Abstract
The constant regeneration of stomach epithelium is driven by long-lived stem cells1,2,3, but the mechanism that regulates their turnover is not well understood. We have recently found that the gastric pathogen Helicobacter pylori can activate gastric stem cells and increase epithelial turnover4, while Wnt signalling is known to be important for stem cell identity and epithelial regeneration in several tissues5. Here we find that antral Wnt signalling, marked by the classic Wnt target gene Axin2, is limited to the base and lower isthmus of gastric glands, where the stem cells reside. Axin2 is expressed by Lgr5+ cells, as well as adjacent, highly proliferative Lgr5− cells that are able to repopulate entire glands, including the base, upon depletion of the Lgr5+ population. Expression of both Axin2 and Lgr5 requires stroma-derived R-spondin 3 produced by gastric myofibroblasts proximal to the stem cell compartment. Exogenous R-spondin administration expands and accelerates proliferation of Axin2+/Lgr5− but not Lgr5+ cells. Consistent with these observations, H. pylori infection increases stromal R-spondin 3 expression and expands the Axin2+ cell pool to cause hyperproliferation and gland hyperplasia. The ability of stromal niche cells to control and adapt epithelial stem cell dynamics constitutes a sophisticated mechanism that orchestrates epithelial regeneration and maintenance of tissue integrity.
Similar content being viewed by others
Main
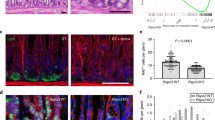
To gain better insight into Wnt signalling in the stomach, we used Axin2 as a marker for active Wnt signalling. Single-molecule in situ hybridization (ISH) showed that the signal is concentrated at the base of glands in the mouse antral mucosa (Fig. 1a), consistent with previous observations1. Lineage tracing of Axin2+ cells after tamoxifen injection in Axin2CreErt2/Rosa26-tdTomato mice verified that they were restricted to the gland base and the lower isthmus at 24 h (Fig. 1b, c). After 7 days, traced cells had repopulated most glands (84%; Fig. 1d, f, g), which remained positive (82%) for more than 4 months (Fig. 1e–g). Similarly, in Rosa26-mTmG reporter mice, which displayed lower reporter activity, the proportion of traced glands remained stable at 7 days, 6 months, and 1 year (Fig. 1h). In addition, Axin2CreErt2/Rosa26-Rainbow mice, which randomly express one of several fluorophores upon tamoxifen treatment, exhibited clonal glandular stripes at 7 days, and units remained monoclonal at 6 weeks post-tracing (Extended Data Fig. 1a–c). Axin2+ cells rarely expressed the mucous neck cell marker GSII and never markers of other differentiated gastric cell types (Fig. 1i), but eventually gave rise to all differentiated antral phenotypes (Fig. 1j). Thus, Axin2 marks a rapidly expanding population of long-lived stem cells in the antrum.
a, Single-molecule ISH for Axin2 in mouse antrum. b–e, Confocal microscopy images of stomach antrum from Axin2CreErt2/Rosa26-tdTomato mice; lineage tracing for 24 h (b, c), 7 days (d), and 120 days (e). f, g, Optical cross-sections through antral glands of Axin2CreErt2/Rosa26-tdTomato mice (f) and quantification of proportions of tdTomato+ clones (g). h, Proportions of GFP+ clones in antrum of Axin2CreErt2/Rosa26-mTmG mice at different time points. i, j, Antrum of Axin2CreErt2/Rosa26-mTmG mice injected with tamoxifen and traced for 24 h (i) or 1 year (j) labelled for GSII, Muc5AC, CGA, or DCAMKL1. Experiments were repeated at least twice in the laboratory. Data represent mean ± s.e.m. from three mice, analysed by Student’s t-test. Scale bar, 100 μm in d, 50 μm in others.
As Lgr5 is known to mark long-lived stem cells in the base of antral glands1, we compared Lgr5 expression to Axin2. Single-molecule ISH revealed that Lgr5 is restricted to the cells at the very base (mainly in positions 1–3), while Axin2 is also expressed directly above this compartment (Fig. 2a, b). Double-ISH confirmed co-expression of Axin2 in the Lgr5+ cells, defining two populations: Axin2+/Lgr5+ cells at the base and Axin2+/Lgr5− cells immediately above in the lower isthmus (Extended Data Fig. 2a). Axin2+ cell-derived clones proliferated rapidly (Extended Data Fig. 2b, c) and repopulated glands within 7 days (Extended Data Fig. 2d). In contrast, repopulation from Lgr5+ cells at that time remained incomplete in most glands (Fig. 2c and Extended Data Fig. 2d), consistent with a turnover time of 10–14 days (ref. 6). Although lineage-labelled Axin2+ cells were evenly distributed in positions 1–7 (Fig. 2d), cells in more distal positions 4–7 were more frequently immunoreactive for the proliferation marker Ki67 (Fig. 2e) and appeared less differentiated (Extended Data Fig. 2e), consistent with a previous model7. This depicts a distinct population of highly proliferative Wnt-responsive stem cells in positions 4–7 of the lower isthmus.
a, Single-molecule ISH for Axin2 and Lgr5 in mouse antrum; arrows indicate cells positive for both markers; arrowheads indicate Axin2+/Lgr5− cells. b, Histogram of the highest cell position of Lgr5 and Axin2 transcripts counted from the gland base (>50 glands analysed). c, Tracing ratio (height between lowest and highest traced cell in the gland divided by gland height) in 7 day lineage-traced Axin2CreErt2/Rosa26-mTmG and Lgr5eGFPCreErt2/Rosa26-mTmG mice. d, e, Axin2CreErt2/Rosa26-mTmG mouse antrum labelled for Ki67 (e) and distribution of Axin2+/Ki67+ and Axin2+/Ki67− cells (d) (>50 glands analysed). f, Antrum of untreated or diphtheria toxin (DT)-treated Lgr5DTR mouse (single dose of diphtheria toxin 24 h before being euthanized). g, Antrum of Lgr5DTR-Axin2CreErt2/Rosa26-mTmG mouse treated with a single dose of diphtheria toxin and tamoxifen 24 h before being euthanized. h, Gastric organoids from untreated and diphtheria toxin-treated Lgr5DTR mice. i, j, Antrum of Lgr5DTR/Axin2CreErt2/Rosa26-mTmG mice treated with diphtheria toxin on 2 consecutive days, with tamoxifen added to the first dose, and euthanized at 7 (i) or 60 days (j). k, Quantification of the proportion of labelled clones in the antrum of Lgr5DTR/Axin2CreErt2/Rosa26-mTmG mice (two mice per time point). Images represent findings reproduced in at least n = 3 biological replicates. Experiments were repeated at least twice in the laboratory. Data represent mean ± s.e.m. from n = 3 mice, analysed by Student’s t-test. Scale bar, 50 μm in e, 100 μm in others.
