Abstract
Histone modifications are fundamental epigenetic regulators that control many crucial cellular processes1. However, whether these marks can be passed on from mammalian gametes to the next generation is a long-standing question that remains unanswered. Here, by developing a highly sensitive approach, STAR ChIP–seq, we provide a panoramic view of the landscape of H3K4me3, a histone hallmark for transcription initiation2, from developing gametes to post-implantation embryos. We find that upon fertilization, extensive reprogramming occurs on the paternal genome, as H3K4me3 peaks are depleted in zygotes but are readily observed after major zygotic genome activation at the late two-cell stage. On the maternal genome, we unexpectedly find a non-canonical form of H3K4me3 (ncH3K4me3) in full-grown and mature oocytes, which exists as broad peaks at promoters and a large number of distal loci. Such broad H3K4me3 peaks are in contrast to the typical sharp H3K4me3 peaks restricted to CpG-rich regions of promoters. Notably, ncH3K4me3 in oocytes overlaps almost exclusively with partially methylated DNA domains. It is then inherited in pre-implantation embryos, before being erased in the late two-cell embryos, when canonical H3K4me3 starts to be established. The removal of ncH3K4me3 requires zygotic transcription but is independent of DNA replication-mediated passive dilution. Finally, downregulation of H3K4me3 in full-grown oocytes by overexpression of the H3K4me3 demethylase KDM5B is associated with defects in genome silencing. Taken together, these data unveil inheritance and highly dynamic reprogramming of the epigenome in early mammalian development.
Similar content being viewed by others
Main
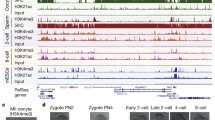
To study the reprogramming of histone modifications in early development, we developed a low-input ChIP–seq method by combining an optimized ChIP protocol that reduces sample loss and TELP, a highly sensitive library preparation method we reported recently3 (Extended Data Fig. 1a; Methods). This method was therefore termed STAR ChIP–seq (small-scale TELP-assisted rapid ChIP–seq). For validation, we performed STAR ChIP–seq for various histone modifications using as few as 200 mouse embryonic stem cells (mES cells) (Extended Data Fig. 1b). These data recapitulated results from those generated using conventional ChIP–seq (Extended Data Fig. 1b–e). We then employed STAR ChIP–seq to profile H3K4me3, a key histone mark for transcription initiation2, in the mouse gametes and pre-implantation embryos by crossing two distinct parental strains, PWK/PhJ (male) and C57BL/6N (female). Genome-wide H3K4me3 was investigated (with two replicates) in metaphase II (MII) oocytes, pronuclear stage 5 (PN5) zygotes, two-cell, four-cell, eight-cell embryos and inner cell masses (ICMs) from blastocysts (Fig. 1a and Extended Data Fig. 2a) (Supplementary Table 1). As minor and major zygotic genome activation (ZGA) occur at the zygote-to-early two-cell and the late two-cell stages, respectively4, we profiled H3K4me3 in both early and late two-cell embryos. As a comparison, we also conducted H3K4me3 ChIP–seq in a somatic tissue—adult mouse cortex (Fig. 1a). RNA-seq was performed using Smart-seq2 (ref. 5) to examine the transcription activities in these samples (Extended Data Fig. 2b). The H3K4me3 STAR ChIP–seq data are highly reproducible between replicates (Extended Data Fig. 2c, d) and are recapitulated using a different antibody (Extended Data Fig. 2e, f). The H3K4me3 peaks in mES cells showed strong overlap with those in ICMs from which mES cells were derived (Extended Data Fig. 2g). We also confirmed that H3K4me3 is correlated with transcription at genes known to be developmentally regulated (Extended Data Fig. 2h). These data demonstrate that we generated high-quality H3K4me3 ChIP–seq data from early embryos.
a, The UCSC browser view shows enrichment of H3K4me3 signals in early embryos (with two replicates for each stage), the cortex, and mES cells (ENCODE19). b, Heat maps showing the expression (FPKM) of all genes (left) and the H3K4me3 enrichment (normalized reads per kilobase per million mapped reads (RPKM)) at their promoters (TSS ± 2.5 kb; middle). Genes are clustered into four major groups with enriched GO terms listed (right).
We then asked if H3K4me3 is associated with transcription genome wide in early embryos. Indeed, promoter H3K4me3 enrichment is generally correlated with RNA levels especially from the late two-cell stage onward when major ZGA occurs (Extended Data Fig. 3a; Supplementary Table 2). As transcripts inherited from oocytes may mask zygotic gene activities, we also analysed ‘non-maternal’ genes that showed no or very low levels of RNA in oocytes (fragments per kilobase of transcript per million mapped reads (FPKM) ≤ 0.5) and obtained similar results (Extended Data Fig. 3b). We further examined the relationship of H3K4me3 and gene expression by grouping all genes into four clusters (Fig. 1b). Notably, for the first cluster (oocyte-specific), the promoters of genes with high levels of transcripts are enriched for H3K4me3 in MII oocytes and zygotes even though these cells are in largely silenced states6, suggesting these marks may reflect previous transcription activities. For the second gene cluster (early-embryo-specific), their promoters showed strong H3K4me3 enrichment from the two-cell stage onward. Intriguingly, lower enrichment of H3K4me3 is also evident before ZGA (Fig. 1b). Therefore, H3K4me3 in MII oocytes and zygotes may mark permissive promoters that are either recently transcribed or poised for future activation. Unexpectedly, H3K4me3 also showed strong enrichment in non-promoter regions in MII oocytes, zygotes and early two-cell embryos (Fig. 1a and Extended Data Fig. 3c). Notably, repeats are highly active in early embryos7 (Extended Data Fig. 3d) and we found H3K4me3 peaks are enriched for repeats in pre-implantation embryos (Extended Data Fig. 3e). These include distal H3K4me3 peaks which frequently overlap repeats such as B1/B2/B4 and ERVL (Extended Data Fig. 3f, g; Methods). However, the enrichment of repeats is generally lower for MII oocytes and zygotes compared to that for late-stage embryos, suggesting that activities of repeats cannot fully explain the widespread distal H3K4me3 peaks in these cells (discussed later). Hence, these data showed distinct H3K4me3 landscapes between early- and late-stage embryos.
