STAT3 activation in serum-starved A549 cells. (c, d) A18 inhibition of STAT3 binding to chromatin in situ as determined using subcellular fractionation and western blotting analysis (c) or ChIP assay of cyclin D1 promoter (d).
To further determine the STAT3-inhibitory activity of A18, we tested whether A18 can inhibit cytokine-induced STAT3 activation and expression of its downstream genes. For this purpose, A549 cells were serum starved and treated with DMSO vehicle or 10 μM A18 followed by interleukin (IL)-6 stimulation and western blotting analysis of phospho-STAT3 (Tyr705) and survivin. As shown in Figure 6b, IL-6 induced activation of STAT3 and expression of survivin in serum-starved cells, and A18 pretreatment did not inhibit IL-6-induced STAT3 activation. However, IL-6-induced survivin expression was inhibited by A18. These data, together with that shown in Figure 6a, suggest that A18 does not affect the constitutive or IL-6-induced STAT3 activation (Tyr705 phosphorylation) but represses STAT3 target gene (for example, survivin) expression.
As shown in Figure 2, A18 along with A26 and A69 all inhibited in vitro DNA-binding activity of STAT3 using EMSA. To determine whether A18 inhibits STAT3 binding to chromatin DNA in situ, we treated A549 and MDA-MB-231 cells with A18 followed by fractionation and western blotting analysis of cytosolic, soluble nuclear and chromatin-bound STAT3. Figure 6c shows that the chromatin-bound STAT3 decreases while soluble nuclear STAT3 increases with the increasing concentration of A18, suggesting that A18 effectively inhibits the binding of STAT3 to its target sequence on chromatin DNA in situ. To confirm this finding, we performed chromatin immunoprecipitation (ChIP) assay of STAT3 bound to the promoter of a STAT3 downstream target gene cyclin D1. As shown in Figure 6d, A18 treatment eliminated STAT3 binding to the promoter of cyclin D1 gene. Thus, we conclude that A18 treatments inhibit STAT3 binding to the promoters of its target genes on chromatin in situ and inhibits the expression of these genes.
A18 inhibits tumor growth, metastasis and expression of STAT3 target genes in vivo
We next evaluated the in vivo efficacy of A18 in a mouse xenograft model of A549 cells. As shown in Figure 7a, A18 treatment via oral dosing significantly reduced tumor growth compared with the vehicle control treatment. However, it had no significant effect on mouse body weight. The final tumor weight in the A18-treated mice is significantly less than that of the control-treated group while there is no significant difference in the wet weight of other organs (Figure 7b). Secondary metastatic tumors were found in the lungs of three of the five mice in the control group (Figure 7c), with extension into the peripancreatic adipose tissues and the adjacent peripancreatic lymph nodes. None of the five A18-treated mice has any signs of lung metastasis. Immunohistochemistry staining analysis of xenograft tumors also revealed that the expression of STAT3 downstream target genes, survivin and VEGF, but not STAT3 itself, were reduced by A18 treatment compared with the vehicle-control-treated group (Figure 7d). These findings suggest that A18 may be effective in suppressing xenograft tumor growth and metastasis with little adverse effect.
A18 suppresses xenograft tumor growth and metastasis in vivo. (a) Volume of xenograft tumors and body weight of mice following implantation and A18 treatments. (*P<0.05, **P<0.01, n=5). (b) Wet weight of final dissected xenograft tumor mass and organs. A single outlier in the treatment group was rejected by Dixon's Q test at 95% confidence. The variance between the two groups are similar. (c) Hematoxylin and eosin-stained paraffin sections of lung tissues of vehicle control and A18-treated mice. The arrow indicates a solitary metastatic tumor. A zoom-in image of the metastatic tumor with adjacent normal tissue is shown. (d) Immunohistochemistry staining of xenograft tumor tissues for the expression of STAT3, survivin and VEGF.