To study the stem cell properties of Axin2+/Lgr5− cells, we depleted all Lgr5+ cells in Lgr5DTR/Axin2CreErt2/Rosa26-mTmG mice by administration of diphtheria toxin (Fig. 2f), which reduced the average number of Axin2+ cells from 1.8 to 1.3 per gland—all of which were confined to the lower isthmus (Fig. 2g and Extended Data Fig. 2f). Organoid-forming capacity was not altered in diphtheria-toxin-treated mice (Fig. 2h). After 7 days, clones emerging from the Axin2+/Lgr5− cells spread to the top of the glands and gave rise to new Lgr5+ cells at the base (Fig. 2i and Extended Data Fig. 2f). The proportion of clones spanning the entire gland remained stable for at least 2 months (Fig. 2j–k), similar to clones in non-Lgr5-depleted Axin2CreErt2/Rosa26-mTmG mice (Fig. 1h). Thus, Axin2+ cells constitute a stem cell pool capable of regenerating entire gastric glands upon Lgr5+ cell depletion.
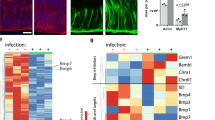
We next examined the distribution of Wnt ligands in the proximity of the stem cell compartment to define areas of intense signalling. ISH revealed Wnt3a, Wnt4, and Wnt5a in the isthmus and mid-glandular compartment and strong expression of Wnt11 (Extended Data Fig. 3). Several members of the Frizzled Wnt receptor family, as well as several Wnt inhibitors, were also expressed in the antrum (Extended Data Fig. 4). However, none was restricted only to the gland base. The Wnt target gene RNF43, which ubiquitinates Frizzled8, showed high expression in the base but was also expressed throughout the gland.
We then analysed R-spondin (Rspo) family members, which enhance Wnt signalling by preventing ubiquitination and turnover of Frizzled9. Indeed, Rspo3 was specifically associated with the stroma adjacent to the stem cell compartment (Fig. 3a and Extended Data Fig. 5a). Double-ISH showed that it is produced by α-smooth muscle actin (SMA)-positive myofibroblasts in the lamina muscularis mucosae directly beneath the glands (Fig. 3b, c). This finding was confirmed in cultured primary antral myofibroblasts (Extended Data Fig. 5d–f), indicating that these cells may drive canonical Wnt signalling in the antrum.
a, Single-molecule ISH for Rspo3 in the antrum; arrows indicate stromal transcripts. b, c, Double-ISH of Rspo3 (blue) and α-SMA (red) in the gastric mucosa, (c) insert from (b); arrows indicate Rspo3 signal in the α-SMA+ cell compartment. d–g, Organoids from antral epithelium grown without (d, f) or in co-culture with (e, g) antral stromal cells with full culture medium (d, e) or medium without Rspo (f, g). h, Co-culture of non-fluorescent antrum epithelium and red fluorescent protein (RFP)-expressing stromal cells. Note that stromal cells do not incorporate into organoids. i, Surface area of organoids grown with or without Rspo either in co-culture or without stromal cells (organoids from n = 3 biological replicates, one-way analysis of variance (ANOVA) and post-hoc analysis). FM, full medium. j–l, Three-dimensional reconstructions from confocal microscopy images of fixed antral organoids from Axin2CreErt2/Rosa26-mTmG mice grown either in full medium (j) or medium without Rspo, either without (k) or with (l) stromal cells. m–o, Axin2CreErt2/Rosa26-tdTomato mice treated with PBS or Rspo1 48 h and tamoxifen 24 h before being euthanized. The number of tdTomato cells per gland section was quantified, (>50 glands). Experiments were repeated at least twice in the laboratory. Data represent mean ± s.e.m. from three mice analysed by Student’s t-test. Scale bar, 100 μm.
Gastric organoids grown in full medium containing the Rspo3 homologue Rspo1 showed no change in size after co-culture with antral stromal cells (Fig. 3d, e). After 4 days in Rspo-free medium, however, their growth was diminished unless stromal cells were present (Fig. 3f–i). Similarly, if organoids from Axin2CreErt2/Rosa26-mTmG mice were grown for 6 days with 4-OH tamoxifen added on day 2 to visualize Wnt-responsive cells, expansion of green fluorescent protein-expressing (GFP+) cells was only observed in the presence of Rspo or in co-culture with stromal cells (Fig. 3j–l). Human organoids also required Rspo to maintain Wnt signalling (Extended Data Fig. 6a–c). Therefore, Rspo from stromal myofibroblasts stimulates Wnt signalling and proliferation of Axin2+ cells in gastric organoids.
To explore how Rspo affects stomach epithelium in vivo, Axin2CreErt2/Rosa26-tdTomato mice were injected with Rspo1 followed by tamoxifen 24 h later to visualize Axin2+ cells. At 48 h, tdTomato expression was expanded in the gland isthmus (Fig. 3m–o). When Rspo was administered 4 or 24 h after tamoxifen, tracing was significantly accelerated at 48 h (Extended Data Fig. 7a, b). A similar response to Rspo3 and Rspo1 was observed for Rosa26-mTmG reporter mice treated with tamoxifen, and 4 h later Rspo, 48 h before being euthanized (Fig. 4a), leading us to conclude that Rspo treatment expands and accelerates tracing of Axin2+ cells.
a, Axin2CreErt2/Rosa26-mTmG mice treated with tamoxifen, and 4 h later PBS, 100 μg Rspo1, or 100 μg Rspo3, 48 h before being euthanized, and quantification of the height from lowest to highest GFP+ cell per gland normalized to total gland height (>50 glands analysed). b, Organoids grown from 250 isolated antral glands from mice pre-treated with either PBS or 100 μg Rspo1 (4 days after plating). c, Organoids from single tdTomato+ cells from Axin2CreErt2/Rosa26-tdTomato mice treated with tamoxifen and either PBS or Rspo3 (100 μg per mouse) 24 h before being euthanized, and quantification of the organoid number that grew from 500 sorted Axin2+ and Axin2− cells per well. d, GSEA analysis showing Rspo-driven enrichment scores for intestinal stem cell gene signature and Reactome gene set for mitosis (n = 2 mice per group). e, qPCR for RNF43 and Sox9 from isolated glands of untreated (n = 4) or Rspo1-treated animals (n = 2). f, Lgr5eGFPCreErt2/Rosa26-mTmG mice treated and quantified as in a. g, Quantification of Ki67 expression in gastric glands and Lgr5+ cells in untreated and Rspo3-treated animals (48 h before being euthanized). h, Antrum tissue from a Myh11CreErt2/Rosa26-mTmG mouse (tamoxifen 7 days before being euthanized). i, qPCR for Rspo3, Axin2, Lgr5 and Sox9 from Myh11CreErt2/Rspo3+/+ (n = 7) and Myh11CreErt2/Rspo3fl/fl (n = 4) mice (tamoxifen 7 days before being euthanized). j, tdTomato signal and height of antral glands from uninfected and 2-month H. pylori-infected Axin2CreErt2/tdTomato mice (tamoxifen 48 h before being euthanized). k, Single-molecule ISH for Rspo3 in the antrum of uninfected and 1-month infected mice, and quantification of total transcripts per image and in the stroma between glands. l, qPCR for Axin2 from uninfected (data from i) or 2-week infected Myh11CreErt2/Rspo3+/+ and Myh11CreErt2/Rspo3fl/+ mice (tamoxifen 7 days before being euthanized). m, n, Confocal microscopy images (m) and colony-forming units (c.f.u.) (n) of H. pylori-infected Myh11CreErt2/Rspo3fl/+ mouse antrum infected 2 weeks before being euthanized (tamoxifen 7 days before being euthanized). Experiments were repeated at least twice in the laboratory except microarray analysis, which was done once on two biological replicates. Except where indicated, data represent mean ± s.e.m. from n = 3 mice, analysed by Student’s t-test. Scale bar, 100 μm.