DNA methylation undergoes allele-specific reprogramming in early development8,9. We asked if this is true for histone modifications by measuring allelic H3K4me3 enrichment in early embryos (Fig. 2a). We also performed H3K4me3 STAR ChIP–seq in sperm, which showed results consistent with published data10 (Extended Data Fig. 4a). As a validation, H3K4me3 near imprinted loci is preferentially enriched on the transcribed allele (Extended Data Fig. 4b) and on the opposite allele of DNA methylation11 (Supplementary Table 3). Intriguingly, the paternal genome, but not the maternal genome, appears to be depleted of H3K4me3 peaks in zygotes (Fig. 2a and Extended Data Fig. 4c). Such parental differences of H3K4me3 in zygotes echoed previous observations for asymmetric H3K4me3 signals using immunofluorescence12. Our clustering analyses revealed that both promoter (Fig. 2b) and global H3K4me3 (Extended Data Fig. 4d) on the paternal allele of zygote are distinct from those of sperm, suggesting extensive reprogramming upon fertilization. Strong paternal H3K4me3 peaks then reappear particularly the late two-cell stage onward (Fig. 2a and Extended Data Fig. 4c). Notably, the enrichment of paternal H3K4me3 is still weaker than the maternal H3K4me3 even in ICMs (Fig. 2c and Extended Data Fig. 4e). By performing H3K4me3 ChIP–seq in epiblast (E6.5; Methods), we found the allelic H3K4me3 enrichment becomes comparable in post-implantation embryos (Fig. 2c). Intriguingly, the paternal H3K4me3 at promoters in zygotes showed basal enrichment that resembles the pattern in the late two-cell embryos (Fig. 2b), raising the possibility that H3K4me3 may poise these promoters for activation13. A careful examination revealed this is caused by low levels but very large domains of H3K4me3 on the paternal allele (Fig. 2a). We confirmed these domains by performing H3K4me3 ChIP–seq in isolated paternal and maternal pronuclei from zygotes (Methods; Fig. 2a and Extended Data Fig. 4f). It remains to be determined if these domains may include any H3K4me3 modified nucleosomes transmitted from sperm. In sum, these data suggest that extensive reprogramming occurs for paternal H3K4me3 after fertilization.
a, The UCSC genome browser view of H3K4me3 enrichment in gametes, each allele of early embryos, and pronuclei from zygotes (top). A magnified view is also shown (bottom). b, Heat map showing the k-means clustering result of the promoter H3K4me3 enrichment (normalized RPKM) in spermatozoa, the paternal alleles of early embryos and paternal pronuclei. c, Box plots showing the differences of read numbers between two alleles (paternal – maternal) normalized by the total number of allelic reads (paternal + maternal) for H3K4me3 peaks (covered by at least 20 reads) across the genome.
We then asked how H3K4me3 is reprogrammed on the maternal genome. As the pattern of H3K4me3 in MII oocytes is distinct from those in late-stage embryos and the cortex (Fig. 1a), we referred to these two patterns as ‘non-canonical H3K4me3’ (ncH3K4me3) and ‘canonical H3K4me3’ patterns, respectively. We also observed very similar ncH3K4me3 patterns on the maternal alleles of zygotes and early two-cell embryos (Fig. 3a), indicating possible inheritance of ncH3K4me3 from oocytes. The maternal ncH3K4me3 pattern is distinct from that on the paternal allele (Fig. 2a) and also does not simply reflect nucleosome enrichment (Extended Data Fig. 5a, b). Notably, ncH3K4me3 in distal regions is significantly reduced in the late two-cell embryos and is nearly erased by the four-cell stage (Fig. 3a, shaded regions). On the other hand, promoter H3K4me3 starts to adopt a canonical form from the late two-cell stage (Fig. 3a). The switch of maternal ncH3K4me3 to canonical patterns at the two-cell stage is confirmed in a global clustering analysis (Extended Data Fig. 5c), indicating possible involvement of both global demethylation and methylation.
a, The UCSC genome browser view of H3K4me3 enrichment in spermatozoa, oocytes of various stages, the maternal alleles of pre-implantation embryos and the cortex. The DNA methylation levels for indicated stages are also shown14,20. PMDs with ncH3K4me3 in MII oocytes are shaded. D, day; w, week. b, Bar chart showing the percentages of the genome covered by H3K4me3 distal peaks for each stage. c, Heat maps showing the enrichment of H3K4me3 (normalized RPKM) around oocyte PMDs in day 14 (D14) oocyte, MII oocyte, ICM and cortex (top). The average DNA methylation levels around oocyte PMDs at each stage are also shown (bottom). d, Heat maps showing the CpG density and normalized H3K4me3 enrichment in MII oocyte and ICM around all TSSs, sorted by promoter CpG density (top). Average signals across all promoters were also shown (bottom). The dashed lines show the H3K4me3 peak summits downstream of TSSs.
To investigate how ncH3K4me3 is established during oogenesis, we conducted H3K4me3 ChIP–seq and RNA-seq in growing oocytes (postnatal days 7, 10 and 14) and full-grown oocytes (8 weeks) (Methods; Extended Data Fig. 5d). We found H3K4me3 largely remains a canonical pattern in growing oocytes (day 7–14), before adopting a non-canonical pattern in full-grown oocytes that resembles that in MII oocytes (Fig. 3a). Consistently, a positive correlation was found between the transcriptome and promoter H3K4me3 in growing oocytes, and the correlation decreased in grown oocytes and MII oocytes (Extended Data Fig. 5e). Notably, H3K4me3 in day 10 and day 14 oocytes appears to be in transition states as revealed by a hierarchical clustering analysis (Extended Data Fig. 5f). Indeed, we observed that weak levels of distal H3K4me3 start to occur in these cells, indicating the possible onset of ncH3K4me3 establishment (Extended Data Fig. 5g). In sum, these data showed that the global ncH3K4me3 pattern is readily established in full-grown oocytes.
We then focused on distal ncH3K4me3 peaks during oocyte development. These peaks cover large fractions of the genome in grown and mature oocytes preferentially in low CG regions (Fig. 3b and Extended Data Fig. 6a). Notably, we found distal H3K4me3 almost exclusively overlaps with DNA hypomethylated regions in MII oocytes14 (Fig. 3a, c). DNA methylome establishment starts in growing oocytes in a transcription-dependent manner, leading to preferential DNA methylation in transcribing gene bodies15. As a result, intergenic regions and non-transcribing gene bodies are poorly methylated, which we referred to as ‘partially methylated domains’ (PMDs)16 (Fig. 3a; Methods). Approximately 91% of distal ncH3K4me3 marked regions are located in oocyte PMDs (Extended Data Fig. 6b). On the other hand, about 60% of PMDs overlap with oocyte H3K4me3, and the rest PMDs are often present in gene deserts (Extended Data Fig. 6c). Notably, the oocyte PMDs persist throughout pre-implantation development (Fig. 3a, c). Distal ncH3K4me3, on the other hand, is nearly absent after the late two-cell stage, suggesting that the maintenance of oocyte PMDs is independent of ncH3K4me3 (Fig. 3c). We then asked if the establishment of oocyte PMDs precedes that of ncH3K4me3, or vice versa. It was reported that DNA methylation establishment in oocytes starts around postnatal day 10 (ref. 17). Through DNA methylome profiling (Methods), we confirmed that PMDs are clearly evident in day 14 growing oocytes (Fig. 3c and Extended Data Fig. 6d). As weak levels of distal H3K4me3 also occur in these cells (Extended Data Fig. 5g), the patterning of PMDs in oocytes appears to be accompanied with gradual establishment of ncH3K4me3. Future experiments are needed to further dissect their possible causal relationship. Taken together, these data showed that distal ncH3K4me3 is highly correlated with PMDs in oocytes.