Discussion
In this study, we successfully identified a potential lead inhibitor, A18, targeting the DBD of STAT3. A18 not only binds to the DBD of STAT3 but also inhibits the DNA-binding activity of STAT3 both in vitro and in situ as well as the expression of STAT3 downstream target genes. However, it does not inhibit constitutive or IL-6-stimulated STAT3 activation in cancer cells. It further suppresses xenograft tumor growth and metastasis possibly by inhibiting STAT3 activity in vivo. Interestingly, A18 is completely soluble in an oral formulation and is likely specific to STAT3. Furthermore, A18 is tolerable by mice up to 200 mg/kg in multiple dosing. These characteristics make A18 a promising potential lead inhibitor for further modification and development.
Domain mapping of the binding site using purified recombinant STAT3 protein and pull-down assay indicate that these compounds likely bind directly to the DBD as originally designed. It also suggests that the improved in silico screening approach in our previous study31 is effective in identifying STAT3-selective inhibitors targeting DBD of STAT3 and offers a new promising approach for drug discovery targeting the ‘undruggable‘ DBD of other oncogenic transcription factors. Although inS3-54 and its active analogs likely bind to the DBD of STAT3 and their binding mode can be predicted using in silico approach,31 how each compound interact with specific residues in the DBD of STAT3 await further investigation using experimental approaches, such as generating co-crystal structures.
The potential core structure of compounds that can bind to the DBD of STAT3 is 5-phenyl-1H-pyrrol-2(3H)-ketone with variation of R1 and R2 (Figure 1b). Based on the predicted binding mode of inS3-54 in a previous study,31 the R2 group binds residues Met331, Val343, Met420, Ile467 and Met470 in STAT3 via hydrophobic interactions while the R1 group stabilizes the binding via interaction with residues Lys340 and Asn466. Active compounds appear to have p-hydroxybenzene, p-carboxybenzene or o-methoxybenzene as R1 and p-nitrobenzene, p-chlorobenzene or benzenamide as R2. However, appropriate combinations of R1 and R2 may also be needed. For example, compound A23 with p-carboxybenzene as R1 and p-chlorobenzene as R2 does not appear to have any significant activity in inhibiting STAT3-dependent luciferase expression. It is currently unclear whether the combination of different R1 and R2 affects their cellular permeability, binding affinity to STAT3 or recognition by cellular detoxification system. However, it is clear that both R1 and R2 groups are critical for specificity of these compounds. For example, inS3-54 with R1 being carboxybenzene is less specific to STAT3 than analog A69 with the same R2 but different R1 (hydroxybenzene) from inS3-54 as determined using in vitro colony-formation assay of STAT3−/− hematopoietic progenitor cells.
It is noteworthy that A18 has an IC50 of ~11 μM in luciferase reporter assay. However, its IC50 in cytotoxicity assay ranges from ~3.2–4.7 μM in cancer to ~7.0–10.5 μM in non-cancer cells. Currently, it is unknown why the IC50 of A18 is lower in cytotoxicity assay than that in luciferase reporter assay. It is possible that the cytotoxicity assay is more sensitive as it measures both proliferation and cell death induced by A18, which is an amplified result of reduced expression of STAT3 target genes. It is also possible that A18 may have off-target effects that can impact on cell survival, making it more effective in suppressing cancer cell survival. Interestingly, a similar but wider difference was also observed with inS3-54 in our previous study. The reduced difference for A18 is consistent with its increased selectivity for STAT3. Clearly, future more in-depth studies are needed to differentiate these possibilities.
Finally, drug discovery and development targeting STAT3 needs to be cautiously performed. Because STAT3 is important for hematopoiesis and innate immunity,32, 33 and as we show herein (Figure 2b) that STAT3 inhibitors can suppress formation of colonies in vitro from mouse STAT3+/+ hematopoietic progenitor cells, chronic use of effective STAT3 inhibitors may cause adverse effect on bone marrow and innate immunity. However, whether multiple dosing of A18 inhibits proliferation of these progenitor cells in vivo remains to be determined. Lack of weight loss, apparent phenotypic changes and change in organ size suggest that multiple dosing of A18 in mice may not cause sever adverse effect and thus A18 warrants further development.