The number of organoids derived from cells with stem cell potential10,11, as determined by cultivating glands (Fig. 4b) or single cells (Extended Data Fig. 6d), was also significantly higher if mice had been pre-treated with Rspo1 for 24 h. About 1% of Axin2+ but only 0.25% of Axin2− cells from Axin2CreErt2 reporter mice generated organoids (Fig. 4c). Rspo administration significantly increased the number of organoids from Axin2+ but not Axin2− cells (Fig. 4c). Consistent with this, microarray gene set enrichment analysis (GSEA) of antral epithelium from mice treated with Rspo1 revealed a significant enrichment of genes involved in cell division and proliferation, as well as ‘intestinal stem cell signature genes’ (Fig. 4d), which are specifically overexpressed in Lgr5+ cells in the intestine12, although Lgr5 itself was not regulated. Quantitative PCR (qPCR) confirmed the upregulation of RNF43 and Sox9 (Fig. 4e). Together, these data indicate that Rspo stimulates expansion of stem cell signalling and proliferation of Axin2+ cells in the stomach.
To examine the effect of Rspo on the Lgr5+ cell compartment, we treated Lgr5CreErt2/Rosa26-mTmG mice with Rspo1 or Rspo3. This caused neither expansion of Lgr5+ clones (Fig. 4f) nor increased expression of Lgr5–GFP (Extended Data Fig. 7c). Ki67 immunostaining confirmed that there was an overall increase in proliferation, but not within the Lgr5+ population (Fig. 4g and Supplementary Fig. 7d). We found that the Lgr5 homologue Lgr4 is expressed throughout the gland epithelium (Extended Data Fig. 8d), and can thus serve as an alternative receptor through which Rspo can modulate Wnt signalling in Lgr5− cells9. The Lgr5 lineage-tracing ratio in mice pre-treated with single or multiple doses of Rspo decreased after 7 days compared with control mice, probably because of preferential expansion of Axin2 cells (Extended Data Fig. 8a, b). However, by day 14, Lgr5+ cell-derived clones had again populated the glands in both groups (Extended Data Fig. 8b), suggesting that the effect of injected Rspo is transient. Accordingly, no expansion of Axin2 cells was observed when lineage tracing was induced 6 days after Rspo treatment (Extended Data Fig. 8c). Together, these data confirm Rspo as a regulator of gastric Wnt signalling that specifically activates gland regeneration through Axin2+/Lgr5− cells.
To deplete endogenous Rspo3, we generated Myh11CreErt2/Rspo3fl/fl mice, in which Rspo3 expression could be turned down in Myh11+ myofibroblasts13 surrounding the glands (Fig. 4h). Conditional depletion of Rspo3 7 days previously showed that Axin2, Lgr5, Sox9, and Rspo3 were significantly downregulated (Fig. 4i), demonstrating that endogenous Rspo3 induces Wnt signalling.
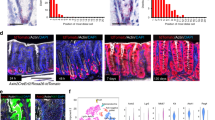
As we have previously found that H. pylori increases stem cell turnover in infected glands4, we asked whether this pathogen-specific response is also driven by Rspo/Wnt signalling. Axin2CreErt2/Rosa26-tdTomato mice were infected with H. pylori for 2 months and injected with tamoxifen 48 h before being euthanized, revealing a twofold increase in the number of Axin2+ cells in the antrum, accompanied by increased proliferation of the cells in the gland base (Extended Data Fig. 9a) and gland hyperplasia (Fig. 4j and ISH confirmation in Extended Data Fig. 9b, c). Only a partial increase in Axin2 expression was seen after infection with an isogenic CagE mutant of H. pylori (Extended Data Fig. 9d, e), indicating that a functional cagPAI type IV secretion system is important for inducing this host response.
Using Myh11CreErt2/Rosa26-mTmG mice, we noticed that H. pylori-induced epithelial gland hyperplasia was accompanied by an expansion of surrounding Myh11+ myofibroblasts (Extended Data Fig. 9f). Single-molecule ISH showed that expression of Rspo3 increased significantly in myofibroblasts beneath and between the glands in mice infected with H. pylori for 2 months (Fig. 4k), but not in CD45+ blood cells or PECAM+ endothelial cells (Extended Data Fig. 9g). Rspo-expressing cells between the glands were present next to the isthmus, in position 5.6 on average, rising to position 8.2 after infection (Extended Data Fig. 9h). Further, in Rspo3fl/+ mice, infection-driven expression of Axin2 was significantly lower after 2 weeks (Fig. 4l), while colonization density was higher (Fig. 4m, n), suggesting that the Rspo-mediated epithelial response influences the degree of infection.
Our results show that Lgr5−/Axin2+ cells constitute a highly proliferative, non-differentiated compartment capable of regenerating entire antral glands and establish Rspo3 as a critical niche-specific regulator that orchestrates stem cell homeostasis and proliferation dynamics. It promotes proliferation of Lgr5−/Axin2+ cells while the less proliferative Lgr5+ cells appear to be silenced. Both cell types contribute to homeostatic gland renewal, the former being reminiscent of the highly proliferative Lgr5+ cells of the intestinal crypt14, and the latter of the more quiescent Troy+ chief cells of the corpus11. Our findings are consistent with previously described differential responses of stem cell subpopulations to environmental changes15,16.
Rspo-driven expansion of the stem cell compartment upon infection with H. pylori may bear increased risk of malignant transformation17. This is supported by data implicating aberrant Rspo signalling in colorectal18 and gastric cancer19. On the other hand, regenerative responses are important for maintaining epithelial integrity and replacing infected or damaged cells. This study provides a basis for a future understanding of how stem cells adaptively respond to bacteria and how these responses contribute to mucosal pathologies.