Notably, even at promoters, the pattern of ncH3K4me3 in MII oocytes is different from canonical H3K4me3. Promoter ncH3K4me3 is preferentially enriched in the low-CpG regions instead of high-CpG regions (Extended Data Fig. 6a). A close examination revealed that the canonical promoter H3K4me3 is primarily enriched in CpG-rich regions centred around the transcription start sites (TSSs), whereas the promoter ncH3K4me3 shows highest enrichment in CpG-poor loci flanking the TSSs (Fig. 3d). As a result, the summit of ncH3K4me3 peak is further away from TSSs compared to canonical H3K4me3 (Fig. 3d, bottom, dashed lines). It is worth noting that the unique promoter ncH3K4me3 pattern can be resulted from a decrease of H3K4me3 centred around TSSs or an increase of H3K4me3 in regions flanking the TSSs, or both. In addition, the relatively weak promoter ncH3K4me3 can be due to the dilution of sequencing reads by abundant distal ncH3K4me3. Thus, both promoter and distal H3K4me3 demonstrate non-canonical patterns in MII oocytes.
As ncH3K4me3 appears to be replaced by canonical H3K4me3 in the late two-cell embryos which coincides with major ZGA, we asked if such reprogramming depends on zygotic transcription. To do so, we inhibited transcription by treating embryos with α-amanitin before major ZGA (starting from the stages of late zygote or early two-cell; Methods). The resulting embryos were arrested at the two-cell stage and we confirmed the blockage of ZGA (Extended Data Fig. 7a). Notably, H3K4me3 ChIP–seq analysis revealed that ncH3K4me3 failed to switch to canonical H3K4me3 patterns in these embryos (Fig. 4a), suggesting that the reprogramming of H3K4me3 at the two-cell stage requires zygotic transcription. We then asked if the loss of ncH3K4me3 is caused by active histone demethylation or passive dilution in cell division. We treated late zygotes/early two-cell embryos with aphidicolin which arrested embryos in the S phase at the two-cell stage, whereas the control group developed beyond the two-cell stage (Methods). ZGA in replication-blocked embryos was largely unaffected4 (Extended Data Fig. 7a). Notably, the loss of ncH3K4me3 still occurred in these embryos (Fig. 4a). This is also supported by a hierarchical clustering analysis and an analysis of the average H3K4me3 patterns in these embryos (Fig. 4b). Similar observations were made for allelic H3K4me3 data (Extended Data Fig. 7b, c). Hence, the erasure of ncH3K4me3 in the two-cell embryos probably requires active demethylation but not passive dilution.
a, The UCSC genome browser view shows H3K4me3 enrichment in early embryos and those treated by alpha-amanitin (α-amanitin) and aphidicolin. b, Hierarchical clustering of embryo stages based on global H3K4me3 enrichment (left). Averaged H3K4me3 enrichment patterns at the promoters (middle) and distal oocyte PMDs (right) are also shown. The dashed lines show the positions of the H3K4me3 peak summits downstream of TSSs (middle) or the boundaries of PMDs (right). c, Immunostaining of H3K4me3 (green) and BrUTP (red) in SN stage oocytes with overexpression of KDM5A or KDM5B by mRNA microinjection. Uninjected oocytes were used as control. DAPI stains for DNA (blue). Scale bar, 10 μm. d, A schematic model showing the reprogramming of H3K4me3 from gametes to the somatic tissue in the next generation. ncH3K4me3 on the paternal and maternal alleles are represented by different colours. D, day; w, week.
Notably, the global occurrence of ncH3K4me3 coincides with genome silencing from mature oocytes to the early two-cell stage, raising a paradoxical possibility that it may contribute to gene repression. In support of this hypothesis, the genome silencing in peri-ovulatory oocytes is impaired in the absence of KMT2B, an H3K4me3 methyltransferase18. To test if this observation is probably due to a direct effect from H3K4me3, we isolated full-grown oocytes where we overexpressed a panel of H3K4me3 demethylases (Methods). Silenced oocytes with characteristic surrounded nucleolus were identified17. Notably, the overexpression of KDM5B, but not KDM5A (Fig. 4c and Extended Data Fig. 7d) or other demethylases (data not shown) led to significant downregulation of H3K4me3 and reactivated transcription (as determined by the BrUTP assay) in a substantial portion of oocytes. These data indicate that ncH3K4me3 may play a role in genome silencing during oogenesis.
Finally, we ask whether ncH3K4me3 is unique to mammals. We found that H3K4me3 in zebrafish oocytes showed a canonical pattern that resembles those at stages after ZGA (Extended Data Fig. 8a–c). The absence of ncH3K4me3 in zebrafish oocytes may be related to the characteristics of their DNA methylomes which generally lack PMDs (Extended Data Fig. 8d–f). The strong correlation between the presence of PMDs and ncH3K4me3 indicates that the reprogramming of histone modifications and DNA methylation is inherently linked.
Using a highly sensitive ChIP–seq method, we investigated genome-wide H3K4me3 in mouse early development at the nucleosome resolution. Our data unveiled surprisingly dynamic chromatin landscapes in gametes and early embryos characterized by the presence of ncH3K4me3 (Fig. 4d). The ncH3K4me3 pattern appears to be inherited from oocytes to early embryos before ZGA. Extensive reprogramming also occurs on the paternal genome in zygotes where H3K4me3 peaks are largely depleted. Instead, very broad H3K4me3 domains at weak levels were observed which could be considered as a type of paternal ncH3K4me3 (Fig. 4d). Surprisingly, the genome silencing in oocytes is impaired when H3K4me3 is depleted. Perhaps distal ncH3K4me3 can work as ‘molecular sponges’ by absorbing and sequestering transcriptional resources to prevent spontaneous and inadvertent transcription. Eliminating ncH3K4me3 may be important for restoring the quiescent genome back to the active state during ZGA. Taken together, our study not only discovered a highly dynamic landscape of modified histones in pre-implantation development, but also shed light on the fundamental mechanisms for inheritance and reprogramming of epigenetic modifications.
Methods
Data reporting
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during outcome assessment.
Cell culture
Mouse ES cells (R1) were cultured without irradiated mouse embryonic fibroblasts (MEFs) in DMEM containing 15% FBS, leukaemia inhibiting factor (LIF), penicillin/streptomycin, l-glutamine, β-mercaptoethanol, and non-essential amino acids.
Mouse gametes and early embryo collection
Pre-implantation embryos were collected from 5 to 6 week-old C57BL/6N females (purchased from Vital River) mated with PWK/PhJ males (purchased from Jackson Laboratory). To induce ovulation, females were administered 5 IU of hCG intraperitoneally, 44–48 h after injection of 5 IU of PMSG (San-Sheng pharmaceutical Co. Ltd). Each set of embryos at a particular stage was flushed from the reproductive tract at defined time periods after hCG administration: 20 h (MII oocyte), 27–28 h (PN5 zygote), 30 h (early two-cell), 43 h (late two-cell), 54–56 h (four-cell), 68–70 h (eight-cell) and 92–94 h (blastocysts) in HEPES-buffered CZB medium. The zona pellucida of embryos selected by cell number or morphology was gently removed by treatment of 10 IU/ml pronase (Sigma P8811) for several minutes. The embryos were then manually picked and treated with the lysis buffer for STAR ChIP–seq or Smart-seq2.