Materials and methods
Materials
Antibodies against STAT3 (sc-482x), STAT1 (sc-346x), matrix metalloproteinase-9 (sc-10737), VEGF (sc-152) and cyclin D1 (sc-246) were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA, USA). Antibodies against pSTAT3 (9145), STAT3 for immunohistochemistry (9139), survivin (2808), histone H2A (2578) and histone H3 (9715) were all from Cell Signaling Technology (Danvers, MA, USA). Antibodies against FLAG tag (F3165), α-tubulin (T9026) and β-actin (A5316) and protease inhibitor cocktail were obtained from Sigma-Aldrich (St Louis, MO, USA). ChIP Assay kit and CpGenome Universal DNA Modification Kit were purchased from EMD Millipore (Billerica, MA, USA). Metafectene Pro transfection reagent, the Easy AT vector, and the Formulation Kit were obtained form Biontex (München, Germany), Promega (Madison, WI, USA), and Pharmatek Laboratories, respectively. The enhanced chemiluminescence regagents, iScript cDNA Synthesis Kit and the StepOnePlus Systems Kit were purchased from GE Healthcare (Uppsala, Sweden), Bio-Rad Laboratories (Hercules, CA, USA) and Applied Biosystems (Warrington, UK), respectively. The Cell Death Detection ELISAPLUS Kit, Matrigel-coated Boyden Chambers and CNBr-activated Sepharose 4B were from Roche (Mannheim, Germany), BD Biosciences (Bedford, MA, USA) and GE Healthcare, respectively. Cell culture media and fetal bovine serum were from Media Tech (Herndon, CA, USA) or Applied Biosystems-Life Technologies (Carlsbad, CA, USA). All other chemicals were purchased from Sigma (St Louis, MO, USA) or Fisher Scientific (Chicago, IL, USA).
Cell lines
Human cancer cell lines A549, MDA-MB-231 and MDA-MB-468 and human lung fibroblast IMR90 were all from ATCC (Manassas, VA, USA) and cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and appropriate antibiotics in a 5% CO2 incubator at 37 °C. H1299 cells were maintained in RPMI 1640 medium containing 10% fetal bovine serum and appropriate antibiotics. Human mammary epithelial cell line MCF10A1 was cultured in Dulbecco’s modified Eagle’s medium/F-12 (50:50) with 10% equine serum, 10 μg/ml insulin, 25 ng/ml epidermal growth factor, 500 ng/ml hydrocortisone and 100 ng/ml cholera toxin. These cell lines were authenticated by short tandem repeat analysis on 21 January 2013.
Resynthesis of A18 and A26
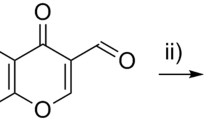
Compounds A18 and A26 were prepared as outlined below in Scheme 1 from commercially available starting materials 4-oxo-4-phenylbutanoic acid (compound 1) and 4-chlorobenzaldehyde (compound 2). Compound 1 reacted with compound 2 in Ac2O with catalytic amount of N,N-Diisopropylethylamine to give the key intermediate 3. Intermediate 3 reacted with corresponding amine at reflux for 4 h, followed by 6 n HCl at reflux for 2 h to give desired compounds A18 and A26.
Engineering recombinant STAT3 constructs
The Flag-tagged STAT3 was engineered in our previous study31 and was used as the template for engineering truncated STAT3 constructs to produce recombinant proteins. Briefly, the different STAT3 deletion mutants were generated using PCR with the same forward primer 5′-CGGATATCCATGGCCCAATGGAATCAGCTACAG-3′ containing a EcoR V site and different reverse primers containing a Xho I site (5′-CGCTCGAGCTATTACCGACAATACTTTCCGAATGCCTC-3′ for ΔTAD-C; 5′-CGCTCGAGCTATTAGTACCCTTCGTTCCAAAG-3′ for ΔSH2-C; and 5′-CGCTCGAGCTATTAACTTTTCATTAAGTTTCTAAACAGC-3′ for ΔDBD-C). The PCR products were first cloned into Easy AT vector (Promega), released by digestion using EcoR V and Xho I and then cloned into pET-28a(+) vector for production of His-tagged proteins. All final constructs were verified by sequencing.
