Abstract
Alzheimer’s disease is a common and devastating disease characterized by aggregation of the amyloid-β peptide. However, we know relatively little about the underlying molecular mechanisms or how to treat patients with Alzheimer’s disease. Here we provide bioinformatic and experimental evidence of a conserved mitochondrial stress response signature present in diseases involving amyloid-β proteotoxicity in human, mouse and Caenorhabditis elegans that involves the mitochondrial unfolded protein response and mitophagy pathways. Using a worm model of amyloid-β proteotoxicity, GMC101, we recapitulated mitochondrial features and confirmed that the induction of this mitochondrial stress response was essential for the maintenance of mitochondrial proteostasis and health. Notably, increasing mitochondrial proteostasis by pharmacologically and genetically targeting mitochondrial translation and mitophagy increases the fitness and lifespan of GMC101 worms and reduces amyloid aggregation in cells, worms and in transgenic mouse models of Alzheimer’s disease. Our data support the relevance of enhancing mitochondrial proteostasis to delay amyloid-β proteotoxic diseases, such as Alzheimer’s disease.
Similar content being viewed by others
Main
Ageing is often accompanied by the onset of proteotoxic degenerative diseases, characterized by the accumulation of unfolded and aggregated proteins. Amyloid diseases are proteotoxic disorders that can affect the nervous system, which is the case in Alzheimer’s disease (AD), the most common form of dementia1. However, amyloid diseases can also affect other organs, such as is seen in amyloidosis-associated kidney disease2 and inclusion body myositis3. To date, no efficient therapy is available for AD4, a disease with a strong component of amyloid-β (Aβ) aggregation1. Clinical trials for AD have focused primarily on counteracting Aβ aggregation in the brain, which is considered the key pathogenic mechanism5. However, AD is a complex, multifactorial disease6 and mitochondrial dysfunction has been shown to be a common pathological hallmark7. Mitochondrial abnormalities in AD include decreased mitochondrial respiration and activity and alterations in mitochondrial morphology7. However, the relevance of other aspects of mitochondrial homeostasis, such as mitochondrial proteostasis, to the pathogenesis of AD is still mostly unknown.
Mitochondrial function pathways are disrupted in AD
We analysed brain expression datasets from patients with AD (GN327, GN328 and GN314) archived in GeneNetwork (http://www.genenetwork.org/webqtl/main.py) to define the mitochondrial signature associated with the disease. Gene set enrichment analysis of datasets from healthy individuals versus patients with AD from prefrontal cortex, primary visual cortex and whole brain showed that downregulation of mitochondrial oxidative phosphorylation (OXPHOS) and disruption of mitochondrial import pathways were hallmarks of AD (Fig. 1a, Extended Data Fig. 1a–c, e, f and Supplementary Tables 1–4). Because these processes are linked with and affect mitochondrial proteostasis8, we used comprehensive gene sets for two major mitochondrial quality control pathways, the mitochondrial unfolded protein response (UPRmt) and mitophagy, to evaluate whether their expression is co-regulated in patients with AD (Supplementary Table 5). Whereas we observed a strong correlation between genes associated with UPRmt and mitophagy in all brain datasets investigated (Fig. 1b and Extended Data Fig. 1d, g), other stress pathways, such as the endoplasmic reticulum stress (UPRER) response and heat shock response (HSR), were co-regulated to a lesser degree.
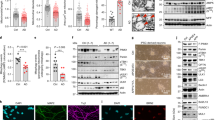
a, Heat map of the expression levels of OXPHOS and mitochondrial import genes in human prefrontal cortex of patients with AD (GN328; healthy, n = 195; AD, n = 388 individuals). b, Correlation plots of mitochondrial stress genes, UPRER and HSR levels in prefrontal cortex from patients with AD (GN328; n as in a). PARK2 is also known as PRKN. See Extended Data Fig. 1 and Supplementary Table 1–5. c, d, Transcript analysis of the MSR signature (c; n = 8 per group) and western blot (d, n = 2 individuals) of mtDNAJ and CLPP in brains of humans with no cognitive impairment (NCI), mild-cognitive impairment (MCI) and mild to moderate AD. e, f, Transcript (e) and immunoblot (f) analysis of MSR gene and protein expression in the cortex of nine-month-old wild-type (WT) and 3×TgAD mice (wild-type, n = 5; 3×TgAD, n = 5 for RNA analysis; wild-type, n = 4; 3×TgAD, n = 4 for western blot analysis, representative of six animals). Hsp60 is also known Hspd1; p62 is also known as Sqstm1; LC3 is also known as Map1lc3a; Cox2 is also known as mt-Co2; Nd1 is also known as mt-Nd1. g, Immunoblot (wild-type, n = 4; 3×TgAD, n = 4, representative of five animals) of mitophagy and autophagy proteins in mitochondrial extracts from cortex tissues of the mice in e, f. Data are mean ± s.e.m. *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001. Overall differences between conditions were assessed by two-way ANOVA. Differences in individual genes or proteins were assessed using two-tailed t-tests (95% confidence interval). All experiments were independently performed twice. See also Extended Data Fig. 2. For uncropped gel source data, see Supplementary Fig. 1. For all individual P values, see Source Data.
Evidence for a mitochondrial stress signature in AD
We next measured UPRmt, mitophagy and OXPHOS transcripts in the human cortex. We have previously reported that several UPRmt genes were upregulated during frank familial or sporadic AD9. Here, we extend that analysis and observe that, compared to subjects with no cognitive impairment, several UPRmt and mitophagy transcripts were upregulated in patients with mild cognitive impairment, a putative prodromal stage of AD10, and in mild and moderate AD (Fig. 1c and Supplementary Table 6), whereas OXPHOS genes were downregulated, consistent with our gene set enrichment analysis. The induction of this mitochondrial stress response was also observed at the protein level in subjects with mild cognitive impairment and AD (Fig. 1d and Extended Data Fig. 1h). The occurrence of this disruption already in patients with mild cognitive impairment suggests that mitochondrial dysfunction contributes to neuron and synapse loss, and that mitochondrial stress pathways may be activated as a protective response during disease progression10. We also analysed cortex samples of wild-type and a transgenic mouse model of AD (3×TgAD mice)11 (Extended Data Fig. 2a), at six and nine months of age. Both mitochondrial quality control and OXPHOS genes were induced in mice with AD (Fig. 1e and Extended Data Fig. 2b), although to a different extent over time. In fact, pairing wild-type and 3×TgAD mice at these two times indicated a marked attenuation of this stress signature during disease progression (Extended Data Fig. 2d–f). Immunoblotting of total lysates from the cortex of wild-type and 3×TgAD mice showed the induction of PINK1, LONP1 and LC3 at both time points (Fig. 1f and Extended Data Fig. 2c). Additional analysis of the nine-month-old mice also indicated a reduction in VDAC, a marked increase in p62 phosphorylation (Fig. 1g) and reduced citrate synthase activity (Extended Data Fig. 2g) in mice with AD, indicative of autophagy and mitophagy. PINK1 and LC3-I, the unmodified form of LC3, were also increased in mitochondrial extracts from cortex samples of mice with AD (Fig. 1g), confirming that these proteins are recruited to the mitochondria to promote mitophagy, as supported by the increased ubiquitylation of mitochondrial proteins (Fig. 1g). For simplicity, we abbreviate the comprehensive mitochondrial stress footprint that we analysed as mitochondrial stress response (MSR).
Identification of a cross-species MSR signature
The functional impact of changes in mitochondrial homeostasis during disease and ageing in mammals can be modelled reliably using the nematode C. elegans12. Worm models of Aβ aggregation have been used extensively to study the basic consequences of proteotoxic stress on conserved biological pathways between worms and mammals13,14. We took advantage of the GMC101 worm model of Aβ proteotoxicity15. GMC101 worms constantly express the human Aβ isoform 1–42 in muscle cells, but adults only develop age-progressive paralysis and amyloid deposition in the body wall muscle after a temperature shift from 20 to 25 °C (ref. 15), whereas the control strain CL2122 does not express the Aβ peptide (Extended Data Fig. 3a).
Transcripts of the worm orthologues of MSR were induced in adult GMC101 worms compared to CL2122 after the temperature shift to 25 °C (Fig. 2a), whereas these were only partially changed at 20 °C (Extended Data Fig. 3b). The basal and maximal respiratory capacity were decreased in GMC101 worms (Fig. 2b and Extended Data Fig. 3c), in contrast to the increased OXPHOS transcript levels (Fig. 2a), suggesting a compensatory induction to ensure respiration. Mitochondrial content was lower in GMC101 worms, as shown by decreased OXPHOS proteins, a reduced mitochondrial:nuclear DNA ratio and decreased citrate synthase activity (Fig. 2c and Extended Data Fig. 3d, e). Notably, GMC101 fitness, measured as spontaneous movement16, was robustly reduced relative to CL2122 worms in line with the muscle disorganization and alteration of the mitochondrial network in the body wall muscle (Extended Data Fig. 3f, g). These data highlight the cross-species conservation of MSR, and show that GMC101 is an excellent proxy for the characterization of the mitochondrial dysfunction and phenotypic impact observed in Aβ diseases in mammals.
a, MSR transcript analysis of day 1 (D1) CL2122 and GMC101 worms (n = 3 biologically independent samples). b, Basal respiration and after carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP) (45 min, 10 μM) of day 1 (D1) and 3 (D3) adult worms (CL2122, n = 8; GMC101, n = 8 biologically independent samples). c, Immunoblot (CL2122, n = 4; GMC101, n = 4, representative of five biological replicates) of OXPHOS proteins in control and GMC101 at D1. d, Respiration assay as in b in CL2122 worms (ev, n = 8; atfs-1, n = 8 biologically independent samples) and GMC101 worms (ev, n = 8; atfs-1, n = 6 biologically independent samples) fed with atfs-1 RNAi. e, Amyloid aggregation in GMC101 worms upon atfs-1 RNAi shown by western blotting of two biological repeats. f, Mobility of GMC101 fed with 50% dilution of atfs-1 RNAi (ev, n = 59; atfs-1 1/2, n = 50 worms). g, MSR transcript analysis of GMC101 worms upon atfs-1 RNAi (n = 3 biologically independent samples). h, Mobility of control and afts-1-overexpressing GMC101 strains (GMC101, n = 61; AUW9, n = 48; AUW10, n = 41 worms). Data are mean ± s.e.m. *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; NS, not significant. Overall differences between conditions were assessed by two-way ANOVA. Differences in individual genes or proteins were assessed using two-tailed t-tests (95% confidence interval). All experiments were performed independently at least twice. ev, scrambled RNAi; AU, arbitrary units. See also Extended Data Figs 3, 4. For uncropped gel source data, see Supplementary Fig. 1. For all individual P values, see Source Data.
