Abstract
Deleted in Breast Cancer 1 (DBC1), a negative regulator of deacetylase SIRT1, has been shown to act as an estrogen receptor α (ER) coactivator that has a key role in ER transcription complex assembly and estrogen-dependent breast cancer cell proliferation. However, little is known about its physiological role and mechanism of action in ER-negative breast cancer cells. Here we report that DBC1 functions as a coactivator for the oncogenic ETS transcription factor PEA3 to promote ER-negative breast cancer progression. DBC1 is required for the expression of PEA3 target genes and for recruitment of PEA3 and RNA polymerase II to PEA3 target promoters. We also demonstrated that acetylation of PEA3 stimulates its DNA binding and association with DBC1 by disrupting the intramolecular interaction of PEA3. The molecular mechanism underlying DBC1 function in PEA3-mediated transcription involves inhibition of SIRT1 interaction with PEA3 and of SIRT1-mediated deacetylation of PEA3. Moreover, DBC1 depletion inhibited the tumorigenic properties of ER-negative breast cancer cells in vitro and in vivo. Importantly, increased DBC1 expression correlated with shorter relapse-free survival of ER-negative breast cancer patients. Our results firmly established DBC1 as a critical coactivator of PEA3 and as a key player in PEA3-mediated breast cancer progression.
Similar content being viewed by others
Introduction
Breast cancer is a heterogeneous disease and is made up of a number of subtypes.1, 2 Among the breast cancer subtypes, estrogen receptor α (ER)-negative breast cancers are more aggressive and metastatic than ER-positive breast cancers and unresponsive to anti-estrogen therapy.3 Elevated expression of human epidermal growth factor receptor 2 (HER2), which activates the mitogen-activated protein kinase (MAPK) signaling pathways, is also associated with aggressiveness and metastasis in breast cancer.4 It has been shown that ER-positive and -negative breast cancers exhibit large differences in their gene expression profiles.1, 2 Unlike ER-positive breast cancer, few risk factors have been identified for ER-negative breast cancer. Thus it remains a major challenge to identify novel regulators of ER-negative breast cancer progression.
The ETS family transcription factors regulate a wide range of cellular processes in both normal and cancer cells, including mammary gland development, growth, tumorigenesis and metastasis.5, 6 ETS transcription factors share a highly conserved ETS DNA-binding domain and control gene expression by binding to ETS-binding sites (EBS) in the promoter of target genes. PEA3, the founding member of the PEA3 subfamily (PEA3/ETV4, ER81/ETV1 and ERM/ETV5), is a downstream target of HER2-MAPK pathway, overexpressed in HER2-positive breast tumors and many metastatic ER-negative breast cancer cells, and often associated with poor prognosis and metastasis in breast cancer.6, 7, 8 The transcriptional activity of PEA3 is enhanced upon activation of the MAPK signaling pathway.9 PEA3 is acetylated by p300, and its acetylation is enhanced by MAPK signaling.10 Although acetylation enhances the transcriptional activity of PEA3, its mechanism in PEA3-mediated transcription is largely unknown.
Deleted in breast cancer (DBC1; also known as CCAR2) is a coactivator for nuclear receptors as well as a negative regulator of epigenetic modifiers, such as deacetylases SIRT1 and HDAC3.11, 12, 13, 14, 15 Recently, we have shown that DBC1 functions as a coactivator for ER and inhibits SIRT1-mediated deacetylation of ER and consequently enhances DNA binding and transcriptional activity of ER.11 In addition, DBC1 is important for estrogen-induced expression of ER target genes and estrogen-dependent growth of breast cancer cells. Although previous studies suggested that DBC1 has the potential to participate in hormone-driven tumorigenesis, whether DBC1 has a role in hormone-independent, ER-negative breast cancer remains unknown. We report here that DBC1 functions as a coactivator for PEA3 to promote ER-negative breast cancer progression.
Results
Identification of DBC1 target genes in ER-negative breast cancer cells
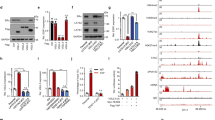
To investigate global gene expression changes induced by depletion of DBC1 and identify DBC1 target genes in ER-negative breast cancer cells, we performed genome-wide gene expression analysis in MDA-MB-231 cells (Figure 1a). DBC1 mRNA and protein levels were specifically reduced in MDA-MB-231/shDBC1 cells infected with lentiviruses expressing DBC1 short hairpin RNA (shRNA) (shDBC1) compared with MDA-MB-231/shNS cells infected with a non-specific shRNA (shNS) (Figure 1b). We then performed microarray analysis and identified 280 DBC1-dependent genes that exhibited a fold change >1.5 between MDA-MB-231/shNS and MDA-MB-231/shDBC1 cells. Among the differentially expressed genes, 221 genes (79%) and 59 genes (21%) were downregulated and upregulated, respectively, in MDA-MB-231/shDBC1 cells relative to control MDA-MB-231/shNS cells (Figure 1c). These differentially expressed genes are depicted in a heat map (Supplementary Figure S1). To gain insight into the functions of the genes changed by DBC1 depletion, we performed gene ontology analyses of the overlapping genes using the PANTHER classification system and observed that several of these genes were involved in the PANTHER Biological Process categories: cell cycle (5.8%), cell adhesion (13.9%), cell communication (35.9%) and metabolic process (44.4%) (Supplementary Figure S2).