Hematopoietic progenitor cell colony-formation assay
Hematopoietic progenitor cell colony-formation assay was performed as previously described.33 Briefly, 5 × 104 bone marrow cells isolated from wild-type (STAT3+/+) and bone-marrow specific STAT3 knockout (STAT3−/−) C57BL/6 mice32 were stimulated in vitro with 1 U/ml recombinant human erythropoietin, 50 ng/ml recombinant mouse stem cell factor and 5% vol/vol pokeweed mitogen mouse spleen cell conditioned medium and 0.1 mM hemin in the presence of Iscove’s Modified Dulbecco’s Medium medium, Iscove’s Modified Dulbecco’s Medium medium plus inS3-54 analogs or DMSO vehicle control. Colonies were scored 7 days after incubation at 37 °C with 5% CO2 and lowered (5%) O2.
DNA-binding activity assays
EMSA was used for testing in vitro DNA-binding activity of STAT3 as previously described,31 Briefly, 20 μg lysate of H1299 cells with transient expression of FLAG-tagged STAT3 or STAT1 was preincubated with compounds or vehicle, specific antibodies to STAT3 or STAT1 or excess cold SIE probe (5′-AGCTTCATTTCCCGTAAATCCCTA-3′) for 30 min at room temperature in binding buffer (0.1 μg/μl poly(dI·dC), 10 mM HEPES–KOH, pH 7.9, 50 mM KCl, 10% glycerol, 0.05 μg/μl bovine serum albumin, 1 mM dithiothreitol and 0.2 mM phenylmethanesulfonylfluoride) before incubating with 4 × 104 c.p.m. [32P]-labeled SIE probe for 20 min at room temperature. The reactions were separated on 6% non-denaturing polyacrylamide gel and signals were detected by autoradiography.
The in situ DNA-binding activity of STAT3 was performed by analyzing chromatin-bound STAT3 also as previously described.31 Briefly, following treatment with compounds or DMSO vehicle control, cells were harvested and subjected to subcellular fractionation to separate cytosol, soluble nuclear and chromatin-bound proteins exactly as we previously described31 followed by western blotting analysis of STAT3 in these fractions.
Western blotting and real-time reverse transcription (RT–PCR) analyses
Both western blotting and real-time RT–PCR analyses were performed as we previously described.31 For western blotting analysis, briefly proteins were separated by SDS–PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) and transferred to polyvinylidene difluoride membranes that were probed using antibodies against different proteins and visualization by exposing to X-ray films or using FluoChem HD2 Imaging System (ProteinSimple, Santa Clara, CA, USA) with enhanced chemiluminescence. For real-time RT–PCR analysis, total mRNAs were reverse-transcribed to cDNA using iScript cDNA Synthesis Kit followed by real-time PCR using StepOnePlus Systems with primers as described in our previous publications.31
Immobilization of A26 and pull-down assay
A26 containing a secondary amine group was immobilized to CNBr-activated Sepharose 4B according to the manufacturer’s instruction. Vehicle-treated and an irrelevant compound-conjugated CNBr-activated Sepharose 4B were also generated in the same way as control beads. As A26 is yellow, conjugation of A26 to CNBr-activated Sepharose 4B was verified by monitoring the change in color of the beads. For pull-down assay, A26-conjugated and control beads equilibrated with binding buffer (20 mM Tris-HCl, pH 8.0, 150 mM KCl, 1 mM EDTA, 15% glycerol, 0.5% NP-40) were blocked with 10% non-fat milk in the binding buffer containing 0.2 mM phenylmethanesulfonylfluoride and 1:1000 diluted protease inhibitor cocktail followed by incubation with 120 μg total lysate of H1299 cells with FLAG-STAT3 or 1 μg recombinant STAT3 protein in the same buffer at 37 °C for 1 h. Unbound proteins were removed by washing beads seven times with the binding buffer, and the bound proteins were separated by SDS–PAGE followed by analysis of FLAG-STAT3 using western blotting or silver staining. For competition analysis, cell lysate was preincubated with DMSO vehicle or different compound for 1 h at 37 °C prior to pull-down assay using A26-conjugated beads.