Methods
Mouse experiments
All procedures involving animals were approved by the institutional and local as well as national legal authorities at Stanford University and the Max Planck Institute for Infection Biology. Wild-type C56BL6 mice were from Charles River; Lgr5–eGFP–IRES-CreERT2 (Lgr5eGFP)14, Axin2CreERT2 (ref. 20), Rosa26-tdTomato16, Rosa26-mTmG21, Lgr5DTR15, as well as Rosa26-Rainbow reporter mice22 were previously described. Rspofl/fl mice13 were a gift from J. Cobb. Myh11CreErt2 mice23 were a gift from S. Offermanns. All animals were maintained in autoclaved microisolator cages and provided with sterile drinking water and chow ad libitum. Male, 6- to 8-week-old mice were used for this study. Littermates were used for the experiments and randomly allocated to experimental groups. Tamoxifen (Sigma) was injected intraperitoneally into mice as a single dose (4 mg per 25 g body weight, diluted in 200 μl corn oil) at the indicated time points to induce lineage tracing. We noticed that the reporter activity of the different Rosa26 reporter mice used in the study for Axin2 lineage tracing showed substantial differences. While the recombination efficiency was high in Rosa26-tdTomato mice, Rosa26-mTmG reporters were much less efficient, with expression of GFP in only a limited proportion of glands. The Rosa26-Rainbow mice showed an even lower efficiency and only a few glands with fluorescent reporter signal could be visualized. Recombinant Rspo1 (Peprotech, 100 μg per mouse) or Rspo3 (Peprotech, 100 μg per mouse) was injected intravenously at indicated time points. Diphtheria toxin (Sigma, 50 μg per kg intraperitoneal) was injected at indicated time points. For short-term analysis, a single dose of diphtheria toxin was injected and mice were euthanized 24 h after injection. A complete loss of GFP+ Lgr5 cells was observed at 24 h. For lineage-tracing analyses, Lgr5DTR/Axin2CreErt2/Rosa26-mTmG mice were treated with tamoxifen and diphtheria toxin on the same day. In addition, follow-up doses of diphtheria toxin were administered at days 2 and 3 after tamoxifen.
At the time of harvest, the forestomach was removed and the glandular stomach was opened along the lesser curvature and laid flat. The stomach contents were removed and the tissue was divided into two halves along the greater curvature. For microscopy analysis, a similar longitudinal section at the midline along the greater curvature was used in all animals to minimize sampling error. Experiments were performed in at least three biological replicates per condition. For all mouse experiments, mice were randomly allocated to experimental groups.
H. pylori infection
Animals were infected with a single oral dose of 108 H. pylori and euthanized at indicated time points. H. pylori strain PMSS1 as well as an isogenic ΔCagE mutant were used for this study, as previously described4. A longitudinal section of stomach tissue was weighed and then mechanically homogenized in brain–heart infusion medium. Serial dilutions of the homogenates were plated for enumeration of colony-forming units and bacterial counts were expressed as colony-forming units per gram of stomach. Experiments were performed in at least three biological replicates per condition. Mice were randomly allocated to experimental groups. The investigator was blinded for analysis of colony-forming units.
Confocal microscopy
Tissue samples were processed for confocal microscopy as previously described, with minor modifications4. Tissue was fixed in 2% paraformaldehyde for 1 h and washed three times with PBS. Tissue was embedded in 4% agarose and longitudinal stomach sections 100–300 μm thick were generated using a vibratome (Leica). Tissue sections were permeabilized in PBS with 3% BSA, 1% saponin, and 1% Triton X-100 before staining. The samples were stained overnight with primary antibodies, followed by 2 h with secondary antibodies, and counterstained with 4′,6-diamidino-2-phenylindole (DAPI) or Hoechst for nucleus visualization and with phalloidin to visualize cell boundaries, if indicated. Samples were imaged with a Zeiss LSM 700 or Leica Sp8 confocal microscope. Rabbit anti-H. pylori antibodies were generated against paraformaldehyde-fixed PMSS1 bacteria (Covance, Princeton, New Jersey, USA) and used at 1:300 dilution. Further, rabbit anti-Ki67 (D3B5 Cell Signaling), mouse anti-E-cadherin (BD, 610181), Alexa Fluor 647-conjugated GSII Lectin (Thermo Scientific), mouse anti-Muc5AC (Invitrogen, 12178), chromogranin A (Abcam, ab15160), rabbit anti-DCAMKL1 (Thermo Scientific, PA5-14046), DAPI (Sigma), and Alexa Fluor 564 and Alexa Fluor 647 fluorophor-conjugated phalloidin (Life Technologies, A12381 and A22287) were used.
To quantify the turnover kinetics within the gland, we measured the gland height and the height between the lowest and highest lineage-traced cell within the gland. The ratio between the distances was defined as the tracing ratio. All antral glands from a longitudinal section through the mouse stomach were used for the analysis.
To assess the proportion of glands with Cre-driven expression of tdTomato in Rosa26-tdTomato mice, antrum wholemount tissue was visualized after removal of the muscular layer in x–z projection and the proportion of tdTomato+ glands was counted and normalized to the total number of glands. Mice were randomly allocated to experimental groups. The investigator was blinded for image analysis.
In addition to analysing antrum tissue, we observed that, in the corpus, Axin2+ did not fully trace glands (Extended Data Fig. 1d, e) and transcripts were sparse (Extended Data Fig. 1f), while in the duodenum it was substantially higher but not restricted to the crypt base (Extended Data Fig. 1g, h), suggesting that Wnt signalling intensities differ in distinct regions of the gastrointestinal tract.
Organoid cultures
Antrum tissue was incubated for 90 min in buffered saline solution containing 0.5 mM DTT/3 mM EDTA to dissociate gastric glands. Isolated glands were washed with PBS and mixed with 25 or 50 μl Matrigel (BD) and plated in 48- or 24-well plates. After polymerization of Matrigel, mouse gastric culture medium (Advanced Dulbecco’s Modified Eagle Medium/F12 supplemented with B27, N2 (Invitrogen), N-acetyl cysteine (Sigma) penicillin/streptomycin containing 50 ng ml−1 epidermal growth factor, 100 ng ml−1 noggin, 100 ng ml−1 fibroblast growth factor 10, 10 mM gastrin, and 500 ng ml−1 R-spondin 1 (Peprotech) or R-spondin 1-conditioned medium and Wnt3A-conditioned medium) was overlaid. The medium supplemented with growth factors was replaced every 2–3 days.
For clonal experiments, single-cell suspension from Axin2CreErt2/Rosa26-mTmG or Rosa26-tdTomato mouse glands was achieved using TrypLE. Viable cells were sorted from isolated gastric glands and seeded into Matrigel at low density (500 or 1,000 cells per well). Organoids from sorted Axin2+ cells were passaged and maintained in culture for 3 months to demonstrate their longevity (Extended Data Fig. 6e).