Blastocysts from E3.5–4.0 were subjected to immunosurgery as previously described21. In brief, blastocysts were incubated in a 1:3 dilution of anti-mouse rabbit serum in DMEM for 20 min, washed in PBS and further incubated for 20 min in a 1:5 dilution of rat serum in DMEM for the complement reaction. The ICM was subsequently cleaned from lysed trophectoderm with a narrow glass pipette.
To isolate the E6.5 epiblast from post-implantation embryos, extraembryonic region was removed firstly with two tungsten needles. Epiblast was then separated from visceral endoderm by trypsin and pancreatic enzyme digestion followed by mechanical dissection as described previously22.
The cortex tissue was isolated from 8-week old male C57BL/6 mice following a protocol that was previously described by ENCODE http://genome.ucsc.edu/ENCODE/protocols/cell/mouse/Cortex_Ren_protocol.pdf.
To inhibit transcription in the early embryos, late zygotes (PN5) and early two-cell embryos were cultured in CZB supplemented with α-amanitin for about 14 h. To block DNA synthesis, late zygotes (PN5) and early two-cell embryos were cultured in CZB medium supplemented with aphidicolin for about 14 h.
The isolation of follicles was performed as described previously23. Briefly, growing oocytes were isolated from 7, 10 and 14-day-old mice by mechanical dissection using a 26 gauge needle, followed by the digestion with 4 mg/ml of collagenase I and 0.02 mg/ml of DNase I in L15 medium for 30 min at 37 °C. To collect germinal vesicle (GV) stage oocytes from 8-week-old mice, the whole ovaries were clipped mechanically with a razor blade. To remove the granulosa cells, the dissociated oocyte-granulosa cell complexes were transferred into PBS (Ca2+-, Mg2+-free) containing 1 mg/ml of collagenase I and were digested for 20 min24. Oocytes were then incubated in lysis buffer after removing the zona pellucida by pronase treatment.
All animal maintenance and experimental procedures used in current study were carried out according to guidelines of Institutional Animal Care and Use Committee (IACUC) of Tsinghua University, Beijing, China.
Isolation of mouse pronuclei
Zygotes at PN3–PN4 stages for pronuclei isolation were collected from C57BL/6N females mated with DBA/2N males. Female pronuclei were distinguished from male pronuclei on their size and distance from the polar bodies and were first harvested by a micromanipulator combined with a Piezo drive unit (Prime Tech). Male pronuclei within cytoplasm were extracted after removal of the zona pellucida and polar bodies. Isolated pronuclei were then lysed for STAR ChIP–seq.
Gene overexpression and immunostaining in SN GV oocytes
The cDNA of complete open reading frame of KDM5A or KDM5B was cloned into the RN3P vector. These plasmids were first linearized with Sfi and mRNA was synthesized with T3 polymerase using the mMessage mMachine kit (Ambion). GV stage oocytes were collected by puncturing the follicles of ovaries from 6–8-week-old C57BL/6 or F1 (C57BL/6 × CBA) female mice. To arrest oocytes in GV stage, Milrinone (Sigma) was added to a final concentration of 20 μM during isolation, microinjection and culture25. Microinjection of mouse KDM5A and KDM5B mRNA was performed as previously described25. After injection, oocytes were cultured in M16 medium supplemented with 5% fetal calf serum (FCS) at 37 °C with 5% CO2. For BrUTP incorporation experiments, oocytes injected with control, KDM5A and KDM5B mRNAs were cultured in M16 medium with Milrinone and 5% FCS for 12 h. Then they were injected with 30 mM BrUTP(B7166, Sigma, in 2 mM Pipes, pH 7.4, 140 mM KCl buffer)26 and cultured in the same medium for another 2 h. For immunostaining, oocytes were treated briefly with acidic Tyrode’s solution to remove zona pellucida. Then they were fixed in 1% PFA in PBS 0.1% Tween20 (PBST), 4 °C overnight, permeabilized with 0.25% Triton X-100 in PBST at room temperature for 20 min and blocked with 3% BSA in PBST at 4 °C overnight. Oocytes were incubated with the H3K4me3 antibody (in-house) and the BrdU/BrUTP antibody (11170376001, Roche). Secondary antibodies used were DyLight 633 conjugated goat anti-rabbit IgG and Alexa Fluor 546 conjugated goat anti-mouse IgG (Thermo). DNA was stained with DAPI. After staining, oocytes were mounted on glass slides in VECTASHIELD Antifade mounting medium (Vector Laboratories). Images were acquired on a Nikon A1Rsi confocal microscope. The same parameters were used for each channel of all groups of samples. The fluorescence intensity of H3K4me3 and BrUTP were quantified using NIS-Elements AR analysis software.
Zebrafish oocyte collection
The wild-type zebrafish line AB was used in this study. Unfertilized oocytes were obtained by squeezing the females gently on the ventral side. Next, the oocytes were activated by adding 3 ml fresh holfreter (0.05 g/l KCl, 0.1 g/l CaCl2, 0.025 g/l NaHCO3, 3.5 g/l NaCl, pH 7.0) in 35-mm dishes, and then followed by standard pronase dechorionation treatment27. In order to de-yolk oocytes (where yolk mass were highly abundant), dechorionated oocytes were transferred into a 1.5 ml tube and washed with PBS for several times, then disrupted with 200-μl tips. The cells of oocytes were spun down at 5,000 r.p.m. for 5 min at 4 °C.