Survival, apoptosis, migration and invasion assays
Survival, apoptosis, migration and invasion assays were performed as previously described.31 Briefly, cells were treated with or without compounds for 72 h followed by sulforhodamine B assay to determine the effects of the compounds on cell survival. Apoptosis induced by STAT3 inhibitors was quantified using Cell Death Detection ELISAPLUS Kit by ELISA according to the manufacturer’s instructions. Wound-healing assay was performed by culturing 1 × 105 cells/well in six-well plates, followed by generating a wound using a pipette tip and continuous culture with photographic recording at different times. Cell invasion assay was performed using Matrigel-coated Boyden Chambers following the manufacturer’s instructions with 1.25 × 105 cells/well.
Chromatin immunoprecipitation
ChIP was performed as previously described.31 Briefly, H1299 cells were treated with 1% formaldehyde for 10 min, and lysates of the cells were sonicated to shear DNA to lengths between 200 and 1000 bases, which were then subjected to ChIP using the Chromatin Immunoprecipitation Assay Kit and normal IgG or STAT3 specific antibody according to the manufacturer’s instructions. The primers for PCR of cyclin D1 promoter are 5′-AACTTGCACAGGGGTTGTGT-3′ (forward) and 5′-GAGACCACGAGAAGGGGTGACTG-3′ (reverse).
Efficacy analysis using xenograft mouse model
The in vivo efficacy study was approved by the institution's IACUC committee at Indiana University. Briefly, 5 × 106 A549 cells were injected subcutaneously in the flanks of 5–6-week-old male NOD/SCID mice. When the tumor volume reached about 50.0 mm3, the mice were randomized using online program into two different groups (6/group) with one group treated by formulation vehicle control and the other by A18 at 200 mg/kg with oral dosing 2–3 times a week for 4 weeks. One mouse in each group died during the study and was eliminated. Tumor volume and body weight of the remaining five mice in each group were measured twice a week without blinding. On the thirty-fifth day after implant, mice were euthanized and the tumor tissues were harvested and weighed. Necropsy was also performed to determine changes in the heart, lungs, kidneys, liver and spleen.
To evaluate the histological alternation, metastasis in the lung and the expression of STAT3 and its downstream proteins, paraffin-embedded tissue sections were stained with hematoxylin and eosin and/or used for immunohistochemistry analysis as we previously described.34, 35 Briefly, the paraffin-embedded tissues were deparaffinized and rehydrated overnight, followed by incubation with 0.3% H2O2 and target retrieval buffer (10 mM citrate acid, 0.05% Tween-20, pH 6.0) at 95–100 °C for 30 min. Following preincubation in phosphate-buffered saline containing 10% fetal bovine serum and 0.1% Tween-20 for 30 min at room temperature, the tissue sections were probed with primary antibody against STAT3, survivin or VEGF at 4 °C overnight followed by further incubation with secondary antibody and streptavidin–horseradish peroxidase substrate. Slides were finally counterstained with hematoxylin and mounted with coverslips, and images were captured under a microscope.
Statistical considerations
The primary outcome of the in vivo efficacy study is the reduction of xenograft tumor size from control. Independent Student’s t-test was used with two-sided P-value <0.05 considered as statistically significant. With n=4 per group, we would have a power level of 0.80 to detect a mean difference of 50% in the treatment group with s.d. at 25%. For cell-based assays, with n=3, we would have a power of level of 0.80 to detect a mean difference of 25% with an s.d. at 10%. In general, continuous outcomes were summarized by mean±s.d. and compared using Student’s t-test.

Synthesis of compounds A18 and A26 ((a) Ac2O, N,N-Diisopropylethylamine (cat.), reflux; (b) 4-aminophenol, toluene, 100 °C; (c) N-(4-aminophenyl)acetamide, toluene, 100 °C; (d) HCl (6 n), reflux).
References
Bromberg J, Darnell JE Jr . The role of STATs in transcriptional control and their impact on cellular function. Oncogene 2000; 19: 2468–2473.
Debnath B, Xu S, Neamati N . Small molecule inhibitors of signal transducer and activator of transcription 3 (Stat3) protein. J Med Chem 2012; 55: 6645–6668.
Lee H, Pal SK, Reckamp K, Figlin RA, Yu H . STAT3: a target to enhance antitumor immune response. Curr Top Microbiol Immunol 2011; 344: 41–59.