Identification of DBC1 target genes by microarray analysis. (a) Flow chart depicting the strategy of cDNA microarray analysis and DBC1-dependent gene selection process. (b) MDA-MB-231 cells were infected with lentiviruses expressing shNS or shDBC1#3. Total RNA was examined by qRT–PCR analysis with primers specific for DBC1. Results shown were normalized to β-actin mRNA levels and are means±s.d. (n=3). Protein levels were monitored by immunoblot using the indicated antibodies. (c) Pie graph shows that 280 genes are differentially expressed by DBC1 depletion. (d) Validation of DBC1-regulated genes. Total RNAs were examined by qRT–PCR analysis with primers specific for the indicated mRNAs. Results shown were normalized to β-actin mRNA levels and are means±s.d. (n=3).
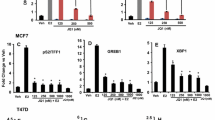
Based on previous reports16, 17, 18 and data analysis using Transcriptional Regulatory Element Database (http://rulai.cshl.edu), we found that the expression of several genes (MMP1, CXCR4, PDGFA, GPR160, LPCAT2, IL8, BMP4, TGFBR2, NRG1, EGLN3, CST1, TIMP3, TLR4 and PECAM1) known as targets of ETS transcription factors, which have overlapping target specificity, was significantly inhibited by depletion of DBC1. These genes were selected for validation by quantitative reverse transcriptase–PCR (qRT–PCR; Figure 1d and Supplementary Figure S3a) and immunoblot (Supplementary Figure S3b), and the results were very similar to those found by the microarray analysis. Among these genes, we focused on matrix metalloproteinase 1 (MMP1) and C-X-C motif chemokine receptor 4 (CXCR4), which are well-characterized PEA3 target genes and well recognized for their roles in breast cancer progression and metastasis.16, 17, 19, 20 Similar results on the expression of MMP1 and CXCR4 were also observed in DBC1-depleted SK-BR-3 and BT-20 cells (Supplementary Figure S4). To exclude the possible off-target effects, we repeated the knockdown experiments using two additional shRNAs targeting other regions of DBC1 mRNA and observed similar results in MDA-MB-231 cells (Supplementary Figures S5a and b). In addition, rescue experiments using shRNA-resistant DBC1 constructs confirmed that the reduction in the expression of MMP1 and CXCR4 caused by the DBC1-specific shRNA was specifically due to the reduction of cellular DBC1 levels (Supplementary Figures S6a and b). Thus our data suggest a role of DBC1 in the regulation of a subset of ETS target gene expression in ER-negative breast cancer cells.
DBC1 functions as a PEA3 coactivator
Given our finding that DBC1 is required for the expression of a subset of ETS target genes and previous findings that ETS transcription factors, especially PEA3, are upregulated and function as oncogenes in breast cancer,7, 8, 21 we hypothesized that DBC1 might act as a coactivator for PEA3. To test this hypothesis, we first examined the association between DBC1 and PEA3 by coimmunoprecipitation assays. Endogenous DBC1 and PEA3 were reciprocally coimmunoprecipitated from MDA-MB-231 cells (Figure 2a and Supplementary Figure S7a), and endogenous interaction between DBC1 and PEA3 was also detected in SK-BR-3, BT-20 and BT-549 cells (Supplementary Figure S7b). Similarly, PEA3 was co-immunoprecipitated specifically with DBC1 from extracts of transiently transfected 293T cells (Supplementary Figure S7c). In vitro GST (glutathione S-transferase) pull-down assays confirmed the interaction between DBC1 and PEA3 (Figure 2b) and showed that DBC1 interacts with the N-terminal region of PEA3 (Supplementary Figures S7d and e), suggesting that DBC1 interacts directly with PEA3.
DBC1 interacts with PEA3 and functions as a PEA3 coactivator. (a) MDA-MB-231 cell lysates were immunoprecipitated with normal IgG or anti-PEA3 antibody. The immunoprecipitates were analyzed by immunoblot with the indicated antibodies. (b) In vitro-translated HA-tagged PEA3 was incubated with recombinant GST-DBC1. Bound proteins were analyzed by immunoblot with an anti-HA antibody. (c) 293T cells were transfected with 5 × EBS-TA-LUC and expression vectors as indicated and harvested for luciferase assays. Data are means±s.d. (n=3). (d and e) MDA-MB-231 cells were transfected with DBC1 siRNA or non-specific (NS) siRNA, as indicated. Seventy-two hours after transfection, protein levels were monitored by immunoblot using the indicated antibodies (d), and luciferase activity was measured (e). Data are means±s.d. (n=3). (f) MDA-MB-231 cells transfected with 5 × EBS-TA-LUC and indicated siRNAs were untreated or treated with 10 nM phorbol myristate acetate (PMA) for 6 h and harvested for luciferase assays. Data are means±s.d. (n=3).
In reporter gene assays using a reporter containing five copies of the ETS-binding sites (5 × EBS-TA-LUC), DBC1 enhanced the transcriptional activity of PEA3 in a dose-dependent and PEA3-dependent manner (Figure 2c). When DBC1 protein levels were specifically reduced in MDA-MB-231 cells by siRNA transfection (Figure 2d), PEA3-induced expression of transiently transfected reporter genes driven by 5 × EBS, MMP1 and CXCR4 gene regulatory regions was inhibited compared with the results using non-specific (NS) siRNA (Figure 2e). In addition, phorbol myristate acetate (a potent activator of the MAPK signaling)-induced PEA3 activity was also inhibited by DBC1 depletion (Figure 2f). These results suggest that DBC1 is required as a coactivator for the expression of PEA3 target genes.