Mitochondrial homeostasis counteracts Aβ proteotoxicity
The control of mitochondrial function and UPRmt during stress in the worm is mostly attributable to the activating transcription factor associated with stress, atfs-1 (refs 17, 18). Depletion of atfs-1 by RNA interference (RNAi) through feeding of GMC101, but not CL2122, worms, caused a severe developmental delay even in the absence of the disease-inducing temperature shift (Extended Data Fig. 3h), phenocopying mitochondrial respiration mutants that rely on atfs-1 for survival and adaption18. Comparative analysis of transcripts involved in cytosolic and nuclear adaptation pathways, such as UPRER, HSR and daf-16, also showed a mild induction of the UPRER and a notable upregulation of the HSR in GMC101 worms (Extended Data Fig. 3i), in line with the role of the HSR as a primary defence against proteotoxic stress in worms19. We therefore evaluated the effect of RNAis targeting key regulators of these pathways, hsf-1 and xbp-1, on the development of GMC101 worms. Only atfs-1 RNAi led to extreme developmental delays in GMC101 worms, whereas no alterations were observed with any of these RNAis in CL2122 worms (Extended Data Fig. 3j).
Notably, basal and maximal respiration in adult GMC101 worms was significantly impaired upon atfs-1 silencing, whereas only the maximal respiration was partially affected in CL2122 worms (Fig. 2d). In addition, atfs-1 RNAi exacerbated aggregation (Fig. 2e and Extended Data Fig. 3k) and paralysis (Fig. 2f) in GMC101 worms, whereas the mobility of CL2122 worms was unaffected (Extended Data Fig. 3l). Furthermore, atfs-1 knockdown in GMC101 worms prominently repressed the MSR signature, including mitophagy effectors (Fig. 2g), whereas in CL2122 worms, pdr-1, dct-1 and OXPHOS transcripts were even induced, despite silencing of atfs-1 in both strains (Extended Data Fig. 3m, n).
Given the induction of the HSR in GMC101 worms, we tested whether hsf-1 repression would have an effect on fitness. Notably, while atfs-1 knockdown paralysed only GMC101 worms, silencing of hsf-1 reduced mobility in both CL2122 and wild-type N2 strains when incubated at 25 °C (Extended Data Fig. 3o), reflecting a general effect of hsf-1 on homeostasis independent of the strain. Worm mobility was similarly impaired in GMC101, but not in CL2122, worms following silencing of atfs-1 with an alternative RNAi that we generated (atfs-1 #2) (Extended Data Fig. 3p, q and Supplementary Table 13), confirming a specific role of atfs-1 in ensuring organismal homeostasis in GMC101 worms. Furthermore, silencing of ubl-5, another positive regulator of the UPRmt (ref. 20), also delayed development and decreased health- and lifespan specifically in GMC101 worms (Extended Data Fig. 4a–c). Interestingly, atfs-1 silencing led to further upregulation of the HSR in GMC101 worms and to its induction in CL2122 worms (Extended Data Fig. 4d, e), while repressing the MSR specifically in GMC101 worms (Fig. 2g and Extended Data Fig. 3n).
Conversely, to increase atfs-1 function, we generated two GMC101-derived strains, AUW9 and AUW10, and one CL2122-derived line (AUW11), overexpressing atfs-1 (Supplementary Tables 12, 13). This resulted in induction of the UPRmt (Extended Data Fig. 4f), a significant increase in fitness and a decrease in paralysis and death scores in the GMC101-derived strains AUW9 and AUW10 (Fig. 2h and Extended Data Fig. 4g, h), while no changes were observed in AUW11 worms (Extended Data Fig. 4g). As an alternative approach to increase the MSR, we crossed GMC101 worms with two long-lived mitochondrial mutants, that is, clk-1 (ref. 21) and nuo-6 (ref. 22) (Supplementary Tables 12, 13). Consistently, GMC101 worms with a mutation in these mitochondrial genes (AUW12, with the clk-1 mutation, and AUW13, mutated for nuo-6) had intermediate phenotypes between GMC101 and the mitochondrial mutants, with an increased health- and lifespan (Extended Data Fig. 4i–l).
Together, these results indicate that atfs-1 and the induction of MSR ensure proteostasis and survival in this worm model of Aβ aggregation, and that mitochondria have an active, rather than passive, role during Aβ proteotoxic stress. This prompted us to investigate the potential of increasing mitochondrial proteostasis to halt the progression of this deleterious phenotype.
Effect of mitochondrial translation on Aβ proteotoxicity
Given the strong association between UPRmt and AD observed above, we investigated the effects of two established strategies to induce UPRmt in C. elegans: genetically, by silencing the expression of the mitochondrial ribosomal protein mrps-5 (ref. 23); and pharmacologically, by using the mitochondrial translation inhibitor doxycycline (DOX)23,24. Both interventions, which increase worm health and lifespan23, markedly induced transcripts of UPRmt, mitophagy and respiration genes in GMC101 worms (Fig. 3a, b), without causing major development and growth delays (Extended Data Fig. 5a). DOX similarly induced these pathways in CL2122 worms (Extended Data Fig. 5b). No changes in UPRER and HSR were observed in GMC101 worms treated with DOX or mrps-5 RNAi (Extended Data Fig. 5c, d), whereas one daf-16 target, the mitochondrial superoxide dismutase sod-3, was induced (Extended Data Fig. 5c, d). The transcriptional induction of the MSR positively affected fitness and lifespan of GMC101 worms (Fig. 3c–e). Furthermore, Aβ aggregation was reduced by mrps-5 RNAi and DOX (Fig. 3f and Extended Data Fig. 5e). Importantly, the improvement in motility and Aβ clearance in GMC101 worms required atfs-1 (Fig. 3g, h), demonstrating the contribution of the UPRmt to the phenotypic improvements. Respirometry on GMC101 worms fed with mrps-5 RNAi at day 3 and day 6 of adulthood (Extended Data Fig. 5f), showed that mrps-5 knockdown prevented the decrease in respiration upon ageing, suggesting a stabilization of mitochondrial function following the MSR-dependent improvement of proteostasis.
a, b, MSR transcript levels in GMC101 worms that were fed mrps-5 RNAi or treated with DOX (a, b, n = 3 biologically independent samples). c, Mobility of GMC101 upon mrps-5 RNAi (n = 35 worms) or DOX treatment (n = 54 worms). The ANOVA P values are reported on the left bottom side of the plots. d, e, Percentage of paralysed (d) and dead (e) D8 adult GMC101 worms after mrps-5 RNAi or DOX treatment (n = 3 independent experiments). f, Western blot of amyloid aggregation in GMC101 upon mrps-5 RNAi or DOX treatment (n = 2 biologically independent samples). g, Mobility of GMC101 worms upon atfs-1 RNAi feeding (ev, n = 54; mrps-5, n = 49; DOX, n = 49 worms). The ANOVA P values are reported on the left bottom side of the plots. h, Amyloid aggregation in DOX-treated GMC101 worms upon atfs-1 RNAi (n = 2 biological replicates). i, Mobility of GMC101 upon dct-1 RNAi (ev, n = 44; DOX, n = 59; mrps-5, n = 66 worms). ***P = 0.0004; ****P ≤ 0.0001. The ANOVA P values are reported on the bottom of the plots. j, Amyloid aggregation in DOX-treated GMC101 worms upon dct-1 RNAi (n = 3 biologically independent samples). k, Confocal images of the SH-SY5Y neuroblastoma cell line stained with the anti-β-amyloid 1–42 antibody, after DOX and, where indicated, ISRIB treatments for 24 h. Scale bar, 10 μm. See Methods for further details. Data are mean ± s.e.m. *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. Overall differences between conditions were assessed by two-way ANOVA. Differences in individual genes or between two groups were assessed using two-tailed t-tests (95% confidence interval). All experiments were performed independently at least twice. ev, scrambled RNAi; DOX, doxycycline; AU, arbitrary units; ISRIB, integrated stress response inhibitor. See also Extended Data Fig. 5. For uncropped gel source data, see Supplementary Fig. 1. For all individual P values, see Source Data.
We then extended our investigation to a human SH-SY5Y neuroblastoma cell line expressing the Swedish K670N/M671L mutation in APP (APPSwe)25. DOX markedly reduced intracellular Aβ deposits, as shown with an Aβ1–42-specific antibody (Fig. 3k and Extended Data Fig. 5g). This improvement was linked to a mitochondrial–nuclear protein imbalance and the induction of components of the MSR (Extended Data Fig. 5h, i). These results add to observations of in vitro26 and in vivo studies in flies27 and patients28 with AD, which have suggested that DOX treatment may ameliorate AD and Aβ aggregation. Recently, the regulation of the mitochondrial stress responses in mammals, including DOX-dependent mitochondrial stress, was shown to rely on the activation of the integrated stress response (ISR) and on the transcription factor ATF4 (refs 29, 30, 31). Pretreating cells with ISRIB, a global ISR inhibitor32, prevented amyloid clearance by DOX and interfered with DOX-mediated induction of canonical ATF4 target genes, such as CHOP and CHAC1 (ref. 29) (Fig. 3k and Extended Data Fig. 5g, j). This suggests that ISR is involved in resolving Aβ proteotoxic stress in mammalian cells.