STAR ChIP–seq library preparation and sequencing
The ChIP procedure was based on a previously described method28 with modifications to minimize experimental steps and sample loss. Briefly, each sample is lysed in 19 μl lysis buffer (0.5% NP-40, 0.5% Tween-20, 0.1% SDS and proteinase inhibitor) before being subjected to MNase digestion by incubating with 19 μl MNase working buffer (100 mM Tris-HCl pH 8.0, 2 mM CaCl2) and 2 μl diluted MNase (Sigma, N3755-200UN) at 37 °C for 5 min. 0.02 unit of MNase was used for all samples except for sperm (see below; optimal concentration may be determined for different batches of MNase). The reaction is terminated by adding 5 μl Stop buffer (110 mM Tris-HCl pH 8.0, 55 mM EDTA). 45 μl cold 2 × RIPA buffer (1% Triton X-100, 280 mM NaCl, 0.1% SDS, 0.2% sodium deoxycholate (DOC), 5 mM EGTA supplemented with proteinase inhibitor) is added. After spinning at max speed in 4 °C for 15 min, the supernatant is transferred to a new tube. Before adding antibody, each chromatin sample is supplemented with 40 μl RIPA buffer (10 mM Tris-HCl pH 8.0, 140 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.1% DOC, 1 mM EDTA). The IP sample is incubated with ~1–1.5 μg H3K4me3 (in-house antibody for most experiments and Millipore 04-745 for second antibody validation), H3K27me3 (Diagenode, pAb-069–050), H3K27Ac (ActiveMotif, 39133), H3 (Abcam, ab1791) overnight with rotation at 4 °C. Both in-house and commercial H3K4me3 antibodies produced very similar data. The in-house antibody was chosen for most experiments as it produced more robust results. The next day, the sample is incubated with 100 μg protein A dynabeads (Life Technologies) for 2 h with rotation at 4 °C. The beads were washed five times with 150 μl RIPA buffer and once with 150 μl LiCl buffer (250 mM LiCl, 10 mM Tris-HCl pH 8.0, 1 mM EDTA, 0.5% NP-40, 0.5% DOC). After washing, tubes were spun briefly to remove the supernatant. For each IP sample, beads were resuspended with 27 μl ddH2O and 1 μl 10× Ex-Taq buffer (TaKaRa). 1 μl proteinase K (Roche, 10910000) is added at 55 °C for 90 min to elute DNA from beads. The supernatant is then transferred to a new tube and the proteinase K is inactivated at 72 °C for 40 min. 1 μl rSAP (NEB, M0371) is then added to dephosphorylate the 3′ end of DNA at 37 °C for 1 h. Inactive rSAP at 65 °C for 10 min. The resulting sample is subjected to TELP library preparation without DNA purification, starting from poly-C tailing as described previously in full detail3 with a slight modification. Briefly, poly-C tailing is conducted on denatured single-strand DNA using dCTP and terminal deoxynucleotidyl transferase (TDT). Biotin-labelled anchor primer containing poly-G is used for second strand DNA extension. A modified anchor primer was used for better performance ([BIO]GTTCAGACGTGTGCTCTTCCGATCTGGGGGGGGG, where [BIO] refers to 5′-end biotin). The products are captured by magnetic streptavidin beads. After an adaptor ligation to the opposite end of poly-C, the double-stranded DNA is amplified by primers containing Illumina adaptor sequences. The resulting DNA is ready for sequencing.
Mouse sperm ChIP–seq was performed as described previously with modifications29. Briefly, 2 × 106 mouse spermatozoa were treated with 50 mM DTT for 2 h at room temperature, then N-ethylmaleimide (NEM) was added for 30 min to quench DTT. Subsequently, mouse spermatozoa were lysed on ice for 10 min in complete buffer 1(15 mM Tris-HCl (pH 7.5), 60 mM KCl, 5 mM MgCl2 and 0.1 mM EGTA, 0.3 M sucrose, 0.5 mM DTT, 0.5% NP-40 and 1% sodium deoxycholate). Same volume of MNase buffer (85 mM Tris-HCl, pH 7.5, 3 mM MgCl2, 2 mM CaCl2 and 0.3 M sucrose) containing 10 units MNase (Sigma, N3755-200UN) were used for chromatin digestion. Digested chromatin was subjected to STAR ChIP–seq protocol as described above.
RNA-seq library preparation and sequencing
The RNA-seq libraries were generated from early mouse embryos using the Smart-seq2 protocol as described previously with minor modification5. Cells were lysed in hypotonic lysis buffer (Amresco, M334), and the polyadenylated mRNAs were captured by the PolyT primers. After ~3–10 min lysis at 72 °C, the Smart-seq2 reverse transcription reactions were performed. After pre-amplification and AMPure XP beads purification, cDNAs were sheared by Covaris and were subject to Illumina TruSeq library preparation. All libraries were sequenced on Illumina HiSeq1500 or 2500 according to the manufacturer’s instruction.
Growing oocyte methylome profiling
The DNA methylome for the 14 day growing oocytes were determined using a newly developed low-input DNA methylome profiling method (manuscript in submission). Briefly, collected oocytes were lysed in lysis buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1 mM EDTA pH 8.0, NP-40 0.5%) with 1 μl protease K (Roche) and lambda DNA (Promega) (with the mass ratio 1/200) spiked-in. The samples were lysed for at least 3 h at 55 °C and then heat-inactivated for 1 h at 72 °C. The released DNA was then digested with dsDNA fragmentase (NEB) for 30 min. The digested DNA was directly treated with bisulphite conversion reagent using EpiTect Fast Bisulphite Conversion Kit (Qiagen) following a modified protocol: denature for 8 min at 98 °C; incubate at 60 °C for 25 min; and repeat the procedure again. The converted DNA was subjected to column purification and desulfonation using MinElute DNA Spin Columns (Qiagen) with carrier RNA (Qiagen) supplemented. Finally, the purified DNA was then directly proceeded to library construction as previously described3.
STAR ChIP–seq data processing
All STAR ChIP–seq reads were aligned to the mouse reference genome (mm9) using Bowtie2 (version 2.2.2)32 with the parameters –t –q –N 1 –L 25. All unmapped reads, non-uniquely mapped reads and PCR duplicates were removed. For downstream analysis, the read counts were normalized by computing the numbers of reads per kilobase of bin per million of reads sequenced (RPKM). To visualize the STAR ChIP–seq signal in the UCSC genome browser, each read was extended by 250 bp and the coverage for each base was counted. Replicates of STAR ChIP–seq were pooled for the downstream analysis. As the numbers of H3K4me3 peaks that can be detected for each stage are also affected by the sequencing depths, we used the same number of reads (30 million) when available randomly selected from samples of each stage for downstream analysis.
RNA-seq data processing
All RNA-seq data were mapped to the mouse reference genome (mm9) by TopHat (version 2.0.11). The gene expression level was calculated by Cufflinks (version 2.0.2)30 using the refFlat database from the UCSC genome browser. To quantify the expression level of repeats in mouse pre-implantation embryos, the reads were mapped to RepBase31 by Bowtie2 (version 2.2.2)32 and only uniquely mapped reads were kept for further analysis.
Comparison between STAR and conventional ChIP–seq and between STAR ChIP–seq replicates
The correlation between STAR ChIP–seq and conventional ChIP–seq or between different STAR ChIP–seq replicates was calculated as following: the RPKM values were generated in each 100-bp bin and were summed within each 2-kb window for the entire genome. Such enrichment was compared between different samples for correlation analysis.
Identification of promoter and distal H3K4me3 peaks
H3K4me3 peaks were called using MACS233 with the parameters –broad –nomodel –nolambda and noisy peaks with very weak signals (summed RPKM <30) were removed in the further analysis. The Refseq gene annotation databases were used to generate a set of annotated promoters. H3K4me3 peaks that are at least 2.5 kb away from annotated promoters were selected as distal STAR ChIP–seq peaks.