DBC1 is required for optimal recruitment of PEA3 to the ETS-binding sites of PEA3 target genes
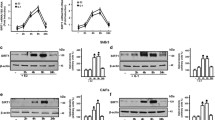
We next performed chromatin immunoprecipitation (ChIP) assays in MDA-MB-231 cells. PEA3, p300 (a known coactivator of PEA3) and RNA Polymerase II (Pol II) were recruited to EBS-containing MMP1 and CXCR4 promoter regions but not to a region lacking PEA3-binding sites (Figure 3a). DBC1 was also recruited to the promoters of MMP1 and CXCR4, indicating that DBC1 is directly involved in the transcriptional regulation of PEA3 target genes. In addition, reciprocal Re-ChIP experiments demonstrated that PEA3 and DBC1 exist in the same transcription complex on the CXCR4 promoter (Figure 3b). To further investigate the role of DBC1 as a PEA3 coactivator, we assessed the effect of DBC1 depletion on transcriptional complex assembly on PEA3 target promoters. DBC1 depletion had no measurable effect on the endogenous levels of PEA3, p300 and Pol II (Figure 3c). However, DBC1 depletion severely affected the recruitment of PEA3 to the MMP1 and CXCR4 promoters, and the recruitment of p300 and Pol II was also greatly reduced (Figure 3d). These results strongly suggest that DBC1 is required for efficient binding of PEA3 to its regulatory regions and facilitates subsequent recruitment of p300 and Pol II to the regulatory regions of PEA3 target genes.
DBC1 is required for optimal association of PEA3 with target promoters. (a) Cross-linked, sheared chromatin from MDA-MB-231 cells was immunoprecipitated with the indicated antibodies. qPCR analyses were performed using primers specific for the indicated regions. A 17-kb upstream region of GREB1 gene was used as a negative control region. The results are shown as the percentage of input and are means±s.d. (n=3). Asterisks indicate statistically significant differences compared with the negative control IgG ChIP (*P<0.05 and **P<0.01; Student’s t-test). (b) Reciprocal Re-ChIP analysis. Soluble chromatin was immunoprecipitated (1° ChIP) with antibodies to PEA3 and DBC1, respectively. Immunocomplexes were eluted and reimmunoprecipitated (2° ChIP) with reciprocal antibodies against DBC1 and PEA3, respectively. qPCR analyses were performed as described in panel (a). Asterisks indicate statistically significant differences (*P<0.05 and **P<0.01; Student’s t-test). (c and d) MDA-MB-231 cells were infected with lentiviruses expressing shNS or shDBC1#3. Protein levels were monitored by immunoblot using the indicated antibodies (c). ChIP assays using the indicated antibodies were performed as described in panel (a) (d). The results are shown as the percentage of input and are means±s.d. (n=3). Asterisks indicate statistically significant differences (*P<0.05 and **P<0.01; Student’s t-test).
Acetylation enhances DNA-binding activity of PEA3 and its interaction with DBC1 by inducing a conformational change
PEA3 is acetylated at multiple sites by p300, and the acetylation is potentiated by MAPK pathway signaling and enhances the transcriptional activity of PEA3.10 To investigate the effect of acetylation on DNA binding of PEA3, we generated K96/K226 (two major acetylation sites) acetylated PEA3 recombinant proteins using a recently developed method that allows the site-specific acetylation of recombinant proteins in bacteria.22 Acetylated PEA3 was purified from Escherichia coli, and acetylation levels were investigated by immunoblot (Figure 4a). In electrophoretic mobility shift assays, recombinant PEA3 showed a weak DNA-binding activity as reported previously (Figure 4b).23, 24 However, acetylation dramatically increased the DNA-binding activity of PEA3, suggesting that acetylation can have a positive role in regulating the DNA-binding activity of PEA3.
Acetylation enhances DNA-binding activity of PEA3 and its interaction with DBC1 by inducing conformational changes. (a) S-tagged recombinant unacetylated (UnAc) and acetylated (Ac) PEA3 proteins were expressed and purified from Escherichia coli. Purified UnAc- and Ac-PEA3 proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Coomassie blue staining and immunoblot with anti-acetyl lysine (AcK) or anti-S-Tag antibodies. (b) The DNA-binding activity of UnAc- and Ac-PEA3 was analyzed by electrophoretic mobility shift assay. The input levels of UnAc- and Ac-PEA3 proteins were analyzed by immunoblot using the indicated antibodies (bottom panels). (c and d) Limited proteolytic digestion of UnAc- and Ac-PEA3 by trypsin (c) and ArgC (d). C-terminally S-tagged fragments were detected by immunoblot using anti-S-Tag antibody. The arrowhead indicates full-length PEA3. (e) Schematic representation of full-length PEA3 and deletion mutants tested in GST pull-down assays. NTD, N-terminal domain; CTD, C-terminal domain; ID, inhibitory domain. (f) Recombinant S-tagged PEA3 fragments were incubated with recombinant GST-PEA3 CTD bound to beads. Bound proteins were analyzed by immunoblot with anti-S-Tag antibody. (g–i) Purified UnAc- and Ac-PEA3 NTD proteins were analyzed by immunoblot with the indicated antibodies (g). UnAc- and Ac-PEA3 NTD proteins were incubated with recombinant GST-PEA3 CTD bound to beads (h) or FLAG-DBC1 bound to M2 agarose (i). Bound proteins were detected by immunoblot using anti-S-Tag antibody.