Given the induction of mitophagy in 3×TgAD mice, human patients with AD and in GMC101 worms, and its further increase upon mrps-5 RNAi and DOX treatments, we also tested the contribution of mitophagy to the homeostasis of GMC101 worms. To achieve this, we silenced, using RNAi, dct-1, an evolutionarily conserved regulator of mitophagy33. dct-1 RNAi already reduced the health- and lifespan of GMC101 worms in basal conditions (Extended Data Fig. 5k–m). Furthermore, it decreased the positive effects of mrps-5 RNAi and DOX on health- and lifespan (Fig. 3i and Extended Data Fig. 5l, m) and proteostasis (Fig. 3j). Conversely, dct-1 knockdown in CL2122 worms affected their movement only during ageing (Extended Data Fig. 5n, o), stressing the relevance of mitophagy in ageing. Taken together, these data show that mitophagy, in addition to UPRmt, is induced and required for the survival of GMC101 worms and for the beneficial effects of the described interventions.
NAD+ boosters attenuate Aβ proteotoxicity
The UPRmt and mitophagy pathways are also potently induced in worms and in various mammalian tissues by NAD+-boosting compounds, such as nicotinamide riboside (NR) and Olaparib (AZD2281 or AZD)34,35,36,37. Similar to DOX treatment and mrps-5 RNAi, treatment of GMC101 worms with NR and AZD induced MSR (Fig. 4a, b), and improved health- and lifespan (Fig. 4c, d and Extended Data Fig. 6a). The NR- and AZD-mediated induction of the MSR was also observed in CL2122 worms (Extended Data Fig. 6b, c); however, these treatments only improved CL2122 fitness during ageing (Extended Data Fig. 6d, e), similar to what has previously been shown in N2 worms34. In addition, treatment of GMC101 worms with NR and AZD reduced proteotoxic stress (Fig. 4e). Notably, the NR-mediated phenotypic and proteostasis benefits required atfs-1 (Fig. 4f–h and Extended Data Fig. 6f) and dct-1 (Fig. 4i–k and Extended Data Fig. 6g). NR has been shown to increase sirtuin activity and activate FOXO (encoded by daf-16 in C. elegans) signalling in mammals and C. elegans34. Therefore, we evaluated the effect of daf-16 and sir-2.1 silencing on development, healthspan and NR-dependent benefits in GMC101 worms. Whereas sir-2.1 knockdown did not delay growth of GMC101 or control CL2122 worms (Extended Data Fig. 6h), it increased paralysis and death in GMC101 worms similar to what was found in GMC101 worms treated with dct-1 and atfs-1 RNAi clones (Extended Data Fig. 6i, k). However, NR still significantly rescued health- and lifespan of GMC101 worms fed with sir-2.1 RNAi, showing that the effects of NR rely mostly on atfs-1 and dct-1 (Extended Data Fig. 6i, k). Instead, feeding GMC101 worms with daf-16 RNAi did not result in major phenotypic changes (Extended Data Fig. 6h, j, k). Furthermore, NR did not affect the expression of daf-16 and its targets (Extended Data Fig. 6l). NR and AZD had only a minimal impact on UPRER and HSR in GMC101 worms, with the induction of hsp-16.41, hsp-16.48 and hsp-16.49 (hsp-16.48/49) (Extended Data Fig. 6l, m). These results indicate that in the GMC101 model, the main mode of action of NAD+ boosting involves the induction of the MSR.
a, b, MSR transcripts in GMC101 worms treated with NR (a) or AZD (b) (a, b, n = 3 biologically independent samples). c, Mobility of GMC101 worms treated with NR (n = 50 worms) or AZD (n = 39 worms). **P ≤0.001 (AZD, P = 0.006); ****P ≤ 0.0001 (NR). d, Percentage of dead D8 adult GMC101 worms after NR or AZD treatment (n = 3 independent experiments). e, Western blot of amyloid aggregation in GMC101 worms after NR or AZD treatment (n = 3 biologically independent samples for all groups). f, Mobility of GMC101 worms treated with NR upon atfs-1 RNAi feeding (ev, n = 52; atfs-1, n = 38; NR, n = 40; NR, atfs-1, n = 41 worms). ***P ≤ 0.001 (atfs-1, P = 0.0006). g, Percentage of dead D8 adult GMC101 worms treated with NR upon atfs-1 RNAi (n = 5 biologically independent samples). h, Amyloid aggregation in NR-treated GMC101 worms upon atfs-1 RNAi feeding (western blot, representative of two biological replicates). i, Mobility of NR-treated GMC101 worms upon dct-1 RNAi (ev, n = 41; dct-1, n = 40; NR, n = 39; NR, dct-1, n = 50 worms). j, Percentage of dead D8 adult GMC101 worms treated with NR upon dct-1 RNAi (n = 5 biologically independent samples). k, Amyloid aggregation immunoblot in NR-treated GMC101 worms upon dct-1 RNAi (n = 3 biologically independent samples). l, Confocal images of APPSwe SH-SY5Y cells stained with anti-β-amyloid 1–42 antibody, after 24 h of NR treatment. Scale bar, 10 μm. Data are mean ± s.e.m. *P < 0.05; **P ≤ 0.01; ***P ≤0.001; ****P ≤ 0.0001; NS, not significant. Overall differences between conditions were assessed by two-way ANOVA. Differences in individual genes or between two groups were assessed using two-tailed t-tests (95% confidence interval). All experiments were performed independently at least twice. ev, scrambled RNAi; AU, arbitrary units. See also Extended Data Fig. 6. For uncropped gel source data, see Supplementary Fig. 1. For all individual P values, see Source Data.
We also assessed the effect of NR on human Aβ-expressing neuroblastoma cells and, consistent with the data from the C. elegans model, we observed a remarkable reduction in intracellular Aβ deposits with NR (Fig. 4l and Extended Data Fig. 6n), accompanied by increased OXPHOS protein (Extended Data Fig. 5h) and MSR transcript levels (Extended Data Fig. 6o).
NR reduces Aβ levels in transgenic mice with AD
We have previously shown that NR exerts beneficial effects on health- and lifespan34,36, but whether it also improves mitochondrial function and proteostasis in vivo in a mouse model of Aβ proteotoxicity remains unknown. We therefore treated APPSwe/PSEN1dE9 mice with AD with NR and assessed the levels of Aβ plaques in brain using thioflavin S, as well as mitochondrial OXPHOS and MSR transcript and protein expression. Consistent with the effects observed in cells and worms, NR robustly reduced Aβ deposits in cortex tissues of the mice with AD (Fig. 5a), and induced the MSR mRNA signature (Fig. 5b) and OXPHOS protein levels (Fig. 5c). NR also increased context-dependent memory in the APPSwe/PSEN1dE9 mice with AD, which was impaired compared to wild-type mice, as assessed by contextual fear conditioning38 (Fig. 5d), suggesting a potential positive impact on cognitive function. We therefore propose that restoring or increasing mitochondrial function and proteostasis induces a conserved repair mechanism, in worms and mice, that leads to decreased Aβ proteotoxicity and improves health- and lifespan.
a, Representative images and corresponding quantification of plaques in cortex samples of APPSwe/PSEN1dE9 mice (APP/PSEN1) with AD following NR treatment, stained using thioflavin S (ThS) (chow diet, n = 5 mice; NR, n = 7 mice; NR treatment: 400 mg kg−1 per day for 10 weeks). Scale bar, 200 μm. *P < 0.05 (relative area, P = 0.041; number, P = 0.014). b, c, MSR transcript (b) and immunoblot (c) analyses of cortex samples of APPSwe/PSEN1dE9 mice following NR treatment (b, n = 5 animals per group; c, n = 4 animals per group). Data are representative of two independent experiments. d, Contextual fear conditioning in wild-type (n = 5 mice) and APPSwe/PSEN1dE9 (n = 7 mice) mice with or without NR treatment, plotted as a function of the time interval (left) or as an average of the total obtained values (right). *P < 0.05 (P = 0.03); ****P ≤ 0.0001. One-tail t-test performed between averaged values of wild-type and APPSwe/PSEN1dE9 mice (P = 0.0574, 95% confidence interval). Data are mean ± s.e.m. *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; NS, not significant. Overall differences between conditions were assessed by two-way ANOVA, unless indicated. Differences in individual genes or between two groups were assessed using two-tailed t-tests (95% confidence interval). See Methods for further details. CD, chow diet. For uncropped gel source data, see Supplementary Fig. 1. For all individual P values, see Source Data.
Discussion
Proteotoxic stress in Aβ diseases, such as AD, is associated with mitochondrial dysfunction, and reduced OXPHOS activity has been considered one of the major hallmarks of these diseases. Here, we identify a cross-species mitochondrial stress response signature that implicates mitochondrial proteostasis as a key mechanism in response to Aβ proteotoxic stress. Notably, we show that Aβ accumulation induces both UPRmt and mitophagy in a manner that is conserved from C. elegans to humans. Currently, the molecular mechanism driving this induction is still unclear. On the basis of our results, we speculate that it involves the alteration of a basic, conserved functional process, such as mitochondrial import, which is linked to the activation of the UPRmt (ref. 18), is disrupted during Aβ proteotoxic stress39 and is downregulated in patients with AD in our analyses. Our work also provides evidence that mitochondria have an active role in the pathogenesis of Aβ diseases, as reducing mitochondrial homeostasis via atfs-1 depletion in GMC101 worms aggravates the hallmarks of the disease (model in Extended Data Fig. 7); conversely, boosting mitochondrial proteostasis by increasing the UPRmt and mitophagy decreases protein aggregation, restores worm fitness and delays disease progression, ultimately translating to increased lifespan. Similarly, in mammals we show that DOX and NR decrease Aβ accumulation in a human SH-SY5Y neuronal cell line and that NR treatment reduces amyloid plaque formation in the brain and improves contextual memory in APPSwe/PSEN1dE9 mice with AD. These findings indicate that boosting mitochondrial function and proteostasis may decrease the formation of detrimental protein aggregates in the context of proteotoxic diseases, which are characterized by reduced mitochondrial activity and loss of proteostasis. Together with initial evidence suggesting potential benefits of targeting dysfunctional mitochondria in AD26,27,28,40,41 and in view of recent findings linking mitochondrial stress to the induction of cytosolic proteostasis mechanisms42,43,44,45, our data support the concept that increasing mitochondrial proteostasis may have potential for the management of pervasive Aβ proteopathies, such as AD.