Analysis of the promoter H3K4me3 enrichment
To ensure that the promoters that we analysed reflect the truly active promoters (as many genes have alternative promoters), we only included genes that contain a single promoter, or multiple promoters that are located within 500 bp, in which case a promoter would be randomly chosen. The ChIP–seq signals at promoters (±2.5 kb) were computed by average signals from each bin within promoter. As the global enrichment of H3K4me3 at promoters differ significantly between ncH3K4me3 and canonical H3K4me3, Z-score normalization was used to allow visualization of all stages in the same heat map or investigation of all stages in the same plot. Z score was obtained using the following formula: for a given promoter i: zi = (xi − μ)/σ, where xi is the RPKM value before normalization, zi is the normalized RPKM value and μ and σ are the mean and standard deviation of all promoter RPKM values for each stage, respectively.
The comparison between promoter H3K4me3 enrichment and gene expression
The Z-score-normalized signals of H3K4me3 at promoter and gene expression levels (FPKM) were used in the analysis. The correlation between H3K4me3 enrichment at promoters and gene expression was showed by a moving average analysis (moving window = 100 genes, step = 1 gene).
Clustering analysis
The k-means clustering of gene expression levels at various stages was conducted using Cluster 3.0 with the parameters –g 7 (Euclidean distance). Heat maps were generated using Java Treeview. The hierarchical clustering analysis for H3K4me3 at various stages was conducted using an R package (ape) based on the Pearson correlation between each pair of stage. The distance between two stages was calculated as: 1 − Pearson correlation.
Distribution of H3K4me3 peaks
To determine the genomic distribution of H3K4me3 peaks, the genome was segmented to TSSs (transcription start sites), exons, introns, TESs (transcription end sites) and intergenic regions using an annotation combining the Refseq, UCSC Known Gene, Ensemble and Gencode databases. H3K4me3 peaks were assigned into each group based on the centre of the peaks.
The comparison between H3K4me3 peaks and repetitive elements
To identify the overlap between repetitive elements and promoter or distal H3K4me3 peaks, we kept the best alignment for non-uniquely mapped reads and called H3K4me3 peaks using MACS2 (ref. 34). These H3K4me3 peaks were compared with the locations of annotated repeats (RepeatMasker) downloaded from the UCSC genome browser. As repeats of different classes vary greatly in numbers, a random set of regions from the same chromosome with identical length for each H3K4me3 peak were used for the same analysis as a control. The numbers of observed peaks that overlap with repeats divided by the numbers of random peaks that overlap with repeats were used and log2 transformed. This is represented as log ratio of ‘observed/random’ enrichment.
Allele assignment of sequencing reads
To minimize the mapping bias introduced by the sequence differences between the two parental alleles, all STAR ChIP–seq reads were aligned to the genomes of the C57BL and PWK strains (mm9) separately using Bowtie2 (version 2.2.2) with the parameters: –t –q –N 1 –L 25 –X 2000–no-mixed –no-discordant. To assign each read to its parental origins, we examined all SNPs in the read that show high quality base calling (Phred score ≥ 30). For paired-end reads, SNP information from both reads in the pair was summed and used. When multiple SNPs were present in a read (or a read pair), the parental origin was determined by votes from all SNPs and the read was assigned to the allele that had at least two thirds of the total votes.
The comparison of H3K4me3 peaks between two alleles
To calculate the paternal to maternal ratio for each development stage, we selected peaks at certain stage covered with at least 20 reads of which the parental origins were identified. We then calculated the differences of read numbers between two alleles (maternal–paternal) normalized by the total number of allelic reads in each peak.
The analysis of allele-specific H3K4me3 enrichment in imprinted control regions
We counted the numbers of H3K4me3 reads that fall into the annotated imprinted control regions on each allele. The significance of allele bias was determined by Fisher’s exact test which varies depending on the numbers of reads and stages. Of 13 regions, 8 have a minimal P value of 0.001 for any stages we examined. The rest include those imprinting control regions that are located in intergenic regions instead of promoters (such as Gpr1/Zbdf2 and Rasgrf1).
Identification of PMDs
PMDs in oocytes were identified as described previously16. Briefly, average DNA methylation levels were calculated for 10-kb bins of the genome and numbers of CpGs covered were also counted. Bins with average DNA methylation level lower than 0.6 and more than 20 CpGs covered were selected and merged into PMDs. Promoter regions (±2.5 kb) were excluded from PMDs.
References
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007)
Ruthenburg, A. J., Allis, C. D. & Wysocka, J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell 25, 15–30 (2007)
Peng, X. et al. TELP, a sensitive and versatile library construction method for next-generation sequencing. Nucleic Acids Res . 43, e35 (2015)
Hamatani, T., Carter, M. G., Sharov, A. A. & Ko, M. S. Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell 6, 117–131 (2004)
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protocols 9, 171–181 (2014)
Lee, M. T., Bonneau, A. R. & Giraldez, A. J. Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 30, 581–613 (2014)
Peaston, A. E. et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell 7, 597–606 (2004)
Smith, Z. D. & Meissner, A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220 (2013)
Wu, H. & Zhang, Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156, 45–68 (2014)
Erkek, S. et al. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat. Struct. Mol. Biol. 20, 868–875 (2013)
Xie, W. et al. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 148, 816–831 (2012)
Lepikhov, K. & Walter, J. Differential dynamics of histone H3 methylation at positions K4 and K9 in the mouse zygote. BMC Dev. Biol. 4, 12 (2004)
Bernstein, B. E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006)
Wang, L. et al. Programming and inheritance of parental DNA methylomes in mammals. Cell 157, 979–991 (2014)
Tomizawa, S., Nowacka-Woszuk, J. & Kelsey, G. DNA methylation establishment during oocyte growth: mechanisms and significance. Int. J. Dev. Biol. 56, 867–875 (2012)
Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009)
Kageyama, S. et al. Alterations in epigenetic modifications during oocyte growth in mice. Reproduction 133, 85–94 (2007)
Andreu-Vieyra, C. V. et al. MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLoS Biol . 8, e1000453 (2010)
Shen, Y. et al. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 (2012)
Hon, G. C. et al. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet. 45, 1198–1206 (2013)
Solter, D. & Knowles, B. B. Immunosurgery of mouse blastocyst. Proc. Natl Acad. Sci. USA 72, 5099–5102 (1975)
Boroviak, T., Loos, R., Bertone, P., Smith, A. & Nichols, J. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat. Cell Biol. 16, 516–528 (2014)