Acetylation often induces conformational changes in transcription factors and enhances their DNA-binding activity.25 To investigate the conformational changes in PEA3 induced by acetylation, C-terminally S-tagged unacetylated and acetylated recombinant PEA3 proteins were subjected to limited proteolytic digestion with various amounts of trypsin. Full-length PEA3 and three major peptide fragments were detected in the tryptic digests of unacetylated PEA3 by immunoblot with S-tag antibodies, and they were partially resistant to trypsin (Figure 4c). In contrast, acetylated PEA3 and its tryptic fragments were much more sensitive to trypsin digestion. We confirmed this result using the arginine-specific protease Arg-C (Figure 4d), demonstrating a conformational change in PEA3 upon acetylation.
Previous studies suggested that DNA binding by PEA3 is tightly inhibited by intramolecular inhibitory regions.23, 24 Indeed, the central inhibitory domain, but not N-terminal activation domain (AD), interacted with the C-terminal region containing ETS domain (Figures 4e and f). Interestingly, acetylation at K96/K226 in the inhibitory region of PEA3 inhibited the interaction between N- and C-terminal regions but enhanced the interaction with DBC1 (Figures 4g–i). Taken together, these results strongly suggest that acetylation converts PEA3 into an active conformation for DNA binding and coactivator recruitment by disrupting the intramolecular interaction.
DBC1 inhibits SIRT1-mediated deacetylation of PEA3 and repression of PEA3 activity
Recent studies reported that DBC1 binds to SIRT1 and inhibits its deacetylase activity.12, 13 We have also reported that DBC1 has an important role in ER transcription complex assembly on target promoters by inhibiting SIRT1-mediated ER deacetylation.11 We next tested the possibility that SIRT1 and DBC1 reciprocally regulate the acetylation status and transcriptional activity of PEA3. In coimmunoprecipitation experiments, SIRT1 was coimmunoprecipitated with PEA3 (Supplementary Figure S8a). To determine the effect of SIRT1 on PEA3 acetylation, 293T cells were transfected with plasmids expressing PEA3, p300 and SIRT1, and the acetylated levels of PEA3 were determined after immunoprecipitation of PEA3 by immunoblots using anti-acetyl lysine antibodies. As reported previously,10 the expression of p300 strongly increased PEA3 acetylation. However, coexpression of SIRT1 dramatically reduced the acetylated level of PEA3 (Supplementary Figure S8b), suggesting that PEA3 is a substrate for SIRT1. However, the deacetylation of PEA3 by SIRT1 was partially reversed by DBC1 expression (Figure 5a). Similar results were observed in in vitro deacetylation assays using purified recombinant proteins (Figure 5b). Furthermore, SIRT1 binding to PEA3 was decreased in the presence of DBC1 in in vitro competitive binding assays (Figure 5c), and DBC1 depletion increased endogenous interaction between SIRT1 and PEA3 in 293T cells (Supplementary Figure S8c). These results indicate that DBC1 inhibits SIRT1 deacetylase activity by blocking the interaction between PEA3 and SIRT1. In reporter gene assays, SIRT1 repressed the transcriptional activity of PEA3 in a dose-dependent manner (Figure 5d), and DBC1 rescued PEA3 activity from SIRT1-mediated repression in a dose-dependent manner (Figure 5e). Consistent with these results, knockdown of SIRT1 or inhibition of SIRT1 deacetylase activity with EX-527, a SIRT1-specific inhibitor, increased the expression of MMP1 and CXCR4 (Supplementary Figure S9), demonstrating that SIRT1 negatively regulates PEA3 target gene expression and that its deacetylase activity is required for target gene repression. Together, these results suggest that DBC1 functions as a positive regulator of PEA3 by inhibiting SIRT1-mediated deacetylation and repression of PEA3 (Figure 5f).
DBC1 inhibits SIRT1-mediated deacetylation and repression of PEA3. (a) 293T cells were transfected with expression vectors as indicated. Cell extracts were immunoprecipitated with anti-PEA3 antibody, and the levels of acetylated PEA3 were determined by immunoblot using anti-acetyl lysine (AcK) antibody. Input and immunoprecipitated proteins were analyzed by immunoblot with the indicated antibodies. (b) Site-specifically acetylated PEA3 was incubated with GST-SIRT1 and GST-DBC1 as indicated. Immunoblots were performed as described in panel (a). (c) GST-PEA3 was incubated with recombinant FLAG-SIRT1 and HA-DBC1 as indicated. Bound proteins were analyzed by immunoblot with the indicated antibodies. (d and e) SIRT1 represses the transcriptional activity of PEA3 (d), and DBC1 reverses SIRT1-mediated repression of PEA3 (e). 293T cells were transfected with 5 × EBS-TA-LUC in combination with various amounts of expression vectors for SIRT1 and DBC1, as indicated, and harvested for luciferase assays. Data are means±s.d. (n=3). (f) A model for the activation of PEA3 by acetylation and proposed role of DBC1 as a PEA3 coactivator. Acetylation induces a conformational change in PEA3 by disrupting the intramolecular interaction between the C-terminal and central domains, thus converting PEA3 into an active conformation for DNA binding and interaction with DBC1. DBC1 inhibits SIRT1-mediated PEA3 deacetylation, thereby increasing DNA binding and transcriptional activity of PEA3.