Methods
Animal experiments
3×TgAD male mice, bearing human mutant APPSwe, PS1M146V and TauP301L transgenes11, and wild-type, hybrid 129/C57BL/6 mouse littermates were transcardially perfused with saline at six and nine months of age (n = 6 per group) and brains from each group were hemisected. One hemisphere was immersion-fixed in 4% paraformaldehyde and 0.1% glutaraldehyde for 24 h and stored in cryoprotectant. From the other hemisphere, the hippocampus, frontoparietal cortex and cerebellum were rapidly dissected and snap-frozen.
APPSwe/PSEN1dE9 male mice (Tg(APPSwe,PSEN1dE9)85Dbo/Mmjax) were purchased from JAX. APPSwe/PSEN1dE9 mice were fed NR pellets (400 mg kg−1 per day) for 10 weeks, starting at the age of 4 months. The pellets were prepared by mixing powdered chow diet (2016S, Harlan Laboratories) with water or with NR dissolved in water. Pellets were dried under a laminar flow hood for 48 h. Mice were housed in groups of two to four animals per cage and randomized to 7–8 animals per experimental group according to their body weight. No blinding was used during the experiment procedures.
Fear conditioning
Fear conditioning was conducted as described previously38, using chambers placed in sound-attenuating boxes (635 × 420 × 445 mm) (Med Associates). Each chamber is equipped with lights (infrared and visible), speakers and the floor of the chamber consisted of a stainless-steel grid connected to a shock generator. A computer running MED PC-IV (Med Associates) controls the presentation of the stimuli (tone and shock). A video camera connected to the MED-VFC-NIR-M automated video-tracking system was used to detect and record movements and freezing behaviour. On the first day (conditioning session), mice were habituated to the testing chamber for 2 min followed by the presentation of an auditory cue for 30 s (5 kHz tone; 80 dB, conditioned stimulus (CS)), co-terminating with a foot shock during the last 2 s (0.7 mA; unconditioned stimulus (US)). Mice received four CS–US pairings with an inter-trial interval of 2 min. The last CS–US pairing was followed by a 2-min resting phase before mice were returned to their home cage. The entire conditioning session lasted 12 min. On the second day, the mouse was re-exposed to the same context with no tone and the freezing response was monitored. Fear was assessed by measuring the percentage of time spent freezing, characterized by a crouching posture and an absence of any visible movement except breathing.
Ethical approval
All experiments were performed in compliance with all relevant ethical regulations. The experiments using post-mortem human samples were authorized by the Michigan State University (MSU) Human Research Protection Program. Informed consent for brain autopsies of Rush Religious Orders Study participants was authorized by the Rush University Medical Center Institutional Review Board. The experiments with 3×TgAD mice were authorized by the MSU Institutional Review Board and Institutional Animal Care and Use Committee. The experiments with APPSwe/PSEN1dE9 mice were authorized by the local animal experimentation committee of the Canton de Vaud under license 3207. The committee that approved the license is the Commission cantonale pour l’expérimentation animale.
Human brain samples
Superior temporal cortex (Brodmann area 22) samples were obtained post mortem from participants in the Religious Orders Study who died with an ante-mortem clinical diagnosis of no cognitive impairment (NCI), mild cognitive impairment (MCI) or AD (n = 8 per group) (Supplementary Table 6). Neuropsychological and clinical examinations, as well as post-mortem diagnostic evaluations, have been described elsewhere46. Demographic, ante-mortem cognitive testing and post-mortem diagnostic variables were compared among the groups using the non-parametric Kruskal–Wallis test with Bonferroni correction for multiple comparisons. Gender and apoE ε4 allele distribution were compared using a Fisher’s exact test with Bonferroni correction.
Bioinformatics analysis
For the in silico analysis of human brain expression datasets, we have used two sets of publicly available RNA-seq data: (1) from the Harvard Brain Tissue Resource Center (HBTRC), for human primary visual cortex (GN accession: GN327) and human prefrontal cortex (GN accession: GN328) and (2) from the Translational Genomics Research Institute, for the whole brain (GN accession: GN314)47. These two datasets are publicly available on GeneNetwork (http://www.genenetwork.org/webqtl/main.py). For correlation analysis, Pearson’s r genetic correlation of the UPRmt, mitophagy, endoplasmic reticulum stress and HSR gene sets was performed to establish the correlation between these pathways and genes that are associated or are causal to the development of AD. Analyses were performed using the hallmarks and canonical pathway gene sets databases. Additional information about the datasets used are provided in Supplementary Information.
Gene expression analyses in C. elegans
A total of approximately 3,000 worms per condition, divided into three biological replicates, was recovered in M9 buffer from NGM plates and lysed in the TriPure RNA reagent. Each experiment was repeated twice. Total RNA was transcribed to cDNA using the QuantiTect Reverse Transcription Kit (Qiagen). Expression of selected genes was analysed using the LightCycler480 system (Roche) and LightCycler 480 SYBR Green I Master reagent (Roche). For C. elegans, two housekeeping genes were used to normalize the expression data, actin (act-1) and peroxisomal membrane protein 3 (pmp-3). See Supplementary Table 7 for the primer list.
Gene expression analyses in mice
Total RNA was extracted from tissues using TriPure RNA isolation reagent (Roche) according to the product manual. Expression of selected genes was analysed using the LightCycler480 system (Roche) and LightCycler 480 SYBR Green I Master reagent (Roche). The β2-microglobulin (B2m) gene was used as housekeeping reference. See Supplementary Table 8 for the primer list.
Gene expression analyses of human samples
Total RNA was extracted using guanidine-isothiocyanate lysis (PureLink, Ambion) from cortex samples, and RNA integrity and concentration were verified using Bioanalysis (Agilent). Samples were randomized based on diagnostic group and assayed in triplicate on a real-time PCR cycler (ABI 7500, Applied Biosystems) in 96-well optical plates. qPCR was performed using Taqman hydrolysis probe primer sets (Applied Biosystems) specific for the following human transcripts (Supplementary Table 10): HSPA9, HSPD1, YME1L1, DNM1L, BECN1, SQSTM1, PARK2 (also known as PRKN), COX5A and CYC1. A primer set specific for human GAPDH was used as a control housekeeping transcript. For the APPSwe-expressing SH-SY5Y cell line, total RNA was extracted using TriPure RNA isolation reagent, and expression of selected transcripts was analysed using the LightCycler480 system (Roche) and LightCycler 480 SYBR Green I Master reagent (Roche). The GAPDH gene was used as housekeeping reference. See Supplementary Table 9 for the primer list. The ΔΔCt method was used to determine relative expression levels of each amplicon. Variance component analyses revealed relatively low levels of within-case variability, and the average value of the triplicate qPCR products from each case was used in subsequent analyses.
C. elegans strains and plasmids generation, and RNAi experiments
C. elegans strains (Supplementary Table 12) were cultured at 20 °C on nematode growth medium (NGM) agar plates seeded with E. coli strain OP50 unless stated otherwise. Strains used in this study were the wild-type Bristol N2, GMC101 (unc-54p::A-beta-1-42::unc-54 3′-UTR + mtl-2p::GFP)15, CL2122 ((pPD30.38) unc-54(vector) + (pCL26) mtl-2::GFP)48, CB4876 (clk-1(e2519)) and MQ1333 (nuo-6(qm200)). Strains were provided by the Caenorhabditis Genetics Center (University of Minnesota). The strain CL2122 was outcrossed three times in the N2 background, and subsequently used in the control experiments reported here. AUW9 and AUW10: (GMC101 + epfEx6(atfs-1p::atfs-1)) and AUW11: (CL2122 + epfEx7(atfs-1p::atfs-1)) overexpression strains, and AUW12: [GMC101 + clk-1(e2519) III), AUW13: (GMC101+ nuo-6(qm200) I) were generated for this study.