Chen Z, Han ZC . STAT3: a critical transcription activator in angiogenesis. Med Res Rev 2008; 28: 185–200.
Devarajan E, Huang S . STAT3 as a central regulator of tumor metastases. Curr Mol Med 2009; 9: 626–633.
Hirano T, Ishihara K, Hibi M . Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene 2000; 19: 2548–2556.
Dolled-Filhart M, Camp RL, Kowalski DP, Smith BL, Rimm DL . Tissue microarray analysis of signal transducers and activators of transcription 3 (Stat3) and phospho-Stat3 (Tyr705) in node-negative breast cancer shows nuclear localization is associated with a better prognosis. Clin Cancer Res 2003; 9: 594–600.
Choi CH, Song SY, Kang H, Lee YY, Kim CJ, Lee JW et al. Prognostic significance of p-STAT3 in patients with bulky cervical carcinoma undergoing neoadjuvant chemotherapy. J Obstet Gynaecol Res 2010; 36: 304–310.
Diaz N, Minton S, Cox C, Bowman T, Gritsko T, Garcia R et al. Activation of stat3 in primary tumors from high-risk breast cancer patients is associated with elevated levels of activated SRC and survivin expression. Clin Cancer Res 2006; 12: 20–28.
Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C et al. Stat3 as an oncogene. Cell 1999; 98: 295–303.
Chiarle R, Simmons WJ, Cai H, Dhall G, Zamo A, Raz R et al. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat Med 2005; 11: 623–629.
Yue P, Turkson J . Targeting STAT3 in cancer: how successful are we? Expert Opin Investig Drugs 2009; 18: 45–56.
Deng J, Grande F, Neamati N . Small molecule inhibitors of Stat3 signaling pathway. Curr Cancer Drug Targets 2007; 7: 91–107.
Ren Z, Cabell LA, Schaefer TS, McMurray JS . Identification of a high-affinity phosphopeptide inhibitor of Stat3. Bioorg Med Chem Lett 2003; 13: 633–636.
Turkson J, Ryan D, Kim JS, Zhang Y, Chen Z, Haura E et al. Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation. J Biol Chem 2001; 276: 45443–45455.
Turkson J, Kim JS, Zhang S, Yuan J, Huang M, Glenn M et al. Novel peptidomimetic inhibitors of signal transducer and activator of transcription 3 dimerization and biological activity. Mol Cancer Ther 2004; 3: 261–269.
Siddiquee KA, Gunning PT, Glenn M, Katt WP, Zhang S, Schrock C et al. An oxazole-based small-molecule Stat3 inhibitor modulates Stat3 stability and processing and induces antitumor cell effects. ACS Chem Biol 2007; 2: 787–798.
Schust J, Berg T . A high-throughput fluorescence polarization assay for signal transducer and activator of transcription 3. Anal Biochem 2004; 330: 114–118.
Schust J, Sperl B, Hollis A, Mayer TU, Berg T . Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol 2006; 13: 1235–1242.
Song H, Wang R, Wang S, Lin J . A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc Natl Acad Sci USA 2005; 102: 4700–4705.
Siddiquee K, Zhang S, Guida WC, Blaskovich MA, Greedy B, Lawrence HR et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci USA. 2007; 104: 7391–7396.
Hao W, Hu Y, Niu C, Huang X, Chang CP, Gibbons J et al. Discovery of the catechol structural moiety as a Stat3 SH2 domain inhibitor by virtual screening. Bioorg Med Chem Lett 2008; 18: 4988–4992.
Fletcher S, Singh J, Zhang X, Yue P, Page BD, Sharmeen S et al. Disruption of transcriptionally active Stat3 dimers with non-phosphorylated, salicylic acid-based small molecules: potent in vitro and tumor cell activities. Chembiochem 2009; 10: 1959–1964.
Lin L, Hutzen B, Li PK, Ball S, Zuo M, DeAngelis S et al. A novel small molecule, LLL12, inhibits STAT3 phosphorylation and activities and exhibits potent growth-suppressive activity in human cancer cells. Neoplasia 2010; 12: 39–50.