DBC1 is required for maximal tumorigenic potential of ER-negative breast cancer cells and its overexpression correlates with a poor outcome in ER-negative breast cancer
We next examined the effect of DBC1 depletion on cell proliferation in ER-negative breast cancer cells. Depletion of DBC1 by shRNA attenuated the proliferation of MDA-MB-231, SK-BR-3 and BT-20 cells (Figure 6a). Similar results were obtained with two additional shRNAs targeting DBC1 in MDA-MB-231 cells (Supplementary Figure S5c). Re-expression of shRNA-resistant DBC1 restored the growth of MDA-MB-231/shDBC1 cells (Supplementary Figure S6c), suggesting a critical role of DBC1 in ER-negative breast cancer cell proliferation. Consistent with these results, cell cycle progression in MDA-MB-231 cells was inhibited by DBC1 depletion (Supplementary Figure S10). In addition, DBC1 depletion inhibited the clonogenic survival, migration and invasion of MDA-MB-231 cells (Figures 6b–d and Supplementary Figures S5d and e), which were rescued by re-expression of shRNA-resistant DBC1 (Supplementary Figures S6d and e), suggesting that DBC1 is required for the tumorigenic and metastatic properties of ER-negative breast cancer cells. To further assess the role of DBC1 in promoting breast tumorigenesis, we examined the effect of DBC1 depletion on the growth of MDA-MB-231 xenograft tumors in nude mice injected with DBC1-depleted or control cells that had been engineered to stably express luciferase. Depletion of DBC1 significantly inhibited the growth of xenograft tumors (Figures 6e and f) and in vivo expression of MMP1 and CXCR4 in xenograft tumors (Supplementary Figure S11), suggesting that DBC1 has a critical role in tumorigenic growth of breast cancer cells and in vivo expression of PEA3 target genes.
DBC1 is required for the tumorigenic properties of ER-negative breast cancer cells. (a) MDA-MB-231, SK-BR-3 and BT-20 cells were infected with lentiviruses expressing shNS or shDBC1#3. Cell viability was determined by 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide assay. Data are means±s.d. (n=6). *P<0.01. (b–d) Representative images of anchorage-dependent colony formation (b), cell migration (c) and invasion (d) assays are shown on the left panel of each figure, and the quantitative analysis (n=3, ±s.d.) is shown on the right panel of each figure. MDA-MB-231 cells were infected with lentiviruses expressing shNS or shDBC1#3. For the colony-formation assay, viable colonies were stained with crystal violet; the dye was extracted and quantified by spectrophotometry (b). Scale bar: 5 mm. Scratch assay was performed on confluent cell monolayers. The white line indicates the migration front of cells (c). Scale bars: 100 μm. Invasion assay was performed using the Transwell chambers with Matrigel (d). Scale bars: 200 μm. *P<0.01. (e and f) MDA-MB-231 cells expressing luciferase and either shNS or shDBC1#3 were injected subcutaneously in athymic female mice. Tumor growth was monitored for 6 weeks. Representative bioluminescence images of tumor-bearing mice and their tumors are shown on the left panel (each index in the ruler represents 1 mm,) and the average signal intensity (n=6, ±s.d.) of regions of interest is shown on the right panel (e). Tumor growth curves are shown (f). *P<0.05. (g and h) Kaplan–Meier analysis of relapse-free survival. Relapse-free survival according to DBC1 expression in ER-negative (g) and ER-negative/HER2-positive (h) breast cancer patients.
To investigate the clinical relevance of DBC1 expression in breast cancer, we searched the Oncomine expression profiling database (http://www.oncomine.org) for clinical tumor sample studies with significant differential expression of DBC1 compared with their corresponding normal tissues. In several studies, we found that DBC1 mRNA levels were significantly higher in invasive breast carcinoma than in normal tissues (Supplementary Figure S12), suggesting a potential role of DBC1 expression in breast tumorigenesis. To further investigate the upregulation and prognostic role of DBC1 in ER-negative breast cancer, we analyzed the DBC1 protein expression by immunohistochemistry in tissue microarrays containing surgical specimens from 207 ER-negative breast cancer patients. Clinical and pathological characteristics of the patients are summarized in Supplementary Table S1. DBC1 immunostaining was mainly nuclear, and tumors were divided into two groups (low and high DBC1 expression) according to DBC1 immunohistochemical scores (Supplementary Figure S13). Kaplan–Meier analysis showed that patients with high levels of DBC1 protein had significantly shorter relapse-free survival than those with low DBC1 expression (Figure 6g). Interestingly, HER2-positive patients with high DBC1 expression had worse relapse-free survival rates (Figure 6h), suggesting that coexpression of DBC1 and HER2 is a significant prognostic factor for ER-negative breast cancer.
Discussion
PEA3 is activated through phosphorylation by MAPK pathways.9 MAPK signaling also enhances SUMOylation and acetylation of PEA3, and these modifications contribute to PEA3 activation.10, 26 Recombinant PEA3 is inactive in DNA binding, but its DNA binding can be activated by N- or C-terminal deletion, antibodies specific for the central region of PEA3, and GST fusion to its N terminus,23, 24 suggesting that the DNA-binding activity of PEA3 is negatively regulated in an intramolecular manner. Consistent with this model, we showed that the central inhibitory domain of PEA3 indeed interacts with the C-terminal ETS domain and, importantly, that acetylation in the N terminus disrupts the intramolecular interaction between the N- and C-terminal domains, thereby leading to a conformational change of PEA3 into an active conformation for DNA binding and association with DBC1. Furthermore, we showed that SIRT1 binds to, deacetylates and represses PEA3. The ability of SIRT1 to deacetylate and repress PEA3 activity mechanistically resembles the effects of SIRT1 on p53 and ER,11, 12, 13 suggesting that SIRT1 functions as a general corepressor for regulating the activity of various transcription factors. Thus our results demonstrated that reversible acetylation has a key role in the regulation of PEA3 function.