Demeestere, I. et al. Effect of preantral follicle isolation technique on in-vitro follicular growth, oocyte maturation and embryo development in mice. Hum. Reprod. 17, 2152–2159 (2002)
Su, Y. Q. et al. MARF1 regulates essential oogenic processes in mice. Science 335, 1496–1499 (2012)
Sharif, B. et al. The chromosome passenger complex is required for fidelity of chromosome transmission and cytokinesis in meiosis of mouse oocytes. J. Cell Sci. 123, 4292–4300 (2010)
Bouniol-Baly, C. et al. Differential transcriptional activity associated with chromatin configuration in fully grown mouse germinal vesicle oocytes. Biol. Reprod. 60, 580–587 (1999)
Whitlock, K. E. & Westerfield, M. The olfactory placodes of the zebrafish form by convergence of cellular fields at the edge of the neural plate. Development 127, 3645–3653 (2000)
Gilfillan, G. D. et al. Limitations and possibilities of low cell number ChIP–seq. BMC Genomics 13, 645 (2012)
Hisano, M. et al. Genome-wide chromatin analysis in mature mouse and human spermatozoa. Nat. Protocols 8, 2449–2470 (2013)
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protocols 7, 562–578 (2012)
Jurka, J. et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110, 462–467 (2005)
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012)
Zhang, Y. et al. Model-based analysis of ChIP–seq (MACS). Genome Biol . 9, R137 (2008)
Feng, J., Liu, T., Qin, B., Zhang, Y. & Liu, X. S. Identifying ChIP–seq enrichment using MACS. Nat. Protocols 7, 1728–1740 (2012)
Yue, F. et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515, 355–364 (2014)
Deng, Q., Ramsköld, D., Reinius, B. & Sandberg, R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343, 193–196 (2014)
Kobayashi, H. et al. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res . 23, 616–627 (2013)
Zhang, Y. et al. Canonical nucleosome organization at promoters forms during genome activation. Genome Res . 24, 260–266 (2014)
Bogdanovic, O. et al. Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res . 22, 2043–2053 (2012)
Potok, M. E., Nix, D. A., Parnell, T. J. & Cairns, B. R. Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell 153, 759–772 (2013)
Acknowledgements
We appreciate help from members of the Xie laboratory for comments during preparation of the manuscript. We thank J. Zhao and G. Qin for helping with pronuclei isolation. We are grateful to the animal core facility, the sequencing core facility, and biocomputing facility at Tsinghua University. This work is supported by the funding provided by the National Basic Research Program of China 2016YFC0900301 (W.X.), 2015CB856201 (W.X.), 2012CB966701 (J.N.), the National Natural Science Foundation of China 31422031 (W.X.), 91519326 (W.X.), 31171381 (J.N.), the Beijing Natural Science Foundation grant 5152014 (J.N.), Tsinghua University Initiative Scientific Research Program 20161080043 (W.X.), the funding from the THU-PKU Center for Life Sciences (W.X., J.N.), and the Youth Thousand Scholar Program of China (W.X.).
Author information
Authors and Affiliations
Contributions
B.Z. developed STAR ChIP–seq and conducted ChIP–seq experiments; B.H., Y.X. and J. Ma collected mouse oocytes and early embryos. B.H. performed RNA-seq; H.Z. performed data analysis; W.L., J. Ming and B.Z. conducted KDM5B experiment. W.L., X.K., Y. Zhao, W.H., C.L. and B.H. collected mouse pronuclei. X.P., B.H. and F.X. advised the development of STAR ChIP–seq. Y. Zhang and Q.Y. performed oocyte DNA methylome profiling. Y.L. and Q.W. conducted high-throughput sequencing; X.W. collected zebrafish oocytes; K.K., A.M., S.G., J.N. and W.X. supervised the project or related experiments. H.Z. and W.X. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
B.Z. and W.X. are co-inventors on a filed patent application for STAR ChIP–seq.
Additional information
Reviewer Information Nature thanks R. Schultz and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Validation of STAR ChIP–seq in mES cells.
a, A brief schematic of STAR ChIP–seq. Based on a previously described ChIP method28, micrococcal nuclease (MNase) is used to fragment native chromatin. Nucleosomes with certain histone modifications are immunoprecipitated using specific antibodies. Proteinase K is added for protein digestion and is heat-inactivated. Shrimp alkaline phosphatase (rSAP) is directly added to the reaction for 3′ DNA end repair, which is required for the poly-C addition in the subsequent TELP library preparation. After heat-inactivation of rSAP, the resulting DNA is subjected to the TELP library preparation as previously described3 without purification. Briefly, poly-C tailing is conducted on denatured single-strand DNA using dCTP and terminal deoxynucleotidyl transferase (TDT). Biotin-labelled anchor primer (indicated by ‘B’) containing poly-G is used for second strand DNA extension. The products are captured by magnetic streptavidin beads. After an adaptor ligation to the opposite end of poly-C, the double-stranded DNA is amplified by primers (P1 and P2) containing Illumina adaptor sequences. The resulting library is ready to be sequenced. b, A snapshot of the UCSC browser view shows enrichment of STAR ChIP–seq for histone modifications H3K4me3, H3K27ac and H3K27me3 using various numbers of mES cells. Data sets from mouse ENCODE are also shown for comparison35. c, The Pearson correlation coefficients showing the comparison between STAR ChIP–seq of H3K4me3 using various numbers of mES cells and conventional ChIP–seq of H3K4me3 from either this study using 1 × 106 mES cells or data from ENCODE35. d, A bar chart showing the percentages of conventional mESC H3K4me3 total/promoter peaks recaptured by STAR ChIP using various numbers of mES cells. e, Box plots showing the H3K4me3 enrichment determined by conventional ChIP–seq at peaks that are either recaptured or missed by STAR ChIP–seq.
Extended Data Figure 2 Validation of H3K4me3 STAR ChIP–seq and RNA-seq in mouse gametes and early embryos.
a, Representative bright-field microscopic images showing the pre-implantation embryos used for STAR ChIP–seq with time points of sample collection indicated. b, The Spearman correlation between the replicates (n = 2) of RNA-seq samples and between RNA-seq in this study and Deng et al.36 at stages when available. c, The Pearson correlation between the replicates (n = 2) of H3K4me3 for mouse pre-implantation embryos at each stage. d, Scatter plots showing the H3K4me3 enrichment levels (2-kb window) between replicates across the genome for mouse pre-implantation embryos at each stage. e, Scatter plot showing the H3K4me3 enrichment levels (2-kb window) in MII oocytes between data sets generated using an in-house antibody or a commercial antibody (Millipore 04-745). f, A snapshot of the UCSC browser view shows H3K4me3 STAR ChIP–seq results using two different H3K4me3 antibodies (in-house or commercial/Millipore 04-745) in mES cells and MII oocytes. Annotated genes and repetitive elements are also shown. g, Venn diagram showing the overlap between mES cell H3K4me3 (conventional ChIP–seq) promoter peaks and ICM H3K4me3 promoter peaks. h, Snapshots of the UCSC browser views showing the H3K4me3 and expression signals near Foxa1 (left), Atp1b1 (middle) and Tbx3 (right).