For co-culture experiments, stromal cells were isolated and cultured. After EDTA-based gland dissociation, the remaining tissue was incubated with type I collagenase (Merck) and accutase (Innovative Culture Technologies) for 1.5 h and vigorously shaken. After washing, dissociated cells were cultured in Advanced DMEM with 5% fetal calf serum (FCS). Cells were passaged after 10 days and showed morphological features of stromal cells. After two passages, the cells were used for experiments. Cells were harvested for reverse transcribed (RT)–PCR or western blot analysis (Western blot for Rspo3 using rabbit anti-Rspo3 antibody (ProSci, 8153)).
For co-culture experiments, the same number of epithelial cells (passage 1) dissociated from organoids and stromal cells was co-cultured in a Matrigel drop. Stromal cells alone did not form organoids. tdTomato-expressing stroma was co-cultured with non-fluorescent epithelium and we observed direct interaction of the epithelium with the stroma but no integration (Fig. 3h). The experiment was performed in three biological replicates. To visualize Axin2 lineages in the organoids, 4OH-tamoxifen was added to cultures from Axin2CreErt2/Rosa26-mTmG mice for 4 days. Cultures were fixed with paraformaldehyde and analysed by confocal microscopy. This experiment was performed twice in independent biological replicates.
Human organoids
Human organoids were isolated and cultured as previously described24. The experiments were approved by the Ethics Committee of the Charité University Medicine, Berlin (EA1-129-12) and samples were obtained after informed consent from the patients. Glands were isolated in chelating solution (distilled H2O with 5.6 mM Na2HPO4, 8.0 mM KH2PO4, 96.2 mM NaCl, 1.6 mM KCl, 43.4 mM sucrose, 54.9 mM d-sorbitol, 0.5 mM dl-dithiothreitol, 2 mM EDTA) for 30 min at 37 °C on a shaking platform. Supernatant was then removed, and tissue fragments were placed in a Petri dish and gently squeezed with a glass slide to isolate the gastric glands. Isolated glands were resuspended in medium containing 10% heat-inactivated FCS (Biochrom), collected in a tube, and allowed to settle for 1 min by gravity before the supernatant containing most of the isolated glands was transferred to a new tube. After five more washes, glands were counted under a microscope and centrifuged (250g, 5 min) followed by three washes with Advanced DMEM/F12 (ADF; Invitrogen).
Isolated gastric glands and fragments of gastric epithelium were mixed with ice-cold Matrigel (growth factor reduced, phenol red free; BD Biosciences) and seeded in pre-warmed 24-well plates at a density of ~300 glands or fragments per 40 μl Matrigel per well. The Matrigel was polymerized for 15 min at 37 °C and overlaid with 500 μl warm expansion medium (ADF, 50% conditioned Wnt3A-medium, 25% conditioned R-spondin1 medium supplemented with 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 1% Glutamax, 2% B27, 1% N2, 20 ng ml−1 human epidermal growth factor (all Invitrogen), 150 ng ml−1 human noggin, 150 ng ml−1 human fibroblast growth factor 10 (both Peprotech), 1.25 mM N-acetyl cysteine, 10 mM nicotinamide, 10 nM human gastrin, 2 μM SB202190 (all Sigma), and 1 μM A83–01 (Calbiochem)). Y-27632 (7.5 μM; Sigma) was added for the first 3 days. Cultures were kept at 37 °C in 5% CO2 in a humidified incubator. Data from three biological replicates were used.
BAT-RED lentiviral construct
Lentivirus construct: replication-deficient lentiviral particles were produced by transfection of mycoplasma-free HEK293T cells (American Type Culture Collection CRL-11268) using 2× HBS (50 mM HEPES, 280 mM NaCl, and 1.5 mM Na2HPO4), 1 M CaCl2, and a mixture of the following vectors: packaging vector psPAX2 (Addgene plasmid 12260), envelope vector pMD2.G (Addgene plasmid 12259), and the TCF/LEF BAT-RED (Addgene plasmid 20674). Two days after transfection, the supernatant containing the lentiviral particles was aspirated, filtered (0.45 μm), and concentrated with Lenti-X Concentrator (Clontech). The lentiviral pellet was dissolved in the ADF medium.
For transduction, organoid cultures were prepared as described for passaging and collected in 250 μl infection medium per sample (expansion medium with ADF containing lentiviral particles instead of normal ADF plus 8 μg ml−1 polybrene (Sigma)). Cells were seeded in a 48-well plate coated with Matrigel and, after overnight incubation, overlaid with a second layer of Matrigel. The culture was passaged after 7 days and RFP-expressing cells were observed. The cultures were then dissociated to single-cell suspension using TrypLE, and RFP+ cells were sorted and re-seeded into Matrigel to enrich for the transfected population. Data represent results from three biological replicates.
Single-molecule RNA ISH
Tissue sections cut at 5 μm thickness were processed for RNA in situ detection using an RNAscope Red Detection Kit according to the manufacturer’s instructions (Advanced Cell Diagnostics, Hayward, California, USA). RNAscope probes used are shown in Extended Data Table 1. Positive and negative control probes were used for each experiment according to the manufacturer’s instructions. We were able to confirm the expression pattern of previously described transcripts in the stomach, for example Lgr5 in the base of the antral glands1 and the absence of the chief cell marker Troy in the antrum11. Rspo1 was not expressed in the antrum, but we noticed some expression in the same slides in the forestomach (Extended Data Fig. 5b). To validate the Rspo probes, we performed ISH on embryonic day 10.5 mouse tissue and confirmed a distinct pattern of expression of the different homologues (Extended Data Fig. 5c), in agreement with wholemount data13.
For quantitative analysis, at least three biological replicates per condition were used. The investigator was blinded for analysis. For Rspo3 and Axin2 quantification, consecutive images of bases of all antral glands in a longitudinal section and the underlying stroma were taken at high magnification (×40 objective). Using ImageJ software, the Fast Red signal was selected by image deconvolution and the number of molecules per image was automatically calculated. To calculate the highest cell position within a gland positive for a specific signal, we measured the distance between the gland base and the highest position of the signal within a gland, and divided by a distance of a single cell.