Cloning and sequencing primers used for C. elegans analyses are listed Supplementary Table 13. The atfs-1p::atfs-1 expression vector was created by amplifying the 1,488-bp sequence upstream of the transcription start site of the atfs-1 coding region by using worm genomic DNA for the promoter region. The PCR product was digested with PciI and AgeI and ligated into the pPD30.38 expression vector containing the gfp coding sequence cloned between inserted AgeI and NotI restriction sites. The atfs-1 coding sequence was instead amplified using C. elegans cDNA, and the PCR product was inserted into pPD30.38 downstream of the promoter region, between AgeI and NheI restriction sites. The correctness of the atfs-1p::atfs-1 construct was assessed by sequencing with the indicated pPD30.38 and atfs-1 sequencing primers. Two independent GMC101 lines (AUW9 and AUW10), and one CL2122 strain (AUW11) carrying the atfs-1p::atfs-1 transgene as extrachromosomal array were analysed in this study. Injection marker myo-2p::gfp was cloned by amplifying the 1,179-bp sequence upstream of the transcription start site of the myo-2 coding region by using C. elegans genomic DNA. The PCR product was digested with PciI and AgeI and ligated into the pPD30.38 expression vector containing atfs-1::gfp to replace the atfs-1 promoter sequence between PciI and AgeI restriction sites. All transgenic strains were created using microinjection. For the generation of the GMC101 lines AUW12 and AUW13, GMC101 males were generated after exposure of L4 worms to 30 °C for 3 h, and allowed to mate with L4 hermaphrodites from clk-1 or nuo-6 mutant strains. The derived progeny was selected for homozygosis of the GMC101 intestinal GFP marker for a few generations, and the homozygosis of the clk-1 or nuo-6 mutant alleles verified by amplifying and sequencing a 500-bp region of the worms genomic DNA encompassing the desired mutation (Supplementary Table 13). Bacterial feeding RNAi experiments were carried out as described49. Clones used were atfs-1 (ZC376.7), ubl-5 (F46F11.4), mrps-5 (E02A10.1), dct-1 (C14F5.1), daf-16 (R13H8.1), sir-2.1 (R11A8.4), xbp-1 (R74.3) and hsf-1 (Y53C10A.12). Clones were purchased from GeneService and verified by sequencing. The novel atfs-1 RNAi construct (atfs-1 #2) used in this study was generated by amplifying 1,400 bp of the atfs-1 genomic DNA sequence, starting from the last exon of atfs-1. Gateway cloning (Thermo Scientfic) was used to insert the PCR product into the gateway vector pL4440gtwy (Addgene 11344), and verified by sequencing. atfs-1 knockdown was verified by qPCR for all the atfs-1 RNAi constructs used here. For phenotypic studies (see below), worms at the L4 larval stage were allowed to reach adulthood and lay eggs on the treatment plates. The derived F1 worms were shifted from 20 °C to 25 °C at the L4 stage to induce amyloid accumulation and paralysis, and phenotypes were assessed over time as indicated. For double RNAi experiments, we used a combination of atfs-1 with mrps-5 RNAi constructs as indicated in the text, with 80% amount of atfs-1 and 20% mrps-5 RNAi. For mRNA analysis, synchronized L1 worms were exposed to the treatment plates, and shifted from 20 °C to 25 °C when reaching the L4 stage. After 24-h incubation at 25 °C, corresponding to day 1 of adulthood, they were collected in M9 for analysis.
Pharmacological treatment of C. elegans
Doxycycline was obtained from Sigma-Aldrich and dissolved in water23. For experiments, a final concentration of 15 μg ml−1 was used. Olaparib (AZD2281) was dissolved in DMSO to experimental concentrations of 300 nM34. Nicotinamide riboside triflate (NR) was custom synthesized by Novalix (http://www.novalix-pharma.com/) and dissolved in water, and used at a final concentration of 1 mM34. Compounds were added just before pouring the plates. For phenotyping experiments, parental F0 L4 worms were allowed to reach adulthood and lay eggs on the treatment plates. The derived F1 worms were therefore exposed to compounds during the full life from eggs until death. For RNA analysis experiments, synchronized L1 worms were exposed to the compounds until collection. To ensure a permanent exposure to the compound, plates were changed twice a week.
Worm phenotypic assays
Mobility
C. elegans movement analysis was performed as described16, starting from day 1 of adulthood, using the Movement Tracker software. The experiments were repeated at least twice.
Development
50 adult worms per condition were transferred to NGM agar plates (10 worms per plate) and allowed to lay eggs for 3 h. They were then removed and the number of eggs per plate was counted. After 72 h, the number of L1–L3 larvae, L4 and adult worms was counted. The experiment was done twice with five individual plates.
Paralysis and death score
5 L4 worms per condition were allowed to reach adulthood and lay eggs on the treatment plates. 45 to 60 F1 worms per condition were manually scored for paralysis after poking, as previously described15. Worms that were unable to respond to any repeated stimulation, were scored as dead. Results are representative of data obtained from at least three independent experiments.
Oxygen-consumption assays
Oxygen consumption was measured using the Seahorse XF96 equipment (Seahorse Bioscience)34,50. Respiration rates were normalized to the number of worms in each individual well and calculated as averaged values of 5–6 repeated measurements. Each experiment was repeated at least twice.
MitoTracker Orange CMTMRos staining
A population of 20 worms at L4 stage were transferred on plates containing MitoTracker Orange CMTMRos (Thermo Scientific) at a final concentration of 2 μg μl−1. The plates were incubated at 25 °C and the worms were collected and washed in 200 μl of M9 in order to remove the residual bacteria after 24 h of treatment. The worms were then incubated for 30 min on regular NGM plates at 25 °C and mounted on an agar pad in M9 buffer for visualization. Mitochondria were observed using confocal laser microscopy.
Phalloidin and DAPI staining
A population of 100 L4 worms was incubated for 24 h at 25 °C. The worms were then washed in M9 and frozen in liquid nitrogen, immediately after this, they were lyophilized using a centrifugal evaporator. Worms were permeabilized using acetone. 2 U of phalloidin (Thermo Scientific) was resuspended in 20 μl of a buffer containing: Na-phosphate pH 7.5 (final concentration 0.2 mM), MgCl2 (final concentration 1 mM), SDS (final concentration 0.004%) and dH2O to volume. The worms were incubated for 1 h in the dark and then washed twice in PBS and incubated in 20 μl of 2 μg ml−1 DAPI in PBS for 5 min. Following the immobilization, worms were observed using confocal laser microscopy.
Real-time PCR for mitochondrial:nuclear DNA ratio
Absolute quantification of the mtDNA copy number in worms was performed by real-time PCR. Relative values for nd-1 and act-3 were compared within each sample to generate a ratio representing the relative level of mitochondrial DNA per nuclear genome. The results obtained were confirmed with a second mitochondrial gene MTCE.26. The average of at least three technical repeats was used for each biological data point. Each experiment was performed on at least five independent biological samples.
Cell culture and treatments
The SH-SY5Y neuroblastoma cell line expressing the APP Swedish K670N/M671L double mutation (APPSwe)25 was a gift from A. Cedazo-Minguez (Karolinska Institute). Cells were grown in DMEM/F-12, supplemented with 10% fetal bovine serum (FBS, Gibco), GlutaMAX (100×, Gibco) and penicillin–streptomycin (1×, Gibco). Cells were selected in 4 μg ml−1 Geneticin Selective Antibiotic (G418 Sulphate, Sigma-Aldrich) and grown for three generations before experiments with cells plated and passaged at 4 × 103 cells per ml and 60% confluence, respectively. Cells were cultured at 37 °C in a 5% CO2 atmosphere and tested for mycoplasma using Mycoprobe (CUL001B, R&D systems) following the manufacturer’s instructions. Cells were treated with 10 μg ml−1 DOX, 1 or 3 mM NR, or 0.5 μM ISRIB (Sigma-Aldrich), as indicated for 24 h before cell collection or fixation. For immunostaining, cells were fixed with 1× Formal-Fixx (Thermo Scientific) for 15 min. After 15 min permeabilization with 0.1% Triton X-100, cells were blocked in PBS supplemented with 5% FBS for 1 h and immunostained overnight at 4 °C with the anti-β-amyloid 1–42 antibody (1:100, Millipore AB5078P). The secondary antibody was coupled to an Alexa-488 fluorochrome (Thermo Scientific) and nuclei were stained with DAPI (Invitrogen, D1306). After washing in PBS, cell slides were mounted with DAKO mounting medium (DAKO, S3023) and analysed with a Zeiss LSM 700 confocal microscope (Carl Zeiss MicroImaging) equipped with a Plan-Apochromat 40×/1.3 NA oil immersion objective lens using a 488 nm laser. Laser power was set at the lowest intensity allowing clear visualization of the signal. Imaging settings were maintained with the same parameters for comparison between different experimental conditions.
Western blot analysis (C. elegans)
Worms were lysed by sonication with RIPA buffer containing protease and phosphatase inhibitors (Roche), and analysed by SDS–PAGE and western blot. The concentration of extracted protein was determined using the Bio-Rad Protein Assay. Proteins were detected using the following antibodies: anti-β-actin (Sigma-Aldrich), anti-tubulin (Santa Cruz), atp-5, ucr-1 (OXPHOS cocktail, Abcam), anti-β-amyloid 1–16 (6E10) (BioLegend). In addition to the housekeeping proteins, loading was monitored by ponceau red to ensure homogeneous loading. Pixel intensity was quantified using ImageJ software. Each immunoblot experiment was repeated at least twice using three biological replicates each containing approximately 1,000 worms.
Western blot analysis (mice)
Frozen cortex tissue samples were lysed by mechanical homogenization with RIPA buffer containing protease and phosphatase inhibitors, and analysed by SDS–PAGE and western blot. Subsequently, the concentration of extracted protein was determined using the Bio-Rad Protein Assay. Proteins were detected using the following antibodies: HSP60 (Enzo Life Science), CLPP (Sigma-Aldrich), GAPDH (14C10) (Cell Signaling), LONP1 (Sigma-Aldrich), PINK1 (Novus Biologicals), LC3 A/B (Cell Signaling), SDHB (OXPHOS cocktail, Abcam), MTCO1 (Abcam), ubiquitin (Enzo), p62 (BD Transduction Laboratories), phopshorylated p62 (Cell Signaling), VDAC (Abcam). In addition to the housekeeping proteins, loading was monitored by ponceau red to ensure homogeneous loading. Antibody detection reactions for all immunoblot experiments were developed by enhanced chemiluminescence (Advansta) and imaged using the c300 imaging system (Azure Biosystems). Pixel intensity was quantified using ImageJ software.