Extended Data Figure 3 Characterization of promoter and distal H3K4me3 in mouse gametes and early embryos.
a, Scatter plots showing the relationship between the expression levels of all genes and their promoter H3K4me3 enrichment (Z-score-normalized) for each development stage. Mouse ES cell data from ENCODE are used as control. Spearman correlation coefficients are also shown. b, Moving average plots (window = 100 genes, moving step = 1 gene) showing the relationship between the expression levels of non-maternal genes and their promoter H3K4me3 enrichment (Z-score-normalized) for each stage after major ZGA. c, The genomic distributions of H3K4me3 peaks are shown for early embryos and mES cells. TSS, transcription start site (±2.5 kb); TES, transcription end site (±2.5 kb). d, A bar chart showing the percentages of all mappable reads that can be aligned to RepBase for mouse embryos of various stages. The error bars denote the standard deviations of two biological replicates of RNA-seq for each stage. e, Enrichment of repeats (log ratio of observed/random) in H3K4me3 promoter and distal peaks compared to that in random peaks at each stage. f, The enrichment (log ratio of observed/random) of top repeat subfamily members in distal H3K4me3 peaks in mouse embryos of various stages and mES cells. g, The snapshots of distal H3K4me3 signals near various types of repeats.
Extended Data Figure 4 Reprogramming of H3K4me3 on the paternal genome.
a, A snapshot of UCSC genome browser shows the enrichment of H3K4me3 in sperm from a published data set10 and this study. b, Snapshots showing the allelic H3K4me3 signals and RNA-seq signals near the imprinted loci Snrpn and Impact. Please note the maternal RNAs in zygotes, early two-cell and late two-cell embryos are presumably inherited from oocytes. The paternal zygotic transcripts appear from the late two-cell stage onward for both genes. c, Bar chart showing the numbers of H3K4me3 peaks identified in sperm (in regions covered by SNPs) and the paternal alleles of early embryos. d, Hierarchical clustering of sperm, early embryos and somatic cortex based on the paternal allele H3K4me3 enrichment. For sperm H3K4me3 data, only regions covered by SNPs were included for analysis. e, A bar chart showing the percentages of raw reads assigned to the maternal or paternal allele for each stage. f, Scatter plots showing the H3K4me3 enrichment levels between SNP-assigned parental alleles from zygotes and pronuclei for both the paternal (left) and maternal genomes (right).
Extended Data Figure 5 Non-canonical H3K4me3 (ncH3K4me3) in oocytes and early embryos
a, A snapshot of the UCSC browser view shows the enrichment of H3K4me3 in MII oocytes, isolated maternal pronuclei, PN5 zygotes (maternal allele), and histone H3 (maternal allele) in PN5 zygotes. b, Scatter plots showing the comparisons between the enrichment of histone H3 (maternal) in PN5 zygotes and H3K4me3 from either MII oocytes (left) or the maternal allele of PN5 zygotes (right). c, Hierarchical clustering of early embryo stages (maternal) based on global H3K4me3 enrichment. d, A bar chart showing the expression levels (log2[FPKM + 1]) of several marker genes for granulosa cells and oocytes, as well as a house-keeping gene (Actb), in oocyte samples collected at different stages. ICMs are included as a non-oocyte control. e, Heat maps showing the expression (FPKM) of all genes and the H3K4me3 enrichment (normalized RPKM) at their promoters (TSS ± 2.5 kb) for oocytes and early embryos. f, Hierarchical clustering of stages of oocytes and early embryos based on global H3K4me3 enrichment. g, A snapshot of UCSC browser shows the H3K4me3 enrichment in oocyte of various stages as well as DNA methylation in MII oocyte. A PMD with ncH3K4me3 enrichment is shaded.
Extended Data Figure 6 Non-canonical distal H3K4me3 in oocytes correlates with DNA hypomethylation.
a, Box plots showing the CpG density of H3K4me3 promoter and distal peaks in early embryos, mES cells, and the cortex. b, Pie charts showing the percentage of the genome covered by PMDs (left) and the percentage of distal H3K4me3 marked regions covered by PMDs (right) in MII oocytes. c, A global view of H3K4me3 promoter and distal peak density of chr12 in MII oocytes (top). Gene densities and PMD densities are also shown (bottom). Three gene desert regions are shaded. d, DNA methylation levels around active genes are shown for those in embryonic day 16.5 (E16.5) female PGCs37, day 14 oocytes, and MII oocytes14.
Extended Data Figure 7 The transition from ncH3K4me3 to canonical H3K4me3 requires zygotic transcription but not DNA replication.
a, Box plots of gene expression levels (log2[FPKM + 1]) show the effects of α-amanitin and aphidicolin on genes activated during ZGA (left). Genes expressed at all stages (middle) and genes repressed at all stages (right) are also shown. b, Hierarchical clustering of two alleles of early embryos including those treated with α-amanitin or aphidicolin based on the global H3K4me3 enrichment. c, A snapshot of the UCSC browser view shows the global view of H3K4me3 enrichment for maternal and paternal alleles in various types of embryos including those treated with α-amanitin or aphidicolin. d, Bar charts showing normalized H3K4me3 and BrUTP fluorescence intensity of individual surrounded nucleolus (SN)-stage oocytes. Each pair of bars for H3K4me3 and BrUTP represents an individual oocyte. Data from three independent experiments were pooled. GV, germinal vesicle.
Extended Data Figure 8 ncH3K4me3 is not found in zebrafish.
a, A snapshot of the UCSC genome browser shows the global view of H3K4me3 enrichment in oocytes (this study), the dome stage embryos38, 48 hours post fertilization (hpf) embryos39 and DNA methylation level40 for various zebrafish developmental stages. b, The genomic distributions are shown for the H3K4me3 peaks in zebrafish oocytes, embryos at the dome stage38 and 48 hpf embryos39. c, Box plots showing the CpG densities of H3K4me3 promoter peaks in zebrafish oocytes, embryos at the dome stage38 and embryos at 48 hpf39. A random set of peaks that match the lengths of H3K4me3 peaks on the same chromosomes were used as a control in the analysis. d, Violin plots showing the distributions of DNA methylation levels from gametes to somatic tissues in zebrafish (left panel) or mice (right panel). e, Box plot showing the lengths of distal PMDs in zebrafish and mouse oocytes, sperms, and somatic tissues. f, Bar chart shows the percentages of the genome covered by distal PMDs in oocytes, sperms and somatic tissues in zebrafish or mouse.
Supplementary information
Supplementary Data
This file contains Supplementary Table 1. (XLSX 39 kb)
Supplementary Data
This file contains Supplementary Table 2. (XLSX 9167 kb)
Supplementary Data
This file contains Supplementary Table 3. (XLSX 43 kb)
Rights and permissions
About this article
Cite this article
Zhang, B., Zheng, H., Huang, B. et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537, 553–557 (2016). https://doi.org/10.1038/nature19361
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature19361
- Springer Nature Limited
This article is cited by
-
Evaluating histone modification analysis of individual preimplantation embryos
BMC Genomics (2024)
-
Emergence of replication timing during early mammalian development
Nature (2024)
-
Dynamic and distinct histone modifications facilitate human trophoblast lineage differentiation
Scientific Reports (2024)
-
Meiotic maturation failure in primary ovarian insufficiency: insights from a bovine model
Journal of Assisted Reproduction and Genetics (2024)
-
Stable maternal proteins underlie distinct transcriptome, translatome, and proteome reprogramming during mouse oocyte-to-embryo transition
Genome Biology (2023)