Microarray analysis
For microarray analysis, mouse glands from Rspo-treated mice and corresponding controls were isolated from the antrum using the isolation procedure described for organoid culture. Total RNA was isolated by the TRIzol (Invitrogen) method following the manufacturer’s protocol. Microarray experiments were performed as independent dual-colour dye-reversal colour-swap hybridizations using two biological replicates. Quality control and quantification of total RNA was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies) and a NanoDrop 1000 UV-Vis spectrophotometer (Kisker). Total RNA was isolated with TRIzol (Life Technologies) according the supplier’s protocol using glycogen as co-precipitant. RNA labelling was performed with a dual-colour Quick-Amp Labelling Kit (Agilent Technologies). In brief, mRNA was reverse transcribed and amplified using an oligo-dT-T7 promoter primer, and the resulting cRNA was labelled with cyanine 3-CTP or cyanine 5-CTP. After precipitation, purification, and quantification, 1.25 μg of each labelled cRNA was fragmented and hybridized to whole-genome mouse 4×44K multipack microarrays (Agilent-014868, whole mouse genome 4×44K microarray kit) according to the supplier’s protocol (Agilent Technologies). Scanning of microarrays was performed at 5 μm resolution using a G2565CA high-resolution laser microarray scanner (Agilent Technologies) with extended dynamic range (XDR). Microarray image data were analysed with Image Analysis/Feature Extraction software G2567AA version A.11.5.1.1 (Agilent Technologies) using default settings and the GE2_1105_Oct12 extraction protocol. The extracted MAGE-ML files were analysed further with Rosetta Resolver Biosoftware, build 7.2.2 SP1.31 (Rosetta Biosoftware). Ratio profiles comprising single hybridizations were combined in an error-weighted fashion to create ratio experiments. A 1.5-fold change expression cutoff for ratio experiments was applied together with anti-correlation of dye-swapped ratio profiles, rendering the microarray analysis highly significant (P < 0.01), robust, and reproducible25. Microarray data have been deposited in the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) of the National Center for Biotechnology Information under accession number GSE79494.
GSEA analysis
We used a gene set of a stem cell signature obtained from Lgr5+ cells in intestinal crypts published previously12 as well as gene sets from collections H, C2, C3, C5BP, and C6 from MSigDB version 5.0, and performed GSEA26 on genes pre-ranked by gene expression-based t-score between gastric antral epithelium isolated from Rspo pre-treated and PBS-treated controls, using standard settings with 1,000 permutations. GSEA2-2.1.0 was used for the analysis. GSEA can be obtained from http://software.broadinstitute.org/gsea/downloads.jsp. R-3.2 was used and can be obtained from https://cran.r-project.org/. R code used to produce the ranked gene list for GSEA from raw microarray files can be accessed at https://gist.github.com/anonymous/0d3a3a6add821557b009d114c8c78667.
Quantitative RT–PCR
Organoids were released from Matrigel with cold DPBS and pelleted by centrifugation (7 min at 4 °C, 300g), followed by RNA isolation using a GeneJET RNA Purification Kit (Fermentas) according to the manufacturer’s protocol. qPCR was performed using a Power SYBR Green RNA-to-CT 1-Step Kit (Applied Biosystems). Reactions were performed in 25 μl containing 50–200 ng RNA, 10 μl SYBR Green mix, 0.16 μl RT mix, and 0.2 μM per primer.
Programme: 30 min at 48 °C; 10 min at 95 °C; followed by 40 cycles of 15 s at 95 °C/60 s at 60 °C. Primers for human tissue used for this study were Axin2 forward CCTGCCACCAAGACCTACAT, reverse CTTCATTCAAGGTGGGGAGA; LGR5 forward CACCTCCTACCTAGACCTCAGT, reverse CGCAAGACGTAACTCCTCCAG; GAPDH forward GGTATCGTGGAAGGACTCATGAC, reverse ATGCCAGTGAGCTTCCCGTTCAG. For primers used for mouse tissue, see Extended Data Table 1. For each oligonucleotide pair and RNA sample, the reaction was performed in triplicate. The amplification plots obtained from the RT–PCR were analysed with the ABI Prism SDS Software package (version 2.2.2; Applied Biosystems). The expression levels of the target genes were normalized to the levels of glyceraldehyde-3-phosphate dehydrogenase gene expression in each individual sample.
Statistics
No statistical methods were used to predetermine sample size. Mouse experiments were performed on n = 3 biological replicates except where stated otherwise. Microarray analysis was performed using samples from two biological replicates. No mice were excluded from experiments. In vitro experiments were performed on n = 3 biological replicates except where stated otherwise. All data are mean ± s.e.m. for the various groups. Statistics are based on ‘n’ biological replicates. For the comparisons of two groups, a t-test or non-parametric test was performed. Non-parametric testing was performed if a normal distribution could not be assumed. All analyses of statistical significance were calculated and displayed compared with the reference control group unless otherwise stated.
For the analysis of antral samples used for immunofluorescence, the data from all glands from consecutive images taken from a longitudinal section through the antrum were used to exclude sampling errors. Statistical analysis used GraphPad Prism software.
Data availability
The microarray data from this manuscript have been deposited in the GEO under accession number GSE79494. Quantitative data supporting the findings of this study are available within the paper and the Supplementary Information. All other data supporting these findings are available from corresponding author upon reasonable request.
Accession codes
References
Barker, N. et al. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36 (2010)
Hayakawa, Y. et al. CCK2R identifies and regulates gastric antral stem cell states and carcinogenesis. Gut 64, 544–553 (2015)
Matsuo, J. et al. Identification of stem cells in the epithelium of the stomach corpus and antrum of mice. Gastroenterology 152, 218–231 (2017)
Sigal, M. et al. Helicobacter pylori activates and expands Lgr5+ stem cells through direct colonization of the gastric glands. Gastroenterology 148, 1392–404.e21 (2015)
Clevers, H., Loh, K. M. & Nusse, R. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346, 1248012 (2014)
Leushacke, M., Ng, A., Galle, J., Loeffler, M. & Barker, N. Lgr5+ gastric stem cells divide symmetrically to effect epithelial homeostasis in the pylorus. Cell Reports 5, 349–356 (2013)
Lee, E. R. & Leblond, C. P. Dynamic histology of the antral epithelium in the mouse stomach: II. Ultrastructure and renewal of isthmal cells. Am. J. Anat. 172, 205–224 (1985)
Koo, B. K. et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 488, 665–669 (2012)
de Lau, W. et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297 (2011)
Barker, N., Bartfeld, S. & Clevers, H. Tissue-resident adult stem cell populations of rapidly self-renewing organs. Cell Stem Cell 7, 656–670 (2010)
Stange, D. E. et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell 155, 357–368 (2013)
Muñoz, J. et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 31, 3079–3091 (2012)
Neufeld, S. et al. A conditional allele of Rspo3 reveals redundant function of R-spondins during mouse limb development. Genesis 50, 741–749 (2012)
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007)
Tian, H. et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478, 255–259 (2011)
Yan, K. S. et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl. Acad. Sci. USA 109, 466–471 (2012)
Tomasetti, C. & Vogelstein, B. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347, 78–81 (2015)
Seshagiri, S. et al. Recurrent R-spondin fusions in colon cancer. Nature 488, 660–664 (2012)
Wilhelm, F. et al. Novel insights into gastric cancer: methylation of R-spondins and regulation of LGR5 by SP1. Mol. Cancer Res. 15, 776–785 (2017)
van Amerongen, R., Bowman, A. N. & Nusse, R. Developmental stage and time dictate the fate of Wnt/β-catenin-responsive stem cells in the mammary gland. Cell Stem Cell 11, 387–400 (2012)
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007)
Ueno, H. & Weissman, I. L. Clonal analysis of mouse development reveals a polyclonal origin for yolk sac blood islands. Dev. Cell 11, 519–533 (2006)
Herring, B. P., Hoggatt, A. M., Burlak, C. & Offermanns, S. Previously differentiated medial vascular smooth muscle cells contribute to neointima formation following vascular injury. Vasc. Cell 6, 21 (2014)
Schlaermann, P. et al. A novel human gastric primary cell culture system for modelling Helicobacter pylori infection in vitro. Gut 65, 202–213 (2016)
Churchill, G. A. Fundamentals of experimental design for cDNA microarrays. Nat. Genet. 32 (Suppl.), 490–495 (2002)
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005)
Acknowledgements
We thank K. Hoffmann, S. Jackisch, D. Frahm, S. Müllerke, C. Fung, and R. Cooper for technical assistance, R. Zietlow for editing the manuscript, and D. Zhao for comments. Financial support for this work was provided by the Deutsche Forschungsgemeinschaft to M.S. (SI 1983/2-1 and SI 1983/3-1) and T.F.M. (ME 705/18-1) and the Innovation Fund of the Max Planck Society to T.F.M. M.S. was supported by the WILL Foundation and is a participant in the Berlin Institute of Health Charité Clinician Scientist Program funded by the Charité University Medicine Berlin and the Berlin Institute of Health.