Western blot analysis (human samples)
Frozen cortex tissue samples were prepared as previously described51. Samples were randomized based on diagnostic group and assayed in triplicate. For CLPP, blots were incubated overnight at 4 °C with a mouse monoclonal antibody against CLPP (1:1,000; clone 2E1D9, ProteinTech) and then incubated for 1 h with a near-infrared-labelled goat anti-mouse IgG secondary antiserum (IRDye 800LT, 1:10,000; Licor) and analysed on an Odyssey imaging system (Licor). After imaging, the membranes were stripped and re-probed with a mouse monoclonal GAPDH antibody (1:10,000; clone 2D9, Origene) overnight followed by a 1-h incubation with near-infrared-labelled goat anti-mouse IgG secondary antiserum and Odyssey imaging. For mtDNAJ (also known as TID1), blots were incubated overnight at 4 °C with both a mouse monoclonal antibody against mtDNAJ (1:500; clone RS13, Cell Signaling) and the GAPDH antibody, followed by goat anti-mouse IgG incubation and Odyssey imaging. Signals for CLPP and mtDNAJ were normalized to GAPDH for quantitative analysis. Details about the antibodies used are listed in Supplementary Table 11.
Citrate synthase activity assay
Citrate synthase enzymatic activity was determined in mouse cortex samples and C. elegans using the citrate synthase assay kit (Sigma-Aldrich). Absorbance at 412 nm was recorded on a Victor X4 (PerkinElmer) with 10 readings over the 1.5-min timespan. These readings were in the linear range of enzymatic activity. The difference between baseline and oxaloacetate-treated samples was obtained and used to calculate total citrate synthase activity according to the formula provided in the manual. The obtained values were normalized to the amount of protein used for each sample.
Extraction of the mitochondrial fraction
Cortexes were placed in homogenizer glass-Teflon potter tubes with 5 ml of a buffer prepared by adding 10 ml of 0.1 M Tris–MOPS and 1 ml of EGTA-Tris to 20 ml of 1 M sucrose and bringing the volume to 100 ml with distilled water. Homogenization was performed at 1,600 r.p.m. for 5 min. The samples were then centrifuged for 10 min at 600g and the supernatants were collected. A second centrifugation at 7,000g for 10 min was performed on the supernatants in order to precipitate the mitochondrial fraction. All steps were performed on ice or at 4 °C.
Histology
Brain hemispheres were collected from anaesthetized mice and immediately frozen in isopentane. 8-μm cryosections were collected and fixed with 4% paraformaldehyde. For immunostaining, sections were stained with 0.01% thioflavin S (Sigma-Aldrich) for 15 min at room temperature, and after washes in ethanol and PBS, stained with Hoechst (Life Technology). After washing in PBS, tissue sections were mounted with DAKO mounting medium. Images were acquired using a Leica DM 5500 (Leica Microsystems) CMOS camera 2900 Colour at the same exposure time. Quantitative analysis of the immunofluorescence data was carried out by histogram analysis of the fluorescence intensity at each pixel across the images using ImageJ (Fiji; National Institutes of Health). Appropriate thresholds were used for all images of each single experiment to eliminate the background signal in the images before histogram analysis. Fluorescence intensity and signal-positive areas were calculated using the integrated ‘analyse particles’ tool of the Fiji software, and statistical analysis was performed using Prism 6 (GraphPad Software).
Statistical analyses
No statistical methods were used to predetermine sample size. Differences between two groups were assessed using two-tailed t-tests. Differences between more than two groups were assessed by using two-way ANOVA. GraphPad Prism 6 (GraphPad Software, Inc.) was used for all statistical analyses. Variability in all plots and graphs is presented as the s.e.m. All P < 0.05 were considered to be significant. *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001 unless stated otherwise. All mouse experiments were performed once. Animals that showed signs of severity, predefined by the animal authorizations were euthanized. These animals, together with those who died spontaneously during the experiments, were excluded from the calculations. These criteria were established before starting the experiments. For motility, fitness and death scoring experiments in C. elegans, sample size was estimated based on the known variability of the assay. All experiments were done non-blinded and repeated at least twice.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information). Source data for uncropped gels are provided in Supplementary Fig. 1 and Source Data for all the individual P values are provided in the online version of the paper. The expression datasets that support the findings of this study are available in GeneNetwork (http://www.genenetwork.org/webqtl/main.py) with identifiers GN327, GN328, GN31447, and from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) with the identifiers GSE3112, GSE3945452,53. The worm strains generated in this study are available upon request from the corresponding author.
References
Alzheimer’s Association. 2016 Alzheimer’s disease facts and figures. Alzheimers Dement. 12, 459–509 (2016)
Dember, L. M. Amyloidosis-associated kidney disease. J. Am. Soc. Nephrol. 17, 3458–3471 (2006)
Askanas, V. & Engel, W. K. Sporadic inclusion-body myositis: conformational multifactorial ageing-related degenerative muscle disease associated with proteasomal and lysosomal inhibition, endoplasmic reticulum stress, and accumulation of amyloid-β42 oligomers and phosphorylated tau. Presse Med. 40, e219–e235 (2011)
Gauthier, S. et al. Why has therapy development for dementia failed in the last two decades? Alzheimers Dement. 12, 60–64 (2016)
Soejitno, A., Tjan, A. & Purwata, T. E. Alzheimer’s disease: lessons learned from amyloidocentric clinical trials. CNS Drugs 29, 487–502 (2015)
Herrup, K. et al. Beyond amyloid: getting real about non-amyloid targets in Alzheimer’s disease. Alzheimers Dement. 9, 452–458 (2013)
Selfridge, J. E., E, L., Lu, J. & Swerdlow, R. H. Role of mitochondrial homeostasis and dynamics in Alzheimer’s disease. Neurobiol. Dis. 51, 3–12 (2013)
Pellegrino, M. W. & Haynes, C. M. Mitophagy and the mitochondrial unfolded protein response in neurodegeneration and bacterial infection. BMC Biol. 13, 22 (2015)
Beck, J. S., Mufson, E. J. & Counts, S. E. Evidence for mitochondrial UPR gene activation in familial and sporadic Alzheimer’s disease. Curr. Alzheimer Res. 13, 610–614 (2016)
Mufson, E. J. et al. Mild cognitive impairment: pathology and mechanisms. Acta Neuropathol. 123, 13–30 (2012)
Oddo, S. et al. Triple-transgenic model of Alzheimer’s disease with plaques and tangles: intracellular Aβ and synaptic dysfunction. Neuron 39, 409–421 (2003)
Jovaisaite, V., Mouchiroud, L. & Auwerx, J. The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. J. Exp. Biol. 217, 137–143 (2014)
Cohen, E., Bieschke, J., Perciavalle, R. M., Kelly, J. W. & Dillin, A. Opposing activities protect against age-onset proteotoxicity. Science 313, 1604–1610 (2006)
Link, C. D. C. elegans models of age-associated neurodegenerative diseases: lessons from transgenic worm models of Alzheimer’s disease. Exp. Gerontol. 41, 1007–1013 (2006)
McColl, G. et al. Utility of an improved model of amyloid-beta (Aβ1–42) toxicity in Caenorhabditis elegans for drug screening for Alzheimer’s disease. Mol. Neurodegener. 7, 57 (2012)
Mouchiroud, L. et al. The movement tracker: a flexible system for automated movement analysis in invertebrate model organisms. Curr. Protoc. Neurosci. 77, 8.37.1–8.37.21 (2016)
Nargund, A. M., Fiorese, C. J., Pellegrino, M. W., Deng, P. & Haynes, C. M. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPRmt. Mol. Cell 58, 123–133 (2015)
Nargund, A. M., Pellegrino, M. W., Fiorese, C. J., Baker, B. M. & Haynes, C. M. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337, 587–590 (2012)
Prahlad, V. & Morimoto, R. I. Integrating the stress response: lessons for neurodegenerative diseases from C. elegans. Trends Cell Biol. 19, 52–61 (2009)
Benedetti, C., Haynes, C. M., Yang, Y., Harding, H. P. & Ron, D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics 174, 229–239 (2006)
Wong, A., Boutis, P. & Hekimi, S. Mutations in the clk-1 gene of Caenorhabditis elegans affect developmental and behavioral timing. Genetics 139, 1247–1259 (1995)
Yang, W. & Hekimi, S. Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans. Aging Cell 9, 433–447 (2010)
Houtkooper, R. H. et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 497, 451–457 (2013)
Moullan, N. et al. Tetracyclines disturb mitochondrial function across eukaryotic models: a call for caution in biomedical research. Cell Rep. 10, 1681–1691 (2015)
Zheng, L. et al. Macroautophagy-generated increase of lysosomal amyloid β-protein mediates oxidant-induced apoptosis of cultured neuroblastoma cells. Autophagy 7, 1528–1545 (2011)
Forloni, G., Colombo, L., Girola, L., Tagliavini, F. & Salmona, M. Anti-amyloidogenic activity of tetracyclines: studies in vitro. FEBS Lett. 487, 404–407 (2001)
Costa, R., Speretta, E., Crowther, D. C. & Cardoso, I. Testing the therapeutic potential of doxycycline in a Drosophila melanogaster model of Alzheimer disease. J. Biol. Chem. 286, 41647–41655 (2011)
Loeb, M. B. et al. A randomized, controlled trial of doxycycline and rifampin for patients with Alzheimer’s disease. J. Am. Geriatr. Soc. 52, 381–387 (2004)
Quirós, P. M. et al. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 216, 2027–2045 (2017)
Münch, C. & Harper, J. W. Mitochondrial unfolded protein response controls matrix pre-RNA processing and translation. Nature 534, 710–713 (2016)
Bao, X. R. et al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. eLife 5, e10575 (2016)
Sidrauski, C., McGeachy, A. M., Ingolia, N. T. & Walter, P. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. eLife 4, e05033 (2015)