Lin L, Hutzen B, Zuo M, Ball S, Deangelis S, Foust E et al. Novel STAT3 phosphorylation inhibitors exhibit potent growth-suppressive activity in pancreatic and breast cancer cells. Cancer Res 2010; 70: 2445–2454.
Yang J, Liao X, Agarwal MK, Barnes L, Auron PE, Stark GR . Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Dev 2007; 21: 1396–1408.
Timofeeva OA, Chasovskikh S, Lonskaya I, Tarasova NI, Khavrutskii L, Tarasov SG et al. Mechanisms of unphosphorylated STAT3 transcription factor binding to DNA. J Biol Chem 2012; 287: 14192–14200.
Nkansah E, Shah R, Collie GW, Parkinson GN, Palmer J, Rahman KM et al. Observation of unphosphorylated STAT3 core protein binding to target dsDNA by PEMSA and X-ray crystallography. FEBS Lett 2013; 587: 833–839.
Caboni L, Lloyd DG . Beyond the ligand-binding pocket: targeting alternate sites in nuclear receptors. Med Res Rev 2012; 33: 1081–1118.
Leung CH, Chan DS, Ma VP, Ma DL . DNA-binding small molecules as inhibitors of transcription factors. Med Res Rev 2013; 33: 823–846.
Huang W, Dong Z, Wang F, Peng H, Liu JY, Zhang JT . A small molecule compound targeting STAT3 DNA-binding domain inhibits cancer cell proliferation, migration, and invasion. ACS Chem Biol 2014; 9: 1188–1196.
Welte T, Zhang SS, Wang T, Zhang Z, Hesslein DG, Yin Z et al. STAT3 deletion during hematopoiesis causes Crohn's disease-like pathogenesis and lethality: a critical role of STAT3 in innate immunity. Proc Natl Acad Sci USA 2003; 100: 1879–1884.
Mantel C, Messina-Graham S, Moh A, Cooper S, Hangoc G, Fu XY et al. Mouse hematopoietic cell-targeted STAT3 deletion: stem/progenitor cell defects, mitochondrial dysfunction, ROS overproduction and a rapid aging-like phenotype. Blood 2012; 120: 2589–2599.
Cheng L, Pan CX, Zhang JT, Zhang S, Kinch MS, Li L et al. Loss of 14-3-3sigma in prostate cancer and its precursors. Clin Cancer Res 2004; 10: 3064–3068.
Yang Y, Liu H, Li Z, Zhao Z, Yip-Schneider M, Fan Q et al. Role of fatty acid synthase in gemcitabine and radiation resistance of pancreatic cancers. Int J Biochem Mol Biol 2011; 2: 89–98.
Acknowledgements
We thank Anthony Sinn in the In Vivo Therapeutics Core and Sheng Zhang in the Chemical Genomics Core for their technical assistance for in vivo studies and chemical synthesis, respectively. Jiayuh Lin at Ohio State University provided MDA-MB-231-STAT3 cells with the STAT3-dependent luciferase reporter while George R Stark at Cleveland Clinic provided STAT1 cDNA. This work was supported in part by the Lung Cancer Working Group at IUSCC.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Huang, W., Dong, Z., Chen, Y. et al. Small-molecule inhibitors targeting the DNA-binding domain of STAT3 suppress tumor growth, metastasis and STAT3 target gene expression in vivo. Oncogene 35, 783–792 (2016). https://doi.org/10.1038/onc.2015.215
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.215
- Springer Nature Limited
This article is cited by
-
Unraveling the complexity of STAT3 in cancer: molecular understanding and drug discovery
Journal of Experimental & Clinical Cancer Research (2024)
-
JAKs and STATs from a Clinical Perspective: Loss-of-Function Mutations, Gain-of-Function Mutations, and Their Multidimensional Consequences
Journal of Clinical Immunology (2023)
-
Small molecule targeting FOXM1 DNA binding domain exhibits anti-tumor activity in ovarian cancer
Cell Death Discovery (2022)
-
eIF3a regulation of mTOR signaling and translational control via HuR in cellular response to DNA damage
Oncogene (2022)
-
Systemic delivery of gemcitabine analogue and STAT3 siRNA promotes antitumor immunity against melanoma
Nano Research (2022)