Author information
Authors and Affiliations
Contributions
The study was designed by M.S., C.Y.L., M.R.A., and T.F.M. M.S. performed most of the experiments. C.Y.L. performed ISH studies. M.K. and M.S. performed lineage-tracing studies and analysis. Data were analysed by M.S. and M.K. H.J.M. performed microarray studies, and H.B. and H.J.M. analysed the microarray data. R.N. and B.W. gave advice for experimental design and data interpretation. The manuscript was written by M.S. and T.F.M.; all the authors provided comments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks M. Hatakeyama, J. Mills, T. Wang, and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Axin2 expression and Wnt signalling intensities differ in distinct regions of the gastrointestinal tract.
a–c, Confocal microscopy images of mouse antral glands from an Axin2CreErt2/Rosa26-Rainbow mouse treated with tamoxifen 7 days (a) or 6 weeks (b) before being euthanized, and number of clones per section (c) (n = 2–3 mice per time point). d, e, In the corpus, Axin2+ does not fully trace glands: Confocal microscopy image from the corpus of an Axin2CreErt2/Rosa26-tdTomato mouse treated with tamoxifen 7 days before being euthanized. e, Confocal microscopy image from the corpus of an Axin2CreErt2/Rosa26-tdTomato mouse treated with tamoxifen 120 days before being euthanized. f–h, Image of ISH for Axin2 in the stomach corpus (f), antrum (g), and duodenum (h). Red dots mark Axin2 mRNA transcripts. Note that expression in the duodenum is substantially higher but not restricted to the crypt base. Experiments were repeated at least twice in the laboratory. Data represent mean ± s.e.m. from two or three mice analysed by Student’s t-test. Scale bar, 100 μm.
Extended Data Figure 2 Co-localization of Lgr5 and Axin2.
a, Double-ISH for Axin2 (red) and Lgr5 (blue) in the mouse antrum. b, Confocal microscopy images of antrum from Axin2CreErt2/Rosa26-mTmG mice treated with tamoxifen for 1, 2, 3, or 4 days, as well as 6 months, before being euthanized. c, Average numbers of Axin2+ cells in different cell positions counted from the gland base at day 1, 2, 3 and 4 after tamoxifen treatment. A rapid expansion of the signal is observed. d, Confocal microscopy images of antrum from Axin2CreErt2/Rosa26-mTmG and Lgr5CreErt2/Rosa26-mTmG mice treated with tamoxifen 7 days before being euthanized. e, Haematoxylin and eosin images of antral glands. The gland base contains mucous cells that are more differentiated (arrowheads), whereas cells in the lower isthmus above appear less differentiated and have larger nuclear to cytoplasmic ratios (arrows). f, Confocal images of antrum from Lgr5DTR/Axin2CreErt2/Rosa26-mTmG mice treated with tamoxifen 24 h before being euthanized (left), tamoxifen and diphtheria toxin 24 h before being euthanized (middle), and tamoxifen and diphtheria toxin 30 days before being euthanized (right). Arrows indicate Axin2+/Lgr5− cells in the isthmus, arrowheads indicate Lgr5+ cells in the base. Images represent findings that were reproduced at least twice in the laboratory in independent biological replicates. Scale bar, 100 μm.
Extended Data Figure 3 Wnt ligand expression in the antrum.
In situ hybridization of Wnt ligands in the mouse antrum. For the probes that detected a signal, a higher magnification image is also displayed. Red dots mark the mRNA of the respective target. Scale bar, 100 μm. Images represent findings that were reproduced at least twice in the laboratory on independent biological replicates. Scale bar, 100 μm.
Extended Data Figure 4 Wnt receptors and inhibitors expression in the antrum.
a–c, In situ hybridization of the Wnt receptors Frizzled (Fz)1–10 (a), Dkk1–4 (b), as well as Troy and RNF-43 (c) in the mouse antrum. Images represent findings that were reproduced at least twice in the laboratory on independent biological replicates. Scale bar, 100 μm.
Extended Data Figure 5 Rspo expression in the antrum.
a, In situ hybridization of Rspo1–4 in the mouse antrum with an inset displaying the Rspo3 signal in the stroma at high magnification. Scale bar, 100 μm. b, Expression of Rspo1 was observed in the forestomach at lower level, but not in the antrum on the same slide. c, ISH for Rspo1–4 in embryonic day 10.5 mouse embryo, displaying distinct patterns for all four probes in the developing brain and limb bud. Pol2 and DapB served as positive and negative controls. d, e, qPCR from cultured myofibroblasts and epithelial organoids isolated from mouse antrum for α-SMA, vimentin, and collagen 1A1 (d) and Rspo homologues (e). f, Western blot for Rspo3 from epithelial organoids and cultured myofibroblasts (Con, positive control, HEK293T cells). Epi, antral epithelial three-dimensional organoids; Str, cultures of antral stroma. Experiments were repeated at least twice in the laboratory. Data represent mean ± s.e.m. from n = 3 biological replicates, analysed by Student’s t-test. Scale bar, 100 μm.