Palikaras, K., Lionaki, E. & Tavernarakis, N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 521, 525–528 (2015)
Mouchiroud, L. et al. The NAD+/sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell 154, 430–441 (2013)
Gariani, K. et al. Eliciting the mitochondrial unfolded protein response by nicotinamide adenine dinucleotide repletion reverses fatty liver disease in mice. Hepatology 63, 1190–1204 (2016)
Zhang, H. et al. NAD+ repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 352, 1436–1443 (2016)
Fang, E. F. et al. NAD+ replenishment improves lifespan and healthspan in ataxia telangiectasia models via mitophagy and DNA repair. Cell Metab. 24, 566–581 (2016)
Corcoran, K. A., Lu, Y., Turner, R. S. & Maren, S. Overexpression of hAPPswe impairs rewarded alternation and contextual fear conditioning in a transgenic mouse model of Alzheimer’s disease. Learn. Mem. 9, 243–252 (2002)
Cenini, G., Rüb, C., Bruderek, M. & Voos, W. Amyloid β-peptides interfere with mitochondrial preprotein import competence by a coaggregation process. Mol. Biol. Cell 27, 3257–3272 (2016)
Gong, B. et al. Nicotinamide riboside restores cognition through an upregulation of proliferator-activated receptor-γ coactivator 1α regulated β-secretase 1 degradation and mitochondrial gene expression in Alzheimer’s mouse models. Neurobiol. Aging 34, 1581–1588 (2013)
Martire, S. et al. Bioenergetic impairment in animal and cellular models of Alzheimer’s disease: PARP-1 inhibition rescues metabolic dysfunctions. J. Alzheimers Dis. 54, 307–324 (2016)
Kim, H. E. et al. Lipid biosynthesis coordinates a mitochondrial-to-cytosolic stress response. Cell 166, 1539–1552 (2016)
Wang, X. & Chen, X. J. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature 524, 481–484 (2015)
Wrobel, L. et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 524, 485–488 (2015)
D’Amico, D., Sorrentino, V. & Auwerx, J. Cytosolic proteostasis networks of the mitochondrial stress response. Trends Biochem. Sci. 42, 712–725 (2017)
Tiernan, C. T. et al. Protein homeostasis gene dysregulation in pretangle-bearing nucleus basalis neurons during the progression of Alzheimer’s disease. Neurobiol. Aging 42, 80–90 (2016)
Liang, W. S. et al. Alzheimer’s disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc. Natl Acad. Sci. USA 105, 4441–4446 (2008)
Fonte, V. et al. Suppression of in vivo β-amyloid peptide toxicity by overexpression of the HSP-16.2 small chaperone protein. J. Biol. Chem. 283, 784–791 (2008)
Kamath, R. S., Martinez-Campos, M., Zipperlen, P., Fraser, A. G. & Ahringer, J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, RESEARCH0002 (2001)
Koopman, M. et al. A screening-based platform for the assessment of cellular respiration in Caenorhabditis elegans. Nat. Protoc. 11, 1798–1816 (2016)
Counts, S. E., Nadeem, M., Lad, S. P., Wuu, J. & Mufson, E. J. Differential expression of synaptic proteins in the frontal and temporal cortex of elderly subjects with mild cognitive impairment. J. Neuropathol. Exp. Neurol. 65, 592–601 (2006)
Greenberg, S. A. et al. Plasma cells in muscle in inclusion body myositis and polymyositis. Neurology 65, 1782–1787 (2005)
Zhu, W. et al. Genomic signatures characterize leukocyte infiltration in myositis muscles. BMC Med. Genomics 5, 53 (2012)
Acknowledgements
We thank P. Gönczy and M. Pierron (EPFL) for sharing reagents, the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), for providing worm strains. V.S. is supported by the ‘EPFL Fellows’ program co-funded by the Marie Skłodowska-Curie, Horizon 2020 Grant agreement (665667). D.D. is supported by a fellowship funded by Associazione Italiana per la Ricerca sul Cancro (AIRC). S.E.C. is supported by NIH grants (P01AG014449, R21AG053581 and P30 AG053760). The research of J.A. is supported by the EPFL, NIH (R01AG043930), Systems X (SySX.ch 2013/153), Velux Stiftung (1019), the Jebsen Foundation and the Swiss National Science Foundation (31003A-140780).
Author information
Authors and Affiliations
Contributions
V.S. and J.A. conceived and designed the project. V.S., L.M., M.R., J.S.B., H.Z., D.D., F.P., N.M., A.W.S., S.R. and S.E.C. performed the experiments. V.S., M.R. and L.M. independently replicated worm experiments in Figs 2, 3, 4. V.S., L.M. and J.A. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Mitochondrial function pathways are disrupted in patients with AD.
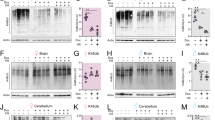
a, Gene set enrichment analysis of OXPHOS (false-discovery rate (FDR) = 0.051, nominal P = 0.008) and mitochondrial import (FDR = 0.295, nominal-P = 0.002) genes in human prefrontal cortex of patients with AD (GN328; healthy, n = 195; AD, n = 388 individuals). b, e, Gene set enrichment analysis of OXPHOS and mitochondrial import genes in human visual cortex (b) (GN327; healthy, n = 195; AD, n = 388) and whole brain (e) (GN314, n = 16 healthy individuals and 33 individuals with AD) of patients with AD. b, FDR = 0.754, P = 0.038 for OXPHOS, FDR = 0.657, P = 0.031 for mitochondrial import. e, FDR = 0.076, P = 0.002 for OXPHOS, FDR = 0.218, P = 0.006 for mitochondrial import. c, f, Heat maps of genes from visual cortex (c) and whole brain (f) datasets. d, g, Correlation plots of mitochondrial stress genes, UPRER and HSR levels in human visual cortex (d) and whole brain (g) from patients with AD. For further information, see Supplementary Table 5. h, Quantification of immunoblots of mtDNAJ and CLPP (n = 8 per group shown in Fig. 1d) from brains of humans with no cognitive impairment (NCI), mild-cognitive impairment (MCI) and mild to moderate AD. This experiment was performed independently twice. Data are mean ± s.e.m. ***P ≤ 0.001. Differences were assessed using two-tailed t-tests (95% confidence interval). Mito., mitochondrial. For all individual P values, see Source Data.
Extended Data Figure 2 MSR analysis and mitochondrial function in 3×TgAD mice.
a, Human APP expression in cortex tissues of wild-type and 3×TgAD mice (n = 4 mice per group). **P = 0.002. b, MSR transcript analysis from cortex tissues of wild-type (n = 4 mice) and 3×TgAD (n = 4 mice) mice at six months of age. c, Immunoblot analysis (wild-type, n = 5; 3×TgAD, n = 6, western blot of four representative mice) and quantification of the samples as in b. *P < 0.05. (P = 0.035, P = 0.029). d–f, MSR transcript analysis from cortex tissues of wild-type (d; 6 months, n = 4 mice; 9 months, n = 5 mice) and 3×TgAD (e; 6 months, n = 4 mice; 9 months, n = 5 mice) mice at six and nine months of age, and corresponding heat maps (f) representing relative variation in gene expression between groups. g, Citrate synthase activity assay in cortex tissues from wild-type and 3×TgAD mice (wild-type, n = 8 mice; 3×TgAD, n = 7 mice). *P = 0.039. Data are mean ± s.e.m. *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001; n.s., not significant. Overall differences between conditions were assessed by two-way ANOVA. Differences in individual genes, proteins or between two groups were assessed using two-tailed t-tests (95% confidence interval). All experiments were performed independently twice. For uncropped gel source data, see Supplementary Fig. 1. For all individual P values, see Source Data.
Extended Data Figure 3 Characterization of Aβ proteotoxicity and stress response pathways in GMC101 worms.
a, Amyloid aggregation in CL2122 and GMC101 worms (n = 3 biologically independent samples) at 20 °C or 25 °C. b, MSR transcript analysis in worms at 20 °C (n = 3 biologically independent samples). c, Respiration assay in CL2122 and GMC101 worms (CL2122, n = 8; GMC101, n = 8 biologically independent samples). d, mitochondrial:nuclear DNA ratio in CL2122 and GMC101 worms (n = 13 animals per group). e, Citrate synthase activity in CL2122 and GMC101 worms on D1 (n = 5 biologically independent samples). **P = 0.004. f, CL2122 and GMC101 mobility (CL2122, n = 48; GMC101, n = 59 worms). g, Confocal images of D1 adult worms showing muscle cell integrity, nuclear morphology and mitochondrial networks. Scale bar, 10 μm. See Methods. h, Representative images and fraction of worms in different stages of development upon atfs-1 RNAi (n = 4 independent experiments). *P < 0.05 (larvae, P = 0.048; adults, P = 0.035). i, Transcript analysis of UPRER, HSR and daf-16 target genes (n = 3 biologically independent samples). j, Representative images and fraction of worms in different stages of development fed with atfs-1, xbp-1 and hsf-1 RNAis at 20 °C (n = 8 per group; xbp-1, n = 3 biologically independent samples). k, Amyloid aggregation upon atfs-1 RNAi (n = 2 biological replicates). l, Mobility of CL2122 fed with 50% dilution of atfs-1 RNAi (ev, n = 48; atfs-1 1/2, n = 47 worms). m, Validation of the efficacy of atfs-1 RNAi in CL2122 and GMC101 worms (n = 3 biologically independent samples). n, MSR transcript analysis of CL2122 worms upon atfs-1 RNAi (n = 3 biologically independent samples). o, Mobility of D1 adult worms fed with atfs-1 or hsf-1 RNAi at 25 °C (CL2122, ev, n = 22; atfs-1, n = 27; hsf-1, n = 28; GMC101, ev, n = 27; atfs-1, n = 21; hsf-1, n = 18; N2, ev, n = 31; atfs-1, n = 38; hsf-1, n = 27 worms). p, Validation of the efficacy of the newly generated atfs-1 #2 RNAi (n = 3 biologically independent samples). For further information, see Methods. q, Worm mobility upon atfs-1 #2 RNAi (CL2122, ev, n = 47; atfs-1 #2, n = 42; GMC101, ev, n = 55; atfs-1 #2, n = 46 worms). ev, scrambled RNAi; A.U., arbitrary units. Data are mean ± s.e.m. *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; n.s., not significant. Overall differences between conditions were assessed by two-way ANOVA. Differences in individual genes or between two groups were assessed using two-tailed t-tests (95% confidence interval). All experiments were performed independently at least twice. For uncropped gel source data, see Supplementary Fig. 1. For all individual P values, see Source Data.