We have previously shown that DBC1 functions as an ER coactivator and is required for estrogen-dependent growth of ER-positive breast cancer cells.11 In this study, we identified PEA3 as a target transcription factor of DBC1 through a microarray screen and extend our previous findings to ER-negative breast cancer by demonstrating that DBC1 contributes to the tumorigenesis and progression of ER-negative breast cancer by serving as a coactivator for PEA3. PEA3 is frequently overexpressed in ER-negative breast cancer cell lines and primary breast tumors and has an important role in onset, progression and metastasis of breast cancer.6, 7, 8, 21, 27 Depletion of DBC1 caused reduction in the expression of a subset of PEA3 target genes, markedly reduced recruitment of PEA3, p300 and Pol II to the ETS-binding sites on PEA3 target gene promoters and attenuated the growth of ER-negative breast cancer cells. Furthermore, DBC1 depletion reduced the tumorigenic and metastatic potential of MDA-MB-231 cells. Our results firmly established DBC1 as a PEA3 coactivator that has an important physiological role in ER-negative breast cancer cell growth, tumorigenesis and invasiveness.
Recent studies identified DBC1 as a negative regulator of several enzymes involved in posttranslational modifications of histones and transcription factors.11, 12, 13, 15 Here, we add another line of evidence supporting the role of DBC1 as a negative regulator of SIRT1: SIRT1-mediated PEA3 deacetylation was inhibited by DBC1; DBC1 blocks the interaction between PEA3 and SIRT1; and the SIRT1-mediated repression of PEA3 was reversed by DBC1 expression. Our ChIP analysis further showed that DBC1 was required for efficient recruitment of PEA3 to its target promoters, probably through protecting PEA3 from SIRT1-mediated deacetylation. These results are consistent with results showing that PEA3 acetylation enhances its DNA-binding activity. However, it is also possible that PEA3 binds to its target DNA more efficiently when complexed with DBC1. In line with the reduced promoter occupancy of PEA3 by DBC1 depletion, the recruitment of p300 and Pol II was greatly reduced, suggesting a role of DBC1 in the assembly of PEA3 transcription complex. ETV1, a member of PEA3 subfamily, is also acetylated by p300, and ETV1 acetylation enhances its transcriptional potential and DNA-binding activity.28 It will be interesting to investigate whether DBC1 and SIRT1 reciprocally regulate the transcriptional activity of other PEA3 family members through a similar mechanism described in this study.
Two recent papers reported that DBC1 expression was significantly associated with distant metastatic relapse and shorter relapse-free survival of breast cancer patients.29, 30 Here, we further demonstrated, using in vitro and in vivo models, that DBC1 confers growth and metastatic advantages to ER-negative breast cancer cells by acting as a coactivator for PEA3. Moreover, importantly, we provided evidence that DBC1 expression can be an independent prognostic predictor of shorter relapse-free survival in ER-negative breast cancer. The results of this study, together with our previous finding that DBC1 has an important physiological role in ER signaling,11 suggest that DBC1 has critical oncogenic roles in both ER-positive and -negative breast cancers. In breast cancer, CXCR4 and MMP1 are associated with metastasis and poor clinical outcomes.19, 20 CXCR4 has been implicated as a major contributor to the cross-talk between cancer cells and microenvironment, and cancer cell–microenvironment interactions promote cancer cell migration and invasion by stimulating the expression of several metalloproteases, including MMP1.31, 32 The critical functions of DBC1 in promoting the expression of CXCR4 and MMP1, together with its upregulation, suggest that DBC1 should be considered as a therapeutic target for breast cancer.
HER2 is overexpressed in 20~30% of human breast cancers,1 and its overexpression leads to the activation of the RAS-RAF-MEK-MAPK signaling pathway.4 The PEA3 subfamily members, downstream targets of HER2 signaling, are required for mammary tumorigenesis in transgenic mice overexpressing HER2.21 In addition, overexpression of PEA3 occurs in the vast majority of breast cancers and in nearly all HER2-positive breast cancers.7, 8 Interestingly, we found that high DBC1 protein expression correlated with poorer relapse-free survival in the HER2-positive subgroup of ER-negative breast cancer patients, indicating a positive role for DBC1 in HER2-mediated breast cancer progression and suggesting that high expression of HER2 and DBC1 may have a synergistic effect on PEA3-mediated breast cancer progression. As PEA3 and its subfamily members have key roles in the progression of multiple cancers and metastasis,6 further elucidation of roles and mechanisms of DBC1 in PEA3-mediated transcription and cancer progression will provide useful information for developing therapeutic drugs for the treatment of multiple cancers as well as breast cancer.
Materials and methods
Gene expression analysis by Affymetrix microarray
The microarrays were performed following the Affymetrix standard protocol as described previously33 (details in Supplementary Information). The data have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE54707).
Cell culture and transient transfection
MDA-MB-231, SK-BR-3, BT-20 and BT-549 were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) or from Korean Cell Line Bank (KCLB, Seoul, Korea). MDA-MB-231, BT-20 and BT-549 cells were cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum. SK-BR-3 cells were cultured in McCoy’s 5A medium with 10% fetal bovine serum. Transient transfections and reporter gene assays were performed as described previously.11, 33, 34, 35 Each experiment was repeated independently at least three times.
RNAi and real-time qRT–PCR
The depletion of DBC1 by siRNA or shRNA was performed according to previously described protocol.11 qRT–PCR was performed with total RNA and Brilliant SYBR Green QRT–PCR Master Mix 1-Step (Stratagene, La Jolla, CA, USA). siRNAs, shRNAs and primers used are listed in Supplementary Information.