Extended Data Figure 6 a
, Human gastric organoids require Rspo for active Wnt signalling: images from human gastric antrum-derived organoids grown from FACS sorted RFP+ cells from a culture infected with BAT-RED lentiviral construct to visualize Wnt signalling. Cells were grown either in full medium (left column), without Rspo (middle column), or without either Rspo or Wnt3a (right column). b, c, Expression of Wnt target genes in organoids requires Rspo qPCR analysis of Axin2 (b) and Lgr5 (c) expression in the human organoids grown in full medium or medium lacking Rspo or Rspo and Wnt3a. d, Organoid growing capacity is higher in antral cells from Rspo1 pre-treated mice plated at low density. Images show cells at passage one from two independent biological replicates per group. e, Single Axin2+ cells generate clonal organoids that can be expanded and passaged over months: GFP+ cells from Axin2CreErt2/Rosa26-mTmG mice, pre-treated with tamoxifen 48 h before being euthanized and seeded at clonal density, generate organoids that can be expanded and grown for several passages. Shown are organoids at passages 2 and 6. Organoids were cultured for 3 months and passaged weekly at a ratio of 1:2 or 1:3. Experiments were repeated at least twice in the laboratory. Data represent mean ± s.e.m. from three independent experiments, analysed by one-way ANOVA.
Extended Data Figure 7 Rspo treatment accelerates Axin2+ cell tracing.
a, Axin2CreErt2/Rosa26-mTmG mice were treated with tamoxifen 4 h before administration of Rspo1 or PBS and euthanized 48 h later. Tile scans and high magnification of antral glands are shown. The tracing ratio (distance between the lowest and highest tdTomato+ cell divided by the gland height) of antral glands was quantified (n = 2 mice per group). b, Axin2CreErt2/Rosa26-mTmG mice were treated with tamoxifen 24 h before Rspo1, Rspo3, or PBS and euthanized 24 h later; shown are antral glands and their tracing ratio (n = 3 mice per group). c, Rspo does not expand the Lgr5eGPF+ cell population. Lgr5eGFP mice were treated with PBS or 100 μg Rspo3 48 h before being euthanized. Right: graph demonstrating the highest position of GFP+ cells per gland in PBS versus Rspo3-treated animals. Data represent mean ± s.e.m. from antral glands analysed in two mice per group. d, Confocal microscopy images of Ki67-labelled antrum of C57Bl6 mice treated either with PBS or Rspo1 48 h before being euthanized (left) and Lgr5-eGFP mice treated either with PBS or Rspo3 48 h before being euthanized (right). Experiments were repeated at least twice in the laboratory. Except where indicated, data represent mean ± s.e.m. from n = 3 mice, analysed by Student’s t-test. Scale bar, 100 μm.
Extended Data Figure 8 Rspo transiently affects turnover dynamics in the gland and could interact through Lgr4 receptors.
a, Confocal microscopy images from the antrum of Lgr5CreErt2/Rosa26-mTmG mice treated with tamoxifen and PBS (top) or tamoxifen and Rspo1 (bottom) 7 days before being euthanized. b, Tracing ratio from all glands with membrane-bound GFP signal in a longitudinal section from mice treated with either Rspo1 or PBS, as well as tamoxifen at 7 and 14 days before being euthanized. In addition, one group received four injections of Rspo1 for 4 consecutive days, as well as tamoxifen together with the last Rspo injection 7 days before being euthanized. Further, one group received tamoxifen at 7 days and Rspo at 5 days before being euthanized. Each dot represents a single gland, Data represent mean ± s.e.m. from antral glands of two mice per group. c, Rspo-driven Axin2 expansion is a transient response. Confocal microscopy image from a mouse antrum of an Axin2CreErt2/Rosa26-tdTomato mouse treated with 100 μg Rspo1 (left) or PBS (right) 8 days and tamoxifen 48 h before being euthanized (n = 2 mice per group). d, Single-molecule ISH for the Lgr homologues Lgr4, Lgr5, and Lgr6 in mouse antrum. Images represent findings that were reproduced at least twice in the laboratory in independent biological replicates.
Extended Data Figure 9 Expression of Axin2 is increased upon H. pylori infection.
a, Quantification of Axin2+/Ki67+ cells and Axin2+/Ki67− cells in glandular positions 1–3 and 4–12 in the antrum of uninfected mice and mice infected with H. pylori for 2 months; n = 3 mice per group. b, Single-molecule ISH for Axin2 in uninfected and 1-month infected mouse antrum and (c) quantification of the ISH images (number of Axin2mRNA transcripts per image, n = 3 mice per group). d, e, Images and quantification of short-term Axin2 lineage tracing (48 h) from uninfected and 2-month infected Axin2CreErt2/Rosa26-mTmG mice infected with either wild type (WT) H. pylori strain PMSS1 or an isogenic strain with a mutation in the CagE gene. f, Images from uninfected and 2-month infected Myh11CreErt2/Rosa26-mTmG mice. Tamoxifen was injected 7 days before mice were euthanized to visualize myofibroblasts/smooth muscle cells in the stomach (red, actin; blue, nuclei; green, Myh11+ myofibroblasts). g, Upon infection with H. pylori, Rspo3 is expressed in α-SMA+ myofibroblasts but not in CD45+ blood cells or in PECAM+ endothelial cells: double-ISH for Rspo3 (blue) and either α-SMA, CD45, or PECAM (red) in mouse antrum infected with H. pylori 2 months before being euthanized. Scale bar, 100 μm. h, Quantification of Rspo3 transcripts between glands detected by ISH. Images represent findings that were reproduced at least twice in the laboratory in independent biological replicates. Data represent mean ± s.e.m. from n = 3 mice, analysed by Student’s t-test.
Supplementary information
Supplementary Information
This file contains uncropped blots. (PDF 675 kb)
Source data
Rights and permissions
About this article
Cite this article
Sigal, M., Logan, C., Kapalczynska, M. et al. Stromal R-spondin orchestrates gastric epithelial stem cells and gland homeostasis. Nature 548, 451–455 (2017). https://doi.org/10.1038/nature23642
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature23642
- Springer Nature Limited
This article is cited by
-
Decoding spatiotemporal transcriptional dynamics and epithelial fibroblast crosstalk during gastroesophageal junction development through single cell analysis
Nature Communications (2024)
-
Increased ONECUT2 induced by Helicobacter pylori promotes gastric cancer cell stemness via an AKT-related pathway
Cell Death & Disease (2024)
-
Impact of media compositions and culture systems on the immunophenotypes of patient-derived breast cancer cells
BMC Cancer (2023)
-
A synergistic partnership between IL-33/ST2 and Wnt pathway through Bcl-xL drives gastric cancer stemness and metastasis
Oncogene (2023)
-
Role of PDGFRA+ cells and a CD55+ PDGFRALo fraction in the gastric mesenchymal niche
Nature Communications (2023)