Extended Data Figure 4 Reliance on ubl-5 and on increased mitochondrial stress response of GMC101 worms.
a, Fraction of worms that reached D1 adulthood fed with ubl-5 RNAi (n = 5 biologically independent samples). b, c, Mobility of worms (b) and percentage of paralysed and dead D8 adult worms (c) upon ubl-5 RNAi (b; CL2122, ev, n = 39; ubl-5, n = 43; GMC101, ev, n = 40; ubl-5, n = 41 worms; c; n = 5 biologically independent samples). d, e, Transcript analysis of UPRER, HSR and daf-16 target genes in GMC101 (d) and CL2122 (e) worms upon atfs-1 RNAi (n = 3 biologically independent samples). f, Validation of the atfs-1-overexpressing strains AUW9, AUW10 and AUW11 (n = 3 biologically independent samples). See Methods. g, Worm mobility in atfs-1-overexpressing CL2122- and GMC101-derived lines (CL2122, n = 40; GMC101, n = 57; AUW9, n = 40; AUW10, n = 38; AUW11, n = 42 worms). h, Percentage of paralysed and dead D6 adult worms (n = 5 biologically independent samples). *P < 0.05 (from left to right, P = 0.019, 0.046, 0.041). i, j, Mobility (i) and percentage of paralysed and dead D8 adult worms (j) of GMC101, clk-1 mutant (CB4876) and AUW12 strains (i; GMC101, n = 35; CB4876, n = 42; AUW12, n = 38 worms; j; n = 5 biologically independent samples). k, l, Mobility (k) and percentage of paralysed and dead D8 adult worms (l) of GMC101, nuo-6 mutant (MQ1333) and AUW13 strains (k; GMC101, n = 46; MQ1333, n = 50; AUW13, n = 47 worms; l; n = 5 biologically independent samples). For further information on all strains, see Methods. ev, scrambled RNAi; A.U., arbitrary units. Data are mean ± s.e.m. *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; n.s., not significant. Overall differences between conditions were assessed by two-way ANOVA. Differences in individual genes or between two groups were assessed using two-tailed t-tests (95% confidence interval). All experiments were performed independently at least twice. For uncropped gel source data, see Supplementary Fig. 1. For all individual P values, see Source Data.
Extended Data Figure 5 Effects of the inhibition of mitochondrial translation and mitophagy in worms, and of compound treatments in mammalian cells.
a, Representative images of GMC101 worms upon mrps-5 RNAi feeding or DOX treatment (15 μg ml−1) from eggs to D1 (n = 2 independent experiments). b, MSR transcript analysis of DOX-treated CL2122 worms (n = 4 biologically independent samples). c, d, Transcript analysis of UPRER, HSR and daf-16 target genes in GMC101 worms fed with mrps-5 RNAi (c) or treated with DOX (15 μg ml−1) (d) (c, d, n = 3 biologically independent samples). e, Amyloid aggregation in worms upon mrps-5 RNAi or DOX treatment (n = 3 biologically independent samples). f, Respiration on D3 and D6 in GMC101 worms fed with mrps-5 RNAi (n = 9; mrps-5 D6, n = 8 biologically independent samples). g, Additional confocal images of the SH-SY5Y(APPSwe) cells stained with the anti-β-amyloid 1–42 antibody, after DOX and ISRIB treatment. Scale bar, 10 μm. h, Immunoblot of OXPHOS proteins in SH-SY5Y(APPSwe) cells showing the effects of NR (1 mM) and DOX (10 μg ml−1) (n = 2 biologically independent samples). i, j, Transcript levels of MSR and OXPHOS genes (i) and ATF4 target genes (j) in APPSwe-expressing cell line after 24 h of DOX (10 μg ml−1; n = 4 biologically independent samples). k, Mobility of GMC101 worms upon dct-1 RNAi (ev, n = 54; dct-1, n = 44 worms). l, Mobility of GMC101 worms fed with dct-1, mrsp-5 or both RNAis (ev, n = 54; dct-1, n = 44; mrps-5, n = 35; mrps-5;dct-1, n = 45 worms). m, Mobility of GMC101 worms treated with DOX and/or fed dct-1 RNAi (ev, n = 54; dct-1, n = 44; DOX, n = 52; DOX and dct-1, n = 54 worms). n, o, Mobility of CL2122 worms fed with dct-1 RNAi (n) from D1 to D4 (ev, n = 44; dct-1, n = 40 worms) or (o) at D8 (n = 38 worms, *P < 0.05 (P = 0.018)). ev, scrambled RNAi; DOX, doxycycline; NR, nicotinamide riboside; ISRIB, integrated stress response inhibitor; A.U. Data are mean ± s.e.m. *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; n.s., not significant. Overall differences between conditions were assessed by two-way ANOVA. Differences in individual genes or between two groups were assessed using two-tailed t-tests (95% confidence interval). All experiments were performed independently at least twice. For uncropped gel source data, see Supplementary Fig. 1. For all individual P values, see Source Data.
Extended Data Figure 6 Effect of NAD+-boosting compounds and sirtuin depletion in worms, and NR treatment in mammalian cells.
a, Percentage of paralysed D8 adult GMC101 worms after NR or AZD treatment (n = 3 independent experiments). b, c, MSR transcript analysis of CL2122 worms treated with NR (b; 1 mM) or AZD (c; 0.3 μM) (b, c, n = 3 biologically independent samples). d, e, Mobility of CL2122 treated with NR (1 mM) or AZD (0.3 μM) from (d) D1 to D4 (vehicle, n = 44; NR, n = 48; AZD, n = 43 worms) or (e) at D8 (vehicle, n = 38; NR, n = 36; AZD, n = 33 worms, *P < 0.05 (P = 0.017); **P ≤ 0.01 (P = 0.004)). f, g, Percentage of paralysed D8 adult GMC101 treated with NR upon atfs-1 RNAi (f) or dct-1 RNAi (g) (n = 5 biologically independent samples). h, Representative images of worms fed with atfs-1, sir-2.1 or daf-16 RNAis (n = 2 independent experiments). i, Mobility of NR-treated GMC101 worms (1 mM) fed with sir-2.1 RNAi (ev, n = 52; sir-2.1, n = 37; NR, n = 40; sir-2.1 and NR, n = 51 worms). j, Mobility of NR-treated GMC101 worms (1 mM) fed with daf-16 RNAi (ev, n = 52; daf-16, n = 43; NR, n = 40; daf-16 and NR, n = 48 worms). k, Percentage of paralysed and dead D8 adult GMC101 worms treated with NR or fed with sir-2.1, daf-16 or atfs-1 RNAis (n = 5 biologically independent samples). l, m, Transcript analysis of UPRER, HSR and daf-16 target genes in GMC101 worms treated with NR (l; 1 mM, *P < 0.05 (hsp-16.41, P = 0.03; hsp-16.48/49, P = 0.008)), or AZD (m; 0.3 μM, *P < 0.05 (P = 0.033); **P ≤ 0.01 (P = 0.0004)) (n = 3 biologically independent samples). n, Additional confocal images of the intracellular amyloid deposits in the SH-SY5Y(APPSwe) cells after 24 h NR treatment. o, Transcript levels of MSR genes in APPSwe-expressing cells after NR (1 mM) (n = 4 biologically independent samples). NR, nicotinamide riboside; ISRIB, integrated stress response inhibitor; AZD, Olaparib; ev, scrambled RNAi; A.U., arbitrary units. Data are mean ± s.e.m. *P < 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; n.s., not significant. Overall differences between conditions were assessed by two-way ANOVA. Differences in individual genes or between two groups were assessed using two-tailed t-tests (95% confidence interval). All experiments were performed independently twice. For all individual P values, see Source Data.
Extended Data Figure 7 Proposed model.
Scheme illustrating the role of mitochondrial proteostasis in Aβ proteopathies based on our studies in the GMC101 model. (1) Accumulation of amyloid aggregates triggers mitochondrial dysfunction, which induces the MSR. (2) atfs-1 depletion results in loss of mitochondrial homeostasis, more pronounced amyloid aggregation and decreased healthspan. (3) Enhancing mitochondrial proteostasis with DOX, mrps-5 RNAi, and NAD+ boosters (NR and Olaparib) increases organismal fitness, delaying the development of Aβ proteotoxicity.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-13, Supplementary Text and Supplementary Figure 1, the uncropped western blots (PDF 2292 kb)
Source data
Rights and permissions
About this article
Cite this article
Sorrentino, V., Romani, M., Mouchiroud, L. et al. Enhancing mitochondrial proteostasis reduces amyloid-β proteotoxicity. Nature 552, 187–193 (2017). https://doi.org/10.1038/nature25143
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25143
- Springer Nature Limited
This article is cited by
-
The concept of resilience to Alzheimer’s Disease: current definitions and cellular and molecular mechanisms
Molecular Neurodegeneration (2024)
-
Mitochondrial quality control in human health and disease
Military Medical Research (2024)
-
The crosstalk between mitochondrial quality control and metal-dependent cell death
Cell Death & Disease (2024)
-
Enhancing mitochondrial proteolysis alleviates alpha-synuclein-mediated cellular toxicity
npj Parkinson's Disease (2024)
-
Mitochondrial dysfunction: mechanisms and advances in therapy
Signal Transduction and Targeted Therapy (2024)