ChIP assays
MDA-MB-231 cells were treated with 10 nM phorbol myristate acetate for 6 h. ChIP and Re-ChIP experiments were performed according to the procedure described previously.11, 33, 34, 35 The immunoprecipitated DNAs were amplified by qPCR. The primers used are listed in Supplementary Information.
Expression and purification of site-specifically acetylated PEA3
Site-specifically acetylated PEA3 was generated by a strategy described previously.22 Briefly, BL21(DE3) cells transformed with pAcKRS-3 and pCDF PylT-1-PEA3 with amber codons at K96 and K226 were grown in LB supplemented with 50 μg/ml kanamycin and 50 μg/ml spectinomycin at 37 °C. At an OD600 of 0.7, the culture was supplemented with 20 mM nicotinamide (NAM) and 10 mM acetyl-lysine. Thirty minutes later, protein expression was induced by addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside, and incubation was continued at 30 °C for 4 h. Cell pellets were resuspended in Ni-NTA lysis buffer (50 mM sodium phosphate, 300 mM NaCl, 20 mM imidazole, pH 8.0) containing 20 mM NAM and sonicated. The acetylated PEA3 was purified using Ni-NTA agarose (Qiagen, Valencia, CA, USA) and eluted with lysis buffer containing 250 mM imidazole.
Xenografts
Mouse xenograft experiments were performed as described previously.33, 35 The detailed method is provided in Supplementary Information. Animal experiments were conducted with the approval of the Institutional Animal Care and Use Committee of Laboratory Animal Research Center at Samsung Biomedical Research Institute.
ER-negative breast tumor samples, tissue microarray and immunohistochemistry
We collected 207 ER-negative breast cancer cases from 1290 primary invasive breast cancer samples collected between January 1995 and December 2002 at the Samsung Medical Center.36 Tissue microarray construction and immunohistochemical staining of DBC1 were performed as described previously.37 More details are provided in Supplementary Information. The human specimen analysis complied with all ethics requirements and was approved by the Samsung Medical Center Institutional Review Board.
A full description of the methods used in this study can be found in Supplementary Information.
References
Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 2001; 98: 10869–10874.
Reis-Filho JS, Pusztai L . Gene expression profiling in breast cancer: classification, prognostication, and prediction. Lancet 2011; 378: 1812–1823.
Barcellos-Hoff MH . Does microenvironment contribute to the etiology of estrogen receptor-negative breast cancer? Clin Cancer Res 2013; 19: 541–548.
Hynes NE, Lane HA . ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 2005; 5: 341–354.
Sharrocks AD . The ETS-domain transcription factor family. Nat Rev Mol Cell Biol 2001; 2: 827–837.
Oh S, Shin S, Janknecht R . ETV1 4 and 5: an oncogenic subfamily of ETS transcription factors. Biochim Biophys Acta 2012; 1826: 1–12.
Myers E, Hill AD, Kelly G, McDermott EW, O'Higgins NJ, Young LS . A positive role for PEA3 in HER2-mediated breast tumour progression. Br J Cancer 2006; 95: 1404–1409.
Benz CC, O'Hagan RC, Richter B, Scott GK, Chang CH, Xiong X et al. HER2/Neu and the Ets transcription activator PEA3 are coordinately upregulated in human breast cancer. Oncogene 1997; 15: 1513–1525.
O'Hagan RC, Tozer RG, Symons M, McCormick F, Hassell JA . The activity of the Ets transcription factor PEA3 is regulated by two distinct MAPK cascades. Oncogene 1996; 13: 1323–1333.
Guo B, Panagiotaki N, Warwood S, Sharrocks AD . Dynamic modification of the ETS transcription factor PEA3 by sumoylation and p300-mediated acetylation. Nucleic Acids Res 2011; 39: 6403–6413.
Yu EJ, Kim SH, Heo K, Ou CY, Stallcup MR, Kim JH . Reciprocal roles of DBC1 and SIRT1 in regulating estrogen receptor α activity and co-activator synergy. Nucleic Acids Res 2011; 39: 6932–6943.
Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W . Negative regulation of the deacetylase SIRT1 by DBC1. Nature 2008; 451: 587–590.
Kim JE, Chen J, Lou Z . DBC1 is a negative regulator of SIRT1. Nature 2008; 451: 583–586.
Fu J, Jiang J, Li J, Wang S, Shi G, Feng Q et al. Deleted in breast cancer 1, a novel androgen receptor (AR) coactivator that promotes AR DNA-binding activity. J Biol Chem 2009; 284: 6832–6840.
Chini CC, Escande C, Nin V, Chini EN . HDAC3 is negatively regulated by the nuclear protein DBC1. J Biol Chem 2010; 285: 40830–40837.
Gu S, Chen L, Hong Q, Yan T, Zhuang Z, Wang Q et al. PEA3 activates CXCR4 transcription in MDA-MB-231 and MCF7 breast cancer cells. Acta Biochim Biophys Sin 2011; 43: 771–778.
Sun Y, Wenger L, Brinckerhoff CE, Misra RR, Cheung HS . Basic calcium phosphate crystals induce matrix metalloproteinase-1 through the Ras/mitogen-activated protein kinase/c-Fos/AP-1/metalloproteinase 1 pathway. Involvement of transcription factor binding sites AP-1 and PEA-3. J Biol Chem 2002; 277: 1544–1552.
Wollenick K, Hu J, Kristiansen G, Schraml P, Rehrauer H, Berchner-Pfannschmidt U et al. Synthetic transactivation screening reveals ETV4 as broad coactivator of hypoxia-inducible factor signaling. Nucleic Acids Res 2012; 40: 1928–1943.
Bohn OL, Nasir I, Brufsky A, Tseng GC, Bhargava R, MacManus K et al. Biomarker profile in breast carcinomas presenting with bone metastasis. Int J Clin Exp Pathol 2009; 3: 139–146.
Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 2003; 3: 537–549.
Shepherd TG, Kockeritz L, Szrajber MR, Muller WJ, Hassell JA . The pea3 subfamily ets genes are required for HER2/Neu-mediated mammary oncogenesis. Curr Biol 2001; 11: 1739–1748.
Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J et al. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell 2009; 36: 153–163.
Greenall A, Willingham N, Cheung E, Boam DS, Sharrocks AD . DNA binding by the ETS-domain transcription factor PEA3 is regulated by intramolecular and intermolecular protein.protein interactions. J Biol Chem 2001; 276: 16207–16215.
Bojovic BB, Hassell JA . The PEA3 Ets transcription factor comprises multiple domains that regulate transactivation and DNA binding. J Biol Chem 2001; 276: 4509–4521.
Gu W, Roeder RG . Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 1997; 90: 595–606.
Guo B, Sharrocks AD . Extracellular signal-regulated kinase mitogen-activated protein kinase signaling initiates a dynamic interplay between sumoylation and ubiquitination to regulate the activity of the transcriptional activator PEA3. Mol Cell Biol 2009; 29: 3204–3218.
Baert JL, Monte D, Musgrove EA, Albagli O, Sutherland RL, de Launoit Y . Expression of the PEA3 group of ETS-related transcription factors in human breast-cancer cells. Int J Cancer 1997; 70: 590–597.
Goel A, Janknecht R . Acetylation-mediated transcriptional activation of the ETS protein ER81 by p300, P/CAF, and HER2/Neu. Mol Cell Biol 2003; 23: 6243–6254.
Lee H, Kim KR, Noh SJ, Park HS, Kwon KS, Park BH et al. Expression of DBC1 and SIRT1 is associated with poor prognosis for breast carcinoma. Hum Pathol 2011; 42: 204–213.
Hiraike H, Wada-Hiraike O, Nakagawa S, Saji S, Maeda D, Miyamoto Y et al. Expression of DBC1 is associated with nuclear grade and HER2 expression in breast cancer. Exp Ther Med 2011; 2: 1105–1109.
Eck SM, Cote AL, Winkelman WD, Brinckerhoff CE . CXCR4 and matrix metalloproteinase-1 are elevated in breast carcinoma-associated fibroblasts and in normal mammary fibroblasts exposed to factors secreted by breast cancer cells. Mol Cancer Res 2009; 7: 1033–1044.
Cojoc M, Peitzsch C, Trautmann F, Polishchuk L, Telegeev GD, Dubrovska A . Emerging targets in cancer management: role of the CXCL12/CXCR4 axis. Onco Targets Ther 2013; 6: 1347–1361.
Seo WY, Jeong BC, Yu EJ, Kim HJ, Kim SH, Lim JE et al. CCAR1 promotes chromatin loading of androgen receptor (AR) transcription complex by stabilizing the association between AR and GATA2. Nucleic Acids Res 2013; 41: 8526–8536.
Kim JH, Yang CK, Heo K, Roeder RG, An W, Stallcup MR . CCAR1, a key regulator of mediator complex recruitment to nuclear receptor transcription complexes. Mol Cell 2008; 31: 510–519.
Yu EJ, Kim SH, Kim MJ, Seo WY, Song KA, Kang MS et al. SUMOylation of ZFP282 potentiates its positive effect on estrogen signaling in breast tumorigenesis. Oncogene 2013; 32: 4160–4168.
Choi YL, Oh E, Park S, Kim Y, Park YH, Song K et al. Triple-negative, basal-like, and quintuple-negative breast cancers: better prediction model for survival. BMC Cancer 2010; 10: 507.
Kim SH, Kim JH, Yu EJ, Lee KW, Park CK . The overexpression of DBC1 in esophageal squamous cell carcinoma correlates with poor prognosis. Histol Histopathol 2012; 27: 49–58.
Acknowledgements
We thank Jason W Chin (MRC Laboratory for Molecular Biology, Cambridge, UK) for providing vectors and Yoon-La Choi (Department of Pathology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea) for providing the tissue microarray slides of primary invasive breast carcinoma specimens. This work was supported by Samsung Biomedical Research Institute (SBRI) grant (SMX1132501) and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (NRF-2013R1A1A2059697) to JHK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, H., Kim, SH., Yu, E. et al. A positive role of DBC1 in PEA3-mediated progression of estrogen receptor-negative breast cancer. Oncogene 34, 4500–4508 (2015). https://doi.org/10.1038/onc.2014.381
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.381
- Springer Nature Limited
This article is cited by
-
DBC1 regulates Wnt/β-catenin-mediated expression of MACC1, a key regulator of cancer progression, in colon cancer
Cell Death & Disease (2018)
-
DBC1 promotes castration-resistant prostate cancer by positively regulating DNA binding and stability of AR-V7
Oncogene (2018)
-
A novel crosstalk between CCAR2 and AKT pathway in the regulation of cancer cell proliferation
Cell Death & Disease (2016)
-
Positive regulation of β-catenin–PROX1 signaling axis by DBC1 in colon cancer progression
Oncogene (2016)
-
DBC1/CCAR2 is involved in the stabilization of androgen receptor and the progression of osteosarcoma
Scientific Reports (2015)